Abstract
Background Peroxisomes are highly dynamic, metabolically active organelles that used to be regarded as a sink for H2O2 generated in different organelles. However, peroxisomes are now considered to have a more complex function, containing different metabolic pathways, and they are an important source of reactive oxygen species (ROS), nitric oxide (NO) and reactive nitrogen species (RNS). Over-accumulation of ROS and RNS can give rise oxidative and nitrosative stress, but when produced at low concentrations they can act as signalling molecules.
Scope This review focuses on the production of ROS and RNS in peroxisomes and their regulation by antioxidants. ROS production is associated with metabolic pathways such as photorespiration and fatty acid β-oxidation, and disturbances in any of these processes can be perceived by the cell as an alarm that triggers defence responses. Genetic and pharmacological studies have shown that photorespiratory H2O2 can affect nuclear gene expression, regulating the response to pathogen infection and light intensity. Proteomic studies have shown that peroxisomal proteins are targets for oxidative modification, S-nitrosylation and nitration and have highlighted the importance of these modifications in regulating peroxisomal metabolism and signalling networks. The morphology, size, number and speed of movement of peroxisomes can also change in response to oxidative stress, meaning that an ROS/redox receptor is required. Information available on the production and detection of NO/RNS in peroxisomes is more limited. Peroxisomal homeostasis is critical for maintaining the cellular redox balance and is regulated by ROS, peroxisomal proteases and autophagic processes.
Conclusions Peroxisomes play a key role in many aspects of plant development and acclimation to stress conditions. These organelles can sense ROS/redox changes in the cell and thus trigger rapid and specific responses to environmental cues involving changes in peroxisomal dynamics as well as ROS- and NO-dependent signalling networks, although the mechanisms involved have not yet been established. Peroxisomes can therefore be regarded as a highly important decision-making platform in the cell, where ROS and RNS play a determining role.
Keywords: Antioxidants, arabidopsis, autophagy, β-oxidation, nitric oxide, nitrosative stress, oxidative stress, photorespiration, peroxisomes, reactive nitrogen species, RNS, reactive oxygen species, ROS, signalling
INTRODUCTION
Peroxisomes are ubiquitous organelles in eukaryotic cells and have a close relationship with chloroplasts and mitochondria (Fig. 1). Initially, these organelles were regarded as involved in removing H2O2 produced by different sources both inside and outside peroxisomes by catalases. The role of peroxisomes was therefore limited to cleaning up reactive oxygen species (ROS) produced by other organelles. However, in recent years biochemical, transcriptomic and proteomic approaches have demonstrated that these organelles are much more complex and perform functions hitherto unknown (Reumann et al., 2009; Hu et al., 2012; Sandalio et al., 2013). Peroxisomes are highly dynamic, metabolically active organelles that participate in different cellular processes involved in development, morphogenesis and cell responses to stress (del Río et al., 1998; Sandalio et al., 2013). Changes in metabolic pathways, number, size, morphology and even speed of movement in peroxisomes can take place depending on cell type, tissue, organism, developmental stage (Hu et al., 2012) or environmental conditions involving herbicides, clofibrate, metals, wounding or nutrient disturbances among others (Romero-Puertas et al., 2004; Rodríguez-Serrano et al., 2009; Sinclair et al., 2009; Hu et al., 2012; McCarthy-Suarez et al., 2011; Sandalio et al., 2013) and in response to pathogens (Koh et al., 2005; Kuzniak and Skłodowska, 2005). Photomorphogenesis and senescence are also associated with changes in the metabolism and dynamics of peroxisomes (del Río et al., 1998; Rosenwasser et al., 2011; Hu et al., 2012). In addition, peroxisomes are an important source of ROS and reactive nitrogen species (RNS) and contain a complex battery of antioxidant defences that regulate the accumulation of ROS and RNS. Overproduction of ROS and RNS can cause severe oxidative and nitrosative damage, respectively (del Río, 2011). However, ROS and RNS can also act as signalling molecules that regulate developmental processes and stress responses (Neill et al., 2008; Mittler et al., 2011).
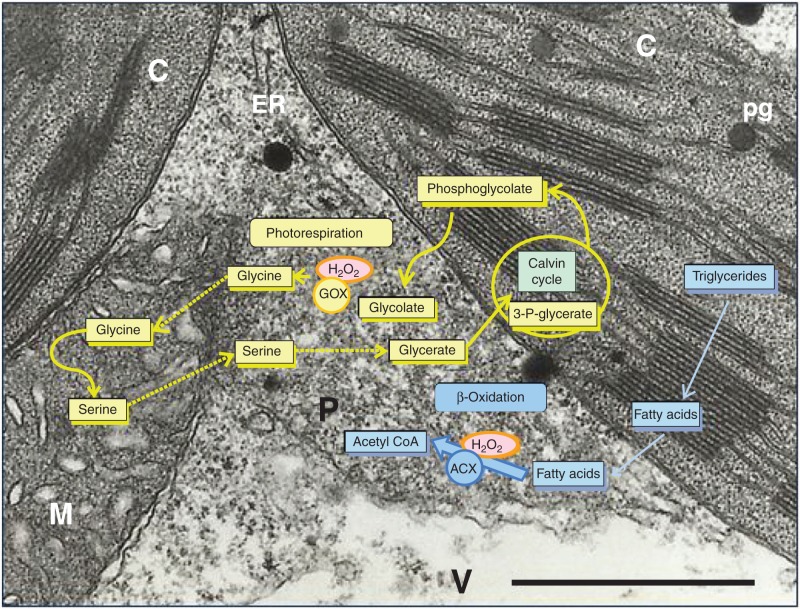
Fig. 1.
Electron micrograph of arabidopsis leaves showing a close relationship between chloroplasts, mitochondria and peroxisomes. Schemes showing H2O2 production in the reaction of glycolate oxidase (GOX) during photorespiration involving chloroplasts, peroxisomes and mitochondria (yellow) and H2O2 production associated with acyl CoA oxidase (ACX) activity from fatty acid β-oxidation (blue; chloroplasts and peroxisomes). C, chloroplast; ER, endoplasmic reticulum; M, mitochondria; P, peroxisome; pg, plastoglobuli; V, vacuole. Scale bar = 1 µm.
Peroxisomes can also play an important role in cellular redox homeostasis, which is, in turn, a key element in the regulation of cell metabolic pathways (Foyer and Noctor, 2003; Yun et al., 2012). It has been demonstrated that peroxisomes can rapidly sense changes in their environment and modify their metabolism and dynamics, all of which is regulated by ROS (Rodríguez-Serrano et al., 2009; Sinclair et al., 2009; Hu et al., 2012). However, the mechanisms involved in the perception of these stimuli have not yet been established. This review focuses on the production of ROS and RNS in peroxisomes and their regulation by antioxidants, and attempts to connect up all the pieces of the puzzle available in order to determine how peroxisomes perceive changes in their environment and trigger cell responses using ROS and RNS as key molecules.
PEROXISOMES ARE AN IMPORTANT SOURCE OF ROS AND RNS
Peroxisomes produce and scavenge different ROS
The term ROS includes oxygen-derivative species obtained through the reduction of oxygen, such as superoxide radicals (O2.–), hydroxyl radicals (·OH), alkoxyl radicals (RO·) and peroxyl radicals (ROO·), and also some non-radical compounds, such as H2O2, the excited oxygen species singlet oxygen (1O2), ozone (O3) and hypochlorous acid (HOCl–) (Halliwell and Gutteridge, 2007). The reactivity of each species differs, although ·OH is considered to be the strongest oxidant, capable of reacting with all types of molecule (Halliwell and Gutteridge, 2007). Different sources of ROS, including H2O2, O2.– and ·OH, have been reported to be produced in peroxisomes (Fig. 2). Superoxide radical production takes place in a short electron chain associated with the NADH/NADPH-driven peroxisomal membrane and also in the peroxisomal matrix associated with xanthine oxidoreductase (XOD/XDH) and uricase, both of which are important enzymes in the catabolism of nucleic acids and ureide metabolism, respectively (Sandalio et al., 2013) (Fig. 2). Recently, sulphite oxidase has been added to the list of peroxisomal enzymes involved in O2.– production associated with sulphite detoxification and assimilatory reduction of sulphate. However, the function of this enzyme in peroxisomes has not been clearly established and may prevent oxidative damage to sulphite (Byrne et al., 2009) (Fig. 2). In animal tissues it has been suggested that nitric oxide synthase (NOS) present in these organelles can also produce significant amounts of O2.– in the absence of an adequate amount of its substrate (Stuehr et al., 2001). However, a gene orthologous to the mammalian gene for NOS, whose identity remains elusive, has not been found in plants (Moreau et al., 2010).
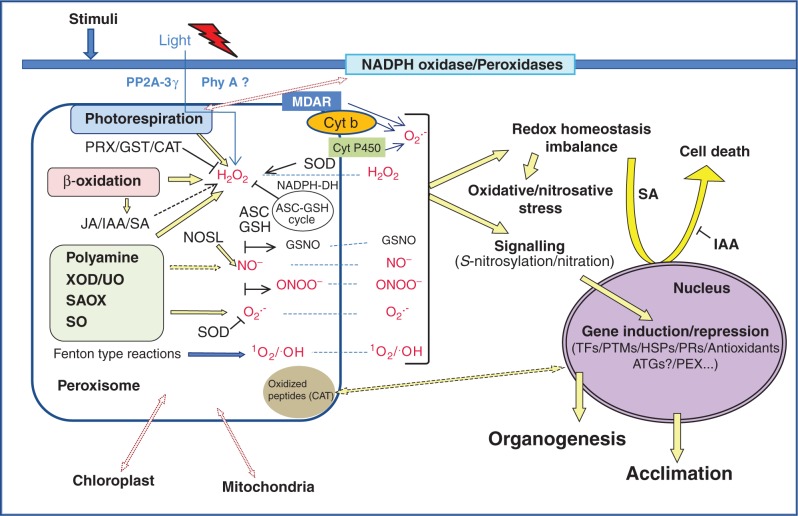
Fig. 2.
Schematic overview of the signalling networks involving ROS and RNS in peroxisomes. H2O2 and O2.– are produced in different metabolic pathways and in a small electron transport chain associated with the membrane and their steady state is regulated by SODs, catalase, the ASC–GSH cycle and probably peroxiredoxin (PRX). ROS, particularly H2O2, can diffuse to the cytosol were they can alter redox homeostasis and regulate gene transcription in the nucleus, giving rise to acclimatization or cell death depending on the stimuli. Light can regulate photorespiratory peroxisome-dependent changes in cytosolic redox homeostasis that cause SA-dependent lesions and induction of pathogenesis-related proteins. This process is dependent on SA and can also be regulated by PP2A-B’γ and Phy A. In addition, peroxisomes are a source of hormones, such as JA and IAA, that participate in the network regulating cell responses to stress (i.e. IAA, which negatively regulates light-dependent cell death). NO is produced in peroxisomes by a NOS-like activity, although other sources can also be involved, such as xanthine oxidase (XOD) and polyamines. ONOO– and GSNO are produced in peroxisomes by reaction of NO with O2.– and GSH, respectively, and can also act as signal molecules regulating gene expression and protein activity throughout post-translational modifications (PTMs) such as nitration and S-nitrosylation. Oxidized peptides may also regulate gene expression and autophagy in order to control homeostasis and peroxisomal quality. Peroxisomal ROS promote cross-talk with mitochondria and chloroplasts, probably through changes in the redox state of each organelle. ATGs, autophagy-related genes; CAT, catalase; Cyt P450, cytochrome P450 reductase; GST, glutathione-S-transferase; HSPs, heat shock proteins; IAA, indoleacetic acid; JA, jasmonic acid; MDAR, monodehydroascorbate reductase; NADPH-DH, NADPH dehydrogenases; NOS-L, NO synthase-like activity; ONOO–, peroxynitrite; PRX, peroxiredoxin; PEX, peroxins; Phy A, phytochrome A; PP2A-B’γ, 2A protein phosphatase subunit; PRs, pathogenesis-related proteins; SAOX, sarcosin oxidase; SO, sulphite oxidase; TFs, transcription factors; UO, urate oxidase.
The production of H2O2 in peroxisomes takes place in association with different metabolic pathways. Photorespiration is a light-dependent process that results in the uptake of O2 and the release of CO2 and is compartmentalized in chloroplasts, peroxisomes and mitochondria (Foyer et al., 2010) (Fig. 1). Glycolate oxidase (GOX) is one of the key enzymes in the photorespiration cycle and is probably the main source of H2O2 in green tissues (Fig. 1). The rate of H2O2 production through this process is 2-fold higher than in chloroplasts and 50-fold higher than in mitochondria (Foyer and Noctor, 2003). Fatty acid β-oxidation is another important source of H2O2 in peroxisomes (Fig. 1). This metabolic pathway is more active in germinating seeds, while being ubiquitous in green tissues and also involved in the metabolism of phytohormones such as jasmonic acid (JA) and auxins (Hooks, 2002). Other functions associated with β-oxidation have recently been described in non-storage tissue, such as benzoic acid, biotin and isoprenoid biosynthesis (Hu et al., 2012; Linka and Theodoulou, 2013; Cassin-Ross and Hu, 2014). Acyl CoA oxidase (ACX) is the principal enzyme in this pathway and catalyses the oxidation of acyl-CoA to trans-2-enoyl-CoA to produce H2O2 as a by-product (Kaur et al., 2009). The spontaneous or enzymatic dismutation of O2.– catalysed by superoxide dismutases (SODs) is another source of H2O2. The presence of Mn-SOD and CuZn-SOD in peroxisomes has been demonstrated in different plant species (Sandalio et al., 2013) (Fig. 2). Sarcosine oxidase has been added to the list of candidates for peroxisomal H2O2 sources in plants and catalyses the oxidation of sarcosine, N-methyl amino acids and l-pipecolate, producing formaldehyde, glycine and H2O2 (Goyer et al., 2004) (Fig. 2). The catabolism of polyamines is further source of H2O2 in peroxisomes, where diamine oxidases and polyamine oxidases have been identified (Kamada-Nobusada et al., 2008; Osno et al., 2012; Planas-Portell et al., 2013). The presence of haem- and non-haem iron-containing proteins and ascorbate in peroxisomes may contribute to ·OH production in these organelles through Fenton-type reactions. In peroxisomes purified from watermelon cotyledons, the production of ·OH was demonstrated by electron spin resonance spectroscopy (Sandalio et al., 1988). Fenton-type reactions may also give rise to the production in peroxisomes of singlet oxygen, an ROS typically associated with the chloroplast, as it has been reported by using the fluorescent probe Singlet Oxygen Sensor Green (SOSG; Mor et al., 2014). Thus, 1O2 could be a new player in the peroxisome-derived signalling network.
The steady-state levels of ROS in peroxisomes are strongly regulated by scavenging systems, including enzymatic and non-enzymatic antioxidants. Plant peroxisomes contain enzymatic defences to cope with different types of ROS (Table 1). Superoxide dismutases are involved in removing superoxide radicals, with the presence of two isoforms of SOD containing Cu and Zn (CuZn-SOD) and Mn (Mn-SOD) being reported in peroxisomes from different plant species (Sandalio et al., 2013) and a Fe-SOD in carnation petals (Droillard and Paulin, 1990). Catalase is the main antioxidant involved in removing H2O2 (Huang et al., 1983; Mhamdi et al., 2012). The ascorbate–glutathione cycle, made up of ascorbate peroxidase (APX), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), glutathione reductase (GR), ascorbate (ASC) and glutathione (GSH) (Jiménez et al., 1997; Romero-Puertas et al., 2006; Mhamdi et al., 2012), is also involved in maintaining H2O2 levels under control in peroxisomes. NADPH, which is necessary for the ASC–GSH cycle, is supplied by isocitrate dehydrogenase (ICDH) and the pentose phosphate cycle present in these organelles (Corpas et al., 1999; Reumann et al., 2009). Peroxisomes can also produce NADPH de novo through NADH phosphorylation (Waller et al., 2010). Other peroxidases may be present in peroxisomes, such as glutathione peroxidase and peroxiredoxins (Dietz, 2003), although neither the proteins nor the genes have yet been characterized. To our knowledge, the presence of thioredoxins in peroxisomes has not yet been demonstrated, although the identification of uricase as a target of thioredoxins suggests the presence of thioredoxins in peroxisomes (Du et al., 2010). The presence of glutathione transferases in peroxisomes from arabidopsis plants has also been reported (Dixon et al., 2009). These enzymes require GSH and participate in the detoxification of xenobiotic compounds and lipid peroxide products in addition to removing H2O2 (Dixon et al., 2009).
Table 1.
Enzymatic antioxidant defences present in plant peroxisomes
| Antioxidant | Substrate | References |
|---|---|---|
|
O2.– | Droillard and Paulin, 1990; Rodríguez-Serrano et al., 2007; Sandalio et al., 2013 |
| Catalase | H2O2 | Huang et al., 1983; Mhamdi et al., 2012 |
| Ascorbate peroxidase | H2O2, organic hydroperoxides | Jiménez et al., 1997; Lisenbee et al., 2003; Leterrier et al., 2005 |
| Glutathione reductase | GSSG | Romero-Puertas et al., 2006 |
| Monodehydroascorbate reductase | Monodehydroascorbate | Jiménez et al., 1997; Lisenbee et al., 2005 |
| Dehydroascorbate reductase | Dehydroascorbate | Jiménez et al., 1997, Reumann et al., 2009 |
| Peroxiredoxin | H2O2, organic hydroperoxides, ONOO– | Verdoucq et al., 1999; Dietz, 2003 |
| Glutathione S-transferase | H2O2, organic hydroperoxides | Dixon et al., 2009 |
|
NADP | Corpas et al., 1999; Reumann et al., 2009; Mhamdi and Noctor, 2015 |
Oxidative stress can lead directly to reversible or irreversible protein modifications. Protein carbonylation is a highly important oxidative-derived modification that consists of carbonyl radical production and is considered to be an irreversible process. The increase in carbonylated proteins in peroxisomes has been observed under different stress conditions associated with an increase in H2O2, such as cadmium toxicity (Romero-Puertas et al., 2002), senescence (Vanaker et al., 2006), treatment with the herbicide 2,4-dichlorophenoxiacetic acid (2,4-D; McCarthy et al., 2011), combined salt and continuous light treatment (Mano et al., 2014) and, in peroxisomes, treatment with metals to induce Fenton-derived ·OH production (Nguyen and Donaldson, 2005). The proteome of carbonylated proteins in peroxisomes has been studied in different plant species, and several proteins prone to carbonylation have been identified, some of which participate in antioxidant defences, fatty acid β-oxidation or cell death (Table 2) (Romero-Puertas et al., 2002; Nguyen and Donaldson, 2005; Mano et al., 2014). Oxidized proteins are dangerous and need to be proteolytically degraded by proteases to preserve peroxisomal functionality. The presence of serine protease activity has been demonstrated in purified peroxisomes from pea leaves using biochemical techniques, with catalase, GOX and glucose 6-P dehydrogenase (G6PDH) being the target of these proteases (Distefano et al., 1999; Romero-Puertas et al., 2002). A cysteine protease, resistant to drought 21A-like 1 (RDL1), has also been shown to be involved in fatty acid β-oxidation and development, although its substrate is still unknown (Quan et al., 2013). However, by using a genetic technique, damaged or obsolete proteins have been demonstrated to be transported outside the organelles for degradation by LON2 proteases with the assistance of the peroxines PEX4, PEX6 and PEX22, involved in protein import into peroxisomes (Farmer et al., 2013). In peroxisomes from yeast, fungi and mammal cells, LON proteases have been shown to be capable of selectively degrading oxidatively damaged peroxisomal matrix proteins and may act as a key player in the coordination of ROS metabolism in peroxisomes (Kumar et al., 2014; Nordgren and Fransen, 2014). A genetic screen for suppressors of the lon2 mutant has demonstrated that the degradation of proteins from the glyoxylate cycle during the functional transition of glyoxysomes to leaf peroxisomes, requiring not only LON2 but also autophagy, is involved in this process (Farmer et al., 2013; Shibata et al., 2014).
Table 2.
Main peroxisomal proteins identified as target of post-translational modifications associated with ROS and RNS
| Protein | Carbonylation | S-nitrosylation | Nitration |
|---|---|---|---|
| Catalase | Romero-Puertas et al., 2002 | Ortega-Galisteo et al., 2012 | |
| Glutathione reductase | Romero-Puertas et al., 2002 | ||
| Mn-superoxide dismutase | Romero-Puertas et al., 2002 | ||
| Glycolate oxidase | Ortega-Galisteo et al., 2012 | Lozano-Juste et al., 2011 | |
| Hydroxypyruvate reductase | Ortega-Galisteo et al., 2012 | Corpas et al., 2013 | |
| Malate synthase | Nguyen and Donaldson, 2005 | ||
| Isocitrate lyase | Nguyen and Donaldson, 2005 | ||
| Malate dehydrogenase | Nguyen and Donaldson, 2005 | Ortega-Galisteo et al., 2012 | Lozano-Juste et al., 2011 |
| Serine-glyoxylate aminotransferase | Ortega-Galisteo et al., 2012 | ||
| Aminotransferase 1 | Ortega-Galisteo et al., 2012 | ||
| Nitrile-specifier protein 5 nitrilase 1 | Mano et al., 2014 | ||
| Nucleoside diphosphate kinase AIM1 protein | Mano et al., 2014 | ||
| Glycine-rich RNA-binding protein 8 | Mano et al., 2014 | ||
| Aldehyde dehydrogenase family 7 member B4 | Mano et al., 2014 |
Peroxisomes produce NO and RNS
In addition to ROS, NO and its derivative RNS can also be produced in peroxisomes. Nitric oxide is a gaseous free radical produced in almost all eukaryotic cells and has attracted considerable attention in the last decade (Yun et al., 2012; Chen et al., 2014). Although NO has emerged as an important plant signalling molecule, our current knowledge of the metabolic sources of NO and the mechanisms involved in NO scavenging in plants is incomplete (Mur et al., 2013). Most NO produced in animal systems is due to the enzyme NOS, and NOS2, the inducible form of this enzyme, displays a dual cytosolic–peroxisomal localization in hepatocytes (Stolz et al., 2002; Pacher et al., 2007). However, in plants, NO can be produced by both enzymatic and non-enzymatic sources (Gupta et al., 2011). A gene orthologous to NOS from animals has not been found in plants, although several attempts to identify plant NOS have been described. However, the data were not reproducible at a later stage or the protein was not directly involved in NO production (Moreau et al., 2010). Several studies, mostly based on the use of NOS inhibitors, have suggested the presence of an NOS-like activity in plants, and the presence of this activity has also been described in peroxisomes (del Río, 2011). Additionally, peroxisomes contain xanthine oxidase (XOD), which is also capable of generating NO (Godber et al., 2000; Antonenkov et al., 2010). Polyamine catabolism has been identified as another source of NO (Wimalasekera et al., 2011) in plants and, because peroxisomes contain polyamine oxidases and amine oxidases, it is possible to speculate that polyamines may contribute to NO production in peroxisomes.
The presence of NO in plant peroxisomes under physiological and stress conditions has been described (Corpas et al., 2004; del Río, 2011). Nitric oxide promotes the formation of peroxynitrite (ONOO–) and nitrosoglutathione (GSNO) in the presence of O2.– and GSH, respectively. Peroxynitrite has recently been detected in peroxisomes from arabidopsis (Corpas and Barroso, 2014), and some peroxisomal proteins, such as GOX, malate dehydrogenase (MDH) and hydroxypyruvate reductase (HPR) (Lozano-Juste et al., 2011; Corpas et al., 2013) (Table 2), have been previously described as targets of nitration. Nitrosoglutathione, considered to be a cellular reservoir of NO bioactivity, has been localized in pea leaf peroxisomes by electron microscopic immunocytochemistry (Ortega-Galisteo et al., 2012). A proteomic study has shown that six peroxisomal proteins are targets of S-nitrosylation involved in β-oxidation (MDH), photorespiration (HPR, GOX, serine-glyoxylate aminotransferase and aminotransferase 1) and the antioxidant system (catalase) (Ortega-Galisteo et al., 2012) (Table 2).
RNS are important signalling molecules that influence many aspects of cell function, including differentiation and cell survival in response to environmental cues. An increase in peroxisomal NO and RNS production has been shown under different stress conditions, such as salinity and cadmium stress (Corpas et al., 2009; Corpas and Barroso, 2014). On the other hand, NO can re-establish catalase activity, which was observed to decrease in cadmium-treated sunflowers plants (Laspina et al., 2005) and to negatively affect the catalase capacity of cadmium-treated cells (De Michele et al., 2009). Additionally, the S-nitrosylation levels of catalase and glycolate oxidase changed under abiotic stresses from cadmium and 2,4-D, suggesting that this modification may be involved in regulating peroxisomal H2O2 levels under physiological and stress conditions (Ortega-Galisteo et al., 2012).
Peroxisomal NO has emerged as a new player in auxin-induced root organogenesis. The spatially and temporally coordinated release of NO and indoleacetic acid (IAA) from peroxisomes has been demonstrated to promote lateral root formation induced by indole-3-butyric acid (Schlicht et al., 2013). It has also been shown that pollen tube growth and re-orientation in Lilium longiflorum are regulated by NO production in peroxisomes (Prado et al., 2008). Peroxisomal NO may also participate in epinasty development induced by the herbicide 2,4-D through the post-transductional modification of actin (Pazmiño et al., 2014; Rodriguez-Serrano et al., 2014). Additionally, NO has been reported to be both a pro-oxidant and an antioxidant in plants, and NO pre-incubation has been shown to delay loss of catalase activity in barley aleurone cells (Beligni et al., 2002).
Some RNS are highly unstable and can cause direct oxidative and nitrosative damage, such as lipid peroxidation (Yin et al., 2011). In this context, the presence in peroxisomes of antioxidants capable of regulating RNS, such as SOD, that can remove superoxide and thus avoid ONOO– formation is essential. Although the presence of peroxiredoxins or thioredoxins in peroxisomes, which may regulate peroxynitrite and S-nitrosothiol levels, is predictable, it still requires experimental confirmation. It is also important to mention that the regulation of NO and RNS levels is an issue that needs further study in plant cells peroxisome.
PEROXISOMES PLAY A KEY ROLE IN MAINTAINING CELLULAR REDOX HOMEOSTASIS
Peroxisomes contain many enzymes involved in producing and degrading ROS/RNS and therefore may modulate the cellular redox balance. Glutathione and ascorbic acid are major non-enzymatic cellular redox buffer systems, with S-nitrosylation emerging as a key redox-based post-translational modification (Yun et al., 2012). To enable peroxisomal membranes to function in cellular redox regulatory networks, they need to be capable of channelling small redox molecules such as GSH to the cytosol and other cell compartments. In mammalian peroxisomes, there is some evidence to show that GSH can freely diffuse from the cytosol to peroxisomes through a non-selective pore-forming peroxisomal membrane (Nordgren and Fransen, 2014), and GSNO may do so using a similar system. However, in plant peroxisomes the mechanism by which GSH is transported into and out of peroxisomes has not yet been established (Noctor et al., 2011).There is some evidence to show that changes in the redox state or accumulation of H2O2 in peroxisomes can alter the redox state in the cytosol (Fig. 2). Hydrogen peroxide produced during photorespiration in catalase-deficient arabidopsis plants (cat2) increases total GSH content, mainly in the oxidized (GSSG) form (Mhamdi et al., 2010). As this change in GSH may be involved in transmitting H2O2 signals generated in peroxisomes, it may therefore be an important component of the redox signalling network (Mhamdi et al., 2012). The accumulation of GSH in catalase-deficient plants mainly takes place in the vacuoles, chloroplasts and, to a lesser extent, the cytosol (Mhamdi et al., 2012). Transcriptomic studies carried out on cat2 mutants suggest that cytosolic components of the ASC–GSH system are upregulated (Mhamdi et al., 2010), probably in order to cope with excess H2O2 that escapes from the peroxisome to the cytosol. In animal cells, inhibition of catalase activity interferes with redox homeostasis and ROS metabolism in mitochondria and impairs cell growth and ageing (Nordgren and Fransen, 2014). Cytosolic redox changes influence the dual targeting of 6-phosphogluconolactonase 3 (PGL3) to chloroplasts and peroxisomes in arabidopsis leaves, a process requiring thioredoxin m2 (Trxm2) (Hölscher et al., 2014). Whether these redox changes are regulated by peroxisomal signals has not been demonstrated.
PEROXISOMES REGULATE CELL RESPONSES UNDER STRESS CONDITIONS AND DURING DEVELOPMENT
Considerable advances have been made in the last 10 years that underpin our current knowledge and understanding of the role of ROS and NO as intracellular and intercellular messengers with a broad spectrum of regulatory functions in many physiological processes and stress responses. By promoting oxidative and nitrosative stress, the accumulation of ROS and RNS can be deleterious, and their role in signalling networks depends on the finely tuned regulation of their synthesis and degradation.
ROS production in peroxisomes is associated with metabolic pathways such as photorespiration, ureide metabolism and fatty acid β-oxidation, with disturbances in any of these processes giving rise to changes in the accumulation of ROS. This situation could be perceived by the cell as an alarm bell triggering a whole chain of events to boost defence responses. The specific accumulation of ROS in a cell compartment produces an ROS signature capable of triggering a specific response (Mittler et al., 2011; Rosenwasser et al., 2011; Sewelam et al., 2014), although the mechanism and components involved in recognizing and transmitting the information to the nucleus are unclear. The role of H2O2 as a signalling molecule is based on the activation of transcription factors and intracellular localization of numerous signalling proteins by redox changes in cysteine or S–Fe clusters (Yun et al., 2011). However, peptides derived from the proteolytic breakdown of oxidatively damaged proteins may also act as secondary ROS messengers regulating specific genes and thus contribute to retrograde ROS signalling (Møller and Sweetlove, 2010), although this hypothesis has not yet been confirmed.
Genetic and pharmacological techniques have demonstrated that peroxisomal H2O2 can affect nuclear gene expression (Takahashi et al., 1997; Gechev et al., 2005; Vanderauwera et al., 2005; Queval et al., 2007). It is possible to speculate that peroxisomal NO also regulates gene expression. However, this is difficult to demonstrate as the peroxisomal enzyme involved in NO production has not yet been identified and no specific inhibitors of peroxisomal NO production are available.
Catalase loss-of-function mutants have proved to be an excellent tool for studying the signalling pathway dependent on peroxisomal H2O2 using transcriptomic analyses, which have identified hundreds of genes regulated by peroxisomal H2O2 associated with photorespiration (Vandenabeele et al., 2004; Vanderauwera et al., 2005; Queval et al., 2007; Inzé et al., 2012). In general, down-regulation of CAT makes cells more sensitive to oxidative stress (Mhamdi et al., 2012), with stress symptoms differing according to photoperiod and CO2 content. Arabidopsis cat2 lines show salicylic acid (SA)-dependent lesions and induction of pathogenesis-related proteins under long-day but not short-day conditions (Chaouch et al., 2010), differences that have been associated with distinct transcriptomic signatures (Queval et al., 2012). Induction of pathogen-associated processes has also been observed in catalase-deficient tobacco plants (Takahashi et al., 1997). Genetic studies using double mutants that affect catalase activity and some components of the ASC–GSH cycle in the cytosol have facilitated the study of the interplay between catalase and other antioxidants. Arabidopsis and tobacco double mutants deficient in CAT and APX are more tolerant to stress than single mutants (Rizhsky et al., 2002; Vanderauwera et al., 2011). In the arabidopsis double mutant deficient in APX1 and CAT2, a specific acclimation response was triggered involving DNA repair activation, cell cycle regulation and anti-programmed cell death mechanisms (Vanderauwera et al., 2011). Transcriptomic analyses of arabidopsis mutants deficient in GR1 and CAT2 showed some similarities in the gene expression profile, while the double mutant showed exacerbated oxidative stress (Mhamdi et al., 2010). However, it is important to note that GR1 encodes a protein targeting cytosol and peroxisomes (Kataya and Reumann, 2010). The cytosolic NADP isocitrate dehydrogenase may modulate responses in cat2, while the peroxisomal NADPH isocitrate dehydrogenase does not appear to make any significant contribution (Mhamdi and Noctor, 2015). This further confirms the important role played by cytosolic redox homeostasis in the regulation of peroxisome-dependent H2O2 signalling. Recently, a new component of the peroxisomal-dependent H2O2 network was identified by using arabidopsis double mutants cat2 and pp2a-b′c (2A protein phosphatase subunit), which demonstrates that PP2A-B’γ is a major player in controlling daylength-dependent responses to oxidative stress (Li et al., 2014). PP2A-B’γ may be involved in regulating ROS production in the plasmalemma while phytochrome A could constitute a nexus between oxidative stress, daylength, protein phosphorylation/dephosphorylation and pathogenesis responses (Li et al., 2014). The existence of a strong relationship between plasmalemma/apoplastic ROS production and peroxisomal H2O2-dependent signalling in response to pathogens has also been demonstrated in the double mutant cat2 AtrbohF (Chaouch et al., 2012). Calcium ions have emerged as an important component of ROS-dependent signalling in peroxisomes. These organelles contain an intra-organelle store of Ca2+, which can contribute to the regulation of H2O2 accumulation in peroxisomes by activating catalases (Costa et al., 2010) and may also regulate NO production in peroxisomes.
Comparison of different microarray data sets profiling the arabidopsis transcriptome under conditions that promote high levels of photorespiratory H2O2 revealed that the expression of a total of 783 transcripts changed. Most of these modifications were associated with biotic and abiotic stress responses, the largest overlap being observed with respect to heat and osmotic stress (Foyer et al., 2010; Inzé et al., 2012). Analysis of the subcellular localization of these H2O2-induced proteins points to the nucleus and cytosol as the principal targets, with some being transcription factors or related to nucleic acid binding (Inzé et al., 2012). Transcriptomic techniques have also demonstrated close interaction between peroxisomal H2O2, oxidative stress and phytohormone-dependent signalling involving ethylene, auxins, JA, SA and abscisic acid, suggesting that redox homeostasis, particularly the GSH/GSSG ratio, may modulate this relationship (Queval et al., 2007; Tognetti et al., 2012; Mhamdi et al., 2010, 2012). Recently, interaction between photorespiratory H2O2 and auxin signalling has been reported (Fig. 2). Thus, the supply of natural or synthetic IAA to arabidopsis cat2 mutants inhibits photorespiration-dependent cell death induced by growing plants under continuous light (Kerchev et al., 2015). Downregulation of xanthine dehydrogenase (XDH) in xdh arabidopsis plants prevents the epinasty phenotype induced by the auxinic herbicide 2,4-D, which suggests interaction between peroxisomal ROS and auxin signalling (Pazmiño et al., 2014; Rodríguez-Serrano et al., 2014).
The use of arabidopsis mutants deficient in GOX has facilitated the study of the contribution of H2O2 from different GOX isoforms to the regulation of cell responses to infection with Pseudomonas (Rojas et al., 2012). Hydrogen peroxide, specifically generated by hydroxy acid oxidase 2 (HAOX2) and GOX3, activated components of the SA signal transduction cascade and also appeared to regulate the JA and ethylene pathways involved in cell responses to infection, while GOX1 and GOX2 only played a secondary or indirect role in defence responses (Rojas et al., 2012). Hydrogen peroxide from GOX may constitute a second oxidative wave after 24 h of inoculation of tobacco and arabidopsis plants and could trigger a defence response different from that regulated by NADPH oxidases (Rojas et al., 2012). The increase in GOX and glyoxylate aminotransferase activities is also associated with the hypersensitive response in Cucumis melo line P1 infected with the oomycete Pseudoperonospora cubensis (Taler et al., 2004).
PEROXISOMES CAN ACT AS SENSORS OF CHANGES IN ROS LEVELS IN THE CELL
Signalling and metabolic networks have usually been studied in terms of changes in metabolites, signal molecules and gene expression without taking into account that organelles can change their dynamic properties and communicate with each other in response to their environment. Organelle movement is necessary for cell functionality, including cross-communication between organelles and for signalling (Rodríguez-Serrano et al., 2009; Suzuki et al., 2012). Peroxisomes are highly dynamic organelles, the number of which can increase under stress conditions induced by ozone, light, xenobiotics, salinity and metals in a process termed proliferation (Oksanen et al., 2003; Mitsuya et al., 2011; Sandalio et al., 2013). Peroxisome proliferation takes place through a complex process involving elongation, constriction and fission (Hu et al., 2012). One of the most challenging topics in peroxisome research is to decipher the signalling pathways governing the regulation of this process under different environmental and metabolic conditions. Some evidence suggests that proliferation of peroxisomes is governed by ROS, given that peroxisomal biogenesis genes are regulated by H2O2 in both plant and animal cells (López-Huertas et al., 2000). This has led to the suggestion that peroxisomal proliferation may be a mechanism of protection against oxidative stress. However, arabidopsis and tobacco mutants showing constitutive proliferation of peroxisomes do not show greater resistance to infection or salt treatment, which is probably due to the imbalance in redox homeostasis (Mitsuya et al., 2011; Valenzuela-Soto et al., 2011). Peroxisomal extensions, or peroxules, can be observed under oxidative stress conditions; although their precise function is unclear, it has been suggested that they are involved in the elongation of peroxisomes (Sinclair et al., 2009). The speed at which peroxisomes move can also change under different conditions. In arabidopsis plants exposed to cadmium, an increase in peroxisomal speed has been reported to be regulated by ROS and Ca2+ (Rodríguez-Serrano et al., 2009). However, the role played by these changes in the dynamics of peroxisomes has not been clearly established, with several functions having been suggested: (1) to improve H2O2 detoxification in different parts of the cell; (2) to aid signalling processes; and (3) to facilitate the import of new proteins needed for defence purposes (Rodríguez-Serrano et al., 2009). Thus, peroxisomes can act as sensors of ROS/redox changes in the cell and probably trigger a rapid and very specific response to environmental cues, although this needs to be studied in greater depth. The presence of peroxisomal ROS/RNS and redox signal receptors has not been reported in plant tissues. A recent study of mammalian cells has shown that a cytosolic protein containing valosin (VCP), belonging to the AAA class of ATPases, can sense H2O2 through redox changes in a cysteine group and interacts with PEX19 (required for peroxisome biogenesis), which regulates the import of catalase into the peroxisome (Murakami et al., 2013). In yeast and human fibroblasts, peroxin 5 (PEX5), a receptor of peroxisomal matrix proteins, has been reported to act as a redox switch regulating the import of these proteins into peroxisomes (Ma et al., 2013; Nordgren and Fransen, 2014). Identification of redox-sensitive proteins involved in peroxisomal protein import and proliferation in plants is a key element in understanding the signalling network that governs the rapid response to environmental changes and the role of ROS/RNS in this process.
In order to maintain cellular homeostasis, the proliferation of peroxisomes needs to be controlled, meaning that excess peroxisomes have to be removed. In mammals and fungi, peroxisomes can be degraded by selective autophagy (pexophagy), which requires sequestration of the organelles in autophagosome compartments and further degradation in the vacuole (Baker and Pudyal, 2014). There is growing evidence to support the idea that autophagy may also play an important role in controlling peroxisomal quality in plants through the selective degradation of oxidized peroxisomes (Shibata et al., 2014), in which peroxisomal protease LON2 plays a key role (Shibata et al., 2013; Baker and Paudyal, 2014). Various genetic studies using the arabidopsis mutant lon2, which is defective in AUTOPHAGY-RELATED (ATG) genes (atg2, atg3, atg5, atg7 and atg8), have demonstrated that autophagy is involved in peroxisomal turnover and can be induced by H2O2 and oxidative damage to peroxisomes (Shibata et al., 2013; Baker and Paudyal, 2014). Catalases have been reported to act as a possible molecular link between ROS and the promotion of autophagy-dependent cell death (Hackenberg et al., 2013). This mechanism operates independently of the degradation of peroxisome-associated proteins described previously, although, LON2 plays an important role in both cases (Baker and Paudyal, 2014). The specific peroxisomal receptor involved in the interaction with ATG8 to initiate pexophagy has not yet been identified in plants. However, in animals, LC3-II (homologous to the plant ATG8) is bound to PEX14, which points to the possibility that AtPEX14 alleles may be involved in this process (Baker and Paudyal, 2014).
CONCLUSIONS
The presence of different sources of ROS and RNS in peroxisomes, some of which are associated with important metabolic pathways, in addition to the complex battery of antioxidants present in these organelles, demonstrates the important role of peroxisomes in cellular oxidative metabolism. Although peroxisomes can be a source of stress when over-accumulation of ROS/RNS takes place, they can also act as sensors of oxidative stress induced by different stimuli and by regulating target genes involved in cell responses. In addition to ROS and RNS, peroxisomes also participate in hormone biosynthesis, which in turn may play an important role in the network regulating gene transcription in response to environmental cues (Fig. 2). Peroxisomes can thus be regarded as not merely refuse-collecting organelles, but also as a highly important decision-making platform in the cell. Future research on peroxisomes should aim to elucidate the molecular mechanisms underlying the way in which peroxisomes perceive different signals and regulate their metabolism, morphology and proliferation, as well as the role of ROS/RNS and post-translational modifications of peroxisomal proteins in these processes. Cross-talk between peroxisomes, mitochondria and chloroplasts and the ROS/RNS-dependent signalling events governing these interactions is another exciting field in cell biology that should be studied in depth in order to understand the regulation of development and cell responses to the environment.
ACKNOWLEDGEMENTS
The authors wish to apologize to those colleagues whose work was not cited in this review due to space limitations. The authors acknowledge English editing by Michael O’Shea. This work was supported by ERDF-co-financed grants from the Spanish Ministry of Science and Innovation (BIO2008-04067 and BIO2012-36742) and the Junta de Andalucía (BIO-337; EX12-BIO-296).
LITERATURE CITED
- Antonenkov UD, Gronan S, Ohlmeier S, Hiltunen JK. 2010. Peroxisomes are oxidative organelles. Antioxidant Redox Signalling 13: 525–537. [DOI] [PubMed] [Google Scholar]
- Baker A, Paudyal R. 2014. The life of the peroxisome: from birth to death. Current Opinion in Plant Biology 22: 39–47. [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiology 129: 1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RS, Hansche R, Mendel RR, Hille R. 2009. Oxidative half-reaction of Arabidopsis thaliana sulfite oxidase: generation of superoxide by a peroxisomal enzyme. Journal of Biological Chemistry 284: 35479–35484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassin-Ross G, Hu J. 2014. Systematic phenotypic screen of arabidopsis peroxisomal mutants identifies proteins involved in β-oxidation. Plant Physiology 166: 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, et al. 2010. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiology 153: 1692–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, et al. 2010. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiology 153: 1692–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G. 2012. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in arabidopsis. Plant Journal 69: 613–627. [DOI] [PubMed] [Google Scholar]
- Chen J, Vandelle E, Bellin D, Delledonne M. 2014. Detection and function of nitric oxide during the hypersensitive response in Arabidopsis thaliana: where there's a will there's a way. Nitric Oxide 43: 81–88. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB. 2014. Peroxisomal plant nitric oxide synthase (NOS) protein is imported by peroxisomal targeting signal type 2 (PTS2) in a process that depends on the cytosolic receptor PEX7 and calmodulin. FEBS Letters 588: 2049–2054. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Sandalio LM, Palma JM, Lupiáñez JA, del Río LA. 1999. Peroxisomal NADP-dependent isocitrate dehydrogenase. Characterization and activity regulation during natural senescence. Plant Physiology 121: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, et al. 2004. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiology 136: 2722–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Hayashi M, Mano S, Nishimura M, Barroso JB. 2009. Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of arabidopsis plants. Plant Physiology 151: 2083–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Leterrier M, Begara-Morales JC, et al. 2013. Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochimica et Biophysica Acta 1830: 4981–4989. [DOI] [PubMed] [Google Scholar]
- Costa A, Drago IO, Behera S, et al. 2010. H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant Journal 62: 760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. 2003. Plant peroxiredoxins. Annual Review of Plant Biology 54: 93–107. [DOI] [PubMed] [Google Scholar]
- Distefano S, Palma JM, McCarthy I, del Río LA. 1999. Proteolytic cleavage of plant proteins by peroxisomal endoproteases from senescent pea leaves. Planta 209: 308–313. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R. 2009. Enzymes activities and subcellular localization of members of the arabidopsis glutathione transferase superfamily. Journal of Experimental Botany 60: 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard MJ, Paulin A. 1990. Isozymes of superoxide dismutase in mitochondria and peroxisomes isolated from petals of carnation (Dianthus caryophyllus) during senescence. Plant Physiology 94: 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Kim S, Nam KH, et al. 2010. Identification of uricase as a potential target of plant thioredoxin: implication in the regulation of nodule development. Biochemical and Biophysical Research Communications 18: 22–26. [DOI] [PubMed] [Google Scholar]
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B. 2013. Disrupting autophagy restores peroxisome function to an arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2003. Redox sensing and signalling associated with reactive oxygen species in chloroplasts, peroxisomes and mitochondria. Physiologia Plantarum 119: 355–364. [Google Scholar]
- Foyer CH, Noctor G. 2011. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology 155: 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. 2010. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annual Review of Plant Biology 60: 455–484. [DOI] [PubMed] [Google Scholar]
- Gechev TS, Minkov IN, Hille J. 2005. Hydrogen peroxide induced cell death in arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. International Union of Biochemistry and Molecular Biology Life 57: 181–188. [DOI] [PubMed] [Google Scholar]
- Godber ELJ, Doel JJ, Sapkota PG, et al. 2000. Reduction of nitrite to nitric oxide catalysed by xanthine oxidoreductase. Journal of Biological Chemistry 275: 7757–7763. [DOI] [PubMed] [Google Scholar]
- Goyer A, Johnson TL, Olsen LJ, et al. 2004. Characterization and metabolic function of a peroxisomal sarcosine and pipecolate oxidase from arabidopsis . Journal of Biological Chemistry 279: 16947–16953. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. 2011. On the origins of nitric oxide. Trends in Plant Science 16: 160–168. [DOI] [PubMed] [Google Scholar]
- Hackenberg T, Juul T, Auzina A, et al. 2013. Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in arabidopsis. Plant Cell 25: 4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. 2007. Free radicals in biology and medicine. Oxford: Oxford University Press. [Google Scholar]
- Hölscher C, Meyer T, von Schaewen A. 2014. Dual-targeting of arabidopsis 6-phosphogluconolactonase 3 (PGL3) to chloroplasts and peroxisomes involves interaction with Trxm2 in the cytosol. Molecular Plant 7: 252–255. [DOI] [PubMed] [Google Scholar]
- Hooks MA. (2002). Molecular biology, enzymology, and physiology of β-oxidation. In: Baker A, Graham IA. eds. Plant peroxisomes. London: Kluwer Academic Publishers, 19–55. [Google Scholar]
- Hu J, Baker A, Bartel B, et al. 2012. Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AHC, Trelease RN, Moore TS., Jr 1983. Plant peroxisomes. New York: Academic Press. [Google Scholar]
- Inzé A, Vanderauwera S, Hoeberichts FA, Vandorpe M, Van Gaever T, Van Breusegem F. 2012. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant, Cell and Environment 35: 308–320. [DOI] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, del Río LA, Sevilla F. 1997. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiology 114: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada-Nobusada T, Hayashi M, Fukazawa M, Sakakibara H, Nishimura M. 2008. A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana . Plant and Cell Physiology 49: 1272–1282. [DOI] [PubMed] [Google Scholar]
- Kataya ARA, Reumann S. 2010. Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signaling and Behavior 5: 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Reumann S, Hu J. 2009. Peroxisome biogenesis and function. In: The arabidopsis book 7: e0123. doi:10.1199/tab.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev P, Mühlenbock P, Denecker J, et al. 2015. Activation of auxin signalling counteracts photorespiratory H2O2-dependent cell death. Plant, Cell and Environment 38: 253–265. [DOI] [PubMed] [Google Scholar]
- Koh S, Andre A, Edwards H, Ehrhardt D, Somerville S. 2005. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant Journal 44: 516–529. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kawałek A, van der Klei IJ. 2014. Peroxisomal quality control mechanisms. Current Opinion in Microbiology 22: 30–37. [DOI] [PubMed] [Google Scholar]
- Kuzniak E, Skłodowska M. 2005. Fungal pathogen-induced changes in the antioxidant systems of leaf peroxisomes from infected tomato plants. Planta 222: 192–200. [DOI] [PubMed] [Google Scholar]
- Laspina NV, Groppa MD, Tomaro ML, Benavides MP. 2005. Nitric oxide protects sunflower leaves against Cd-induced oxidative stress. Plant Science 169: 323–330. [Google Scholar]
- Leterrier M, Corpas FJ, Barroso JB, Sandalio LM, del Río LA. 2005. Peroxisomal monodehydroascorbate reductase genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiology 138: 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mhamdi A, Trotta A, Kangasjärvi S, Noctor G. 2014. The protein phosphatase subunit PP2A-B′c is required to suppress day length-dependent pathogenesis responses triggered by intracellular oxidative stress. New Phytologist 202: 145–160. [DOI] [PubMed] [Google Scholar]
- Linka N, Theodoulou FL. 2013. Metabolite transporters of the plant peroxisomal membrane: known and unknown. Sub-cellular Biochemistry 69: 169–194. [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Heinze M, Trelease RN. 2003. Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type arabidopsis cells. Plant Physiology 132: 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee CS, Lingard MJ, Trelease RN. 2005. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant Journal 43: 900–914. [DOI] [PubMed] [Google Scholar]
- López-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A. 2000. Stress induces peroxisome biogenesis genes. EMBO Journal 19: 6770–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-Moreno R, León J. 2011. In vivo protein tyrosine nitration in Arabidopsis thaliana. Journal of Experimental Botany 62: 3501–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Hagstrom D, Polley SG, Subramani S. 2013. Redox-regulated cargo binding and release by the peroxiosomal targeting signal receptor, Pex5. Journal of Biological Chemistry 288: 27220–27231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano J, Nagata M, Okamura S, Shiraya T, Mitsui T. 2014. Identification of oxidatively modified proteins in salt-stressed arabidopsis: a carbonyl-targeted proteomics approach. Plant and Cell Physiology 55: 1233–1244. [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Noctor G. 2015. Analysis of the roles of the arabidopsis peroxisomal isocitrate dehydrogenase in leaf metabolism and oxidative stress. Environmental and Experimental Botany 114: 22–29. [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, et al. 2010. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiology 153: 1144–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Noctor G, Baker A. 2012. Plant catalases: peroxisomal redox guardians. Archives of Biochemistry and Biophysics 525: 181–194. [DOI] [PubMed] [Google Scholar]
- De Michele R, Vurro E, Rigo C, et al. 2009. Nitric oxide is involved in cadmium-induced programmed cell death in arabidopsis suspension cultures. Plant Physiology 150: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, et al. 2011. ROS signaling: the new wave? Trends in Plant Science 16: 300–309. [DOI] [PubMed] [Google Scholar]
- McCarthy-Suárez I, Gómez M, del Río LA, Palma JM. 2011. Role of peroxisomes in the oxidative injury induced by the auxin herbicide 2,4-D in leaves of pea plants. Biologia Plantarum 55: 485–492. [Google Scholar]
- Mitsuya S, El Shami M, Sparkes IA, et al. 2011. Salt stress causes peroxisome proliferation, but inducing peroxisome proliferation does not improve NaCl tolerance in Arabidopsis thaliana. PLoS One 5: e9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Sweetlove LJ. 2010. ROS signalling-specificity is required. Trends in Plant Science 15: 370–374. [DOI] [PubMed] [Google Scholar]
- Mor A, Koh E, Weiner L, Rosenwasser S, Sibony-Benyamini H, Fluhr R. 2014. Singlet oxygen signatures are detected independent of light or chloroplasts in response to multiple stresses. Plant Physiology 165: 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Lindermayr C, Durner J, Klessig DK. 2010. NO synthesis and signaling in plants – where do we stand? Physiologia Plantarum 138: 372–383. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Hebelstrup KH, Gupta KJ. 2013. Striking a balance: does nitrate uptake and metabolism regulate both NO generation and scavenging? Frontiers in Plant Science 4: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Ichinohe Y, Koike M. 2013. VCP is an integral component of a novel feedback mechanism that controls intracellular localization of catalase and H2O2 levels. PLoS One 8: e56012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Bright J, Desikan R, Hancock J, Harrison J, Wilson I. 2008. Nitric oxide evolution and perception. Journal of Experimental Botany 59: 25–35. [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer C. 2011. Glutathione. The arabidopsis book 9: 1–32. doi:10.1199/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Donaldson RP. 2005. Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Archives in Biochemistry and Biophysics 439: 25–31. [DOI] [PubMed] [Google Scholar]
- Nordgren M, Fransen M. 2014. Peroxisomal metabolism and oxidative stress. Biochimie 98: 56–62. [DOI] [PubMed] [Google Scholar]
- Oksanen E, Haikio E, Sober J, Karnosky DF. 2003. Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytologist 161: 791–799. [DOI] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC. 2012. S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. Journal of Experimental Botany 63: 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osno Y, Kim DW, Watanabe K, et al. 2012. Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 42: 867–876. [DOI] [PubMed] [Google Scholar]
- Pacher P, Beckman JS, Liaudet L. 2007. Nitric oxide and peroxynitrite in health and disease. Physiological Review 87: 315–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazmiño MD, Rodríguez-Serrano M, Sanz M, Romero-Puertas MC, Sandalio LM. 2014. Regulation of epinasty induced by 2,4-dichlorophenoxyacetic acid in pea and arabidopsis plants. Plant Biology 16: 809–818. [DOI] [PubMed] [Google Scholar]
- Planas-Portell J, Gallart M, Tiburcio AF, Altabella T. 2013. Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana . BMC Plant Biology 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA. 2008. Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714. [DOI] [PubMed] [Google Scholar]
- Quan S, Yang P, Cassin-Ross G, et al. 2013. Proteome analysis of peroxisomes from etiolated arabidopsis seedlings identifies a peroxisomal protease involved in beta-oxidation and development. Plant Physiology 163: 1518–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, et al. 2007. Conditional oxidative stress responses in the arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant Journal 52: 640–657. [DOI] [PubMed] [Google Scholar]
- Queval G, Neukermans J, Vanderauwera S, Van Breusegem F, Noctor G. 2012. Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in arabidopsis. Plant, Cell and Environment 35: 374–387. [DOI] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, et al. 2009. In-depth proteome analysis of arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiology 150: 125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA. 2011. Peroxisomes as a cellular source of reactive nitrogen species signal molecules. Archives of Biochemistry and Biophysics 506: 1–11. [DOI] [PubMed] [Google Scholar]
- del Río LA, Pastori GM, Palma JM, et al. 1998. The activated oxygen role of peroxisomes in senescence. Plant Physiology 116: 1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Hallak-Herr E, Van Breusegem F, et al. 2002. Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant Journal 32: 329–342. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Pastori GM, et al. 2007. Peroxisomal membrane manganese superoxide dismutase: characterization of the isozyme from watermelon. Journal of Experimental Botany 58: 2417–2427. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Sparkes I, Hawes C, del Río LA, Sandalio LM. 2009. Peroxisome dynamics in arabidopsis plants under oxidative stress induced by cadmium. Free Radicals in Biology and Medicine 47: 1632–1639. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Pazmiño DM, Sparkes I, et al. 2014. 2,4-Dichlorophenoxyacetic acid promotes S-nitrosylation and oxidation of actin affecting cytoskeleton and peroxisomal Dynamics. Journal of Experimental Botany 65: 4783–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Wang K, Ryu C-M, Kaundal A, Mysore K. 2012. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during no host resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Palma JM, Gómez M, del Río LA, Sandalio LM. 2002. Cadmium causes the oxidative modification of proteins in pea plants. Plant, Cell and Environment 25: 677–686. [Google Scholar]
- Romero-Puertas MC, Rodríguez Serrano M, Corpas FJ, Gómez M, del Río LA, Sandalio LM. 2004. Cadmium-induced subcellular accumulation of O2.- and H2O2 in pea leaves. Plant, Cell and Environment 27: 1122–1134. [Google Scholar]
- Romero-Puertas MC, Corpas FJ, Sandalio LM, et al. 2006. Glutathione reductase from pea leaves: response to abiotic stress and characterization of the peroxisomal isozyme. New Phytologist 170: 43–52. [DOI] [PubMed] [Google Scholar]
- Rosenwasser S, Rot I, Sollner E, et al. 2011. Organelles contribute differentially to reactive oxygen species-related events during extended darkness. Plant Physiology 156: 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Fernández VM, Rupérez FL, del Río LA. 1988. Superoxide free radicals are produced in glyoxysomes. Plant Physiology 87: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Rodríguez-Serrano M, Romero-Puertas MC, del Río LA. 2013. Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules. Sub-cellular Biochemistry 69: 231–255. [DOI] [PubMed] [Google Scholar]
- Schlicht M, Ludwig-Müller J, Burbach C, Volkmann D, Baluska F. 2013. Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytologist 200: 473–482. [DOI] [PubMed] [Google Scholar]
- Sewelam N, Jasperta N, Van Der Kelenc K, et al. 2014. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Molecular Plant 7: 1191–1210. [DOI] [PubMed] [Google Scholar]
- Shibata M, Oikawa K, Yoshimoto K, et al. 2013. Highly oxidized peroxisomes are selectively degraded via autophagy in arabidopsis. Plant Cell 25: 4967–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Oikawa K, Yoshimoto K, et al. 2014. Plant autophagy is responsible for peroxisomal transition and plays an important role in the maintenance of peroxisomal quality. Autophagy 10: 936–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J. 2009. Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress . Plant Journal 59: 231–242. [DOI] [PubMed] [Google Scholar]
- Stolz DB, Zamora R, Vodovotz Y. 2002. Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology 36: 81–93. [DOI] [PubMed] [Google Scholar]
- Stuehr D, Pou S, Rosen GM. 2001. Oxygen reduction by nitric oxide synthases . Journal of Biological Chemistry 276: 14533–14536. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. 2012. ROS and redox signalling in the response of plants to abiotic stress. Plant, Cell and Environment 35: 259–270. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Chen Z, Du H, Liu Y, Klessig DF. 1997. Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant Journal 11: 993–1005. [DOI] [PubMed] [Google Scholar]
- Taler D, Galperin M, Benjamion I, Cohen Y, Kenigsbuch D. 2004. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16: 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Mühlenbock P, Van Breusegem F. 2012. Stress homeostasis – the redox and auxin perspective. Plant, Cell and Environment 35: 321–333. [DOI] [PubMed] [Google Scholar]
- Valenzuela-Soto JH, Iruegas-Bocardo F, Martínez-Gallardo NA, Molina-Torres J, Gómez-Lim MA, Délano-Frier JP. 2011. Transformed tobacco (Nicotiana tabacum) plants over-expressing a peroxisome proliferator-activated receptor gene from Xenopus laevis (xPPARα) show increased susceptibility to infection by virulent Pseudomonas syringae pathogens. Planta 233: 507–521. [DOI] [PubMed] [Google Scholar]
- Vanaker H, Sandalio LM, Jiménez A, et al. 2006. Roles for redox regulation in leaf senescence of pea plants grown on different sources of nitrogen nutrition. Journal of Experimental Botany 57: 1735–1746. [DOI] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, et al. 2004. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant Journal 39: 45–58. [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, et al. 2005. Genome-wide analysis of hydrogen peroxide-regulated gene expression in arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology 139: 806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Suzuki N, Miller G, et al. 2011. Extranuclear protection of chromosomal DNA from oxidative stress. Proceedings of the National Academy of Sciences of the USA 108: 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoucq L, Vignols F, Jacquot JP, Chartier Y, Meyer Y. 1999. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. Journal of Biological Chemistry 274: 19714–19722. [DOI] [PubMed] [Google Scholar]
- Waller JC, Dhanoa PK, Schumann U, Mullen RT, Snedden WA. 2010. Subcellular and tissue localization of NAD kinases from arabidopsis: compartmentalization of de novo NADP biosynthesis. Planta 231: 305–317. [DOI] [PubMed] [Google Scholar]
- Wimalasekera R, Tebartz F, Scherer GH. 2011. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Science 181: 593–603. [DOI] [PubMed] [Google Scholar]
- Yin H, Xu L, Porter NA. 2011. Free radical lipid peroxidation: mechanism and analysis. Chemical Reviews 111: 5944–5972. [DOI] [PubMed] [Google Scholar]
- Yun B-W, Spoel SH, Loake GJ. 2012. Synthesis of and signalling by small, redox active molecules in the plant immune response. Biochimica et Biophysica Acta 1820: 770–776. [DOI] [PubMed] [Google Scholar]