Abstract
Background and Aims Despite their importance in many aspects of plant physiology, information about the function of oxidative and, particularly, of nitrosative signalling in fruit biology is limited. This study examined the possible implications of O3 and sodium nitroprusside (SNP) in kiwifruit ripening, and their interacting effects. It also aimed to investigate changes in the kiwifruit proteome in response to SNP and O3 treatments, together with selected transcript analysis, as a way to enhance our understanding of the fruit ripening syndrome.
Methods Kiwifruits following harvest were pre-treated with 100 μm SNP, then cold-stored (0 °C, relative humidity 95 %) for either 2 or 6 months in the absence or in the presence of O3 (0·3 μL L–1), and subsequently were allowed to ripen at 20 °C. The ripening behaviour of fruit was characterized using several approaches: together with ethylene production, several genes, enzymes and metabolites involved in ethylene biosynthesis were analysed. Kiwifruit proteins were identified using 2-D electrophoresis coupled with nanoliquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Expression patterns of kiwifruit ripening-related genes were also analysed using real-time quantitative reverse transcription–PCR (RT–qPCR).
Key Results O3 treatment markedly delayed fruit softening and depressed the ethylene biosynthetic mechanism. Although SNP alone was relatively ineffective in regulating ripening, SNP treatment prior to O3 exposure attenuated the O3-induced ripening inhibition. Proteomic analysis revealed a considerable overlap between proteins affected by both SNP and O3. Consistent with this, the temporal dynamics in the expression of selected kiwifruit ripening-related genes were noticeably different between individual O3 and combined SNP and O3 treatments.
Conclusions This study demonstrates that O3-induced ripening inhibition could be reversed by SNP and provides insights into the interaction between oxidative and nitrosative signalling in climacteric fruit ripening.
Keywords: Actinidia deliciosa, ethylene, fruit softening, gene expression, kiwifruit, mass spectrometry, nitrosative signalling, oxidative signalling, ozone, post-harvest, proteomics, reactive nitrogen species, reactive oxygen species, RNS, ROS, ripening, sodium nitroprusside
INTRODUCTION
Fruit ripening is a sophisticatedly orchestrated developmental process, unique to plants, that is affected by various signalling pathways (Klie et al., 2014). Ethylene, a gaseous plant hormone, is one of the most important ripening-promoting factors for climacteric fruits (Yang and Hoffman, 1984). Ethylene biosynthesis is well documented; however, the interaction of ethylene with other signalling pathways is not fully elucidated (Oracz et al., 2008). For instance, we have previously shown that the post-harvest application of ozone (O3), which is a powerful oxidant compound, was able to inhibit ethylene production in kiwifruit (Minas et al., 2012). In contrast, previous studies documented that O3 exposure provoked ethylene emission and induced ethylene-related genes in leaves of different plant species (Ederli et al., 2006; Pasqualini et al., 2007, 2012; Ahlfors et al., 2009a), indicating downstream differences in ethylene metabolism when fruit and leaves are challenged with O3. Although significant progress has been made in characterizing the molecular mechanisms of O3 signalling in plants using global changes in gene and protein expression analysis (Vainonen and Kangasjärvi, 2014; Vanzo et al., 2014), targets of O3 have not been identified in fruit as yet.
Nitric oxide (NO) is an important signal molecule that is involved in various physiological processes in plants (Neill et al., 2008), including fruit ripening (Zaharah and Singh, 2011; Manjunatha et al., 2012; Singh et al., 2013). Recently, it has been reported that ripening is associated with a higher nitration of proteins and is prevented by treatment of fruits with exogenous NO (Chaki et al., 2015). It has also been shown that NO signalling is integrated with reactive oxygen species (ROS) signalling networks in plants (Lindermayr, 2015). For example, there is a robust interaction between NO and O3 signals in the foliage of various plants. Induction of alternative oxidase, AOX1a, by O3 in tobacco leaves was blocked by an NO scavenger (Ederli et al., 2006). Treatment of arabidopsis plants with the NO donor sodium nitroprusside (SNP) induced the expression of defence-related genes that significantly overlapped with the genes induced by O3 exposure. However, the combined treatment of arabidopsis with O3 and SNP led to attenuated expression of ethylene biosynthesis and defence-related genes in comparison with gene expression under O3 exposure (Ahlfors et al., 2009a, b). Although these data suggest that NO is able to modulate the O3 action in green tissues of plants, no evidence for NO and O3 interaction in fruit biology has been reported yet.
Kiwifruit (Actinidia deliciosa cv. ‘Hayward’) represent an excellent model for investigating fruit ripening physiology. Indeed, kiwifruit is harvested at the physiologically mature stage; however, it is inedible at this stage due to its high flesh firmness. The induction of kiwifruit ripening after harvest is stimulated by exposure of fruit to exogenous ethylene for some hours [or alternatively by chilling exposure (0 °C) for some days] (Antunes and Sfakiotakis, 2002). Kiwifruit can be successfully stored at 0 °C for 6 months in the absence of ethylene since very low ethylene levels induce premature ripening, thus limiting kiwifruit post-harvest life (Wills et al., 2001). In addition, kiwifruit does not produce endogenous ethylene at temperatures <10 °C (e.g. during cold storage), while it produces significant amounts of ethylene at room temperature (e.g. during ripening at 20 °C following cold storage). A recent metabolic profiling in A. deliciosa has shown that kiwifruit metabolism is different from that of other fruit species since carbon is mainly stored as starch (Nardozza et al., 2013). In addition, using oligonucleotide microarray analysis, a surprisingly large number of genes of unknown function was identified in ripe kiwifruit, testifying to the complexity of kiwifruit ripening (Atkinson et al., 2011).
On the basis of the evidence summarized above, in this study, post-harvest treatment with SNP and/or O3 was used as a tool to elicit ripening responses in kiwifruit (cv. ‘Hayward’). Thus, the purpose of this work was first to investigate the possible implications of O3 and SNP in kiwifruit ripening, and their interacting effects. This study also aimed to investigate, for the first time, the kiwifruit proteome and its changes due to SNP and O3 treatments along with selected transcript analysis, as a way to enhance our understanding of the fruit ripening syndrome.
MATERIALS AND METHODS
Fruit material and experimental design
Kiwifruit (Actinidia deliciosa ‘Hayward’) were harvested from a commercial orchard (Meliki, Northern Greece) at the physiologically mature stage [mean weight, 95 ± 5 g; tissue firmness, 6·2 ± 0·2 kg; soluble solids content (SSC) 7·5 ± 0·1 % (w/v)]. ‘Hayward’ kiwifruit was selected as the experimental material since it is the most widely planted and marketed cultivar worldwide (Ferguson and Ferguson, 2003). Fruits were randomly divided into 18 lots of 30 fruits each and the lots were sub-divided into two groups. The fruits of the first group were incubated in containers with SNP (100 μm) for 10 min in the dark, whereas fruit of the second group were incubated in water (control). Fruits were air-dried for 10 h and then cold stored (0 °C, 95 % relative humidity) in the absence or in the presence of continuously supplied gaseous O3 (0·3 μL L–1). It is noted that O3 application within the cold storage rooms was conducted according to the commercially accepted conditions for long-term kiwifruit storage (Minas et al., 2012, 2014). Thus, the following treatments were studied. Control, fruits incubated in H2O and then cold stored in the absence of O3; SNP, fruits incubated in SNP and then cold stored in the absence of O3; O3, fruits incubated in H2O and then cold stored under O3; and SNP + O3, fruits incubated in SNP and then cold stored under O3. In all cases, a Swing Therm catalytic reactor (model BS 500, Fruit Control Equipments, Milano, Italy) was used to control the ethylene concentration at the desired level (<10 nL L–1) in the cold storage rooms. Fruits were removed from cold storage after 2 or 6 months and subsequently were transferred to 20 °C for ripening. The experimental procedure is described schematically in Supplementary Data Fig. S1.
Cylindrical pieces of outer pericarp flesh samples (4·0 g), after removal of the peel, were collected from the equatorial region of each fruit, resulting in triplicate sub-lots (three samples of tissue from ten fruits) per treatment and time point at 20 °C. The first sub-lot was used for direct assessment of quality/ripening parameters; the second and third sub-lots were frozen with liquid nitrogen and stored at –80 °C for proteomic and gene expression analysis, as described below.
Fruit ripening parameters
Tissue firmness, SSC and titratable acidity (TA) were measured as described (Minas et al., 2012). Ethylene measurement was performed with a gas chromatograph (Varian Analytical Instruments, Walnut Creek, CA, USA). Respiration rate was measured by an infrared gas analyser (Combo 280, David Bishop Instruments, UK) (Minas et al., 2012). Data (means of three biological replications) were subjected to analysis of variance (ANOVA) and least significant differences (LSD) at the 5 % level for comparison of means.
Analysis of metabolites and enzyme activities of ethylene biosynthesis
The extraction procedure and determination of aminocyclopropane-1-carboxylic acid (ACC) and 1-malonyl-aminocyclopropane-l-carboxylic acid (MACC) contents as well as of the activities of ACC synthase (ACS) and ACC oxidase (ACO) were performed as described elsewhere (Bulens et al., 2011). Statistical analysis was performed as described above
Preparation of protein extracts and two-dimensional gel electrophoresis (2-DE-PAGE)
Fruit flesh was ground in liquid nitrogen and soluble proteins were extracted (Giraldo et al., 2012) and analysed by 2-DE-PAGE as described by Tanou et al. (2010). Following silver nitrate staining, 2-DE gels were scanned with a Bio-Rad GS-800 Calibrated Densitometer equipped with PDQuest Advanced 2-D Gel Analysis Software. Statistical analysis was done by one-way ANOVA significance (P < 0·05), and individual means were compared using Student’s t-test (significance level 95 %). The statistically significant differences were further combined by a quantitative 1·5-fold change of spot volume.
NanoLC-MS/MS analysis, database searching and data analysis
Gels stained with the Silver stain plus kit (Biorad) and selected spots were analysed by LTQ-Velos-Orbitrap (Thermo Fisher Scientific, Bremen, Germany) online with a nanoliquid chromatography (LC) Ultimate 3000 system (Dionex, Sunnyvale, CA, USA). For protein identification, tandem mass spectrometry (MS/MS) experiments were performed as reported (Vu Hai et al., 2013). Searches were done against the Cornell University kiwifruit protein database (http://bioinfo.bti.cornell.edu/cgi-bin/kiwi/download.cgi) containing 39 004 protein sequences using MASCOT software. Significant differences were analysed through the two-way hierarchical clustering using Permut Matrix software (Meunier et al., 2007). The row-by-row normalization of data was performed using the zero-mean and unit-standard deviation technique. Pearson’s distance and Ward’s algorithm were used for the analysis. Among the positive matches, only protein identifications based on at least two different peptide sequences of >6 amino acids with an individual score >20 were accepted (score >52 for search in NCBI nr, score >22 for search in KIWIFRUIT GENOME and score >72 when using the EST database); in some cases, the protein sequences obtained were BLASTed manually against the current databases. All peptide sequences, accession numbers, database sources, matching criteria, Mascot scores and sequence coverage are provided in Supplementary Data Table S2. When presented, identifications based on single peptide additional information are provided (Supplementary Data ‘Mascot Search Results’).
Gene expression analysis via real-time RT–qPCR
Total RNA was isolated and treated with DNase I (Invitrogen, Paisley, UK) (Chang et al., 1993). First-strand cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen). Target cDNAs were amplified using gene-specific primers (Supplementary Data Table S3) designed with Primer3web version 4.0.0. Quantitative reverse tanscription–PCRs (RT–PCRs) were performed on the Stratagene MX3005P using the KAPA SYBR FAST qPCR Kit (KAPA Biosystems). PCR cycling started with the initial polymerase activation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 58 °C for 20 s and 72 °C for 11 s. An A. deliciosa actin gene was used as a reference gene. Relative transcript levels of the gene of interest (X) were calculated as the ratio to the actin gene transcripts (A), as (1 + E)–ΔCt, where ΔCt was calculated as (CtX – CtA). The PCR efficiency (E) was calculated employing the linear regression method on the Log (Fluorescence) per cycle number data, using the LinRegPCR software (Ramakers et al., 2003).
Estimation of the nitrosative and oxidative status of kiwifruit
Immunoblot analysis of tyrosine (Tyr)-nitrated proteins and quantification of NO2-Tyr signals were determined exactly as described previously (Tanou et al., 2012). Biotin labelling of S-nitrosylated proteins and quantification was conducted according to Tanou et al. (2009). ROS (O2·− and H2O2) imaging in the outer flesh was performed using the fluorescent probes dihydroethidium (DHE) and 2',7'-dichlorofluorescein diacetate (DCF-DA) (Yang et al., 2011; Tanou et al., 2012). Fluorescent signals were visualized by a Nikon D-Eclipse C1 confocal laser-scanning microscope and quantified by IMAGEJ software. In addition, the steady-state level of O2·− and H2O2 was determined as described (Tanou et al., 2012).
RESULTS
The impact of SNP and O3 in kiwifruit ripening physiology
In previous work we documented that O3 exposure during cold storage could modulate ripening-associated changes in kiwifruit, even after kiwifruit removal from the O3-enriched atmosphere (Minas et al., 2012, 2014). Based on these data, we set up an experimental methodology based on preliminary experiments in which kiwifruit were pre-treated with SNP before applying O3 exposure. This initial approach revealed that SNP pre-treatment had the ability to exert a long-term profound impact on the O3-derived ripening effect, encouraging further study. For these reasons, in the present work, we used this combined experimental model (SNP pre-treatment and then cold storage under an O3-enriched atmosphere) in order to analyse the direct impact of these nitro-/oxy- chemical treatments on kiwifruit ripening physiology following relative short-term (2 months) and long-term (6 months) cold storage.
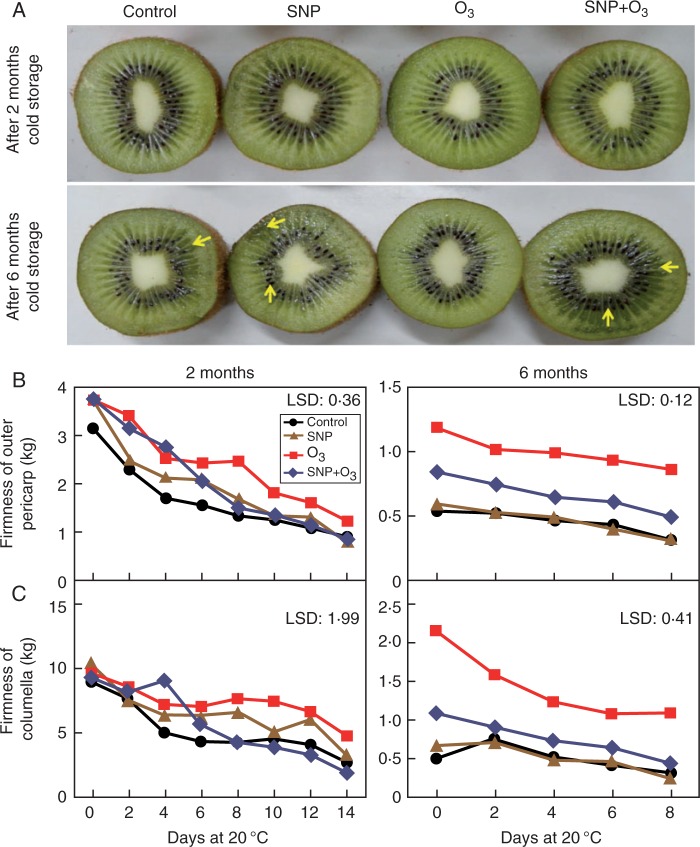
Over-ripening symptoms, expressed as visual discoloration, were observed in control and SNP-treated fruits. Exposure to O3 considerably reduced such symptoms, whereas this O3-mediated effect was remarkably reversed by SNP, especially following O3 exposure for 6 months (Fig. 1A; Supplementary Data Fig. S2). Outer pericarp firmness was decreased during fruit ripening at 20 °C after 2 months of cold storage for all treatments (Fig. 1B), indicative of the kiwifruit ripening status. However, in comparison with the control, fruit exposed to O3 or to SNP + O3 retained higher firmness until 6 or 8 d (Fig. 1D). This higher firmness retention in SNP + O3- and O3-treated fruits was even more evident following 6 months cold storage. Fruits exposed to O3 for 6 months showed higher pericarp firmness than the control during the whole ripening period. The pericarp firmness of kiwifruit exposed to SNP + O3 for 6 months was also higher compared with the control; however, it remained lower with respect to treatment with O3 alone (Fig. 1B). Following 6 months cold storage, O3-treated kiwifruit also retained higher columella tissue firmness during ripening (Fig. 1C), while fruit in the other treatments displayed the same trend as the control fruit.
Fig. 1.
Kiwifruit ripening was markedly affected by SNP and O3. Following harvest, kiwifruit (cv. ‘Hayward’) were treated with SNP (100 μm for 10 min) or water (control) and then cold stored (0 °C) in the absence or in the presence of O3 (0·3 μL L–1) for 2 or 6 months. All experimental fruit were then allowed to ripen at room temperature (20 °C). The arrow indicates over-ripening symptoms (visual discoloration). (A) Effects of SNP and O3 on the kiwifruit phenotype following 2 or 6 months cold storage plus 14 or 8 d ripening at 20 °C, respectively. (B, C) Changes of fruit firmness of the outer pericarp (B) and columella (C) in kiwifruit during ripening at 20 °C for 14 or 8 d following 2 or 6 months cold storage, respectively. Each value represents the mean of three biological replications of ten fruits analysed at each ripening stage. The least significant difference (LSD, P = 0·05) is given in each individual figure.
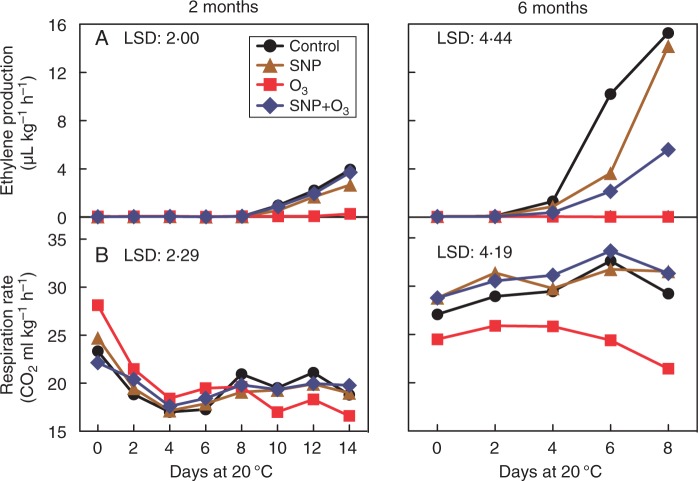
In control fruit that was cold stored for 2 or 6 months, ethylene production initiated after 10 or 4 d at 20 °C, respectively, and then exhibited a climacteric pattern (Fig. 2A). Ethylene production in fruit stored for 2 months and subsequently transferred at 20 °C was similar among control and both SNP treatments; however, the climacteric ethylene production was delayed in SNP-treated fruit after 6 months of storage. Notably, ethylene biosynthesis was almost completely suppressed by O3 treatment throughout ripening, whereas this strong effect of O3 was partially reversed by SNP (Fig. 2A). Meanwhile, O3 application for 6 months reduced the respiration rate only in the absence of SNP compared with the other treatments (Fig. 2B).
Fig. 2.
The impact of SNP and O3 on the climacteric features of kiwifruit undergoing ripening. (A) Ethylene production and (B) respiration rate in kiwifruit following 2 or 6 months cold storage plus 14 or 8 d ripening at 20 °C, respectively. The LSD (P = 0·05) is given in each individual figure (n = 5).
To provide compelling evidence of the reasons for the depression of ethylene production observed after 6 months of cold storage (Fig. 2A), we additionally determined the levels of intermediate metabolites (ACC and MACC) and enzymatic activities (ACS and ACO) of ethylene biosynthesis during kiwifruit ripening. The ACC and MACC contents in both control and SNP-treated kiwifruit were increased after 6 and 4 d of ripening respectively, and reached a peak value at 8 d, whereas SNP + O3 and particularly O3 exposure inhibited the accumulation of ethylene-related metabolites throughout ripening (Fig. 3A, B). Similarly, both ACS and ACO activities were increased in control fruits and those treated with SNP alone after 4 d and 6 d ripening, respectively, whereas this increase was greatly inhibited by O3 in the absence of SNP (Fig. 3C, D). Kiwifruit exposed to SNP + O3 exhibited lower ACS and ACO activities in comparison with control and SNP, but higher compared activities compared with O3 treatment (Fig. 3C, D).
Fig. 3.
The impact of SNP and O3 on ethylene biosynthesis in kiwifruit undergoing ripening at 20 °C following 6 months of cold storage. (A) ACC and (B) MACC content, and (C) ACS and (D) ACO enzyme activities. Each value represents the mean of three biological replications of ten fruits analysed at each ripening stage. (E) ACS and (F) ACO transcript accumulation analysed by quantitative real-time PCR. Bars represent the means (s.e.) of three biological replications. Upper and lower case letters indicate significant differences according to Duncan’s range test (P < 0·05) among treatments and ripening times, respectively.
Based on the ethylene production rate, the accumulation of ACC and MACC and the pattern of ethylene-related enzymatic activities following 6 months of cold storage (Fig. 2A and 3A–D), the time points of 0 d (onset of ripening), 4 d (initiation of ethylene production in control fruit) and 8 d (highest ethylene production level in control fruit) were chosen to analyse ACS and ACO gene expression. In control kiwifruit, the induction of ACS (Fig. 3E) and ACO (Fig. 3F) during ripening was related to increased enzymatic activities (Fig. 3C, D). In line with the enzymatic activities, both ACS and ACO transcripts were unaffected by O3 in the absence of SNP (Fig. 3E, F). Notably, ACO and ACS expression was induced in kiwifruit exposed to SNP plus O3 and was comparable with or even higher then control or SNP-alone treatment (Fig. 3E, F), indicating a differential ripening regulation by SNP in the presence of O3.
Characterization of proteins affected by SNP and O3 during kiwifruit ripening
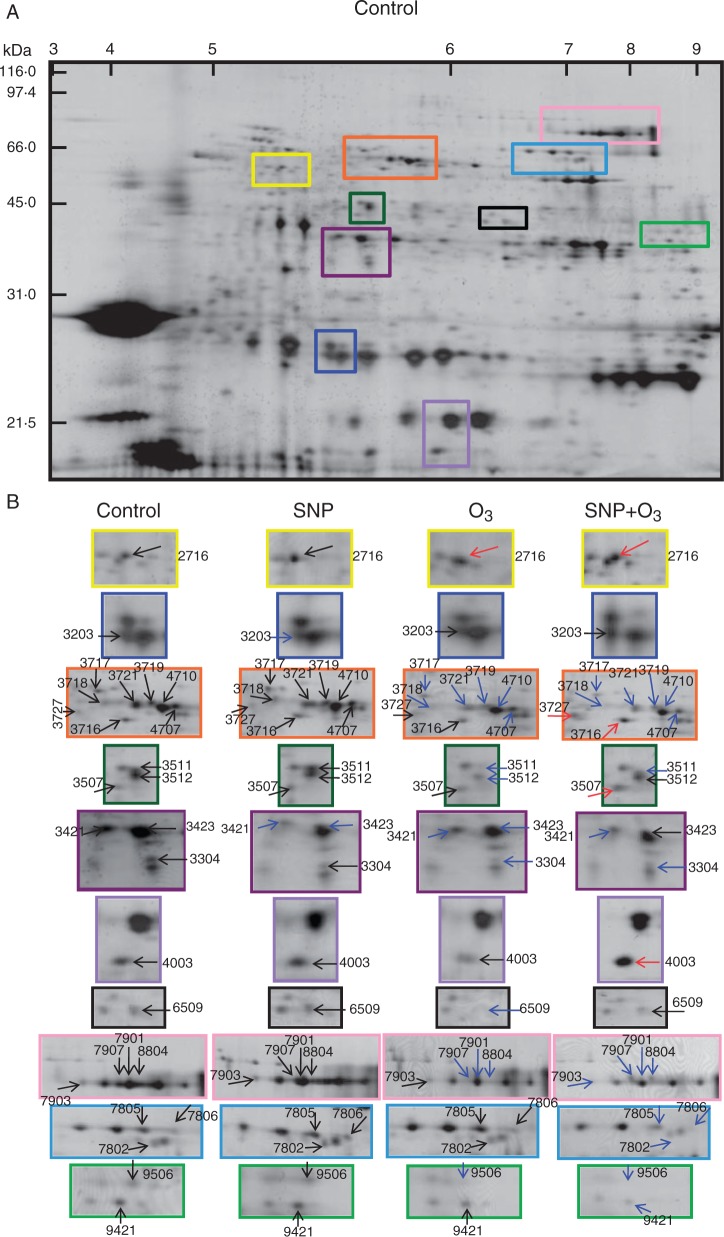
Based on the overall phenotypic and physiological data (Figs 1–3), fruit that were cold stored for 6 months and ripened for 8 d at 20 °C were selected to characterize the proteins that were affected by SNP and O3 through a proteomic approach. Kiwifruit protein identification was an analytical challenge because of the lack of complete genome sequence information for kiwifruit. The 2-DE analysis allowed the detection of 274 spots (Supplementary Data Fig. S3), 52 of which were modified under our experimental conditions based on the Student’s t-test and further validated by the 1·5-fold change threshold (Fig. 4; Supplementary Data Table S2). Following mass spectrum analysis, 48 proteins were identified that were classified into functional classes (Bevan et al., 1998). The complete information regarding protein identification is provided in Table 1 and Table S2.
Fig. 4.
Influence of SNP and O3 on the proteome of kiwifruit (‘Hayward’). Treatments with SNP and O3 were performed as shown in Fig. 1 and fruit were analysed following 6 months cold storage plus 8 d ripening at 20 °C. An equal amount (50 μg) of total protein extracts was loaded on each gel. (A) Representative silver-stained 2-DΕ map of total proteins from control kiwifruit. (B) Close-up views of selected areas of the 2-DΕ map as shown in (A) for the control and for kiwifruit exposed to SNP, O3 and SNP + O3. For each treatment, 2-DE maps were made at least in triplicate and for a minimum of three independent extractions. The numbers assigned to the proteins spots correspond to those listed in Supplementary Data Table S2. Red and blue arrows indicate protein spots that were upregulated or downregulated, respectively, in kiwifruit exposed to post-harvest treatments compared with control fruit.
Table 1.
List of 52 kiwifruit proteins identified by nanoLC-MS/MS
| Spot no.* | Protein name† | Accession no.‡ | SNP§ | Responsiveness to SNP C/U/D¶ | O3** | Responsiveness to O3 C/U/D†† | SNP + O3‡‡ | Responsiveness to SNP + O3 C/U/D§§ | Sub-cellular localization¶¶ | Functional categories*** |
|---|---|---|---|---|---|---|---|---|---|---|
| 1204 | Kiwellin | gi|85701136 | N | C | N | C | Y | D | Extracellular | 11·02-Disease/defence/Defence-related |
| 1705 | HSP70 | gi|255573627 | Y | U | N | C | N | C | Endoplasmic reticulum | 06·01-Protein destination and storage/Folding and stability |
| 1810 | HSP0 luminal binding | gi|168057678 | N | C | N | C | Y | U | Endoplasmic reticulum | 06·01-Protein destination and storage/Folding and stability |
| 2607 | Glutamate decarboxylase | gi|383745688 | Y | U | N | C | N | C | Chloroplast | 01·01-Metabolism/Amino acid |
| 2711 | Chaperonin CPN60 | gi|356534856 | Y | U | N | C | N | C | Mitochondrion | 06·01-Protein destination and storage/Folding and stability |
| 2716 | Leucine aminopeptidase | Achn369241 | N | C | Y | U | Y | U | Chloroplast | 06·13-Protein destination and storage/Proteolysis |
| 3116 | Kiwellin | gi|85701136 | N | C | Y | U | N | C | Extracellular | 11·02-Disease/defence/Defence-related |
| 3203 | Kiwellin | gi|85701136 | Y | D | N | C | N | C | Extracellular | 11·02-Disease/defence/Defence-related |
| 3207 | Aldehyde dehydrogenase | gi|57471048 | Y | D | N | C | Y | D | Mitochondrion | 11·06-Disease/Defence/Detoxification |
| 3304 | Glutelin type-A | gi|195225381 | N | C | Y | D | Y | D | Cytoplasm | 06·20-Protein destination and storage/Storage proteins |
| 3421 | Kiwellin | gi|85701136 | Y | D | Y | D | Y | D | Extracellular | 11·02-Disease/defence/Defence-related |
| 3423 | Polyphenoloxidase | gi|195241520 | Y | D | Y | D | N | C | Chloroplast | 20·1-Secondary metabolism/Phenylpropanoids/Phenolics |
| 3507 | Enolase | Achn246351 | N | C | N | C | Y | U | Plasma membrane | 02·01-Energy/Glycolysis |
| 3511 | Polygalacturonase | gi|195226843 | N | C | Y | D | Y | D | Cell Wall | 09·01-Cell structure/Cell wall |
| 3512 | Enolase | gi|14423687 | N | C | Y | D | N | C | Cytoplasm | 02·01-Energy/Glycolysis |
| 3715 | ATP-citrate synthase | Achn228301 | N | C | Y | U | N | C | Cytosol | 01·06-Metabolism/Lipid and sterol |
| 3716 | F1-ATPase alpha subunit | gi|404481694 | N | C | N | C | Y | U | Mitochondrion | 07·22-Transporters/Transport ATPases |
| 3717 | Glutelin type-A | gi|195225381 | N | C | Y | D | Y | D | Cytoplasm | 06·20-Protein destination and storage/Storage proteins |
| 3718 | Sucrose synthase | gi|398363593 | N | C | Y | D | Y | D | Cytoplasm | 01·05-Metabolism/Sugars and polysaccharides |
| 3719 | Viral A-type inclusion protein repeat containing protein expressed | gi|356510118 | N | C | Y | D | Y | D | Nucleus | 10·04-Signal transduction |
| 3721 | Viral A-type inclusion protein repeat containing protein expressed | gi|356510118 | N | C | Y | D | Y | D | Nucleus | 10·04-Signal transduction |
| 3727 | HSP70 | gi|190442576 | N | C | N | C | Y | U | Cell wall | 06·01-Protein destination and storage/Folding and stability |
| 4003 | Bet v 1 related allergen | gi|281552896 | N | C | N | C | Y | U | Cytoplasm | 11·02-Disease/defence/Defence-related |
| 4408 | Malate dehydrogenase | Achn221601 | Y | D | N | C | N | C | Mitochondrion | 02·10-Energy/TCA pathway |
| 4707 | Phosphoenolpyruvate carboxykinase | gi|195203610 | N | C | Y | D | Y | D | Cytoplasm | 02·02-Energy/Gluconeogenesis |
| 4710 | Pyruvate decarboxylase | gi|51587336 | N | C | Y | D | Y | D | Chloroplast | 11·05-Disease/Defence/Stress responses |
| 5710 | Phosphoenolpyruvate carboxykinase | Achn041831 | N | C | N | C | Y | D | Cytoplasm | 02·02-Energy/Gluconeogenesis |
| 5712 | Non-identified | N | C | N | C | Y | D | 13-Unclassified | ||
| 6207 | Kiwellin | gi|587936047 | N | C | Y | U | N | C | Extracellular | 11·02-Disease/defence/Defence-related |
| 6209 | Kiwellin | gi|85701136 | N | C | Y | U | N | C | Extracellular | 11·02-Disease/defence/Defence-related |
| 6309 | Fructose-bisphosphate aldolase | Achn044851 | N | C | N | C | Y | D | Cytoplasm | 02·01-Energy/Glycolysis |
| 6411 | Triosphosphate isomerase | gi|195201084 | N | C | N | C | Y | U | Cytosol | 02·01-Energy/Glycolysis |
| 6509 | Non-identified | N | C | Y | D | N | C | 13-Unclassified | ||
| 6510 | Non-identified | N | C | N | C | Y | D | 13-Unclassified | ||
| 7417 | Fructose-bisphosphate aldolase | gi|587847389 | N | C | N | C | Y | D | Cytoplasm | 02·01-Energy/Glycolysis |
| 7605 | Non-identified | N | C | N | C | Y | D | 13-Unclassified | ||
| 7714 | NADP-dependent malic enzyme | Achn312431 | N | C | N | C | Y | D | Chloroplast | 02·10-Energy/TCA pathway |
| 7802 | Remorin | Achn227731 | N | C | N | C | Y | D | Nucleus | 10·04-Signal transduction |
| 7805 | NADP-dependent malic enzyme | Achn312431 | N | C | N | C | Y | D | Chloroplast | 02·10-Energy/TCA pathway |
| 7806 | Remorin | Achn227731 | N | C | N | C | Y | D | Nucleus | 10·04-Signal transduction |
| 7807 | NADP-dependent malic enzyme | Achn312431 | Y | U | N | C | Y | D | Chloroplast | 02·10-Energy/TCA pathway |
| 7901 | Beta-d-galactosidase | gi|318136780 | N | C | Y | D | Y | D | Cell wall | 09·01-Cell structure/Cell wall |
| 7903 | Beta-d-galactosidase | gi|318136780 | N | C | N | C | Y | D | Cell wall | 09·01-Cell structure/Cell wall |
| 7907 | Beta-d-galactosidase | gi|318136780 | N | C | Y | D | Y | D | Cell wall | 09·01-Cell structure/Cell wall |
| 8208 | Thaumatin | gi|146737976 | Y | U | N | C | N | C | Extracellular | 11·02-Disease/defence/Defence-related |
| 8804 | Beta-d-galactosidase | gi|318136780 | N | C | Y | D | Y | D | Cell wall | 09·01-Cell structure/Cell wall |
| 9310 | Annexin | Achn387941 | N | C | N | C | Y | D | Cytoplasm | 11·05-Disease/Defence/Stress responses |
| 9316 | Annexin | Achn387941 | N | C | N | C | Y | D | Cytoplasm | 11·05-Disease/Defence/Stress responses |
| 9319 | Endochitinase | gi|195305994 | N | C | N | C | Y | D | Cell Wall | 11·02-Disease/defence/Defence-related |
| 9411 | Endochitinase | gi|195199630 | N | C | Y | D | Y | D | Extracellular | 11·02-Disease/defence/Defence-related |
| 9421 | GAPDHC | gi|166864050 | N | C | N | C | Y | D | Cytoplasm | 02·01-Energy/Glycolysis |
| 9506 | Pectinacetylesterase precursor | Achn130871 | N | C | Y | D | Y | D | Cell Wall | 09·01-Cell structure/Cell wall |
*Spot no, spot label on the reference gel maps presented in Supplementary Data Fig. S3.
†Protein name, identified peptide names.
‡Accession no., accession number in NCBI or the Kiwifruit Genome database.
§SNP, protein whose accumulation level varied under SNP treatment Y, yes; N, no.
¶Responsiveness to SNP, pattern of accumulation; C, protein whose accumulation level was constant; U, upregulated protein; D, downregulated protein.
**O3, protein whose accumulation level varied under O3 treatment Y, yes; N, no.
††Responsiveness to O3 pattern of accumulation; C, protein whose accumulation level was constant; U, upegulated protein; D, downregulated protein.
‡‡SNP + O3, protein whose accumulation level varied under SNP + O3 treatment Y, yes; N, no.
§§Responsiveness to SNP + O3 pattern of accumulation; C, protein whose accumulation level was constant; U, upregulated protein; D, downregulated protein.
¶¶Sub-cellular localization, sub-cellular localization was assigned based on database searches.
***Functional category, proteins ontologically classified into functional categories proposed by Bevan et al. (1998).
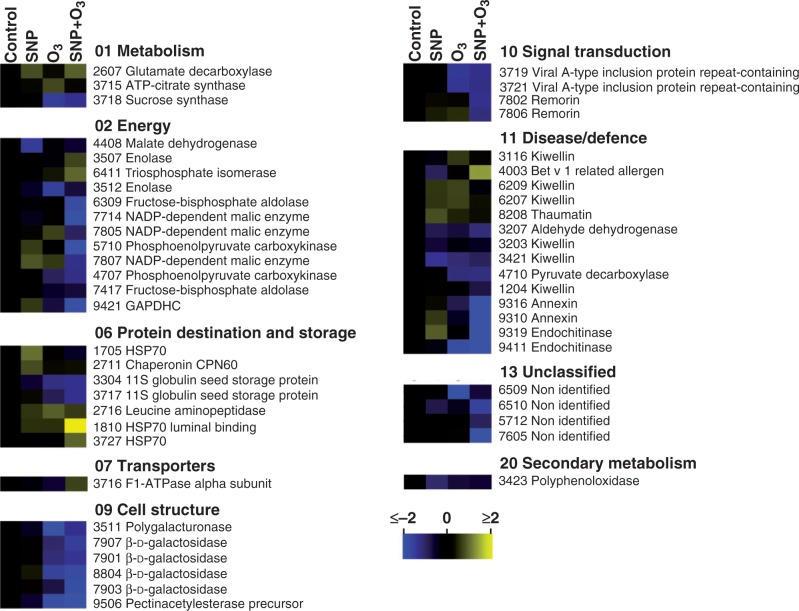
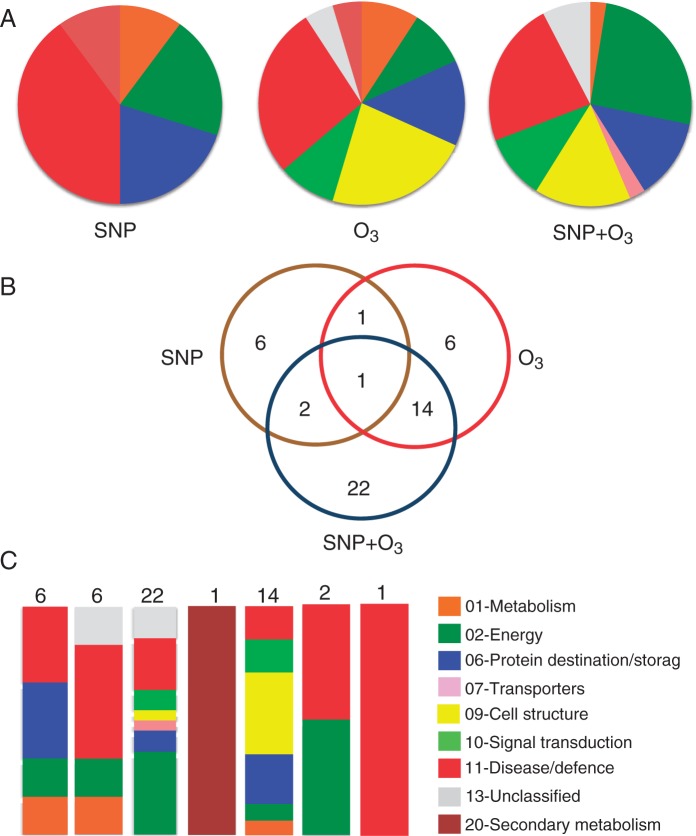
In response to post-harvest treatments, the abundance of many proteins was changed (increased or decreased) as presented by a heat map profile in Fig. 5. Compared with the control, three sets of proteins can be defined: (1) a set of ten proteins whose abundance was changed by SNP (SNP-affected proteins); (2) a set of 22 proteins whose abundance was changed by O3 (O3-affected proteins); and (3) a major set comprising 39 proteins whose abundance was altered following both SNP and O3 application (SNP + O3-affected proteins) (Fig. 6B). Functional analysis disclosed that the SNP-affected proteins were mainly associated with disease/defence (40·0 %) followed by energy (20·0 %) and protein destination/storage (20·0 %) (Fig. 6A). The largest functional category of O3-targeted proteins was involved in disease/defence (27·3 %) and cell structure (22·7 %). The SNP + O3-targeted proteins mainly participate in energy (25·6 %), disease/defence (23·1 %) and cell structure (15·4 %) (Fig. 6A).
Fig. 5.
Protein changes shown as heat map profiles in kiwifruit tissue exposed to SNP and O3. Each coloured cell represents the averaged spot quantity, according to the colour scale at the bottom of the figure. Proteins were grouped according to their known functional role as proposed (Bevan et al., 1998).
Fig. 6.
Distinct and common kiwifruit proteins targeted by SNP and O3. (A) Functional classification of all identified proteins by LC-MS/MS in kiwifruit exposed to post-harvest treatments. The colour code represents the functional classification as presented in Fig. 5 and labelled in (C). (B) Venn diagram of proteins identified in the three post-harvest treatments. The total number in each unique or overlapping set of proteins is shown. (C) Graphical representation of the functional categories for each set of unique or overlapping proteins presented in (B).
The presented Venn diagram provides information concerning the distinct and common proteins that are targeted by SNP and O3 (Fig. 6B). In the group of SNP-affected proteins, six proteins were exclusively modulated in SNP-treated fruit, one and two proteins were also modulated by O3 and SNP + O3 treatments, respectively, whereas one protein was modulated by all treatments. Amongst the 22 O3-affected proteins, six proteins were specifically changed in response to O3 treatment, while one and 14 proteins were also changed in response to either SNP or SNP + O3. Additionally, 22 proteins were exclusively affected by SNP + O3, whereas two and 14 SNP + O3-responsive proteins overlapped with the proteins modulated by SNP and O3 treatments, respectively (Fig. 6B).
The functional distribution of the unique or overlapping group of proteins presented in the Venn diagram was further analysed (Fig. 6C). Proteins associated with protein destination/storage (20·0 %) and disease/defence (20·0 %) were predominant in the group of proteins that were changed only due to SNP treatment (n = 6). Proteins that were specifically affected by O3 (n = 6) are mainly involved in disease/defence (50·0 %). Certain functional categories, including energy (36·4 %) and disease/defence (22·7 %), were represented in the group of proteins that were affected exclusively by SNP + O3 treatment (n = 22). Proteins that were commonly targeted by O3 and SNP + O3 treatments (n = 14) are involved in cell structure (35·7 %), protein destination/storage (21·4 %) and signal transduction (14·3 %). The overlap between SNP and O3 treatment was low and comprised only one protein [polyphenol oxidase (PPO)]. In addition, only one protein (kiwellin) was commonly affected by all treatments (Fig. 6C).
Expression patterns of ripening-related genes in kiwifruit exposed to SNP and O3
RT–qPCR analysis was performed to monitor the expression of transcript levels for selected kiwifruit proteins [e.g. PPO, polygalacturonase (PG), sucrose synthase (SuSy), malate dehydrogenase (MDH) and bet v 1 related allergen] that were remarkably affected by the post-harvest treatments. In addition, genes involved in ripening [e.g. 3 hydroxy-3-methylglutaryl CoA reductase (HMGR), lipoxygenase (LOX) and geranylgeranyl diphosphate synthase (GGP)] were also analysed (Zhang et al., 2006).
PPO expression decreased throughout ripening in all treatments. Between treatments, fruit exposed to SNP retained higher expression levels at 4 and 8 d compared with control treatment (Fig. 7; Supplementary Data Fig. S4). The transcript of the PG gene was induced by ripening. Notably, this induction was significantly restricted in O3-treated fruit (Fig. 7; Supplementary Data Fig. S4). SuSy expression was diminished during ripening, except for SNP +s O3 treatment (Fig. 7; Supplementary Data Fig. S4). SuSy expression was the highest in the control at 0 d; these high levels of expression were also reached in fruit treated with SNP + O3 at 8 d. Transcript levels of MDH diminished during ripening in controls and following individual SNP or O3 application; however, they displayed no significant changes in response to SNP + O3 treatment (Fig. 7; Supplementary Data Fig. S4). The expression of bet v 1 related allergen was suppressed by ripening. Interestingly, in all treated fruit, the expression of this gene remained at relatively higher levels compared with control fruit (Fig. 7; Supplementary Data Fig. S4).
Fig. 7.
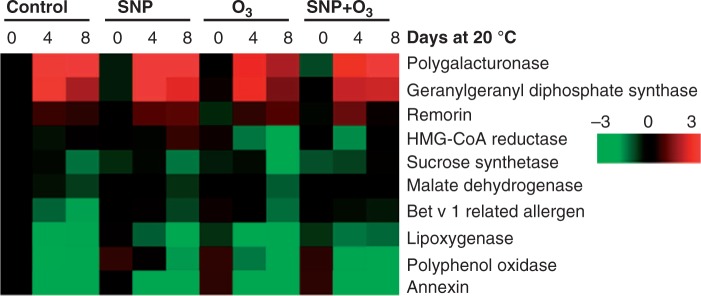
Temporal expression pattern of kiwifruit genes at different ripening stages. Heat diagram showing the expression profiles of selected genes at different ripening times following 6 months cold storage. The relative mRNA abundance was analysed by quantitative real-time PCR using three biological replications. Upregulation is indicated in red; downregulation is indicated in green. A scale of colour intensity is presented. Actual data are provided in Supplementary Data Fig. S4.
The HMGR transcript abundance was unaffected by ripening in the control, whereas it was significantly decreased by O3 treatment; however, the effect of O3 was only temporary in the presence of SNP (Fig. 7; Supplementary Fig. S4). Expression of LOX was reduced by ripening, irrespectively of the treatment applied (Fig. 7; Fig. S4). However, SNP and particularly O3 depressed LOX transcripts whereas SNP + O3 treatment stimulated LOX expression at 8 d. Furthermore, ripening induced GGP expression at 4 d in all treatments; at 8 d, GGP transcripts declined to the initial levels, except in SNP + O3 treatments, in which they were retained at higher levels (Fig. S4). Expression of remorin was induced by ripening but this effect was transient in control and SNP + O3 treatments (Fig. 7; Fig. S4). Also, transcripts of annexin were strongly down-regulated during ripening. The expression of annexin exhibited a similar pattern in control and SNP treatments, whereas O3-treated fruit displayed the lowest rate of decline in annexin expression (Fig. 7; Fig. S4).
DISCUSSION
O3-induced ripening inhibition was partially reversed by SNP
Ethylene production is the hallmark of climacteric fruit ripening (Yang and Hoffman, 1984); therefore, altering its biosynthesis could be an important means to delay the ripening process (Manjunatha et al., 2012). In the present study, untreated control fruit following transfer from low-temperature storage conditions (0 °C) to ripening temperature (20 °C) exhibited a burst in ethylene production (Fig. 2A), which is consistent with the typical climacteric behaviour of kiwifruit (Ritenour et al., 1999). The fact that ethylene production and the respiration rate during ripening at 20 °C was highly dependent on the duration of cold storage (Ritenour et al., 1999; Ilina et al., 2010) can explain the higher ethylene and respiration level observed in kiwifruit that were cold stored for 6 months compared with those stored for 2 months (Fig. 2A, B). Moreover, the patterns of ethylene biosynthesis for each ripening stage clearly suggest that O3 completely suppressed ethylene output (Fig. 2A) through inhibition of ACS expression (Fig. 3E) and ACS activity (Fig. 3C), which concomitantly reduces the ACC content (Fig. 3A). Previous findings indicated that SNP provoked ethylene emission by ripe fruits (Zaharah and Singh, 2011; Manjunatha et al., 2012). Nevertheless, the effect of SNP on ethylene biosynthesis of kiwifruit was minor compared with the effect of O3 (Figs 2 and 3), showing that the impact of SNP in ethylene production is more sophisticated than previously recognized and raises questions about the current knowledge on the autonomous action of SNP in climacteric ripening. Remarkably, ethylene production was suppressed by the co-application of SNP and O3 following a 6 month cold storage period but was not inhibited completely (Fig. 2A), indicating that O3-associated ripening responses in kiwifruit were differentially regulated by SNP. A clear example of this assumption represents the strong induction of ACS expression at the end of the ripening period in fruits exposed to SNP + O3 treatment (Fig. 3E) that was accompanied by enhanced ACS activity (Fig. 3C), ACC content (Fig. 3A) and ethylene biosynthesis (Fig. 2A) compared with fruit exposed only to O3. The current data also showed that O3 reduced the respiration rate following extended cold storage (Fig. 2B), which could be explained by the absence of climacteric ethylene production (Fig. 2A), as previously suggested (Antunes and Sfakiotakis, 2002). However, SNP application prior to O3 exposure reversed the reduction of respiration induced by O3 to the level of the control (Fig. 2B), again indicating that there is considerable interaction between SNP and O3 during ripening. In support of this assumption, kiwifruit exposed to O3 for 6 months exhibited higher firmness retention in both the outer pericarp and columella, whereas SNP pre-exposure suppressed this O3 ripening effect (Fig. 1B, C). Collectively, these data suggest that O3-induced ripening inhibition was partially reversed by SNP. Consequently, this interaction provides scientific challenges for characterizing the targets of SNP and O3 that could significantly improve our understanding of how fruit cells respond to these molecules.
Proteomic-based identification of potential regulators and pathways involved in SNP and O3 function during kiwifruit ripening
The current proteomic analysis revealed that the abundance of ten or 22 proteins was changed in response to SNP or to O3, respectively, while the abundance of 39 proteins was altered following both SNP and O3 treatments (Fig. 6B). These findings provided clear evidence for the interaction of SNP and O3 at the proteome level that could be linked to the physiological data.
Among the six proteins exclusively regulated by SNP, four were upregulated and two were downregulated (Fig. 5; Supplementary Data Table S2). Some of them (e.g. heat shock protein 70 and chaperonin CPN60) act as chaperones and their increase in abundance has been suggested to prevent accumulation of misfolded proteins (Nakamura and Lipton, 2007). The abundance of specific forms of kiwellin and thaumatin was also altered by SNP, indicating that allergenic proteins were selectively targeted by SNP. In particular, the upregulation of thaumatin by SNP could influence kiwifruit flavour since this protein is 1600 times sweeter than sucrose (Liu et al., 2010).
The current proteomic analysis further disclosed that protein repression is essential for O3-inhibited ripening since a widespread downregulation of protein abundance in response to O3 was observed (Fig. 5; Supplementary Data Table S2). Also, six kiwifruit proteins were exclusively affected by O3 (Fig. 6B) with four upregulated and two downregulated (Fig. 5; Table S2). Among them, ATP-citrate lyase that participates in the alternative citrate breakdown pathway that cleaves citrate into oxaloacetate and acetyl-CoA was identified. Thus, it is likely that citrate catabolism via this pathway is activated in O3-treated kiwifruit to control the fruit acidity. This is particularly important since, despite the ripening inhibition, O3-treated kiwifruit was considered to be in the ‘eating-ripe window’ for consumers after ripening at 20 °C based on the SSC/TA level (Supplementary Data Fig. S1). Furthermore, the observed upregulation of three isoforms of kiwellin by O3 together with the fact that several other kiwellin isoforms were downregulated in fruit exposed to all chemical treatments (Fig. 5; Supplementary Data Table S2) suggests that allergen proteins have important biological function during kiwifruit ripening.
The presented proteomic analysis further uncovered an overlap in protein expression regulated by O3 and SNP + O3 since 14 proteins were found to be sensitive to both treatments (Fig. 6B). Many of these commonly affected proteins are involved in cell wall metabolism (Supplementary Data Table S2). Indeed, among those reduced in abundance, polygalacturonase, β-d-galactosidase and pectinacetylesterase precursor were identified, consistent with the higher firmness retention in O3-treated kiwifruit (Fig. 1B, C). Recently, it has been shown that polygalacturonase activity was remarkably suppressed by O3 in kiwifruit during ripening (Minas et al., 2014), which lent support to the analysis of the presented protein characterization.
Of particular interest is the fact that out of the 22 proteins that were affected by O3, 15 proteins were also modulated in the same manner by SNP + O3 (Fig. 5; Supplementary Data Table S2). This fact along with the 22 proteins that were exclusively targeted by SNP + O3 treatment could provide an explanation for the reversing action of SNP towards O3-delayed ripening. The data indicate that the majority of enzymes involved in energy management, including glycolysis and the tricarboxylic acid (TCA) cycle, were significantly modulated in fruit exposed SNP + O3 (Fig. 5; Table S2), indicating that this treatment may alter kiwifruit energy production. In particular, the upregulation of F1-ATPase α-subunit in response to combined SNP and O3 application suggests that the interaction between the two molecules exerts a positive effect on the glycolytic pathway, inducing energy generation in the fruit cell. The regulatory role of oxy-/nitro- signals in glycolysis is also supported by the fact that the abundance of enolase, of two isoforms of fructose bisphosphate aldolase, phosphoenolpyruvate carboxykinase and of glyceraldehyde 3-phosphate dehydrogenase, which are related to glycolysis, was also changed in kiwifruit challenged with both SNP and O3 (Fig. 5; Table S2). Predominantly, the down-regulation of triose-phosphate isomerase would contribute to decreasing the 3-phosphoglyceraldehyde levels for pyruvate formation, which would be used in the TCA cycle by pyruvate. The TCA cycle maintains a cyclic flux in order to generate reducing NADH and FADH2, facilitating ATP synthesis by oxidative phosphorylation. Beyond the maintenance of a cyclic flux, the TCA cycle also provides carbon skeletons for biosynthetic pathways as well as for metabolizing organic acids generated from other pathways (Sweetlove et al., 2010). Thus, the reduced expression of kiwi fruit proteins involved in the TCA cycle, such as two isoforms of NADP-dependent malic enzyme, suggests a non-cyclic flux controlled by the influx of citrate and malate from the vacuole into the cytosol for its use in the TCA cycle.
Another important point of this analysis was the downregulation of two isoforms of annexin and remorin exclusively identified in kiwifruit treated with both chemicals (Fig. 5; Supplementary Data Table S2). Annexins are a large family of ubiquitous, Ca2+- and membrane-binding proteins that are involved in cell expansion (Konopka-Postupolska, 2007), and may be regulated by ripening, when massive structural remodelling of the cell wall takes place (Proust et al., 1996). In kiwifruit treated with both SNP and O3, annexin could serve as a signal to regulate the ripening programme or be involved in exocytosis of cell wall-degrading enzymes, acting to sequester Ca2+ released from the degrading cell wall matrix, as previously proposed in other fruit undergoing ripening (Bianco et al., 2009). On the other hand, remorins are plant-specific plasma membrane lipid raft-associated proteins that are gaining importance with respect to their role in plant–microbe interactions. The downregulation of two remorin isoforms following SNP + O3 application is of special interest because it has recently been established that remorins are stimulated by phytohormones (Checker and Khurana, 2013). There are several possible roles for remorins in ripening, ranging from provoking cell maturation to acting as scaffold proteins during signalling events (Jarsch and Ott, 2011), aspects that require further research.
Ripening-related gene expression profiling in kiwifruit challenged with SNP and O3
Transcriptional analysis revealed that the expression of only a few of the selected transcripts was induced by ripening (Fig. 7, boxed red; Supplementary Data Fig. S4), while the majority of them were downregulated at the end of the ripening period (8 d) (Fig. 7, boxed green; Fig. S4). The different transcriptional patterns observed in response to post-harvest treatments may have a profound impact on kiwifruit ripening. For example, the completely different regulation of expression of HMG, that encodes the rate-controlling enzyme of the mevalonate pathway (Aharoni et al., 2005), following individual SNP or O3 treatment could result in the channelling of mevalonic acid to specific end-products of the isoprenoid pathway, including sterols, carotenoids, phytoalexins and other specialized terpenoids and hormones (Rodriguez-Concepcion, 1999), thus globally affecting the ripening process.
The analysis of selected genes based on 2-DE-expressed proteins indicated that, along with the protein abundance changes, ripening also induced alterations in the transcripts of PPO, PG, SuSy, MDH, remorin and annexin under all post-harvest conditions (Fig. 7). Nevertheless, we found divergent patterns between transcript and protein profiles, particularly following O3 exposure, providing evidence for post-transcriptional regulation. Remarkably, the transcript corresponding to bet v 1 related allergen displayed a higher level of expression in kiwifruit exposed to both SNP and O3 (Fig. 7), whereas the putative corresponding protein spots also showed an increase in abundance in response to this treatment (Fig. 5; Supplementary Data Table S2). These observations, together with the fact that this protein has been documented to be targeted by oxy-/nitro- agents (Reinmuth-Selzle et al., 2014), suggests that bet v 1 related allergen plays a critical role in SNP and O3 interaction during ripening. An interesting finding that emerged from this work is the fact that there were considerable differences between O3 alone and the co-operative action of SNP + O3 in the majority of transcripts analysed (e.g. ACS, ACO, PG, Susy, MDH, bet v 1 related allergen, HMGR and LOX) (Fig. 7), thus supporting the interaction of SNP and O3 at the transcript level. Taken together, these data, and those of the proteomic analysis described above, suggest that SNP can modify O3-associated gene and protein expression in kiwifruit during ripening.
Possible aspect of the SNP- and O3-dependent ripening regulation
It is worth noting that SNP is a well-acknowledged NO donor, whereas it also releases CN–. In aqueous solutions, SNP in the first 30 min releases approx. 200 times more NO than CN, whereas it is fully degraded within 6 h (Arnold et al., 1984). Therefore, the ripening behaviour of SNP-treated kiwifruit may be associated with a ‘memory’ SNP function (Molassiotis et al., 2010). A typical example of SNP-associated ‘memory’ action is the enhanced acclimation of citrus plants to long-term salinity stress following pre-treatment with SNP (Tanou et al., 2009, 2012). Similarly, in this work, the short duration SNP exposure episodes might represent a ripening signal in kiwifruit, and possibly also in other fruit species, which can be perceived as a ‘memory’ imprint at the post-climacteric ripening stage. This conclusion is endorsed by the induced nitrosative status of SNP-treated kiwifruit at the end of the ripening period, as inferred by the stimulation of NO-sensitive protein post-translation modification, namely Tyr-nitration and S-nitrosylation (Supplementary Data Fig. S5). In addition, the fact that the ripening inhibition observed in O3-treated kiwifruit during the ripening period occurs under O3-free conditions indicates that O3 exerts residual effects in the fruit. This kind of ‘memory’ effect of O3 has been reported in Arabidopsis thaliana, supporting that O3 treatment, as mentioned above for SNP, could also modulate subsequent long-lasting responses (Evans et al., 2005).
However, it remains elusive how SNP can exert its ripening effect only in the presence of O3. It has been proposed that plant cell priming involves accumulation of latent signalling components that are not used until challenged by exposure to stress (Beckers and Conrath, 2007). Hence, a prior exposure to SNP may act as a priming agent capable of rendering kiwifruit more tolerant to subsequent oxidative conditions induced by O3 (Supplementary Data Fig. S6), thereby lowering the effectiveness of O3 and thus its anti-ripening activity. The current state of knowledge in defining the role of SNP in fruit ripening warrants further investigation, including the potential application of synthetic inhibitors of NO biosynthesis.
The current data suggest that kiwifruit ripening is strongly inhibited by application of O3 alone. However, when SNP and O3 are co-applied, a new ripening response is generated, suggesting that the ripening syndromes were also regulated by the interplay of these chemical molecules. Proteomic information along with transcriptomic data disclosed a number of candidate genes and proteins that were triggered by SNP and O3. Overall, this study, using kiwifruit as a ripening model, provides insight into SNP and O3 function in ripening regulation that help us to understand climacteric fruit ripening syndromes.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org amd consist of the following. Table S1: quantitative data for kiwifruit protein spot volumes on 2-DE gels. Table S2: list of kiwifruit proteins identified by nanoLC-MS/MS. Table S3: oligonucleotides used as primers for real-time RT–qPCR. Figure S1: schematic view of the experimental design. Figure S2: content of soluble solids and titratable acidity in kiwifruit during ripening. Figure S3: reference map for kiwifruit proteins. Figure S4: gene expression data in kiwifruit exposed to post-harvest treatments. Figure S5: immunoblot analysis of Tyr-nitrated proteins and biotin labelling of S-nitrosylated proteins. Figure S6: steady-state level and ROS imaging by confocal laser-scanning microscopy. ‘Mascot Search Results’: results for identifications based on single peptide sequences (protein spots 3207, 3775, 3727, 5710 and 6309).
ACKNOWLEDGEMENTS
G.T. was supported by the Greek ‘Education and Lifelong Learning’ Operational Program funded bythe EU-European Social Fund and GSRT (LS9-2703 to K.K.P.).
LITERATURE CITED
- Aharoni A, Jongsma MA, Bouwmeester HJ. 2005. Volatile science? Metabolic engineering of terpenoids in plants. Trends in Plant Science 10: 594–602. [DOI] [PubMed] [Google Scholar]
- Ahlfors R, Brosché M, Kangasjär J. 2009a. Ozone and nitric oxide interaction in Arabidopsis thaliana, a role for ethylene? Plant Signaling and Behavior 4: 878–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfors R, Brosché M, Kollist H, Kangasjärvi J. 2009b. Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. The Plant Journal 58: 1–12. [DOI] [PubMed] [Google Scholar]
- Antunes MDC, Sfakiotakis EM. 2002. Chilling induced ethylene biosynthesis in ‘Hayward’ kiwifruit following storage. Scientia Horticulturae 92: 29–39. [Google Scholar]
- Arnold WP, Longnecker DE, Epstein RM. 1984. Photodegradation of sodium nitroprusside: biologic activity and cyanide release. Anesthesiology 61: 254–260. [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Gunaseelan K, Wang MY, et al. 2011. Dissecting the role of climacteric ethylene in kiwifruit (Actinidia chinensis) ripening using a 1-aminocyclopropane-1-carboxylic acid oxidase knockdown line. Journal of Experimental Botany 62: 3821–3835. [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Conrath U. 2007. Priming for stress resistance: from the lab to the field. Current Opinion in Plant Biology 10: 425–431. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. 1998. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391: 485–488. [DOI] [PubMed] [Google Scholar]
- Bianco L, Lopez L, Scalone AG, et al. 2009. Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. Journal of Proteomics 72: 586–607. [DOI] [PubMed] [Google Scholar]
- Bulens I, Van de Poel B, Hertog ML, et al. 2011. Protocol: An updated integrated methodology for analysis of metabolites and enzyme activities of ethylene biosynthesis. Plant Methods 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Alvarez de Morales P, Ruiz C, et al. 2015. Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Annals of Botany 16: 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11: 113–116. [Google Scholar]
- Checker VG, Khurana P. 2013. Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Reports 32: 1729–1741. [DOI] [PubMed] [Google Scholar]
- Ederli L, Morettini R, Borgogni A, et al. 2006. Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiology 142: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM, Knight MR. 2005. ROS perception in Arabidopsis thaliana: the ozone-induced calcium response. The Plant Journal 41: 615–626. [DOI] [PubMed] [Google Scholar]
- Ferguson AR, Ferguson LR. 2003. Are kiwifruit really good for you? Acta Horticulturae 610: 131–138. [Google Scholar]
- Giraldo E, Díaz A, Corral JM, García A. 2012. Applicability of 2-DE to assess differences in the protein profile between cold storage and not cold storage in nectarine fruits. Journal of Proteomics 75: 5774–5782. [DOI] [PubMed] [Google Scholar]
- Ilina N, Alem HJ, Pagano EA, Sozzi GO. 2010. Suppression of ethylene perception after exposure to cooling conditions delays the progress of softening in ‘Hayward’ kiwifruit. Postharvest Biology and Technology 55: 160–168. [Google Scholar]
- Jarsch IK, Ott T. 2011. Perspectives on remorin proteins, membrane rafts, and their role during plant–microbe interactions. Molecular Plant-Microbe Interactions 24: 7–12. [DOI] [PubMed] [Google Scholar]
- Klie S, Osorio S, Tohge T, et al. 2014. Conserved changes in the dynamics of metabolic processes during fruit development and ripening across species. Plant Physiology 164: 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka-Postupolska D. 2007. Annexins: putative linkers in dynamic membrane–cytoskeleton interactions in plant cells. Protoplasma 230: 203–215. [DOI] [PubMed] [Google Scholar]
- Lindermayr C. 2015. Interplay of reactive oxygen species and nitric oxide: nitric oxide coordinates reactive oxygen species homeostasis. Plant Physiology 167: 1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Sturrock R, Ekramoddoullah AKM. 2010. The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Reports 29: 419–436. [DOI] [PubMed] [Google Scholar]
- Manjunatha G, Gupta KJ, Lokesh V, Mur LAJ, Neelwarne B. 2012. Nitric oxide counters ethylene effects on ripening fruits. Plant Signaling and Behavior 7: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier B, Dumas E, Piec I, Béchet D, Hébraud M, Hocquette J-F. 2007. Assessment of hierarchical clustering methodologies for proteomic data mining. Journal of Proteome Research 6: 358–366. [DOI] [PubMed] [Google Scholar]
- Minas IS, Tanou G, Belghazi M, et al. 2012. Physiological and proteomic approaches to address the active role of ozone in kiwifruit post-harvest ripening. Journal of Experimental Botany 63: 2449–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas IS, Vicente AR, Dhanapal AP, et al. 2014. Ozone-induced kiwifruit ripening delay is mediated by ethylene biosynthesis inhibition and cell wall dismantling regulation. Plant Science 229: 76–85. [DOI] [PubMed] [Google Scholar]
- Molassiotis A, Tanou G, Diamantidis G. 2010. NO says more than ‘YES’ to salt tolerance. Plant Signaling and Behavior 5: 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Lipton SA. 2007. Molecular mechanisms of nitrosative stress-mediated protein misfolding in neurodegenerative diseases. Cellular and Molecular Life Sciences 64: 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozza S, Boldingh HL, Osorio S, et al. 2013. Metabolic analysis of kiwifruit (Actinidia deliciosa) berries from extreme genotypes reveals hallmarks for fruit starch metabolism. Journal of Experimental Botany 64: 5049–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, et al. 2008. Nitric oxide, stomatal closure, and abiotic stress. Journal of Experimental Botany 59: 165–176. [DOI] [PubMed] [Google Scholar]
- Oracz K, El-Maarouf-Bouteau H, Bogatek R, Corbineau F, Bailly C. 2008. Release of sunflower seed dormancy by cyanide: cross-talk with ethylene signalling pathway . Journal of Experimental Botany 59: 2241–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini S, Paolocci F, Borgogni A, Morettini R, Ederli L. 2007. The overexpression of an alternative oxidase gene triggers ozone sensitivity in tobacco plants. Plant, Cell and Environment 30: 1545–1556. [DOI] [PubMed] [Google Scholar]
- Pasqualini S, Reale L, Calderini O, Pagiotti R, Ederli L. 2012. Involvement of protein kinases and calcium in the NO-signalling cascade for defence-gene induction in ozonated tobacco plants. Journal of Experimental Botany 63: 4485–4496. [DOI] [PubMed] [Google Scholar]
- Proust J, Houlné G, Schantz M-L, Schantz R. 1996. Characterization and gene expression of an annexin during fruit development in Capsicum annuum. FEBS Letters 383: 208–212. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–66. [DOI] [PubMed] [Google Scholar]
- Reinmuth-Selzle K, Ackaert C, Kampf CJ, et al. 2014. Nitration of the birch pollen allergen Bet v 1.0101: efficiency and site-selectivity of liquid and gaseous nitrating agents. Journal of Proteome Research 13: 1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritenour MA, Crisosto CH, Garner DT, Cheng GW, Zoffoli JP. 1999. Temperature, length of cold storage and maturity influence the ripening rate of ethylene-preconditioned kiwifruit. Postharvest Biology and Technology 15: 107–115. [Google Scholar]
- Rodriguez-Concepcion M. 1999. Arachidonic acid alters tomato HMG expression and fruit growth and induces 3-hydroxy-3-methylglutaryl coenzyme A reductase-independent lycopene accumulation. Plant Physiology 119: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Z, Khan AS, Zhu S, Payne AD. 2013. Nitric oxide in the regulation of fruit ripening: challenges and thrusts. Stewart Postharvest Review 9: 1–11. [Google Scholar]
- Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. 2010. Not just a circle: flux modes in the plant TCA cycle. Trends in Plant Science 15: 462–470. [DOI] [PubMed] [Google Scholar]
- Tanou G, Job C, Rajjou L, Arc E, et al. 2009. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. The Plant Journal 60: 795–804. [DOI] [PubMed] [Google Scholar]
- Tanou G, Job C, Belghazi M, Molassiotis A, Diamantidis G, Job D. 2010. Proteomic signatures uncover hydrogen peroxide and nitric oxide cross-talk signaling network in citrus plants. Journal of Proteome Research 9: 5994–6006. [DOI] [PubMed] [Google Scholar]
- Tanou G, Filippou P, Belghazi M, et al. 2012. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. The Plant Journal 72: 585–599. [DOI] [PubMed] [Google Scholar]
- Vainonen JP, Kangasjärvi J. 2014. Plant signalling in acute ozone exposure. Plant, Cell and Environment 38: 240–252. [DOI] [PubMed] [Google Scholar]
- Vanzo E, Ghirardo A, Merl-Pham J, et al. 2014. S-nitroso-proteome in poplar leaves in response to acute ozone stress. PLoS OneE 9: e106886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu Hai V, Pages F, Boulanger N, Audebert S, Parola P, Almeras L. 2013. Immunoproteomic identification of antigenic salivary biomarkers detected by Ixodes ricinus-exposed rabbit sera. Ticks and Tick-borne Diseases 4: 459–468. [DOI] [PubMed] [Google Scholar]
- Wills RBH, Warton MA, Mussa DMDN, Chew LP. 2001. Ripening of climacteric fruits initiated at low ethylene levels. Australian Journal of Experimental Agriculture 41: 89–92. [Google Scholar]
- Yang H, Wu F, Cheng J. 2011. Reduced chilling injury in cucumber by nitric oxide and the antioxidant response. Food Chemistry 127: 1237–1242. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology 35: 155–189. [Google Scholar]
- Zaharah SS, Singh Z. 2011. Mode of action of nitric oxide in inhibiting ethylene biosynthesis and fruit softening during ripening and cool storage of ‘Kensington Pride’ mango. Postharvest Biology and Technology 62: 258–266. [Google Scholar]
- Zhang B, Chen K, Bowen J, et al. 2006. Differential expression within the LOX gene family in ripening kiwifruit. Journal of Experimental Botany 57: 3825–3836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.