Abstract
Background:
In spite of modern therapies for non-small-cell lung cancer (NSCLC), prognosis for many patients is still poor and survival rates are low. Immunotherapy is the possibility to improve the lung immune response surrounding the tumour. However, this approach requires detailed understanding of the local immune-responses of NSCLC patients.
Methods:
We analysed samples from three different regions within the lungs of NSCLC patients, whereas we distinguished between patients suffering from adenocarcinoma and squamous cell carcinoma. Expression of type 1 T helper (Th1)/type 1 cytotoxic (Tc1) factors was assessed by quantitative real-time PCR, western blot analyses or immunohistochemistry. Cytotoxic cell activity of CD8+ T cells was determined via co-culture with autologous tumour cells and apoptosis assay.
Results:
We found decreased levels of the transcription factor T-box expressed in T cells (T-bet or Tbx21) and of the downstream activated IFN-γ-dependent pSTAT1α isoform in the lung tumour areas of patients with NSCLC as compared with tumour-free control regions. In these patients, reduced T-bet and pSTAT1α levels were found associated with increased immunosuppressive markers like cytotoxic T lymphocyte-associated protein 4, programmed cell death 1 and with a suppression of the Th1 cell cytokine production and Tc1 cell activity.
Conclusions:
These findings confirm a central role of T-bet in targeted immunotherapy for patients with NSCLC.
Keywords: lung T cells, T-bet, lung tumour, NSCLC, perforin, IFN-γ
Lung cancer is the leading cause of cancer deaths worldwide (Byrne et al, 2011). Although lung cancer is predominantly preventable (the main cause being nicotine), it causes more than one million cancer-related deaths per year worldwide. Unfortunately, only 15% of lung cancer patients survive for more than 5 years from the time of diagnosis (Travis et al, 1995; Greenlee et al, 2001).
The most common type of lung cancer (about 85%) is the non-small-cell lung cancer (NSCLC) and the main histological subtypes among NSCLC are adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma, respectively. Squamous cell carcinoma represents 20–30% of NSCLC and is strongly associated with cigarette smoking. However, lung ADC is the most common subtype of NSCLC (40%) in many countries. It has an increased incidence in smokers, but it is also the most common type of lung cancer in non-smokers and women (Herbst et al, 2008).
Interferons are a group of pleiotropic cytokines that have an important role during innate and acquired immune responses and host defence against viral and mycobacterial infections as well as in tumour immune surveillance (Pestka et al, 2004). Some well-described anti-tumour effects of IFN-γ are upregulation of MHC class I molecules and thus antigen presentation, leading to increased tumour cell recognition and elimination. This effect has been attributed to immune responses mediated by type 1 T helper (Th1) cells and type 1 cytotoxic (Tc1) cells or cytotoxic T lymphocytes (CTLs), which both secrete IFN-γ. T-bet drives IFN-γ production in Th1 and Tc1 cells and is also able to redirect Th2 cells into the Th1 pathway (Szabo et al, 2000). We previously described that T-bet-deficient mice have increased lung tumour load as compared with wild-type littermates (Reppert et al, 2011).
In this project, we analysed the signature markers of Th1 and Tc1 cells as well as their function in NSCLC in lung regions near and distant from the tumour cells.
Materials and methods
Human subjects and study population
This study was performed at the Friedrich-Alexander-University of Erlangen in Germany and was approved by the ethics review board of the University of Erlangen (Re-No: 56_12B; DRKS-ID: DRKS00005376). Thirty-seven patients, who were suffering from NSCLC, underwent surgery and gave their approval to being enrolled in this study. Patients' confidentiality was maintained.
The diagnosis of lung cancer was based on pathological confirmation. The histological types of lung cancer were classified according to the classification of the World Health Organization, formulated in 2004. The staging of lung cancer was based on the Cancer TNM Staging Manual, formulated by the International Association for the Study of Lung Cancer in 2010.
Tissue samples were taken from the tumoral area (solid tumour tissue), the peri-tumoral area (up to 2 cm away from the solid tumour) as well as from the tumour-free control area (at least 5 cm away from the solid tumour) of the surgically removed lung material.
Total cell isolation
Samples were cut into very small pieces using scalpels (1–3 mm2). The shredded material was then subjected to collagenase/DNase digestion (20 ml solution per 1 g of material) and incubated overnight at 37 °C under constant shaking (300 r.p.m). The digestion solution contained 4.8 μg ml−1 collagenase (Liberase TM Research Grade REF 05 401 119 001, Roche Diagnostics, Mannheim, Germany) and 15 μg ml−1 DNase (Roche Diagnostics), dissolved in PBS. To get a single-cell suspension, the digested material was sieved through a cell strainer (70 μm). The cell suspension was centrifuged (10 min, 1000 r.p.m., 4 °C) and the supernatant was discarded. The cells were resuspended in 10 ml of hypotonic solution (Ammonium-Chloride-Potassium buffer) and immediately centrifuged again (5 min, 1000 r.p.m., 4 °C) to quickly remove the Ammonium-Chloride-Potassium lysis buffer. This was followed by cell resuspension in 10 ml of PBS+EDTA+1% penicilin/streptomycin and centrifugation (10 min, 1000 r.p.m., 4 °C). Cells were finally resuspended in PBS+EDTA+1% penicilin/streptomycin+5% FCS.
Quantitative real-time PCR
Total RNA was extracted from frozen tissue samples or from cell suspension samples, using peqGold RNA Pure (Peqlab, Erlangen, Germany) according to the manufacturer's instructions. One microgram of the resulting RNA was reverse-transcribed into cDNA via the RevertAid First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany) according to the manufacturer's protocol. Each quantitative real-time PCR reaction mix contained 15 ng of cDNA, 300 nM transcript-specific forward and reverse primer and 2 × SoFast EvaGreen Supermix (Bio-Rad, Munich, Germany) in a total volume of 20 μl. Quantitative real-time PCR primers were purchased from Eurofins-MWG-Operon (Ebersberg, Germany). The primer sequences are given for HPRT (fw: 5′-TGACACTGGCAAAACAATGCA-3′ rev: 5′- GGTCCTTTTCACCAGCAAGCT-3′), TBX21 (fw: 5′- CAGAATGCCGAGATTACTCAG-3′ rev: 5′- GGTTGGGTAGGAGAGGAGAG-3′), IFNG (fw: 5′- CATGTATTGCTTTGCGTTGG-3′ rev: 5′-TGACCAGAGCATCCAAAAGA-3′), PRF1 (fw: 5′-GGACCAGTACAGCTTCAGCACTG- 3′ rev: 5′- AGTCAGGGTGCAGCGGG-3′), STAT1A (fw: 5′-CACCAGAGCCAATGGAACTT-3′ rev: 5′-ACAGAGCCCACTATCCGAGA-3′), STAT1B (fw: 5′-CTTTCCCTGACATCATTCGCA -3′ rev: 5′-AAGGCTGGCTTGAGGTTTGTA-3′), EOMES (fw: 5′-ACTGGTTCCCACTGGATGAG-3′ rev: 5′-CCACGCCATCCTCTGTAACT-3′), PD-1 (fw: 5′-CAGTTCCAAACCCTGGTGGT-3′ rev: 5′-GGCTCCTATTGTCCCTCGTG-3′), PD-L1 (fw: 5′-AGCAAAGTGATACACATTTGGAG-3′ rev: 5′-CCCCGATGAACCCCTAAACC-3′), IDO (fw: 5′-TGCTGTTCCTTACTGCCAAC-3′ rev: 5′-CGTCCATGTTCTCATAAGTCAGG-3′) and CTLA4 (fw: 5′-CTCTGGATCCTTGCAGCAGT-3′ rev: 5′-GCCTCAGCTCTTGGAAATTG-3′). Reactions were performed for 50 cycles with an initial activation for 2 min at 98 °C, denaturation for 5 min at 95 °C and hybridisation and elongation for 10 min at 60 °C. Quantitative real-time PCR reactions were performed using the CFX-96 Real-Time PCR Detection System (Bio-Rad), and analysed via the CFX Manager Software (Bio-Rad Laboratories GmbH, Munchen, Germany). The relative expression level of specific transcripts was calculated with respect to the internal standard (HPRT). Expression levels were normalised to control lung tissues using ΔΔCT calculation. For comparison of the expression of different genes, efficiency-corrected calculation of relative gene expression, normalised with HPRT, was performed using the relative standard curve method.
Western blot analyses
Western blots were performed with 30–50 μg of protein. Polyacrylamide gel electrophoresis was performed at 80 V for 1.5 h. Proteins were then transferred to a nitrocellulose membrane (PAGEr Ex Gels, Lonza, Rockland, USA) at 200 mA for 45 min. After a washing step and blocking of the membrane for 1 h (5% milk powder, 0.1% Tween-20 in PBS) at room temperature, the primary antibody against pSTAT1 (Cell Signaling Technology, Danvers, MA< USA) was applied and incubated overnight at 4 °C. The next day the membrane was washed and incubated with the HRP-conjugated secondary antibody, solved in blocking buffer, for 1 h at room temperature. After final washing steps, detection was performed using the Western Bright Quantum Western Blotting detection kit (Biozym Scientific, Oldendorf, Germany). For the visualisation of the western blot results, the Fluor Chem FC2 (Biozym Scientific) detection system was used.
Immunohistochemistry of CD8 on paraffin-embedded tissue sections
Immunohistochemistry was performed on paraffin-embedded sections. Before staining, paraffin was removed from the slides by incubation at 72 °C for 30 min and treatment with Roti-Histol (Carl Roth, Karlsruhe, Germany) three times for 10 min. The tissue sections were then rehydrated by immersion in ethanol-series with descending concentrations (100%, 95%, 70%) for 3 min each and in deionised water for 1 min. In the next step, a heat-induced antigen retrieval was performed. Slides were placed into a rack, containing 50 ml of 1 × Tris-EDTA and incubated at 120 °C for 5 min, at 85 °C for 10 s and at RT for 20 min using a pressure cooker. The slides were then washed in deionised water for 1 min. The tissue was surrounded with a hydrophobic barrier using a barrier pen (Dako, Hamburg, Germany). Peroxidase blocking solution (Dako) was then applied on the sections and incubated for 5 min at RT. Slides were washed with washing buffer for 5 min, incubated with primary antibody for 2 h at RT and washed with washing buffer for 5 min. Afterwards, slides were incubated with labelled polymer-HRP for 30 min at RT and washed with washing buffer for 5 min. again. Tissue sections were treated with DAB+Substrate and DAB+Chromogen solution for 10 min at RT. Slides were rinsed with Aqua dest carefully and treated with haematoxylin and eosin solution for 2 min. Slides were rinsed in Aqua dest for 1 min and two times in deionised water for 3 min. each. Sections were dehydrated by immersion in ethanol-series with increasing concentrations (70%, 95%, 100%) for 3 min each and treated with Roti-Histol two times for 5 min. Tissue sections were covered with cover plates using 1 or 2 drops of mounting media (Entellan, Merck, Darmstadt, Germany).
Immunohistochemistry of signal transducer and activator of transcription 1 (STAT1) on paraffin-embedded tissue sections
Deparaffination, rehydration and antigen retrieval was performed as described above. Afterwards, blocking solution (Zytomed Systems, Berlin, Germany) was applied on the sections and incubated for 5 min at RT. Slides were washed with washing buffer for 5 min and incubated with primary antibody to STAT1 (Santa Cruz, Dallas, TX, USA) overnight at RT. The next day slides were washed with washing buffer for 5 min, slides were incubated with Post Block reagent (Zytomed Systems) for 30 min at RT and washed with washing buffer for 5 min again. Tissue sections were treated with AP-Polymer reagent (Zytomed Systems) for 30 min at RT, washed with washing buffer for 5 min and incubated with Permanent AP Red (Zytomed Systems) for 10 min at RT. Slides were then carefully rinsed with Aqua dest and treated with haematoxylin and eosin solution for 1 min. Slides were rinsed in Aqua dest for 1 min and in deionised water for 5 min. Tissue sections were covered with coverslips using Aquatex (Merck).
Immunohistochemistry of STAT1 on cytospins
Cytospins were prepared using 100.000 A549 ADC cells, which were subsequently fixed by incubation with cold aceton for 10 min. The cytospins were surrounded with a hydrophobic barrier using a barrier pen (Dako). Afterwards, Zytomed blocking solution (Zytomed Systems) was applied on the sections and incubated for 5 min at RT. Slides were washed with washing buffer for 5 min, and incubated with primary antibody (Santa Cruz) for 2 h at RT and washed with washing buffer for 5 min. Afterwards, slides were incubated with Post Block Reagent (Zytomed Systems) for 30 min at RT and washed with washing buffer for 5 min again. Tissue sections were treated with AP-Polymer reagent (Zytomed Systems) for 30 min at RT, washed with washing buffer for 5 min and incubated with Permanent AP Red (Zytomed Systems) for 10 min at RT. Slides were then carefully rinsed with Aqua dest and nuclei were stained with haematoxylin solution for 1 min. Slides were rinsed in Aqua dest for 1 min and in deionised water two times for 3 min each. Tissue sections were covered with coverslips using Aquatex (Merck).
CD8+ T cell isolation
CD8+ T cell isolation was performed using magnetic anti-human CD8 Microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany), according to the manufacturer's instructions or by FACS-sorting in cooperation with a facility service of the University of Erlangen (Core Unit Cell Sorting and Immunomonitoring, SFB 643). For that purpose, total cell suspensions were stained with anti-human CD8 PE antibody (BD Biosciences, Heidelberg, Germany) according to the surface staining protocol described below.
Flow cytometry analyses (FACS)
Flow cytometry analyses were performed with 1 × 106 total cells per sample and staining. The cells were washed with PBS, followed by incubation (30 min., 4 °C, dark) with the respective mix of antibodies against surface proteins, solved in PBS (ad 80 μl). After an additional washing step, the cells were fixed and permeabilised with Fixation/Permeabilisation solutions according to the manufacturer's protocol (eBioscience, San Diego, CA, USA) and incubated (30 min, 4 °C, dark) with antibodies against intracellular proteins, dissolved in Permeabilisation buffer (eBioscience; ad 80 μl). After a final washing step with Permeabilisation buffer (eBioscience), the cells were resuspended in PBS+EDTA (Lonza, Cologne, Germany). For FACS analyses of surface markers alone, the cells were washed with PBS and resuspended in PBS+EDTA after incubation with the antibodies directed against antigen expressed on the cell membrane surface. No fixation and permeabilisation were performed in that case. The subsequent flow cytometry measurements were performed via FACS Calibur and analysed using Cell Quest Pro version 4.02 (BD Biosciences). The following antibodies were used during this study: anti-human CD4 FITC (eBioscience), anti-human CD8 Alexa Fluor 488 (eBioscience), anti-human CD11b FITC (eBioscience) and anti-human/mouse T-bet PE (BD Bioscience).
Flow cytometry analyses of phosphorylated STAT1
Single-cell suspensions were generated from a tumoral, peri-tumoral and a control lung sample of a patient with NSCLC. Total lung cells were carefully resuspended in RPMI media, containing FCS, Pen/Strep, αCD3 (2.5 μ ml−1)±IFN-γ (50 ng ml−1) and incubated at 37 °C for 20 min. Flow cytometry staining was performed with 1–1.5 × 106 total cells per sample and staining. The cells were washed with PBS, followed by incubation (30 min., 4 °C, dark) with the respective master mix of antibodies against surface proteins, solved in PBS (ad 50 μl). After an additional washing step, the cells were fixed by incubation with 300 μl 2% PFA (30 min., 4 °C, dark). Afterwards, the samples were centrifuged, the PFA-solution was aspirated and the cells were incubated with 300 μl of cold 90% methanol (30 min, 4 °C, dark). After another cell centrifugation and the aspiration of methanol solution, the cells were incubated with the PE-conjugated pSTAT1 antibody (BD Biosciences), solved in 50 μl Permeabilisation buffer (BD Biosciences; 30 min., 4 °C, dark). After a final washing step with 200 μl of Permeabilisation buffer (BD Biosciences), the cells were resuspended in PBS+EDTA (Lonza). The subsequent flow cytometry measurements were performed via FACS Calibur and analysed using Cell Quest Pro version 4.02 (BD Biosciences).
Co-culture and apoptosis assay
A small piece of the tissue sample from the tumoral region was resected and transferred into culturing medium overnight (RPMI+10% FCS+1% Penicilin/Strepomycin; no digestion or shredding were applied to the sample). The cellular outgrowth of this tumour sample was treated with collagenase seeded in new plate and grown overnight. The rest of the sample was subjected to collagenase digestion, together with the peri-tumoral sample and the control sample as well as a regional lymph node, for subsequent isolation of total cells and CD8+ T cells (in accordance to the procedure described above). The following day, tumoral cells were expanded from the piece of tumour tissue on the cell plate. The expanded tumour cells were harvested, counted and co-cultured with CD8+ T cells, isolated from the tumoral, peri-tumoral and the control area, respectively. 5 × 104 tumour cells and 1 × 105 CD8+ T cells from the respective region were cultured with 5 μg ml−1 soluble anti-CD3 (BD Bioscience) and 2 μg ml−1 anti-CD28 (BD Bioscience) for 24 h. Apoptosis assay was performed on the following day by staining the cells with APC Annexin V (BD Bioscience), propidium iodide (PI; BD Bioscience) and flow cytometry analysis were performed according to the manufacturer's protocol.
Statistical analysis
Differences were evaluated for significance (P<0.05) by the Student's two-tailed t-test for independent events (Excel, PC). Data are given as mean values±s.e.m.
Results
Decreased T-bet expression in the tumoral regions of patients with NSCLC
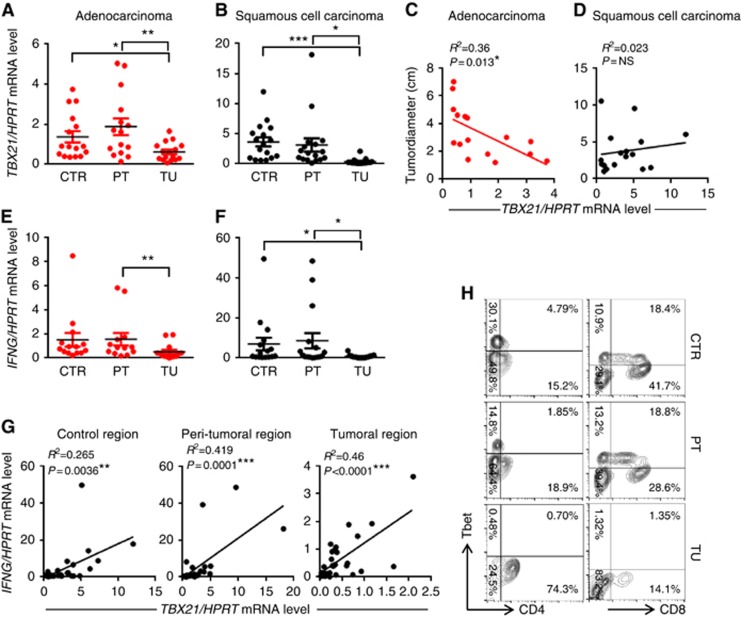
Here we isolated mRNA from the tumoral, peri-tumoral (2 cm around the tumour area) and the control lung region (taken at least 5 cm far away from the tumour border) of patients with ADC as well as SCC and analysed the expression of TBX21 by real-time PCR. TBX21 mRNA expression level was found downregulated in the tumoral region as compared with the peri-tumoral and control region of both ADC and SCC (Figure 1A and B). To examine the influence of T-bet downregulation on the progression of lung cancer, we correlated the mRNA expression of TBX21 with the diameter of the lung tumour from the individual patients, finding a negative correlation between TBX21 expression and the tumour size in patients with adenocarcinoma but not with SCC (Figure 1C and D). In confirmation of this, we found that the number of CD4+ and CD8+ T-bet+ T cells decreased in the tumoral area as compared with those found in the control area (Figure 1H).
Figure 1.
Reduced TBX21 and IFNG mRNA expression in the tumoral region of NSCLC. (A and B) qPCR-based expression analysis of TBX21 mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with adenocarcinoma (ADC) or squamous cell carcinoma (SCC; ADC: NControl=16, NPeri-tumoral=15, NTumoral=16; SCC: NControl=17, NPeri-tumoral=17, NTumoral=17). (C and D) Correlation between the tumour diameter and the TBX21 mRNA expression in the control area of patients with ADC and SCC. (E and F) qPCR-based expression analysis of IFNG mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=14, NPeri-tumoral=13, NTumoral=14; SCC: NControl=16, NPeri-tumoral=16, NTumoral=17). (G) Correlation between TBX21 and IFNG mRNA levels in the control as well as in the peri-tumoral and the tumoral area of patients with non-small-cell lung cancer (NSCLC). (H) Flow cytometry analysis of T-bet+CD4+ and T-bet+CD8+ T cells gated on lymphocytes from the control, the peri-tumoral and the tumoral lung region of one representative patient with NSCLC. Data are shown as mean values±s.e.m. using Student's t-test *P=0.05; **P=0.01; ***P=0.001.
IL-12 is a cytokine inducing IFN-γ and Th1 cells (Leonard et al, 1997). We therefore also analysed IL-12 production in the BALF of patients with NSCLC (Supplementary Table 1). However, here we could not detect this cytokine. This indicates that the Th1 defect could be secondary to a defect of IL-12 in patients with NSCLC.
Decreased IFN-γ in the tumoral region of patients with NSCLC
T-bet directly promotes the transcription of IFNG in Th1 and Tc1 cells and is in turn induced by IFN-γ via a positive feedback loop (Szabo et al, 2000; Lighvani et al, 2001). To determine whether impaired expression of T-bet was associated with reduced expression of IFNG in the tumoral area of patients with NSCLC, we assessed mRNA expression levels of the Th1 cytokine IFN-γ. We observed a significant suppression of IFNG mRNA levels in the tumoral regions of patients with ADC and SCC as compared with the peri-tumoral and control regions (Figure 1E and F). The association of T-bet and IFN-γ expression in the lung tissue of patients with NSCLC was further confirmed by a significant positive correlation of TBX21 and IFNG mRNA levels in all three analysed lung areas (Figure 1G).
Decreased perforin expression in the tumoral region of patients with NSCLC
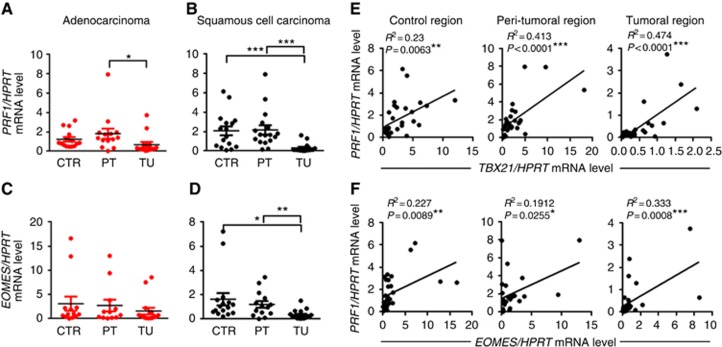
Perforin is expressed by cytotoxic lymphocytes and released from secretory granules to form pores in target cells that allow serine protease granzymes to enter the cell and induce cell death (Metkar et al, 2002; Trapani and Smyth, 2002). Besides IFN-γ expression, T-bet also controls the cytotoxicity of CD8+ T cells and is involved in the regulation of Perforin production (Cruz-Guilloty et al, 2009; Lazarevic et al, 2013). Therefore, we determined the mRNA expression of Perforin (PRF1) in tumoral, peri-tumoral and control lung tissue from patients with ADC and SCC. Comparable to our results for TBX21 and IFNG expression, PRF1 mRNA levels were significantly decreased in the tumoral area of ADC and SCC, suggesting impaired Tc1 responses (Figure 2A and B). Analysing the correlation between PRF1 mRNA expression and the tumour diameter in patients with NSCLC, we found that although there is a tendency, suggesting that low expression of PRF1 is associated with increased tumour diameters in patients with ADC similar to TBX21, the correlation is not statistically significant (Supplementary Figure 1A and B). Finally, we also found a significant positive correlation between TBX21 and PRF1 mRNA expression in the control as well as in the peri-tumoral and the tumoral region of patients with NSCLC (Figure 2E).
Figure 2.
Reduced PRF1 and EOMES mRNA expression levels in the tumoral region of NSCLC. (A and B) qPCR-based expression analysis of PRF1 mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=15, NPeri-tumoral=14, NTumoral=14; SCC: NControl=16, NPeri-tumoral=17, NTumoral=16). (C and D) qPCR-based expression analysis of EOMES mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=13, NPeri-tumoral=12, NTumoral=15; SCC: NControl=16, NPeri-tumoral=14, NTumoral=17). (E) Correlation between TBX21 and PRF1 mRNA levels in the control as well as in the peri-tumoral and the tumoral area of patients with NSCLC. (F) Correlation between EOMES and PRF1 mRNA levels in the control as well as in the peri-tumoral and the tumoral area of patients with NSCLC. Data are shown as mean values±s.e.m. using Student's t-test *P=0.05; **P=0.01; ***P=0.001.
Decreased eomesodermin (EOMES) in the tumoral region of patients with SCC
Eomesodermin also known as T-box brain protein 2 encodes a transcription factor that is involved in the regulation of different developmental processes (Kimura et al, 1999). Besides T-bet, EOMES is also important for the development of effector CD8+ T cells (Intlekofer et al, 2005). Here we found EOMES mRNA significantly decreased in the tumoral region of patients with SCC (Figure 2C and D). Similar to T-bet, EOMES positively correlated with perforin (PRF1) mRNA expression in the control as well as in the peri-tumoral and the tumoral region of patients with NSCLC (Figure 2F). However, EOMES did not inversely correlate with the tumour load (Supplementary Figure 1C and D), neither did EOMES positively correlate with IFNG mRNA expression (Supplementary Figure 1E) in these patients with NSCLC.
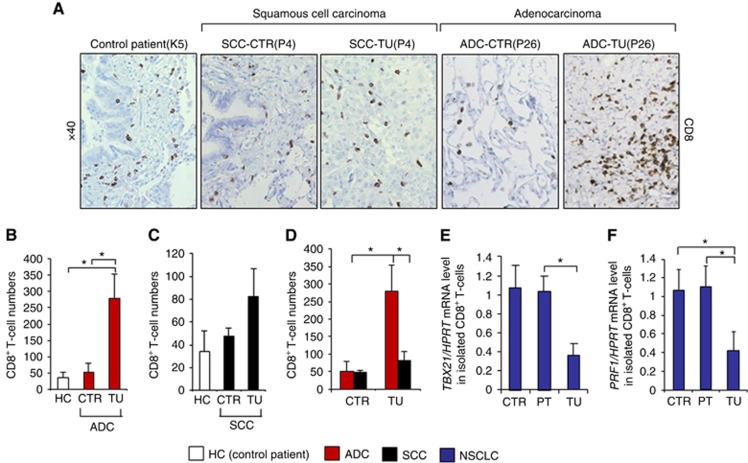
Increased CD8+ T cell numbers with decreased Tc1 gene expression in the tumoral region of NSCLC
We next wondered if the overall numbers of CD8+ T cells are reduced in the tumoral region of patients with NSCLC. Therefore, we performed immunohistochemistry for CD8 on lung tissue histo-arrays from the control and tumoral region of four patients with lung ADC, four patients with SCC and from three control subjects without lung cancer (Table 1). Here we found a higher density of CD8+ T cells in the tumoral region of patients with lung ADC as compared with the control region as well as with samples from control subjects and with the tumoral region of SCC (Figure 3A–D). This result might indicate that the lung ADC is more immunogenic as compared with squamous carcinoma. We next studied the relative mRNA expression of TBX21 and PRF1 in CD8+ T cells, isolated from the lung tissue of NSCLC patients. We analysed CD8+ T cells infiltrating the tumoral, the peri-tumoral and the control region, finding that mRNA expression of these Tc1 genes was significantly decreased in CD8+ T cells isolated from the tumoral region as compared with the peri-tumoral and control region of patients with lung cancer (Figure 3E and F).
Table 1. Clinical data of patient cohort.
| Patient No. | Sample ID | Histological classification | Maximal tumour diameter (cm) | Grading | T | N | M | TNM stadium | Gender | Age (years) | Average smoking (P/Y) | Post-OP development of metastases |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3-MP | ADC | 5 | G3 | 2a | 0 | 0 | IB | Male | 79 | 60 | No |
| 2 | 9-MP | ADC | 2.7 | G2 | 1b | 2 | 0 | IIIA | Female | 84 | 0 | No |
| 3 | 15-MP | ADC | 2.5 | G3 | 1b | 0 | 0 | IA | Male | 63 | 100 | Yes |
| 4 | 16-MP | ADC | 4.6 | G3 | 3 | 0 | 0 | II B | Female | 70 | 15 | No |
| 5 | 17-MP | ADC | 2.6 | G2 | 2 | 0 | 0 | IB | Male | 74 | 70 | No |
| 6 | 19-MP | ADC | 6.5 | # | 2b | 0 | 0 | IIA | Female | 55 | 30 | No |
| 7 | 20-MP | ADC | 2.8 | G3 | 1b | 0 | 0 | IA | Male | 65 | 60 | No |
| 8 | 22-MP | ADC | 7 | G3 | 2b | 1 | 0 | IIB | Male | 68 | 82 | No |
| 9 | 23-MP | ADC | 4.5 | G2 | 2a | 0 | 0 | IB | Male | 73 | 75 | No |
| 10 | 26-MP | ADC | 1.3 | G3 | 1a | 0 | 1 | IV | Female | 52 | 50 | # |
| 11 | 27-MP | ADC | 1.4 | G3 | 1a | 0 | 0 | IA | Female | 70 | 50 | No |
| 12 | 28-MP | ADC | 1.2 | G3 | 1a | 0 | 0 | IA | Male | 76 | 60 | No |
| 13 | 32-MP | ADC | 4.4 | G3 | 2a | 2 | 0 | IIIA | Female | 60 | 30 | Yes |
| 14 | 34-MP | ADC | 1.8 | G3 | 1 | 0 | 0 | I A | Female | 51 | 45 | No |
| 15 | 35-MP | ADC | 3 | G3 | 1b | 0 | 0 | IA | Female | 72 | 0 | No |
| 16 | 39-MP | ADC | 6 | G3 | 2b | 0 | 0 | IIA | Male | 65 | 42 | No |
| 17 | 40-MP | ADC | 1.8 | G3 | 1a | 1 | 0 | IIA | Male | 82 | 100 | No |
| 18 | 43-MP | ADC | 4 | G3 | 2a | 2 | 0 | IIIA | Female | 72 | 0 | # |
| 19 | 44-MP | ADC | 1.5 | # | 1a | 0 | 0 | IA | Male | 53 | 70 | No |
| 20 | 45-MP | ADC | 2.3 | G1 | 1b | 0 | 0 | IA | Male | 78 | 0 | No |
| 21 | 1-MP | SCC | 1.3 | G3 | 1b | 0 | 0 | IA | Male | 80 | 40 | No |
| 22 | 2-MP | SCC | 5.1 | G3 | 1a | 0 | 0 | 0 | Male | 57 | 40 | No |
| 23 | 4-MP | SCC | 2 | G3 | 1b | 1 | 0 | IIA | Female | 53 | 25 | # |
| 24 | 5-MP | SCC | 10.5 | G2 | 3 | 0 | 0 | IIB | Female | 67 | 50 | No |
| 25 | 8-MP | SCC | 5.5 | G3 | 3 | 0 | 0 | IIB | Male | 66 | 30 | Yes |
| 26 | 13-MP | SCC | 3 | G3 | 1b | 0 | 0 | IA | Male | 69 | 50 | No |
| 27 | 14-MP | SCC | 1.9 | G2 | 1a | 0 | 0 | IA | Female | 58 | 30 | No |
| 28 | 21-MP | SCC | 2.5 | G1 | 1b | 0 | 0 | IA | Male | 41 | 10 | No |
| 29 | 29-MP | SCC | 3.7 | G3 | 1b | 0 | 0 | IIB | Male | 74 | 100 | No |
| 30 | 30-MP | SCC | 1.8 | G3 | 1a | 0 | 0 | IA | Female | 70 | 30 | No |
| 31 | 36-MP | SCC | 3.5 | G3 | 2a | 1 | 0 | IB | Male | 74 | 40 | No |
| 32 | 37-MP | SCC | 3.3 | G2 | 2a | 1 | 0 | IIA | Male | 60 | 45 | No |
| 33 | 41-MP | SCC | 1.3 | G3 | 3 | 1 | 0 | IIIA | Male | 70 | 42 | No |
| 34 | 42-MP | SCC | 1.5 | G3 | 2b | 1 | 0 | IIB | Male | 74 | 40 | Yes |
| 35 | 46-MP | SCC | 9.5 | G3 | 3 | 1 | 0 | IIIA | Male | 60 | 30 | # |
| 36 | 47-MP | SCC | 6 | G3 | 2b | 0 | 0 | IIA | Male | 64 | 80 | No |
| 37 | 48-MP | SCC | 5 | G2 | 2b | 0 | 0 | IIA | Male | 55 | 20 | Yes |
| 38 | K5 | Pneumotorax | 0 | / | / | / | / | / | Male | 45 | # | / |
| 39 | K10 | Pneumotorax | 0 | / | / | / | / | / | Male | 22 | # | / |
| 40 | K12 | Pneumotorax | 0 | / | / | / | / | / | Female | 21 | # | / |
Abbreviations: ADC=adenocarcinoma; SCC=squamous cell carcinoma.
#No information available.
T-primary tumour: 0: No evidence of primary tumour; 1a: Tumour 2 cm or less in greatest dimension; 1b: Tumour more than 2 cm but not more than 3 cm in greatest dimension; 2a: Tumour more than 3 cm but not more than 5 cm in greatest dimension; 2b: Tumour more than 5 cm but not more than 7 cm in greatest dimension; 3: Tumour more than 7 cm. N-regional lymph nodes: 0: No regional lymph node metastasis; 1: Metastasis in ipsilateral peribronchial and/or ipsilateral hilar lymph nodes and intrapulmonary nodes, including involvement by direct extension; 2: Metastasis in ipsilateral mediastinal and/or subcarinal lymph node(s).
M-distant metastasis: 0: No distant metastasis; 1: Distant metastasis.
Histopathological grading: G1: well differentiated; G2: moderately differentiated; G3: poorly differentiated.
Figure 3.
Increased numbers of CD8+ T cells in the tumoral lung region of patients with adenocarcinoma. (A) IHC staining of CD8+ cells was performed on paraffin-embedded tissue sections from the control and the tumoral region of the lungs of patients with squamous cell carcinoma (n=4) and adenocarcinoma (n=4) as well as from the lungs of control subjects without lung tumour (n=3). (B–D) Bar charts represent CD8+ cell numbers counted on the tissue sections from the respective NSCLC patients as well as healthy controls. (E) qPCR-based expression analysis of TBX21 mRNA in CD8+ T cells, isolated from lung tissue samples of the tumoral, peri-tumoral and control region of patients with NSCLC (NControl=4, NPeri-tumoral=4, NTumoral=4). (F) qPCR-based expression analysis of PRF1 mRNA in CD8+ T cells, isolated from lung tissue samples of the tumoral, peri-tumoral and control region of patients with NSCLC (NControl=3, NPeri-tumoral=3, NTumoral=3). Data are shown as mean values±s.e.m. using Student's t-test *P=0.05.
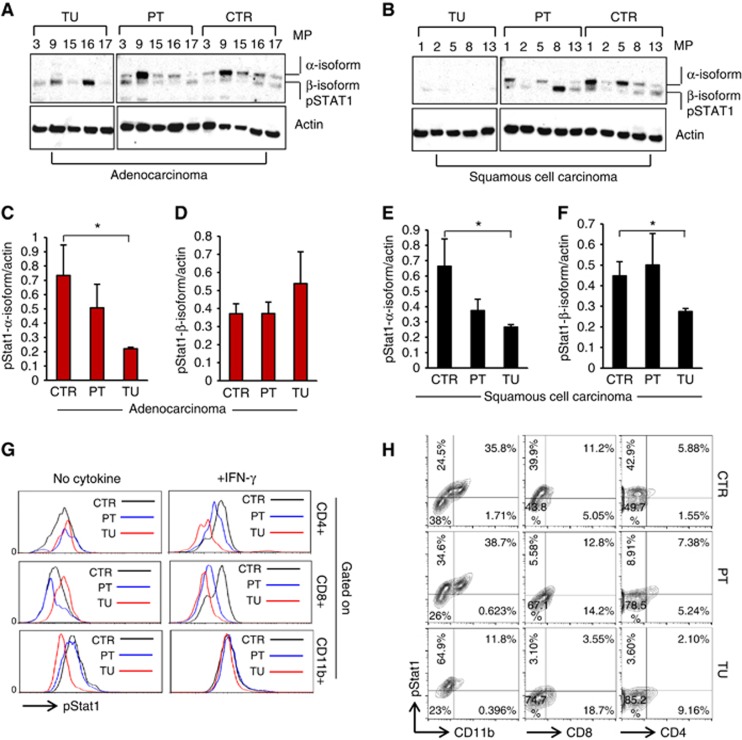
Decreased pSTAT1α in the tumoral region of patients with NSCLC
According to previous studies, TBX21 expression can be induced by IFN-γ in a STAT1-dependent manner (Lighvani et al, 2001; Afkarian et al, 2002). To further analyse the putative alterations of the Th1 signalling pathway in NSCLC, we thus next assessed the protein levels of phosphorylated STAT1 in the tumoral, peri-tumoral and control lung areas of patients with ADC and SCC (Figure 4A and B).
Figure 4.
Reduced STAT1 phosphorylation in the tumoral region of NSCLC. (A–F) Western blot analysis of pSTAT1 and actin expression in lung tissue samples from the tumoral, peri-tumoral and control region of patients with adenocarcinoma (ADC) (NControl=5; NPeri-tumoral=5, NTumoral=5). Bar charts show mean values of the protein expression levels of pSTAT1 α-Isoform (C and D) and pSTAT1 β-Isoform (E and F) relative to actin levels in ADC and in SCC, respectively. (G) Flow cytometry analysis of pSTAT1+ cells in the tumoral, the peri-tumoral and the control lung region of one representative patient with NSCLC. pSTAT1 staining was performed with the lung cell suspensions after 20 min of incubation at 37 °C in the presence or absence of IFN-γ. pSTAT1+ cells were gated on CD4+, CD8+ or CD11b+ cells, respectively. (H) Flow cytometry analysis of pSTAT1+CD4+ and pSTAT1+CD8+ T cells gated on lymphocytes as well as pSTAT1+CD11b+ cells gated on big cells in the control, the peri-tumoral and the tumoral lung region of one representative patient with NSCLC. Data are shown as mean values±s.e.m. using Student's t-test *P=0.05.
In the control and peri-tumoral region of the lungs, we observed two bands, corresponding to the two isoforms of STAT1, which are generated by alternative splicing, STAT1α (91 kDa) and the C-terminally truncated STAT1β (84 kDa; Schindler et al, 1992; Muller et al, 1993). Both isoforms are able to form homo (α/α or β/β) as well as heterodimers (α/β), to translocate to the nucleus and bind to IFN-γ-activated sites. However, STAT1β overexpression has been shown to inhibit STAT1α function and is thus suggested to negatively regulate the expression of IFN-γ responsive genes (Najjar et al, 2008). Here we found that the protein level of phosphorylated STAT1α-isoform was barely detectable in the tumoral lung region of both ADC and SCC as compared with the peri-tumoral as well as the control region (Figure 4A, C and E). By contrast, phosphorylated STAT1β was present in the tumoral, peri-tumoral and the control area of patients with ADC, whereas it could not be detected in the tumoral region of patients with SCC (Figure 4B, D and F).
Cellular distribution of pSTAT1 in the lung of patients with NSCLC
In the next step, we tried to understand the cellular localisation of pSTAT1 in the tumour microenvironment. To this aim, we performed FACS analysis using an antibody against pSTAT1 in tumoral, peri-tumoral and control lung cells isolated from a patient with NSCLC. Before staining, the cell suspensions were incubated at 37 °C in the presence or absence of IFN-γ for 20 min in order to induce STAT1 phosphorylation. We aimed to investigate phosphorylated STAT1 in different immunological cell subsets, such as CD4+ Th1 cells, CD8+ Tc1 cells and CD11b+ macrophages. CD4+pSTAT1+ cells and CD8+pSTAT1+ cells were gated on lymphocytes, whereas CD11b+pSTAT1+ cells were gated on big cells. Generally, we observed that the majority of pSTAT1+ cells in our experiment were CD11b+ macrophages, whereas only a small fraction of pSTAT1+ cells was CD4+ or CD8+ in the control, peri-tumoral and tumoral region of the lung (Figure 4H). Nevertheless, we found that the fraction of pSTAT1-expressing macrophages (CD11b) as well as CD4+ and CD8+ T cells decreased in the tumoral region as compared with the control as well as the peri-tumoral region of the lung of this patient with NSCLC (Figure 4H). By observing the pSTAT1+CD11b− cells gated on big cells, we saw that their number increased in the tumoral region of this lung. We conclude that also tumour cells could contain pSTAT1 (Figure 4H).
We next assessed the mean fluorescence intensity of pSTAT1 gated on CD4+, CD8+ or CD11b+ cells from the tumoral, the peri-tumoral and the control lung area in the presence or absence of IFN-γ. With this approach, we observed a higher pSTAT1 level in CD4+ and CD8+ cells from the control lung area as compared with the peri-tumoral and the tumoral lung area of the NSCLC patient upon IFN-γ treatment, whereas this difference could not be observed in the absence of IFN-γ (Figure 4G). This allows the conclusion that CD4 and CD8+ T cells from the control lung area of patients with NSCLC may be more responsive to IFN-γ signalling, inducing STAT1 phosphorylation.
Taken together these data suggest that pSTAT1+ macrophages as well as IFNγ-responsive Th1 and Tc1 cells are mainly localised in the control and peri-tumoral region of these patients with NSCLC. Moreover, we suggest that the tumour cells themselves can contain pSTAT1.
STAT1 mRNA and protein levels in the lungs of patients with NSCLC
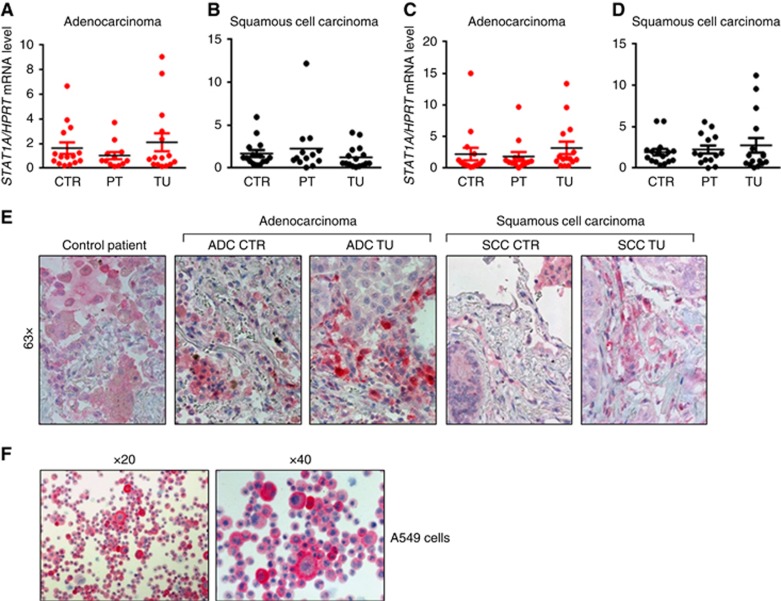
We wondered if only the phosphorylation of STAT1 is changed in the tumoral area of NSCLC or if the two isoforms of STAT1 are differentially expressed in the three analysed regions of the tumour-bearing lungs. Therefore, we next assessed the mRNA levels of STAT1A and STAT1B in the tumoral, the peri-tumoral and the control region of patients with ADC and SCC, finding that there is no difference in the expression levels of STAT1A and STAT1B between the tumoral, the peri-tumoral and the control lung area of patients with ADC and SCC (Figure 5A–D). This observation was further confirmed by immunohistochemical (IHC) staining for STAT1 in a tissue array enclosing the tumoral and control region of most of the patients analysed by real-time PCR. After counting the STAT1-positive cells, no difference in STAT1+ cell numbers was observed between tissue sections from the control and the tumoral lung area of NSCLC patients (Figure 5E and Supplementary Figure 1N and O). Moreover, by observing the IHC staining for STAT1 at higher magnification, we observed that macrophages in the lung of patients with NSCLC as well as in control lungs were an important source of STAT1 (Figure 5E). We also observed that the tumour cells themselves expressed STAT1 and epithelial cells (Figure 5E). This observation was confirmed by IHC for STAT1 on the lung tumour ADC cell line A549 (Figure 5F).
Figure 5.
STAT1 RNA and protein expression in NSCLC lung samples. (A and B) qPCR-based analysis of STAT1A mRNA expression in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC and SCC (ADC: NControl=16, NPeri-tumoral=15, NTumoral=16; SCC: NControl=17, NPeri-tumoral=17, NTumoral=17). (C and D) qPCR-based analysis of STAT1B mRNA expression in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC and SCC (ADC: NControl=15, NPeri-tumoral=13, NTumoral=15; SCC: NControl=16, NPeri-tumoral=14, NTumoral=16). (E) IHC staining of STAT1+ cells was performed on paraffin-embedded tissue sections from the control and the tumoral region of the lungs of patients with ADC and SCC as well as from the lungs of control subjects without lung tumour. (F) IHC staining was performed on cryospins of A549 human adenocarcinoma cell line. Data are shown as mean values±s.e.m. using Student's t-test *P=0.05.
In conclusion, the expression of STAT1 in tumour cells could explain why there is no overall decrease in STAT1 expression between the control versus the tumoral region, although the number of pSTAT1+CD11b+, pSTAT1+CD4+and CD8+ T cells decreased in the tumoral region of the lung of patients with NSCLC.
Programmed cell death 1 (PD-1), PD-L1 expression in the tumoral region of ADC as compared with SCC
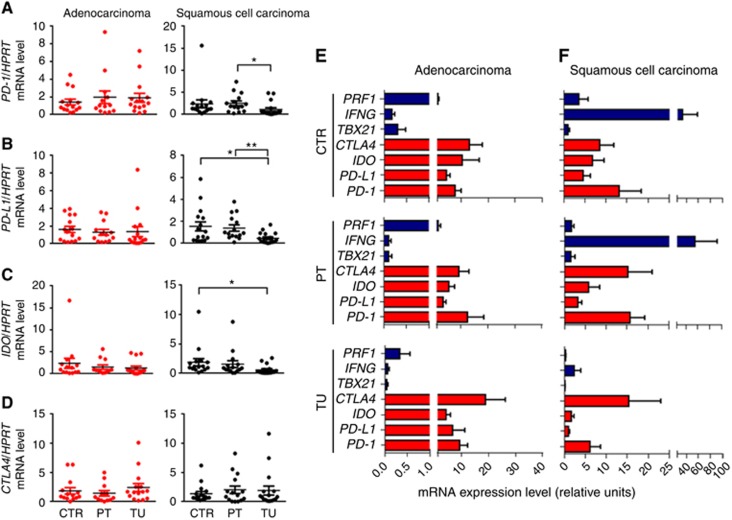
The above data collectively indicate a decrease of Th1 and Tc1 markers in the tumoral region of patients with NSCLC. Programmed cell death 1 and its ligand PD-L1 are necessary for the regulation of the balance between T-cell activation and tolerance. The PD-1/PD-L1 pathway has previously been shown to contribute to tumour-mediated immune suppression and is thus recently being considered as an interesting target for clinical trials (Keir et al, 2008). As it has also been shown that Th1 and Tc1 cells are targets for PD-1/PD-L1-mediated immune suppression (Li et al, 2015), we wanted to see if this pathway is differentially regulated in lung samples from patients with ADC and SCC. The overall expression of these two genes was not found differentially regulated in the different lung regions of patients with ADC. By contrast, in the tumoral region of SCC, unexpectedly, PD-1 and PD-L1 mRNA was found downregulated as compared with the control region (Figure 6A and B).
Figure 6.
mRNA expression levels of T-regulatory cell signature genes in the lungs of patients with NSCLC. (A) qPCR-based expression analysis of PD-1 mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=15, NPeri-tumoral=13, NTumoral=15; SCC: NControl=16, NPeri-tumoral=15, NTumoral=17). (B) qPCR-based expression analysis of PD-L1 mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=15, NPeri-tumoral=13, NTumoral=15; SCC: NControl=16, NPeri-tumoral=15, NTumoral=17). (C) qPCR-based expression analysis of IDO mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=14, NPeri-tumoral=12, NTumoral=15; SCC: NControl=16, NPeri-tumoral=15, NTumoral=17). (D) qPCR-based expression analysis of CTLA-4 mRNA in lung tissue samples from the tumoral, peri-tumoral and control region of patients with ADC or SCC (ADC: NControl=14, NPeri-tumoral=12, NTumoral=15; SCC: NControl=16, NPeri-tumoral=15, NTumoral=17). (E and F) qPCR-based analysis of PRF1, IFNG, TBX21, CTLA-4, IDO, PD-L1, PD-1 mRNA levels in the tumoral, peri-tumoral and control lung regions of patients with ADC or SCC. Calculation of the efficiency corrected relative expression values for the different genes was performed using the relative standard curve method. Data are shown as mean values±s.e.m. using Student's t-test *P=0.05; **P=0.01.
IDO mRNA expression in the lungs of patients with NSCLC
Indoleamine 2, 3-dioxygenase (IDO) is another mediator of tumour immune tolerance, which causes T cell suppression. However, IDO is also known to be up-regulated in tumour cells in an IFNγ and STAT1-dependent manner (Chon et al, 1996). We thus next analysed IDO expression in NSCLC. Here we found decreased expression of IDO mRNA only in the tumoral region of SCC but not in the ADC (Figure 6C). This could imply that the ADC tumour cells express more IDO as they express STAT1.
CTLA-4 mRNA expression in the lungs of patients with NSCLC
Cytotoxic T lymphocyte-associated protein 4 (CTLA-4) is an inhibitory immunoglobulin that is expressed on the surface of helper T cells. Moreover, CTLA-4 has been shown to be upregulated by regulatory T cells and has a role for their suppressive function (Domagala-Kulawik, 2015). The tumour microenvironment is characterised by high levels of inhibitory cytokines, such as IL-10 and TGF-β, which are able to induce regulatory T cells (Mocellin et al, 2005; Jeon and Jen, 2010). We therefore next analysed the mRNA expression of the Treg surface marker CTLA-4 in NSCLC lung samples. However, we could not detect any significant differences at the mRNA level between the tumoral, the peri-tumoral and the control areas of patients with ADC or SCC (Figure 6D).
Relative increase of immunosuppressive genes PD-1, PD-L1, IDO and CTLA-4 in the tumoral region of NSCLC
As we did not observe an upregulation of immunosuppressive genes in the tumoral region of NSCLC lung samples, we wondered how strong the expression of these genes is in relation to the mRNA levels of pro-inflammatory factors. To this aim, we did an efficiency corrected semi-quantitative PCR gene comparison. In patients with ADC, we found that the mRNA levels of the anti-inflammatory genes PD-1, PD-L1, IDO and CTLA-4 are higher as compared with the pro-inflammatory genes TBX21, IFNG and PRF1 in all three analysed lung regions. However, in the tumoral region of ADC, the balance of the expression level is further shifted towards the immunosuppressive genes. In patients with SCC, the mRNA levels of CTLA-4 and PD-1 are higher as compared with the pro-inflammatory genes TBX21, IFNG and PRF1 only in the tumoral lung area, whereas in the control and the peri-tumoral area of SCC the relative expression of IFNG is higher than that of the tested immunosuppressive genes (Figure 6E and F).
Decreased cytotoxic activity of CD8+ T cells isolated from the tumoral region of lung carcinoma
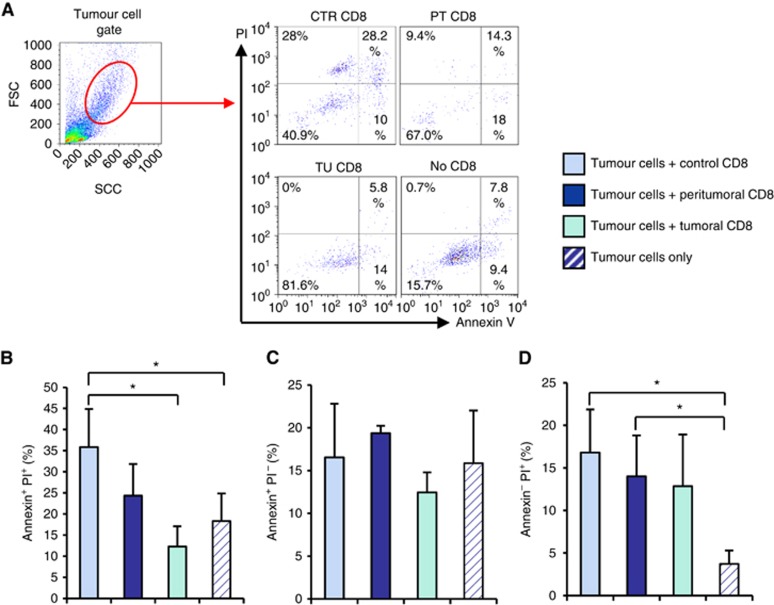
We next asked whether the decreased expression of PRF1 in the tumoral region would affect the cytotoxic activity of local CD8+ T cells. To address this issue, we isolated CD8+ T cells from the different lung carcinoma areas and co-cultured them with tumour cells from the respective patient. Afterwards, we assessed the numbers of apoptotic (Annexin V+) and necrotic (PI+) tumour cells by using an Annexin V/PI assay. We noted that CD8+ T cells from the control and peri- tumoral areas are able to induce tumour cell death in co-culture (PI+ cells), whereas no significant induction of cell death was detected after co-culture with CD8+ T cells from the tumoral area (Figure 7A–D). These findings indicated an impaired cytotoxic activity of intra-tumoral lung CD8+ T cells in patients with NSCLC, which can be relevant for therapeutic settings.
Figure 7.
Increased killing capability of CD8+ T cells from the control area. CD8+ T cells were isolated from the tumoral, peri-tumoral and control region of tissue samples from the lung of lung cancer patients. CD8+ cells were co-cultured with total cells from the tumoral region of the respective patient (tumour cells). Apoptosis and necrosis levels were evaluated by flow cytometry. Cells were double-stained with antibodies against Annexin V and PI. (A) Tumour cell gate and a representative flow cytometry plot of Annexin V versus PI staining is depicted. (B–D) Bar graphs show the mean percentages of Annexin V or PI single positive as well as Annexin V and PI double positive cells. Data are shown as mean values±s.e.m. using Student's t-test *P=0.05.
Discussion
Current treatment modalities for lung cancer are limited and the identification of novel targets for therapy is of high relevance. In the present study, we analysed the local distribution and regulation of tumour-infiltrating immune cell subsets and the expression of their hallmark genes in lung tissue samples of patients suffering from NSCLC.
We thus analysed the expression of specific transcription factors and markers controlling the Th1 and Tc1 differentiation in the tumoral, the peri-tumoral as well as in the control region of patients with NSCLC. It is believed that the most potent antitumour effector cells are CD8+ CTLs producing IFN-γ. Thus, a majority of clinical cancer vaccination studies focusses on the activation of tumour-specific CD8+ T cells (Jager et al, 2000).
Two major effector molecules of the granule-mediated cytolytic pathway, which are thought to be involved in tumour suppression, are granzyme B and Perforin. Perforin creates a membrane channel through which granzyme B is able to enter the target cell and to evolve its cytolytic function (Hodge et al, 2014). Consistently, we detected a downregulation of the mRNA levels of the Th1/Tc1 transcription factor, T-bet as well as of the Th1 and CTL effector molecules IFN-γ and Perforin in the tumoral region of the lungs of NSCLC patients, although the overall numbers of CD8+ T cells were increased in the tumoral area of patients with ADC and were not significantly changed in patients with SCC. This is in line with previous work that demonstrates a relationship between higher numbers of T-bet-positive intra-tumoral lymphocytes and a positive prognosis for patients with breast cancer (Ladoire et al, 2011; Yu and Yang et al, 2014) and the fact that T-bet was found to be required for anti-tumour immune responses in a murine model of lung cancer partially due to its ability to control intra-tumoral development of regulatory T cells (Reppert et al, 2011). Eomesodermin is, like T-bet, another transcription factor involved in CD8+ T cell development. Eomesodermin was found downregulated in the tumoral region of patients with SCC but not ADC. The finding that EOMES neither correlates with IFNG nor inversely correlates with the tumour size, although it does correlate with Perforin, in these patients, questions its relevance in the patho-mechanism of lung cancer as compared with T-bet. In fact, here we found a negative correlation between the size of the tumour and the mRNA level of TBX21 in patients with ADC, which underlines the importance of T-bet-mediated immune responses for anti-tumour effects.
In accordance with these findings, we found that CD8+ T cells isolated from the tumoral region induce less tumour cell death as compared with those isolated from the control and peri-tumoral region of the same patient. We suppose that the increased cytotoxic activity of the CD8+ T cells, isolated from the control area was due to T-bet-induced Perforin expression, both found increased in CD8+ T cells sorted out from the control lung area, as compared with those from the tumoral area. These findings suggest an accumulation of Th1 and Tc1 cells with anti-tumour potential in the control lung region, surrounding the solid tumour of lung cancer patients. Based on these results, a possible approach for a potential therapeutical anti-tumour application could be an in vitro expansion of CD8+ T cells, isolated from the control lung area or from the PBMC of the individual patient, followed by adoptive transfer of the expanded cytotoxic T cells back into the patient, post surgery. Taken together, we identified characteristic alterations of the local immune landscape in tumour tissue of NSCLC patients that were characterised by an impairment of T-bet-associated Th1 and Tc1 immune responses.
In this study, we analysed two of the NSCLC subtypes separately to identify putative differences concerning the immune responses in patients with ADC and SCC. In a previously published work, we were already able to demonstrate that genes, which are associated with the Th17 cell type, such as IL-6R, BATF and RORC are differentially expressed in patients suffering from ADC and SCC (Balabko et al, 2014). Consistently, we could identify some differences between these two types of lung cancer in this study as well. Thus, we found increased numbers of CD8+ T cells in the tumoral area of patients with ADC as compared with SCC. Besides that, we found an association between the tumour size and the expression of TBX21 and PRF1 mRNA in patients with adeno- but not with SCC. Finally, we found that only the phosphorylation of STAT1α, but not STAT1β, was reduced in the tumoral area of patients with ADC, whereas we could not detect phosphorylation of neither of the two STAT1 isoforms in the tumoral region of SCC. Moreover, we observed decreased phosphorylation and thus activation of STAT1α in the tumoral lung region of patients with adeno- as well as with SCC, which is necessary for the transcription of Th1/Tc1 genes downstream of IFN-γ. Tumour infiltrating macrophages (TAMs) have previously been shown to stimulate tumour growth and metastasis in different kinds of cancer (Van Overmeire et al, 2014). Here we identified macrophages infiltrating the lung of these patients as the major source of STAT1. STAT1 has been described as the transcription factor regulating colony-stimulating factor 1 (CSF-1) production. CSF-1 in turn induces TAM via CSF-1 receptor (CSFR) engagement. These data therefore support a central role of STAT1 in lung tumour especially with regard to TAM induction that need to be further investigated. Beside macrophages, also the tumour cells were found to express STAT1. Although we could not find any differential regulation of IDO, PD-1 and PD-L1 mRNA in the different regions of the ADC lung tissues, in the SCC IDO, PD-1 and PD-L1 were found significantly downregulated in the tumoral region, suggesting that these immunosuppressive factors could represent a better possible target for the therapy of the ADC subgroup. Nevertheless, semi-quantitative expression comparison between mRNA levels of Th1/Tc1 genes like TBX21 and suppressive genes like CTLA-4 and PD-1 suggested a shift of the balance towards the suppressive genes in the tumoral lung region of both adeno- and SCC. The reason for the differential regulation of these genes in ADC and SCC still needs further investigation.
Altogether, these results indicate differences in the regulation of anti-tumour immune responses in patients with adeno- and SCC, meaning that in case of the development of an immunotherapy this fact should be considered and patients with these two types of lung cancer would eventually require different treatment.
Acknowledgments
We thank the team at the Department of Molecular Pneumology especially Sonja Trump and Susanne Mittler for their superb technical help, the team at the Department of Thoracic Surgery and at the Institute of Pathology at the Friedrich-Alexander-University of Erlangen-Nürnberg for their help and support to this work. We thank Markus Neurath for critically reading the manuscript. This work was financially supported by the Molecular Pneumology Department, a grant from the SFB 643 (Strategies of cellular immune intervention) (B12), by a grant from the IZKF (Interdisciplinary Center for Clinical Research at the medical Faculties) (A59) and by the Comprehensive Cancer Center at the Friedrich-Alexander University Erlangen-Nürnberg.
Author contributions
KA and DIT were the principal investigators of this study. SF did the protocol design, contributed to the data interpretation and wrote the manuscript with KA. DIT and HS recruited and took care of the patients, provided the clinical data of the patients and did the surgery. RR and AH did the pathological diagnosis as well as the histological analyses and determined the histological score of the patients. JS performed intraoperative bronchoscopy and obtained the BALF from operated patients. All other authors reviewed and approved the final version of the manuscript.
‘The present work was performed in (partial) fulfillment of the requirements for obtaining the degree ‘Dr med. Denis Iulian Trufa'
The authors declared no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM (2002) T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol 3(6): 549–557. [DOI] [PubMed] [Google Scholar]
- Balabko L, Andreev K, Burmann N, Schubert M, Mathews M, Trufa DI, Reppert S, Rau T, Schicht M, Sirbu H, Hartmann A, Finotto S (2014) Increased expression of the Th17-IL-6R/pSTAT3/BATF/Rorgamma T-axis in the tumoural region of adenocarcinoma as compared to squamous cell carcinoma of the lung. Sci Rep 4: 7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne WL, Mills KH, Lederer JA, O'Sullivan GC (2011) Targeting regulatory T cells in cancer. Cancer Res 71(22): 6915–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon SY, Hassanain HH, Gupta SL (1996) Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-gamma-inducible expression of human indoleamine 2,3-dioxygenase gene. J Biol Chem 271(29): 17247–17252. [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A (2009) Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 206(1): 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagala-Kulawik J (2015) The role of the immune system in non-small cell lung carcinoma and potential for therapeutic intervention. Transl Lung Cancer Res 4(2): 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee RT, Hill-Harmon MB, Murray T, Thun M (2001) Cancer statistics, 2001. CA Cancer J Clin 51(1): 15–36. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM (2008) Lung cancer. N Engl J Med 359(13): 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge G, Barnawi J, Jurisevic C, Moffat D, Holmes M, Reynolds PN, Jersmann H, Hodge S (2014) Lung cancer is associated with decreased expression of perforin, granzyme B and interferon (IFN)-gamma by infiltrating lung tissue T cells, natural killer (NK) T-like and N Clin Exp Immunol 178(1): 79–85. [DOI] [PMC free article] [PubMed]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL (2005) Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6(12): 1236–1244. [DOI] [PubMed] [Google Scholar]
- Jager E, Gnjatic S, Nagata Y, Stockert E, Jager D, Karbach J, Neumann A, Rieckenberg J, Chen YT, Ritter G, Hoffman E, Arand M, Old LJ, Knuth A (2000) Induction of primary NY-ESO-1 immunity: CD8+ T lymphocyte and antibody responses in peptide-vaccinated patients with NY-ESO-1+ cancers. Proc Natl Acad Sci USA 97(22): 12198–12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HS, Jen J (2010) TGF-beta signaling and the role of inhibitory Smads in non-small cell lung cancer. J Thorac Oncol 5(4): 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH (2008) PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 26: 677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Nakashima K, Ueno M, Kiyama H, Taga T (1999) A novel mammalian T-box-containing gene, Tbr2, expressed in mouse developing brain. Brain Res Dev Brain Res 115(2): 183–193. [DOI] [PubMed] [Google Scholar]
- Ladoire S, Arnould L, Mignot G, Apetoh L, Rébé C, Martin F, Fumoleau P, Coudert B, Ghiringhelli F (2011) T-bet expression in intratumoral lymphoid structures after neoadjuvant trastuzumab plus docetaxel for HER2-overexpressing breast carcinoma predicts survival. Br J Cancer 105(3): 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Glimcher LH, Lord GM (2013) T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol 13(11): 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JP, Sherman ML, Fisher GL, Buchanan LJ, Larsen G, Atkins MB, Sosman JA, Dutcher JP, Vogelzang NJ, Ryan JL (1997) Effects of single-dose interleukin-12 exposure on interleukin-12-associated toxicity and interferon-gamma production. Blood 90(7): 2541–2548. [PubMed] [Google Scholar]
- Li J, Jie HB, Lei Y, Gildener-Leapman N, Trivedi S, Green T, Kane LP, Ferris RL (2015) PD-1/SHP-2 inhibits Tc1/Th1 phenotypic responses and the activation of T cells in the tumor microenvironment. Cancer Res 75(3): 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O'Shea JJ (2001) T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA 98(26): 15137–15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar SS, Wang B, Aguilar-Santelises M, Raja SM, Uhlin-Hansen L, Podack E, Trapani JA, Froelich CJ (2002) Cytotoxic cell granule-mediated apoptosis: perforin delivers granzyme B-serglycin complexes into target cells without plasma membrane pore formation. Immunity 16(3): 417–428. [DOI] [PubMed] [Google Scholar]
- Mocellin S, Marincola FM, Young HA (2005) Interleukin-10 and the immune response against cancer: a counterpoint. J Leukoc Biol 78(5): 1043–1051. [DOI] [PubMed] [Google Scholar]
- Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE Jr, Stark GR, Kerr IM (1993) Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J 12(11): 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar I, Schischmanoff PO, Baran-Marszak F, Deglesne PA, Youlyouz-Marfak I, Pampin M, Feuillard J, Bornkamm GW, Chelbi-Alix MK, Fagard R (2008) Novel function of STAT1beta in B cells: induction of cell death by a mechanism different from that of STAT1alpha. J Leukoc Biol 84(6): 1604–1612. [DOI] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Walter MR (2004) Interferons, interferon-like cytokines, and their receptors. Immunol Rev 202: 8–32. [DOI] [PubMed] [Google Scholar]
- Reppert S, Boross I, Koslowski M, Tureci O, Koch S, Lehr HA, Finotto S (2011) A role for T-bet-mediated tumour immune surveillance in anti-IL-17A treatment of lung cancer. Nat Commun 2: 600. [DOI] [PubMed] [Google Scholar]
- Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr (1992) Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA 89(16): 7836–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH (2000) A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100(6): 655–669. [DOI] [PubMed] [Google Scholar]
- Trapani JA, Smyth MJ (2002) Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol 2(10): 735–747. [DOI] [PubMed] [Google Scholar]
- Travis WD, Travis LB, Devesa SS (1995) Lung cancer. Cancer 75(1 Suppl): 191–202. [DOI] [PubMed] [Google Scholar]
- Van Overmeire E, Laoui D, Keirsse J, Bonelli S, Lahmar Q, Van Ginderachter JA (2014) STAT of the union: dynamics of distinct tumor-associated macrophage subsets governed by STAT1. Eur J Immunol 44(8): 2238–2242. [DOI] [PubMed] [Google Scholar]
- Yu H, Yang J, Jiao S, Li Y, Zhang W, Wang J (2014) T-box transcription factor 21 expression in breast cancer and its relationship with prognosis. Int J Clin Exp Pathol 7(10): 6906–6913. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.