Abstract
Background and Aims Auxin is the main phytohormone controlling root development in plants. This study uses pharmacological and genetic approaches to examine the role of auxin and nitric oxide (NO) in the activation of NADPH-dependent thioredoxin reductase (NTR), and the effect that this activity has on root growth responses in Arabidopsis thaliana.
Methods Arabidopsis seedlings were treated with auxin with or without the NTR inhibitors auranofin (ANF) and 1-chloro-2, 4-dinitrobenzene (DNCB). NTR activity, lateral root (LR) formation and S-nitrosothiol content were measured in roots. Protein S-nitrosylation was analysed by the biotin switch method in wild-type arabidopsis and in the double mutant ntra ntrb.
Key Results The auxin-mediated induction of NTR activity is inhibited by the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (CPTIO), suggesting that NO is downstream of auxin in this regulatory pathway. The NTR inhibitors ANF and DNCB prevent auxin-mediated activation of NTR and LR formation. Moreover, ANF and DNCB also inhibit auxin-induced DR5 : : GUS and BA3 : : GUS gene expression, suggesting that the auxin signalling pathway is compromised without full NTR activity. Treatment of roots with ANF and DNCB increases total nitrosothiols (SNO) content and protein S-nitrosylation, suggesting a role of the NTR-thioredoxin (Trx)-redox system in protein denitrosylation. In agreement with these results, the level of S-nitrosylated proteins is increased in the arabidopsis double mutant ntra ntrb as compared with the wild-type.
Conclusions The results support for the idea that NTR is involved in protein denitrosylation during auxin-mediated root development. The fact that a high NO concentration induces NTR activity suggests that a feedback mechanism to control massive and unregulated protein S-nitrosylation could be operating in plant cells.
Keywords: Arabidopsis thaliana, auxin, denitrosylation, nitric oxide, NTR, reactive nitrogen species, root growth, thioredoxin reductase
INTRODUCTION
Nitric oxide (NO) is a free radical molecule, produced enzymatically and non-enzymatically in plants, which is involved in different signalling cascades of phytohormones, as well as in biotic and abiotic stress responses (Lamattina et al., 2003). One of the more important modes of action of NO is the S-nitrosylation of proteins. S-nitrosylation has emerged as a relevant post-translational modification (PTM) of proteins given its high reactivity and specificity. It occurs under physiological conditions, influencing the function and activity of a large number of proteins. S-nitrosylation consists of the incorporation of NO moieties by covalent bonding to reactive Cys residues in proteins (Lindermayr and Durner, 2009). Many attempts have been made to establish a consensus sequence of S-nitrosylation in proteins without success. Structural analysis of identified S-nitrosylated proteins showed that Cys residues prone to nitrosylation are flanked by acidic or basic residues. Moreover, a hydrophobic environment, low pKa values and solvent exposure of Cys residues are thought to favour S-nitrosylation (Marino and Gladyshev, 2010). In animals, the transnitrosylase complex shuttles NO from nitric oxide synthase (NOS) isoforms to target proteins via direct NOS–substrate interaction and sequence motif recognition (Kim et al., 2005; Jia et al., 2014). In contrast, studies of the enzymatic transnitrosylation complex have not been reported in plants.
Denitrosylation has been demonstrated to be an enzymatic and selective modification. Elucidation of the molecular mechanisms of denitrosylation in plants is important to understand how signal transduction pathways are regulated by and how plants modulate nitrosative stress. Two systems have emerged as physiologically relevant denitrosylases in plant cells. The first one is the nitrosoglutathione reductase (GSNOR) system conserved in bacteria, plants and humans (Sakamoto et al., 2002; Díaz et al., 2003). When synthesized, NO rapidly reacts with glutathione (GSH) to form nitrosoglutathione (GSNO), a potent transnitrosylating agent that can act also as a mobile reservoir of NO. GSNOR catalyses the NADPH-dependent reduction of GSNO to form glutathione sulfonamide as a major product (Jensen et al., 1998). Therefore, GSNOR indirectly should affect the level of S-nitrosylation in proteins. Arabidopsis thaliana overexpressing GSNOR displays lower levels of nitrosothiols (SNO), suggesting that regulation of GSNO levels modulates S-nitrosylation (Lee et al., 2008). GSNOR is responsive to pathogen attack, wounding, cadmium stress, and salicylic and jasmonic acids (Díaz et al., 2003; Feechan et al., 2005; Barroso et al., 2006; Rustérucci et al., 2007). The redox system formed by NADPH-dependent thioredoxin reductase (NTR) and thioredoxin (Trx), the NTR–Trx system, is an additional mechanism that controls the S-nitrosylation process. Trxs are small ubiquitous proteins with a disulfide active-site that maintains the reduction of Cys residues in target proteins (Meyer et al., 2008). Progression in the redox-based modification of Cys thiols in proteins involves SNO, sulfenic acid (SOH), disulfide (S–S), and sulfinic (SO2–) to the irreversible sulfonic acid (SO3–). Trxs are able to catalyse the denitrosylation of low-molecular-mass molecules and protein-SNO (Sengupta et al., 2007). While in animals, yeast and bacteria there are only two Trxs, plants contain several isoforms, which are classified into f, h, m, o, x, y and z types. Arabidopsis has at least 20 Trx isoforms, distributed in the cytosol (type h), chloroplast (types f, m, x, z and y) and mitochondria (type o) (Meyer et al., 2008). Furthermore, 20 other Trx-related proteins have been identified (Meyer et al., 2008). In contrast, only three genes encoding NTR have been identified in the Arabidopsis genome, termed NTRA, NTRB and NTRC (Serrato et al., 2004). NTRA and NTRB show dual localization in mitochondria and cytosol, although NTRA is the predominant isoform in cytosol and NTRB in mitochondria (Reichheld et al., 2005). NTRC, an NTR with a joint Trx domain at the C terminus, which was first reported as a chloroplast-localized enzyme (Serrato et al., 2004), is also present in plastids from non-photosynthetic tissues (Kirchsteiger et al., 2012).
As stated above, auxin is the main phytohormone involved in plant root development. Auxin promotes the production of NO in roots, which is required for auxin-mediated root organogenesis (Pagnussat et al., 2002; Correa-Aragunde et al., 2004). Moreover, participation of the NTR–Trx system and GSH in auxin signalling and transport was demonstrated using the triple arabidopsis mutant ntra ntrb cad2 (Bashandy et al., 2010). Recently, cytosolic ascorbate peroxidase1 (APX1) was shown to be denitrosylated and partially inactivated by auxin in arabidopsis roots, leading to an increase in H2O2 concentration and modulation of root branching (Correa-Aragunde et al., 2013). In this study, the participation of the NTR–Trx system in auxin-mediated root development and its participation in denitrosylating activity were studied.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana Columbia (Col-0) ecotype seeds were surface-sterilized in 30 % bleach and 0·02 % Triton-X100 for 15 min, rinsed three times with sterile water and stratified at 4 °C for 2 d in the dark. The Arabidopsis thaliana mutant ntra ntrb was generated in the Columbia ecotype. Seeds were placed on ATS medium supplemented with 1 % (w/v) sucrose and 0·8 % (w/v) agar (Wilson et al., 1990) and placed vertically in a growth chamber at 25 °C with a light intensity of 100 µmol photons m–2 s–1 and 16 : 8-h (light–dark) cycles. For root growth measurements, plants were grown for 4 d and then transferred to auxin naphtyl acetic acid (NAA) or sodium nitroprusside (SNP) provided by Sigma (St Louis, MO, USA) for another 4 d. Lateral root (LR) formation was quantified manually using magnifying glasses. For NTR activity assays, plants were grown for 7 d and then subjected to the different treatments for 1 d.
GUS staining
Histochemical assays for GUS activity were performed as described by Jefferson et al. (1987). Five days after germination BA : GUS or DR5 : GUS seedlings were treated with NAA and the NTR inhibitors auranofin (ANF) (Alexis Biochemicals, San Diego, CA, USA) or 1-chloro-2,4-dinitrobenzene (DNCB) (Fluka AG, Buchs, Switzerland) and then submerged in GUS staining buffer containing 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid in 100 mm sodium phosphate (pH 7·5). Tissues were incubated at 37 °C for 3–5 h. Images were taken under a magnifier.
Determination of S-nitrosothiol content
SNO content was measured using the Saville–Griess assay (Park and Kostka, 1997) with modifications. Briefly, Arabidopsis roots were powdered in a mortar in liquid nitrogen and then homogenized in 100 mm phosphate buffer, pH 7·4, containing 1 mm PMSF and incubated on ice for 20 min. After centrifugation (10 000 g, 15 min, 4 °C), 50 µL of supernatant (approximately 80 µg of protein) was incubated with the same volume of 1 % (w/v) sulfanilamide with or without the addition of 4 mm HgCl2 for 15 min in the dark. Then, 50 µL of 0·1 % (w/v) N-1-napthylethylenediamine dihydrochloride (NED) was added and incubated for 10 min. SNO content was measured at an absorbance of 540 nm. SNO content was calculated by using a nitrite standard curve.
NTR activity assay
NTR activity was measured with the thioredoxin reductase assay kit according to the manufacturer’s instructions (Sigma). Root protein extracts, approx. 100 µg of protein, were assayed in 100 mm phosphate buffer, pH 7·0, 10 mm EDTA and 200 µm NADPH. Reactions were initiated by the addition of 300 µm of the substrate 5,5′-dithiobis(2-nitrobenzoic) acid (DTNB; Sigma). The assay is based on the reduction of DTNB with NADPH to 5-thio-2-nitrobenzoic acid (TNB), which can be measured by absorbance at 412 nm. One mole of Trx reduces 1 mol DTNB to produce 2 mol TNB with a molar extinction coefficient of 13 600 m–1 cm–1. Thus, a molar extinction coefficient of 27 200 m–1 cm–1 was applied for quantification of consumed DTNB (Shahpiri et al., 2008). NTR activity results from total reductase activity minus activity with the addition of the inhibitor ANF. For NTR inhibition, 5 µm ANF was used and samples were incubated for 15 min before measurement of activity.
Biotin switch
Arabidopsis roots were homogenized in HEN buffer (25 mm Hepes, pH 7·7, 1 mm EDTA, 0·1 mm neocuproine) containing complete protease inhibitor cocktail (Sigma). Samples were centrifuged at 4 °C for 20 min and protein concentration in the supernatant was measured using the Bradford method. For in vitro S-nitrosylation, protein extracts were incubated with 500 µm of the nitrosylating agent nitrosocysteine (CysNO) in the dark for 30 min with frequent vortexing. Protein extracts were incubated with 30 mm methyl-methanethiosulfate (MMTS; Sigma) and 3·3 % SDS in HEN buffer at 50 °C for 20 min to block free Cys. Proteins were precipitated with 2 volumes of cold acetone and resuspended in HEN buffer with 1 % SDS. After the addition of 20 mm ascorbic acid and 1 mm biotin-HPDP (Pierce Chemical), the mixture was incubated for 1 h at room temperature in the dark. Proteins were then subjected to immunoblot analysis using an anti-biotin antibody.
RESULTS
NO and auxin induce NTR activity in roots
To test the effect of auxin and NO on NTR activity in roots, 7-d-old arabidopsis plants were treated with the NO donor SNP or with the synthetic auxin NAA for 1 d. Total reductase activity was measured in arabidopsis root extracts using DTNB as substrate. Specific NTR activity was calculated from the difference between total reductase activities and the activity in the presence of the NTR inhibitor ANF, as previously used for measurement of NTR activity in plants (Alkhalfioui et al., 2008; Shahpiri et al., 2008; Smiri et al., 2013). The inhibitory effect of ANF on root NTR activity was previously analysed in arabidopsis. ANF inhibits almost 50 % of total reductase activity in roots (Supplementary Data Figure S1). Figure 1A and B show that auxin and NO induce NTR activity in a dose-dependent manner. NTR activity increases two-fold in roots treated with 10 µm NAA withrespect to untreated roots (Fig. 1A). Western blot analysis showed that NTR protein levels in roots were not significantly altered by the NAA treatment (Fig. S2). Treatment with 10 µm of the NO donor SNP triggers, in turn, a stronger response, reaching up to seven-fold induction of NTR activity in roots (Fig. 1B). The effect of SNP is due to the release of NO as the NO scavenger 2 -(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-l-oxyl-3-oxide (CPTIO) abolished the induction of NTR activity by SNP (Fig. 1C). CPTIO was also able to repress the NAA-promoted induction of NTR activity, suggesting that NO is required for the auxin-mediated induction of NTR activity (Fig. 1C).
Fig. 1.
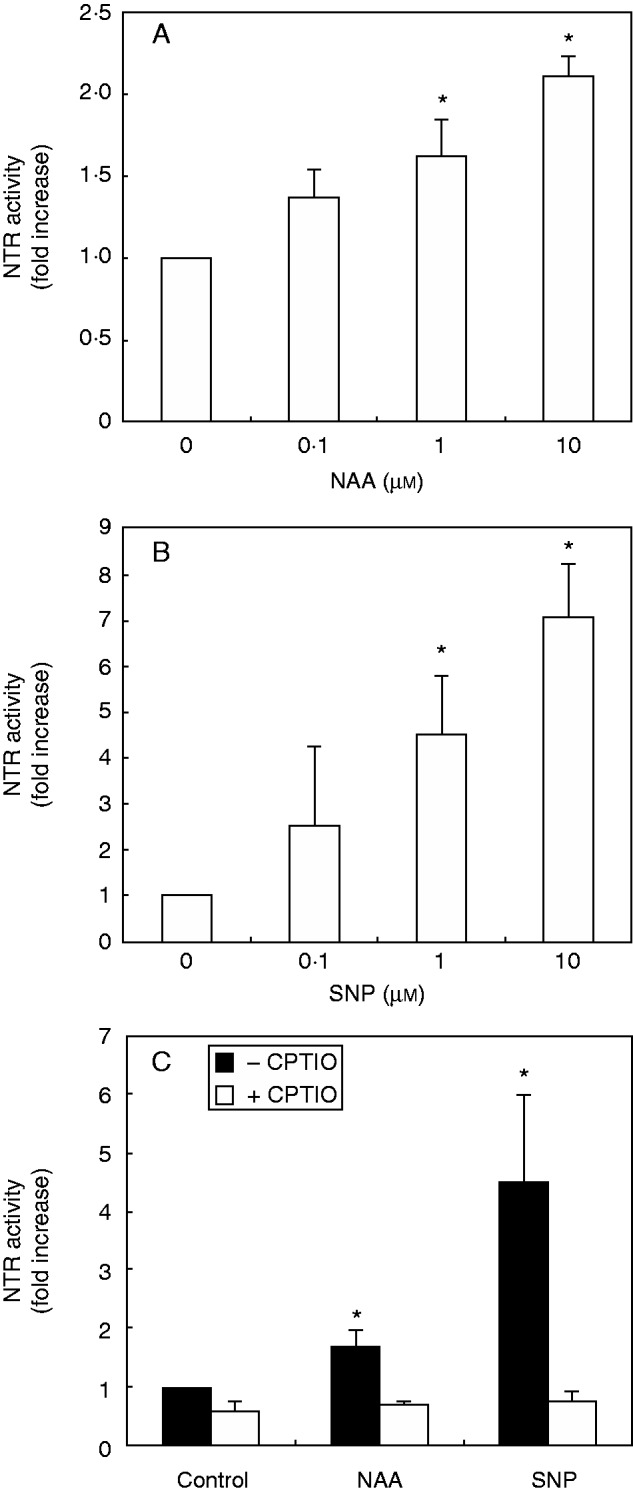
NADPH thioredoxin reductase (NTR) activity is induced by auxin and NO in arabidopsis roots. Arabidopsis plants were grown in ATS-agar for 7 d and then transferred to plates containing ATS-agar with different concentrations of the auxin NAA (A) or the NO donor SNP (B) for an additional day. NTR activity was measured in root extracts using the substrate DTNB in the absence or presence of 5 µm of the NTR inhibitor ANF. (C) NTR activity in roots treated or not (control) with 1 µm NAA or 1 µm SNP in the absence or presence of 0·5 mm CPTIO. Values are the difference between the total activity minus the activity in the presence of 5 µm ANF and expressed as fold increase with respect to control. The actual value of NTR activity in control root extracts without ANF is 6·92 ± 1·03 µm DTNB consumed min–1 mg–1. Asterisks indicate a statistical difference with respect to control (t-test, P > 0·05).
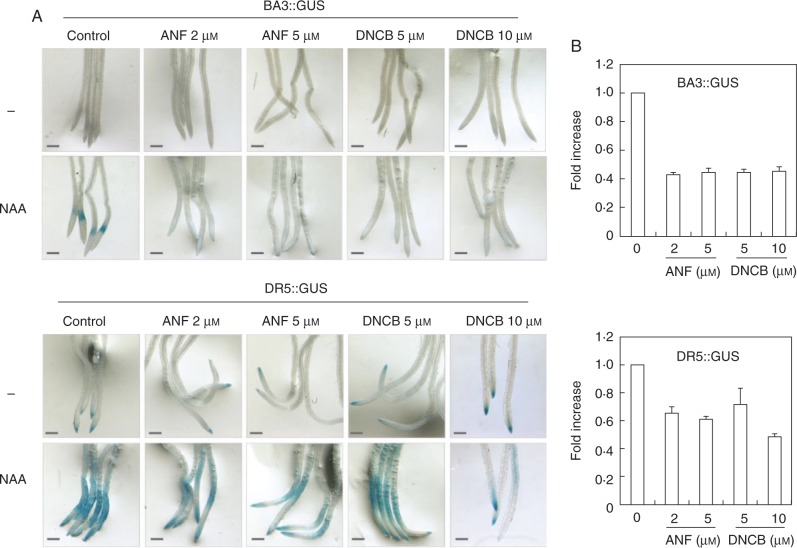
With the aim of dissecting the connection between auxin, NO and cellular reductases during root branching, arabidopsis seedlings were incubated with NAA or SNP in the presence or absence of the NTR inhibitors ANF and DNCB. In mammalian systems, ANF and DNCB have been widely used as inhibitors of NTR (Ishikawa et al., 1999; Rigobello et al., 2004). Figure 2A shows the induction of LR formation by NAA treatment, as was extensively reported elsewhere (Bhalerao et al., 2002; Marchant et al., 2002). Both ANF and DNCB were able to block, in a dose-dependent manner, the effect of NAA on LR formation (Fig. 2). We further examined the effect of NTR inhibitors on auxin signalling through analysis of the arabidopsis auxin responsive reporter lines BA3 : GUS and DR5 : GUS. Figure 3 shows that, as reported, NAA induces the expression of β-glucuronidase (GUS) under the auxin response promoters BA3 or DR5. However, NAA-induced GUS expression was affected in the presence of either of the NTR inhibitors ANF or DNCB (Fig. 3). Together, these results suggest strongly for the participation of NTR activity in auxin signalling during LR development in arabidopsis.
Fig. 2.
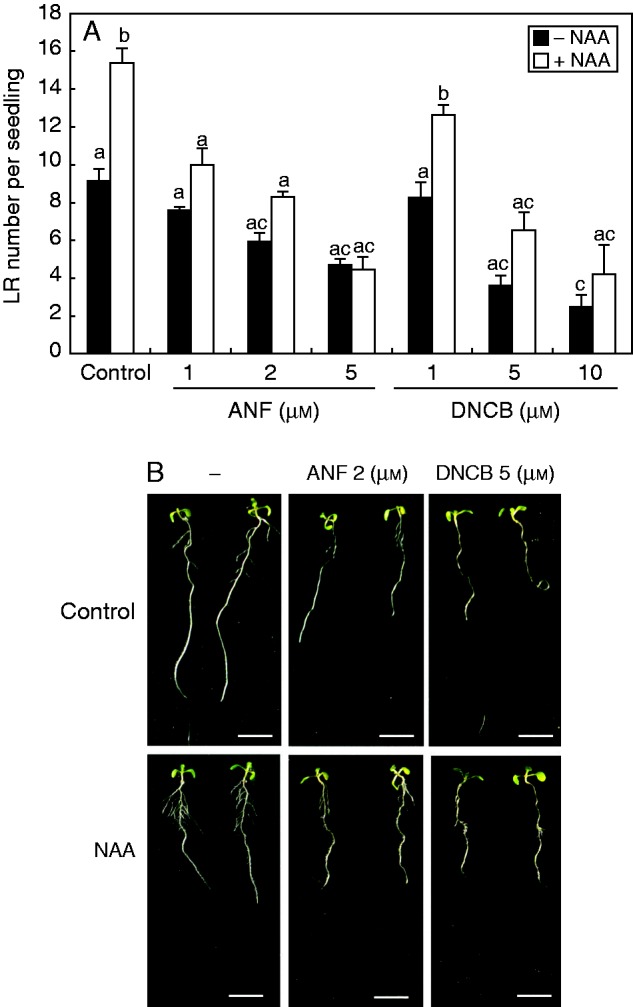
NTR activity inhibition provokes impairment of lateral root (LR) formation. (A) Arabidopsis plants were grown for 4 d in ATS-agar and transferred to treatments with 0·1 µm NAA with or without (control) different concentrations of ANF or DNCB, as indicated, for a further 4 d. LR number was counted with a magnifier. Values are mean and bars are standard error of five independent experiments (n = 5). (B) Photographs of seedlings after 4 d of treatment. Scale bars = 1 cm.
Fig. 3.
Inhibitors of NTR activity interfere with auxin response. Arabidopsis thaliana transgenic plants expressing the BA3 : : GUS and DR5 : : GUS genes were grown in ATS-agar for 4 d. Plants were incubated in ATS solution containing 0·5 µm NAA or not (−) and with or without (control) the NTR inhibitors either ANF or DNCB, at the indicated concentrations, for 18 h. Plants were incubated at 37 °C in the presence of the substrate X-Glu for 2 h. Pictures were taken under a magnifier. Scale bars = 2 mm. (B) Densitometric analysis of roots treated with NAA with or without the inhibitors ANF and DNCB using Image J software. Bars denote standard error of at least two independent experiments (n = 5).
NTR activity influences the level of nitrosylated proteins
Studies in animals have shown that Trx is the major denitrosylating agent and, together with NTR, controls the level of protein nitrosylation in cells (Benhar et al., 2008; Holmgren, 2008). Thus, it was hypothesized that NTR is involved in protein denitrosylation in plants and thereby that treatment with NTR inhibitors would enhance the level of nitrosylated proteins. The Griess–Saville method was used to detect S-nitrosothiol content in roots of arabidopsis plants treated with 1 µm NAA with and without ANF. Figure 4A shows that the treatment with NAA did not significantly change the SNO content in roots. The treatment with ANF, however, increased SNO content in roots in a dose-dependent manner and independently of the auxin treatment.
Fig. 4.
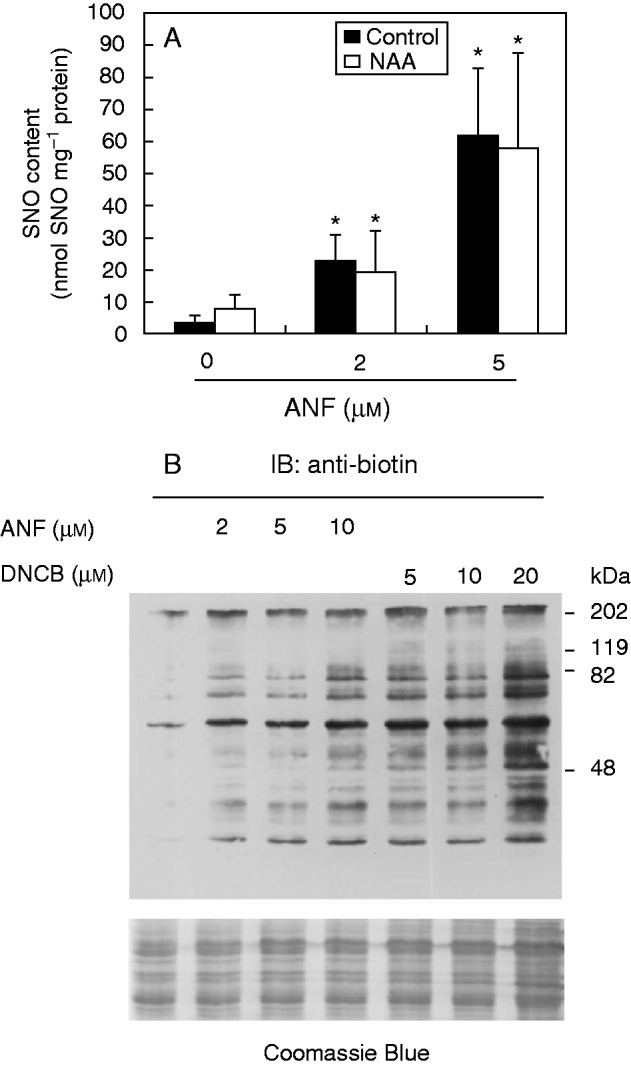
NTR inhibition produces an increase in the content of S-nitrosothiol and in vitro protein S-nitrosylation. (A) Arabidopsis plants were grown in ATS-agar for 7 d and then transferred to plates containing ATS-agar supplemented or not (control) with 1 µm NAA in the absence or presence of auranofin (ANF) at the indicated concentrations. S-nitrosothiol content in root extracts were measured by the Saville–Griess method. Bars indicate standard error (n = 3). Asterisks indicate significant differences for the ANF treatment with respect to untreated (t-test, P < 0·05). (B) Arabidopsis root extracts (60 µg) were treated with different concentrations of the NTR inhibitors ANF and DNCB for 2 h, and then in vitro S-nitrosylated with 500 µm CysNO for 30 min. S-nitrosylated proteins were detected by the biotin switch method using anti-biotin antibody. Molecular weight markers, in kDa, are indicated on the right.
The biotin switch is a three-step method that allows labelling with biotin of the NO-modified Cys residues in proteins (Jaffrey and Snyder, 2001). Arabidopsis root extracts were treated with either ANF or DNCB for 2 h and then in vitro nitrosylated with CysNO for 30 min. The nitrosylated proteins were detected by the biotin switch method followed by immunoblot with the anti-biotin antibody. Figure 4B shows that treatment with the inhibitors ANF or DNCB increases the amount of the in vitro S-nitrosylated proteins in a dose-dependent manner, supporting that NTR functions in protein denitrosylation in arabidopsis roots.
The level of S-nitrosylated proteins is impaired in the arabidopsis mutant ntra ntrb
A genetic approach with the arabidopsis ntra ntrb double mutant was used to analyse the role of NTR in the regulation of the level of S-nitrosylated proteins. The ntra ntrb plants showed a 50 % reduction of total reductase activity with respect to wild-type seedlings (Supplementary Data Fig. S3). Root extracts from wild-type and mutant seedlings were in vitro S-nitrosylated with CysNO for 30 min and the level of S-nitrosylated proteins was detected by the biotin switch method using ascorbate as a specific reductant of S-nitrosothiols. Figure 5A shows that the double mutant ntra ntrb displays an increased level of S-nitrosylated proteins with respect to the wild-type roots. The difference of signal between the wild-type and ntra ntrb mutant was not detected when dithiothreitol was used as reductant, which is able to reduce oxidized Cys (disulfides, sulfenic acid and S-nitrosothiols) (Fig. 5B). This genetic approach further supports the participation of the NTR–Trx system in modulation of the S-nitrosylation pattern in root proteins.
Fig. 5.
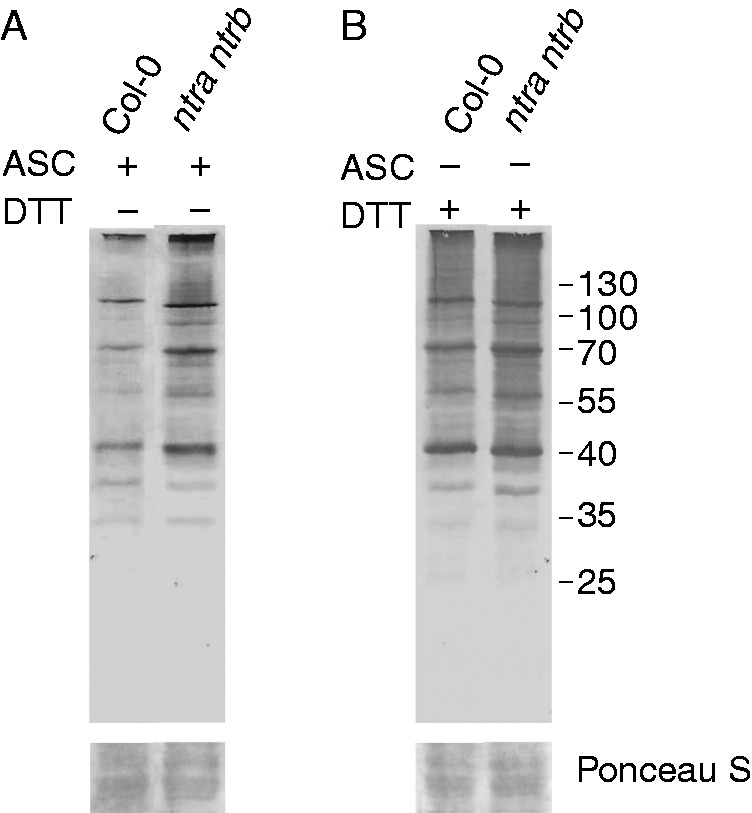
The arabidopsis mutant ntra ntrb has increased levels of S-nitrosylated proteins. Arabidopsis root extracts of wild-type or ntra ntrb mutants were in vitro S-nitrosylated with 500 µm CysNO for 30 min. (A) S-nitrosylated proteins were detected by the biotin switch method using 10 mm ascorbate (ASC) as a specific reductant of S-nitrosothiols. (B) Total oxidized Cys were detected by the biotin switch method using 20 mm dithiothreitol (DTT). Biotinylated Cys were detected by an anti-biotin antibody.
DISCUSSION
The NTR–Trx redox system in plants has been proposed to function in oxidative stress physiology, playing an antioxidant role (Vieira Dos and Rey, 2006). Indeed, in nuclei from wheat seeds cells that suffer high oxidative stress, this system supports the peroxiredoxin-dependent scavenging of H2O2 (Pulido et al., 2009). In addition, the NTR–Trx system has an important role as a modulator of plant growth and development, although this function remains poorly known. Given the complexity of the h-type Trx isoforms in plants, the role of the NTR–Trx system in growth and developmental processes was addressed by analysing NTR-deficient mutants as arabidopsis contains only two genes encoding non-plastidial NTRs, termed NTRA and NTRB (Serrato et al., 2004). Although the arabidopsis ntra ntrb mutant shows an almost wild-type phenotype, a more detailed characterization revealed decreased pollen fertility, slower plant growth and wrinkled seed phenotype (Reichheld et al., 2007). Furthermore, the GSH deficiency in the ntra ntrb mutant background resulted in an altered root meristem, indicating the coordinated action of both redox systems, GSH and NTR–Trx, in this developmental process (Reichheld et al., 2007). The triple mutant ntra ntrb cad2, which combines the deficiency of NTR with the deficiency of γ-glutamylcysteine synthetase, the first enzyme of GSH synthesis, shows loss of apical dominance and reduced LR number, processes in which auxin plays an important role. Indeed, the ntra ntrb cad2 mutant has altered auxin signalling, decreased auxin levels and impaired auxin transport capacities (Bashandy et al., 2010), thus explaining the impairment of developmental processes controlled by auxin. These results suggest participation of the NTR–Trx and GSH systems in auxin metabolism or signalling, although the redox-sensitive targets of this pathway are not known. Here we have addressed this question using both biochemical and genetic approaches. The results show that auxin treatment induces NTR activity in roots of 7-d-old arabidopsis plants. The NTR activity inhibitors ANF and DNCB blocked auxin-induced LR formation and gene expression, supporting a role of NTR in the auxin signal transduction pathway. Moreover, our results show that the NO donor SNP is also able to induce NTR activity, while depletion of endogenous NO results in blockage of the auxin-induced NTR activity, suggesting that NO is required for the auxin-mediated increase in NTR activity.
In animal cells, one of the physiological functions proposed for the NTR–Trx system is protein denitrosylation (Nikitovic and Holmgren, 1996; Benhar et al., 2008, 2010; Wu et al., 2011). Treatment with NTR inhibitors increases the level of S-nitrosylated proteins in murine macrophage cells, thus lending support to the function of the NTR–Trx system in the control of S-nitrosylation (Tello et al., 2009). In plants, participation of the NTR–Trx system in protein denitrosylation was first described in the salicylic acid (SA)-dependent defence response against pathogen attack. In response to SA, the non-expressor of pathogenesis-related 1 protein (NPR1) is denitrosylated by Trx, which favours an oligomer to monomer conversion that promotes NPR1 translocation to the nucleus (Tada et al., 2008). Kneeshaw et al. (2014) showed that cytosolic Trxh5 exhibits protein denitrosylating activity during SA-dependent plant immunity, discriminating among different SNO protein targets. In another study, the use of the NTR inhibitor DNCB in plant protoplasts resulted in an increase of NO donor-induced protein SNO content while no effect was observed with ANF (Kneeshaw et al., 2014). Here we show that the treatment of arabidopsis roots with ANF and DNCB increases SNO content and protein S-nitrosylation independently of auxin treatment. Moreover, the arabidopsis ntra ntrb mutant shows an increase of S-nitrosylated proteins in roots, as compared with the wild-type. Together, these results suggest that NTR activity is able to promote protein denitrosylation and, as a consequence, is an active component modulating the level of the S-nitrosylated proteins in plant roots. In mammalian systems, ANF was shown to inhibit NTR at nanomolar concentrations. At higher concentrations (1–50 µm) ANF inhibits also glutathione reductase (GR) enzyme (Gromer et al., 1998). Thus, although these drugs are commonly used as NTR inhibitors we cannot discard a effect on GSH metabolism or other cellular reductase activities in plants.
The auxin-dependent induction of NTR activity may promote denitrosylation/reduction of specific target proteins required for the hormone signal transduction pathway. The specific Trxs involved in this pathway remain unknown. Recent results have demonstrated that cytosolic APX1 is denitrosylated and partially inhibited by auxin treatment. Denitrosylated APX1 is less active than the S-nitrosylated form of the enzyme, and therefore denitrosylation probably contributes to the increase in H2O2 concentration in auxin-treated arabidopsis roots (Correa-Aragunde et al., 2013). Treatment of roots with ANF resulted in increased levels of S-nitrosylated APX1 and activity, suggesting that the NTR–Trx system is involved in the denitrosylation of APX1 and modulation of its activity (Correa-Aragunde et al., 2013). Accordingly, APX1 was shown to be a target of h-type Trxs (Marchand et al., 2004; Yamazaki et al., 2004; Gelhaye et al., 2006). Therefore, the NTR–Trx redox system seems to play an important function in the modulation of auxin response in plants, in agreement with previous reports (Bashandy et al., 2010), but new proteomic studies are required to identify more targets of Trx-mediated protein denitrosylation/reduction during auxin signalling, including transport and control of hormone levels.
Our results indicate that auxin and NO induce NTR activity and that this activity correlates with protein denitrosylation. Auxin promotes the increase of NO levels in roots enzymatically through nitrate reductase (NR) and NOS-like activities (Kolbert et al., 2007; Flores et al., 2008; Mendez-Bravo et al., 2010; Wang et al., 2010). Moreover, the auxin-mediated increase of NO concentration could be derived from protein denitrosylation. In animal systems, this was described as a novel cellular mechanism of NO production generated from intracellular S-nitrosothiol storage. Chvanov et al. (2006) described the rapid Ca2+-dependent NO release from S-nitrosothiols in isolated pancreatic acinar cells stimulated by acetylcholine. In addition, extracellular ATP stimulates production of NO in human monocytes from intracellular S-nitrosothiol groups rather than de novo synthesis (Hewinson et al., 2008). In plants, the critical role played by the S-nitrosothiol pool in the regulation of N assimilation in arabidopsis has been reported (Frungillo et al., 2014). Thus, S-nitrosothiols, in fact, provide a rapid source of NO available in cells, rather than de novo NO biosynthesis by the action of NOS or NR. The auxin-mediated increase of NO in roots may promote S-nitrosylation of several targets required for hormone response, such as the auxin receptor transport inhibitor response 1 (TIR1). Terrile et al. (2012) showed that TIR1 S-nitrosylation enhances TIR1–Aux/indole-3-acetic acid (IAA) interaction, facilitating Aux/IAA degradation and subsequently promoting auxin-dependent gene expression. In addition, the fact that high NO concentration induces NTR activity suggests that a feedback mechanism to control massive and unregulated protein S-nitrosylation could be operating in plant cells.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, PICT 2383 to L.L. and PICT 1099 to N.C-A.), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP112 20110100903 to L.L. and N.C-A.) and Universidad Nacional de Mar del Plata (UNMdP), Argentina.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: effect of auranofin on thioredoxin reductase activity in arabidopsis roots. Figure S2: NTR protein levels in auxin-treated arabidopsis roots. Figure S3: western blot and NTR activity in the ntra ntrb mutant.
LITERATURE CITED
- Alkhalfioui F, Renard M, Frendo P, et al. 2008. A novel type of thioredoxin dedicated to symbiosis in legumes. Plant Physiology 148: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, et al. 2006. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. Journal of Experimental Botany 57: 1785–1793. [DOI] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, et al. 2010. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. The Plant Cell 22: 376–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Hess DT, Stamler JS. 2008. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320: 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Thompson JW, Moseley MA, Stamler JS. 2010. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry 49: 6963–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. The Plant Journal 29: 325–332. [DOI] [PubMed] [Google Scholar]
- Chvanov M, Gerasimenko OV, Petersen OH, Tepikin AV. 2006. Calcium-dependent release of NO from intracellular S-nitrosothiols. EMBO Journal 25: 3024–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. 2004. Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218: 900–905. [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Foresi N, Delledonne M, Lamattina L. 2013. Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. Journal of Experimental Botany 64: 3339–3349. [DOI] [PubMed] [Google Scholar]
- Díaz M, Achkor H, Titarenko E, Martínez MC. 2003. The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Letters 543: 136–139. [DOI] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. 2005. A central role for S-nitrosothiols in plant disease resistance. Proceedings of the National Academy of Science USA 102: 8054–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores T, Todd CD, Tovar-Mendez A, et al. 2008. Arginase-negative mutants of Arabidopsis exhibit increased nitric oxide signaling in root development. Plant Physiology 147: 1936–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frungillo L, Skelly MJ, Loake GJ, Spoel SH, Salgado I. 2014. S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nature Communications 5: 5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Navrot N, Macdonald IK, Rouhier N, Raven EL, Jacquot JP. 2006. Ascorbate peroxidase-thioredoxin interaction. Photosynthesis Research 89: 193–200. [DOI] [PubMed] [Google Scholar]
- Gromer S, Arscott LD, Williams CH, Schirmer RH, Becker K. 1998. Human placenta thioredoxin reductase isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. Journal of Biological Chemistry 273: 20096–20101. [DOI] [PubMed] [Google Scholar]
- Hewinson J, Moore SF, Glover C, Watts AG, MacKenzie AB. 2008. A key role for redox signaling in rapid P2X7 receptor-induced IL-1 beta processing in human monocytes. Journal of Immunology 180: 8410–8420. [DOI] [PubMed] [Google Scholar]
- Holmgren A. 2008. Biochemistry. SNO removal. Science 320: 1019–1020. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Kubota Y, Murayama T, Nomura Y. 1999. Cell death by 1-chloro-2,4-dinitrobenzene, an inhibitor of thioredoxin reductase and its dual regulation by nitric oxide in rats. Neuroscience Letters 277: 99–102. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. 2001. The biotin switch method for the detection of S-nitrosylated proteins. Science STKE 86: pl1. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D, Belka G, Du Bois G. 1998. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochemical Journal 331: 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Arif A, Terenzi F, Willard B, Plow E, Hazen S, Fox P. 2014. Target-selective protein S-nitrosylation by sequence motif recognition. Cell 159: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SF, Huri DA, Snyder SH. 2005. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science 310: 1966–1970. [DOI] [PubMed] [Google Scholar]
- Kirchsteiger K, Ferrández J, Pascual MB, González M, Cejudo FJ. 2012. NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. The Plant Cell 24: 1534–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneeshaw S, Gelineau SR, Tada Y, Loake GJ, Spoel SH. 2014. Selective protein denitrosylation activity of thioredoxin h 5 modulates plant immunity. Molecular Cell 56: 153–162. [DOI] [PubMed] [Google Scholar]
- Kolbert Z, Bartha B, Erdei L. 2007. Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. Journal of Plant Physiology 165: 967–975. [DOI] [PubMed] [Google Scholar]
- Lamattina L, Garcia-Mata C, Graziano M, Pagnussat G. 2003. Nitric oxide: the versatility of an extensive signal molecule. Annual Review of Plant Biology 54: 109–136. [DOI] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. 2008. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. The Plant Cell 20: 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Durner J. 2009. S-Nitrosylation in plants: pattern and function. Journal of Proteomics 73: 1–9. [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, et al. 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. The Plant Cell 14: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C, Le MP, Meyer Y, Miginiac-Maslow M, Issakidis-Bourguet E, Decottignies P. 2004. New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4: 2696–2706. [DOI] [PubMed] [Google Scholar]
- Marino SM, Gladyshev VN. 2010. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. Journal of Molecular Biology 395: 844–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Bravo A, Raya-Gonzalez J, Herrera-Estrella L, Lopez-Bucio J. 2010. Nitric oxide is involved in alkamide-induced lateral root development in Arabidopsis. Plant Cell Physiology 51: 1612–1626. [DOI] [PubMed] [Google Scholar]
- Meyer Y, Siala W, Bashandy T, Riondet C, Vignols F, Reichheld JP. 2008. Glutaredoxins and thioredoxins in plants. Biochimica et Biophysica Acta 1783: 589–600. [DOI] [PubMed] [Google Scholar]
- Nikitovic D, Holmgren A. 1996. S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. Journal of Biological Chemistry 271: 19180–19185. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L. 2002. Nitric oxide is required for root organogenesis. Plant Physiology 129: 954–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Kostka P. 1997. Fluorometric detection of biological S-nitrosothiols. Analytical Biochemistry 249: 61–66. [DOI] [PubMed] [Google Scholar]
- Pulido P, Cazalis R, Cejudo FJ. 2009. An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. The Plant Journal 57: 132–145. [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Meyer E, Khafif M, Bonnard G, Meyer Y. 2005. AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Letters 579: 337–342. [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y. 2007. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. The Plant Cell 19: 1851–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigobello MP, Scutari G, Folda A, Bindoli A. 2004. Mitochondrial thioredoxin reductase inhibition by gold(I) compounds and concurrent stimulation of permeability transition and release of cytochrome c. Biochemical Pharmacology 67: 689–696. [DOI] [PubMed] [Google Scholar]
- Rustérucci C, Espunya MC, Díaz M, Chabannes M, Martínez MC. 2007. S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiology 143: 1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Ueda M, Morikawa H. 2002. Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Letters 515: 20–24. [DOI] [PubMed] [Google Scholar]
- Sengupta R, Ryter SW, Zuckerbraun BS, Tzeng E, Billiar TR, Stoyanovsky DA. 2007. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry 46: 8472–8483. [DOI] [PubMed] [Google Scholar]
- Serrato AJS, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. 2004. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. Journal of Biological Chemistry 279: 43821–43827. [DOI] [PubMed] [Google Scholar]
- Shahpiri A, Svensson B, Finnie C. 2008. The NADPH-dependent thioredoxin reductase/thioredoxin system in germinating barley seeds: gene expression, protein profiles, and interactions between isoforms of thioredoxin h and thioredoxin reductase. Plant Physiology 146: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiri M, Nahida Jelalib N, Ghoul JE. 2013. Cadmium affects the NADP-thioredoxin reductase/thioredoxin system in germinating pea seeds. Journal of Plant Interactions 8: 125–133. [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, et al. 2008. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello D, Tarín C, Ahicart P, Bretón-Romero R, Lamas S, Martínez-Ruiz A. 2009. A fluorescence switch technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics 9: 5359–5370. [DOI] [PubMed] [Google Scholar]
- Terrile MC, Paris R, Calderon-Villalobos LI, et al. 2012. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. The Plant Journal 70: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Dos SC, Rey P. 2006. Plant thioredoxins are key actors in the oxidative stress response. Trends in Plant Science 11: 329–334. [DOI] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP. 2010. Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. The Plant Cell 22: 2981–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. 1990. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Molecular and General Genetics 22: 377–383. [DOI] [PubMed] [Google Scholar]
- Wu C, Parrott AM, Liu T, et al. 2011. Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach. Journal of Proteomics 74: 2498–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. 2004. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell and Physiology 45: 18–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.