Abstract
The aim of the present meta-analysis was to investigate the correlation of promoter methylation of the p16 and Ras association domain family 1 isoform A (RASSF1A) genes with the risk of the development of papillary thyroid cancer (PTC). A number of electronic databases were searched without language restrictions as follows: Medline (1966–2013), the Cochrane Library database (Issue 12, 2013), Embase (1980–2013), CINAHL (1982–2013), Web of Science (1945–2013) and the Chinese Biomedical Database (CBM; 1982–2013). A meta-analysis was performed with the use of Stata statistical software. The odds ratios (ORs), ratio differences (RDs) and 95% confidence intervals (95% CIs) were calculated. In the present meta-analysis, eleven clinical cohort studies with a total of 734 patients with PTC were included. The results of the current meta-analysis indicated that the frequency of promoter methylation of p16 in cancer tissues was significantly higher compared with that in normal, adjacent and benign tissues (cancer tissues vs. normal tissues: OR=7.14; 95% CI, 3.30–15.47; P<0.001; cancer tissues vs. adjacent tissues: OR=11.90; 95% CI, 5.55–25.52; P<0.001; cancer tissues vs. benign tissues: OR=2.25; 95% CI, 1.67–3.03; P<0.001, respectively). The results also suggest that RASSF1A promoter methylation may be implicated in the pathogenesis of PTC (cancer tissues vs. normal tissues: RD=0.53; 95% CI, 0.42–0.64; P<0.001; cancer tissues vs. adjacent tissues: RD=0.39; 95% CI, 0.31–0.48; P<0.001; cancer tissues vs. benign tissues: RD=0.39; 95% CI, 0.31–0.47; P<0.001; respectively). Thus, the present meta-analysis indicates that aberrant promoter methylation of p16 and RASSF1A genes may play a crucial role in the pathogenesis of PTC.
Keywords: Ras association domain family 1 isoform A, p16, papillary thyroid cancer, meta-analysis
Introduction
Papillary thyroid cancer (PTC) is a common thyroid malignancy originating from the thyroid follicular cells. It accounts for ~85% of all thyroid cancer cases with an estimated 66,200 new cases in the United States in 2013 (1,2). PTC is more prevalent in females and although it may occur at any age, the peak incidence of PTC occurs between 30 and 50 years (3,4). Despite being considered a low grade malignancy, PTC poses a substantial threat to global health due to the marked rise in the incidence rate of PTC, with an estimated 2.9-fold increase from 2.7 to 7.7 cases per 100,000 individuals in the United States over the past three decades (5,6). Although the exact etiology of PTC remains to be fully elucidated, related studies have proposed an association between the risk of developing PTC and exposure to radiation (4,7). Furthermore, with the rapid development of molecular biology, genetic factors have been demonstrated to play a critical role in increasing susceptibility to PTC (8). Therefore, in recent years, an increasing number of studies have attempted to investigate the exact cellular and molecular mechanisms that lead to the development of PTC. Epigenetic inactivation of tumor suppressor genes, including Ras association domain family 1 isoform A (RASSF1A) and p16, has been suggested to be implicated in PTC development (9,10).
The p16 protein, also known as multiple tumor suppressor 1 or cyclin-dependent kinase inhibitor 2A, is a cell cycle regulator protein belonging to a class of cyclin-dependent kinase (CDK) inhibitory proteins that control two critical cell-cycle control pathways, namely the p16-Rb and p14-p53 pathways (11). It plays an important role in cell cycle regulation by decelerating cell progression from the G1 to the S phase, specifically binding to CDK4, and preventing the phosphorylation and subsequent inactivation of the retinoblastoma protein (pRb) (12). Furthermore, p16 is a tumor suppressor protein that regulates cellular proliferation and growth by acting as a CDK4 inhibitor, and is implicated in the prevention of numerous types of cancer, including breast and pancreatic cancers (13,14). The human p16 gene is located on chromosome 9p21, and consists of 3 exons and 2 introns (15). There is evidence to suggest that inactivation and the aberrant methylation status of p16 is implicated in the pathogenesis and development of PTC (9). It has been widely accepted that methylation is a major etiology for p16 inactivation in PTC (9). Furthermore, the aberrant methylation of the p16 gene may give rise to the lack of p16 protein expression and affect its function, which may in turn provide a selective growth advantage for tumor cells, thereby contributing to an increased risk of PTC (16).
Ras association domain family 1 isoform A (RASSF1A) is a microtubule-binding protein that regulates the activation of RAS effector pathways and has been suggested to contribute to maintaining genomic stability, stabilizing microtubules, as well as regulating cell cycle arrest and mitotic progression (17–19). The RASSF1A gene is located on the small arm of the human chromosome 3p21.3 within an area of common heterozygous and homozygous deletions, which occur frequently in a variety of human tumors (20,21). Clinically, reduced expression or inactivation of RASSF1A has been associated with the pathogenesis of various human cancers, including hepatocellular, gastric and bladder carcinomas (22,23). It should be noted that the inactivation of RASSF1A has been found to be associated with the methylation of its CpG-island promoter region (24). Furthermore, RASSF1A CpG-island methylation may lead to transcriptional silencing and thus inhibit its function of tumor suppression, resulting in the development of PTC (9,25).
Previous studies have indicated that promoter methylation of the p16 and RASSF1A genes may play an important role in the pathogenesis of PTC (9,25). However, other studies have obtained contradictory results regarding the association of aberrant promoter methylation of the p16 and RASSF1A genes with PTC development (26,27). Therefore, the present study attempted to perform a meta-analysis of all eligible studies to provide insights into the role of p16 and RASSF1A promoter methylation on the pathogenesis of PTC.
Materials and methods
Literature search and selection criteria
A range of electronic databases were searched without language restrictions, as follows: Medline (1966–2013), the Cochrane Library database (Issue 12, 2013), Embase (1980–2013), CINAHL (1982–2013), Web of Science (1945–2013) and the Chinese Biomedical Database (CBM) (1982–2013). The following keywords and Medical Subject Headings (MeSH) were used, in conjunction with a highly sensitive search strategy: ‘methylation’, ‘DNA methylation’, ‘demethylation’, ‘hypermethylation’ or ‘promoter methylation’; and ‘thyroid cancer, papillary’, ‘papillary thyroid carcinoma’, ‘PTC’, ‘papillary thyroid cancer’ or ‘papillary thyroid neoplasms’; and ‘cyclin-dependent kinase inhibitor p16’, ‘CDKN2 protein’, ‘p16INK4A protein’, ‘RASSF1 protein, human’ or ‘RASSF1’. A manual search was also conducted to identify other potential studies based on the references listed in the individual studies.
The following criteria were used to determine the eligibility of the included studies: i) the study design was a clinical cohort study that focused on the association between promoter methylation of p16 and RASSF1A and the pathogenesis of PTC; ii) all patients met the diagnostic criteria for PTC; and iii) the study provided sufficient information about the frequency of methylation. Studies that did not meet the inclusion criteria were excluded. The most recently published studies or those with the largest sample size were included when the authors had published several studies using the same subjects.
Data extraction and methodological assessment
Data were systematically extracted by two authors from each included study using a standardized form. The form used for data extraction documented the most relevant items including: study language, publication year, first author's surname, geographical location, design of the study, sample size, the source of the subjects, protein expression levels, source of samples and protein detection method.
Methodological quality was evaluated separately by two observers using the Newcastle-Ottawa Scale (NOS) criteria (28). The NOS criteria included three scores: i) subject selection, 0–4; ii) comparability of subjects, 0–2; and iii) clinical outcome, 0–3. NOS scores ranged from 0–9 and a score ≥7 indicated a study of good quality.
Statistical analysis
Meta-analysis was performed using Stata statistical software (version 12.0; Stata Corporation, College Station, TX, USA). The odds ratios (ORs), ratio differences (RDs) and 95% confidence intervals (95% CIs) were calculated as estimates of the associations. The Z-test was used to estimate the statistical significance of pooled ORs. Heterogeneity amongst the studies was estimated by the Cochran's Q and I2 tests (29). If the Q-test revealed a value of P<0.05 or the I2 test exhibited a value >50%, which indicates significant heterogeneity, a random-effects model was conducted, or else a fixed-effects model was used. The reasons for the heterogeneity were also explored using meta-regression and subgroup analyses. In order to evaluate the effect of single studies on the overall estimate, a sensitivity analysis was performed. Funnel plots and Egger's linear regression test were applied to investigate publication bias (30).
Results
Study selection and characteristics of included studies
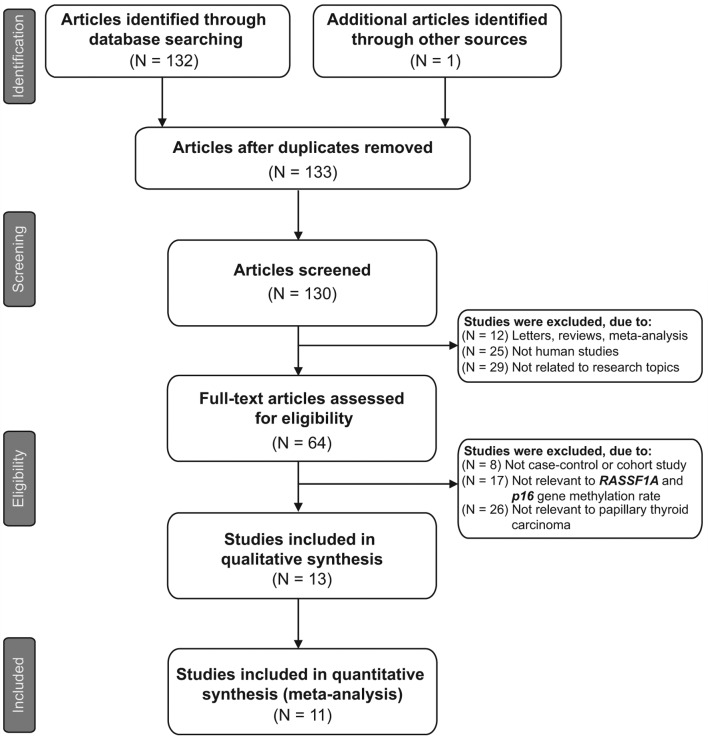
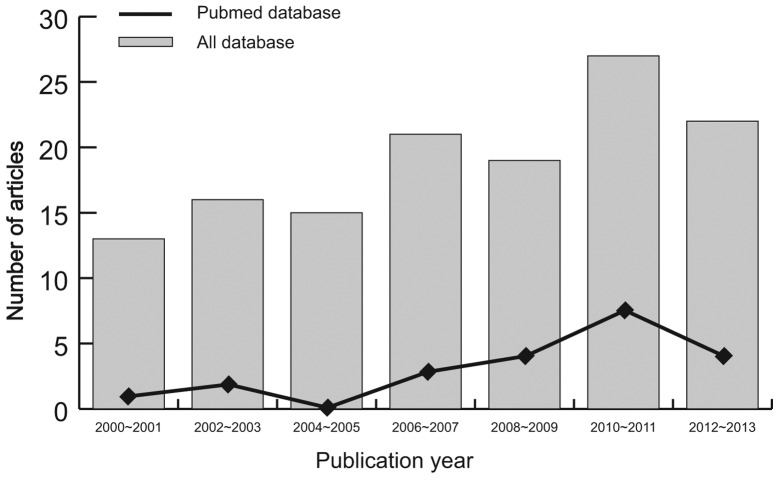
Initially, the highly sensitive search strategy identified 133 studies, of which three were duplicates. The titles and abstracts of all studies were reviewed and 66 studies were excluded. The full texts were subsequently reviewed and a further 51 studies were excluded. Fig. 1 shows the selection process used to identify eligible studies. A total of 11 clinical cohort studies with 734 patients with PTC met the inclusion criteria for qualitative data analysis (9,16,26,27,31–37). Publication years of the eligible studies ranged from 2002–2013. The distribution of the numbers of topic-related studies in electronic databases during the last decade is shown in Fig. 2. Overall, 10 studies were conducted among Asian populations and one study among the Caucasian population. The methylation-specific polymerase chain reaction (PCR; MSP) method was used in 10 studies and one study used the combined bisulfite restriction analysis (COBRA) method. The study characteristics and methodological quality are summarized in Table I.
Figure 1.
Flow chart showing the study selection procedure. A total of 11 cohort studies for quantitative analysis were included in the present meta-analysis.
Figure 2.
Distribution of the number of topic-related studies in the electronic databases during the last decade.
Table I.
Baseline characteristics and methodological quality of all included studies.
| Authors | Year | Country | Language | Ethnicity | No. of cases | Gender (M/F) | Age (years) | Gene | Sample | Method | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Li et al (32) | 2013 | China | Chinese | Asian | 63 | 14/60 | 41±11.7a, 52±11.8b | p16 | Tissue | nMSP | 8 |
| Wang et al (16) | 2013 | China | English | Asian | 95 | - | - | p16 | Tissue | MSP | 6 |
| Mohammadi-asl et al (9) | 2011 | Iran | English | Asian | 50 | - | 56b | p16/RASSF1A | Tissue | COBRA | 7 |
| Wang et al (36) | 2009 | China | Chinese | Asian | 187 | - | - | p16 | Blood | MSP | 6 |
| Peng et al (33) | 2006 | China | Chinese | Asian | 34 | - | - | p16 | Tissue | MSP | 5 |
| Huang et al (26) | 2006 | China | Chinese | Asian | 60 | - | - | p16 | Tissue | MSP | 6 |
| Schagdarsurengin et al (27) | 2002 | USA | English | Caucasian | 18 | - | - | p16 | Tissue | MSP | 5 |
| Qu and Xue (34) | 2012 | China | Chinese | Asian | 28 | 6/22 | 43 (28–68) | RASSF1A | Blood | MSP | 6 |
| Dai et al (31) | 2011 | China | Chinese | Asian | 82 | 15/35 | 40.0±12.0 | RASSF1A | Tissue | MSP | 8 |
| Tang and Su (35) | 2010 | China | Chinese | Asian | 34 | 5/29 | 41 (20–79) | RASSF1A | Blood | MSP | 6 |
| Wang et al (37) | 2009 | China | Chinese | Asian | 83 | - | - | RASSF1A | Tissue | MSP | 7 |
Methylated
unmethylated; MSP, methylation-specific polymerase chain reaction; COBRA, combined bisulfite restriction analysis; NOS, Newcastle-Ottawa scale; nMSP, nested MSP.
Quantitative data synthesis
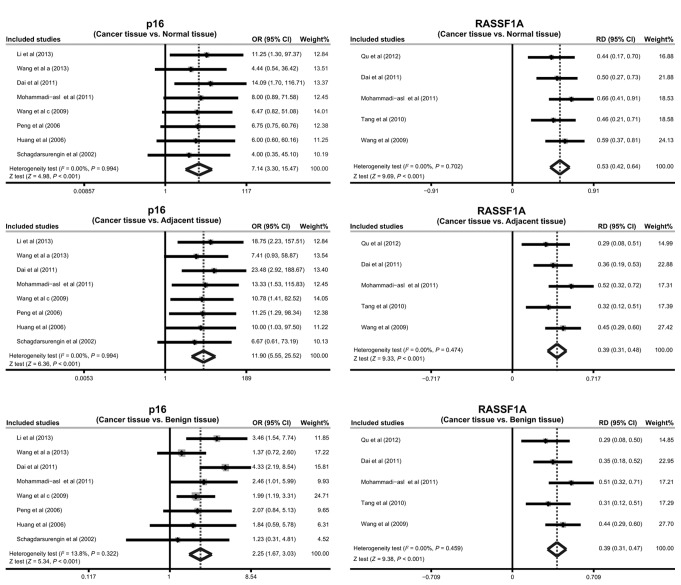
The results of the present meta-analysis indicate that the frequency of promoter methylation of p16 in the cancer tissues was significantly higher compared with that in the normal, adjacent and benign tissues (cancer tissues vs. normal tissues: OR=7.14; 95% CI, 3.30–15.47; P<0.001; cancer tissues vs. adjacent tissues: OR=11.90; 95% CI, 5.55–25.52; P<0.001; cancer tissues vs. benign tissues: OR=2.25, 95% CI: ~1.67–3.03, P<0.001; respectively; Fig. 3). The results also suggest that RASSF1A promoter methylation may be implicated in the pathogenesis of PTC (cancer tissues vs. normal tissues: RD=0.53; 95% CI, 0.42–0.64; P<0.001; cancer tissues vs. adjacent tissues: RD=0.39; 95% CI, 0.31–0.48; P<0.001; cancer tissues vs. benign tissues: RD=0.39; 95% CI, 0.31–0.47; P<0.001; respectively; Fig. 3).
Figure 3.
Forest plots for the correlation between p16 and ras association domain family 1 isoform A (RASSF1A) promoter methylation and susceptibility to papillary thyroid cancer. CI, confidence interval.
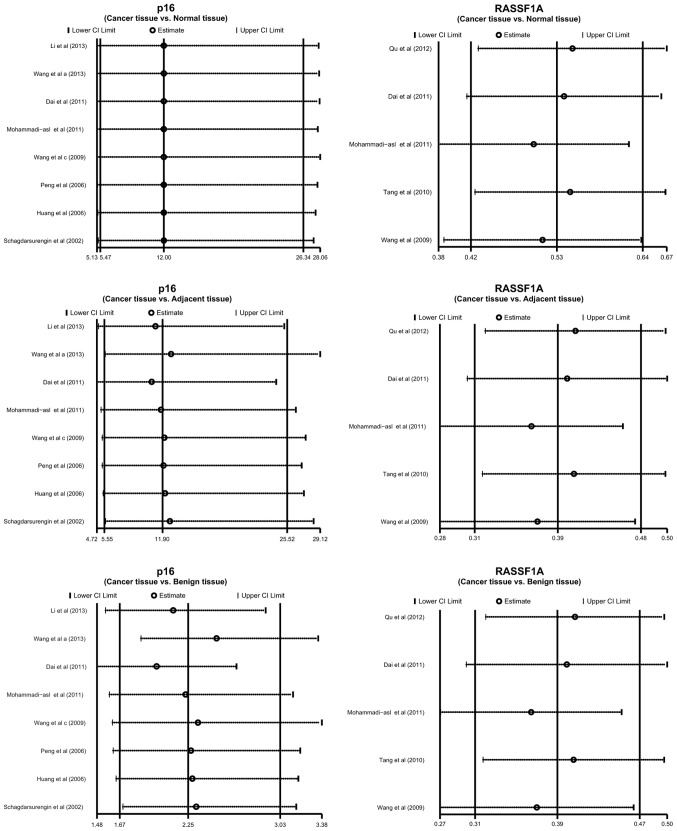
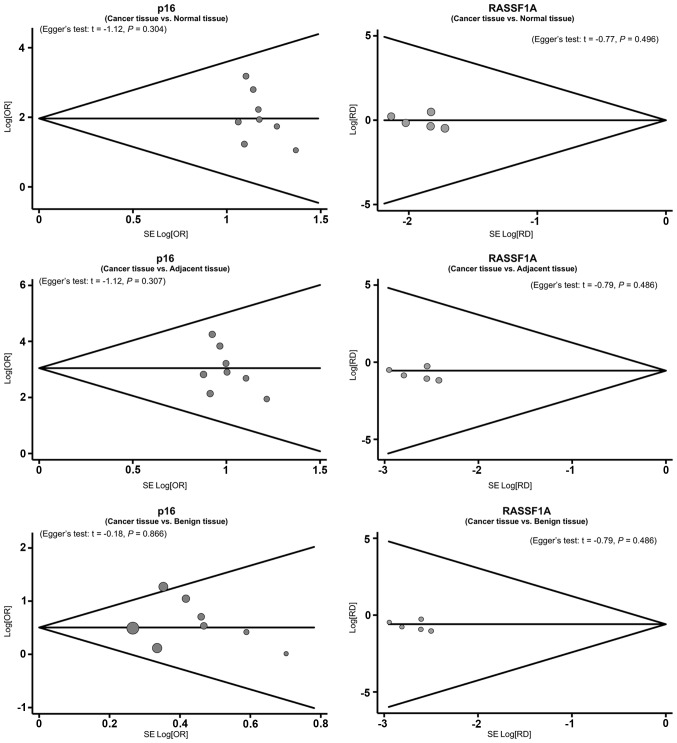
Sensitivity analysis revealed that no single study was able to influence the pooled ORs and RDs (Fig. 4). The funnel plots also demonstrated no evidence of clear asymmetry (Fig. 5). Furthermore, Egger's test did not exhibit strong statistical evidence for publication bias (all P>0.05).
Figure 4.
Sensitivity analysis of the odds ratio coefficients of the correlation between p16 and ras association domain family 1 isoform A (RASSF1A) promoter methylation and susceptibility to papillary thyroid cancer. CI, confidence interval.
Figure 5.
Funnel plot of publication bias on the correlation between p16 and ras association domain family 1 isoform A (RASSF1A) promoter methylation and susceptibility to papillary thyroid cancer. SE, standard error.
Discussion
The present meta-analysis was carried out to determine whether promoter methylation of p16 and RASSF1A is an alternative mechanism for the development and progression of PTC. In the current meta-analysis, promoter methylation of the p16 gene was a frequent and early occurrence in benign to low/high aggressive tumors in patients with PTC, indicating that the methylation profile of p16 may play a significant role in the pathogenesis of PTC. A potential explanation for this is that promoter methylation of p16, an important tumor suppressor gene, may result in transcriptional silencing and consequently, the loss of p16 protein function which is believed to modulate the activity of CDK (38). The CDK inhibitor p16 acts as a negative cell cycle regulator through binding to CDK4 and CDK6, as well as inducing G1 phase arrest in the molecular machinery of the cell cycle by interfering with cyclin D-CDK4 complexes (39). Therefore, it is plausible that p16 promoter methylation may lead to its impaired transcription, contributing to the loss of a designated regulatory mechanism to block cell cycle progression and leading to the uncontrolled growth of genetically damaged cells. The current results are consistent with a previous study reporting that functional repression of the p16 gene, resulting from promoter methylation, plays a vital role in neoplastic progression and tumor dedifferentiation in PTC (40).
The results of the current meta-analysis also demonstrated that the frequency of promoter methylation of RASSF1A in cancer tissues was significantly higher compared with that in normal, adjacent and benign tissues. This implies that RASSF1A promoter methylation may play a causative role in the malignancy of PTC. Despite the precise role of RASSF1A promoter methylation in the development and progression of PTC, its mechanism remains unclear. It is hypothesized that the promoter methylation of RASSF1A may contribute to its silencing and inactivation (41). RASSF1A regulates the activation of RAS effector pathways and has been suggested to have a significant function in the maintenance of genomic stability, cell cycle control, the modulation of apoptosis and cell motility and invasion (19,42). Furthermore, absent RASSF1A expression is considered to be associated with a change in the functional pathway through which RASSF1A may inhibit tumorigenesis (22). The results of a previous association study also demonstrated that the RASSF1A promoter region was methylated in 71% of patients with PTC (27). Thus, the results of the present study are in accordance with previous studies in that they indicate that promoter methylation of the p16 and RASSF1A genes may be implicated in the pathogenesis of PTC. This suggests that p16 and RASSF1A promoter methylation may be a candidate biomarker for the diagnosis and prognosis of PTC.
The current meta-analysis has several limitations that should be acknowledged. First, the results lacked sufficient statistical power to assess the association between p16 and RASSF1A promoter methylation and the pathogenesis of PTC due to the small number of studies included. Since certain studies had small sample sizes and standard deviations, it meant that the current meta-analysis induced fairly wide confidence intervals that restrained the confidence in the conclusions drawn. Furthermore, the small number of studies may constrain the general applicability of the results obtained and consequently the cognitive function of the present meta-analysis should be regarded as preliminary. Secondly, the meta-analysis was a retrospective study that may have led to subject selection bias and thus had an impact on the reliability of the results. Thirdly, the meta-analysis failed to obtain original data from the included studies, which may have limited the further evaluation of the potential role of p16 and RASSF1A promoter methylation in the development of PTC. Although the present study has several limitations, to the best of our knowledge, it is the first meta-analysis focusing on the association between p16 and RASSF1A promoter methylation and the development of PTC. Furthermore, a highly sensitive literature search strategy was performed of the electronic databases. A manual search of the reference lists from the relevant studies was also conducted to identify other potential studies. The selection process to select eligible studies was based on strict inclusion and exclusion criteria. Importantly, rigorous statistical analysis of single nucleotide polymorphism (SNP) data provided a basis for the pooling of information from individual studies.
In conclusion, promoter methylation of p16 and RASSF1A genes may be implicated in the pathogenesis of PTC, suggesting that the promoter methylation of p16 and RASSF1A may be a candidate biomarker for the diagnosis and prognosis of PTC. However, due to the limitations acknowledged above, further studies with larger sample sizes and more detailed information are required in order for a more representative and precise statistical analysis to be possible.
Acknowledgements
The authors would like to acknowledge the reviewers for their helpful comments on this study.
References
- 1.Sipos JA, Mazzaferri EL. Thyroid cancer epidemiology and prognostic variables. Clin Oncol (R Coll Radiol) 2010;22:395–404. doi: 10.1016/j.clon.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Swierniak M, Wojcicka A, Czetwertynska M, et al. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E1401–E1409. doi: 10.1210/jc.2013-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonklaas J, NoguerasGonzalez G, Munsell M, et al. The impact of age and gender on papillary thyroid cancer survival. J Clin Endocrinol Metab. 2012;97:E878–E887. doi: 10.1210/jc.2011-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LiVolsi VA. Papillary thyroid carcinoma: an update. Mod Pathol. 2011;24(Suppl 2):S1–S9. doi: 10.1038/modpathol.2010.129. [DOI] [PubMed] [Google Scholar]
- 5.Hughes DT, Haymart MR, Miller BS, Gauger PG, Doherty GM. The most commonly occurring papillary thyroid cancer in the United States is now a microcarcinoma in a patient older than 45 years. Thyroid. 2011;21:231–236. doi: 10.1089/thy.2010.0137. [DOI] [PubMed] [Google Scholar]
- 6.Enewold L, Zhu K, Ron E, et al. Rising thyroid cancer incidence in the United States by demographic and tumor characteristics, 1980–2005. Cancer Epidemiol Biomarkers Prev. 2009;18:784–791. doi: 10.1158/1055-9965.EPI-08-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Detours V, Delys L, Libert F, et al. Genome-wide gene expression profiling suggests distinct radiation susceptibilities in sporadic and post-Chernobyl papillary thyroid cancers. Br J Cancer. 2007;97:818–825. doi: 10.1038/sj.bjc.6603938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing M. Prognostic utility of BRAF mutation in papillary thyroid cancer. Mol Cell Endocrinol. 2010;321:86–93. doi: 10.1016/j.mce.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammadiasl J, Larijani B, Khorgami Z, et al. Qualitative and quantitative promoter hypermethylation patterns of the p16, TSHR, RASSF1A and RARβ2 genes in papillary thyroid carcinoma. Med Oncol. 2011;28:1123–1128. doi: 10.1007/s12032-010-9587-z. [DOI] [PubMed] [Google Scholar]
- 10.Kunstman JW, Korah R, Healy JM, Prasad M, Carling T. Quantitative assessment of RASSF1A methylation as a putative molecular marker in papillary thyroid carcinoma. Surgery. 2013;154:1255–1261. doi: 10.1016/j.surg.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 11.AbouZeid AA, Azzam AZ, Kamel NA. Methylation status of the gene promoter of cyclin-dependent kinase inhibitor 2A (CDKN2A) in ovarian cancer. Scand J Clin Lab Invest. 2011;71:542–547. doi: 10.3109/00365513.2011.590224. [DOI] [PubMed] [Google Scholar]
- 12.Shidham VB, Mehrotra R, Varsegi G, et al. p16INK4a immunocytochemistry on cell blocks as an adjunct to cervical cytology: Potential reflex testing on specially prepared cell blocks from residual liquid-based cytology specimens. Cytojournal. 2011;8:1. doi: 10.4103/1742-6413.76379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LutfulKabir FM, Agarwal P, Deinnocentes P, et al. Novel frameshift mutation in the p16/INK4A tumor suppressor gene in canine breast cancer alters expression from the p16/INK4A/p14ARF locus. J Cell Biochem. 2013;114:56–66. doi: 10.1002/jcb.24300. [DOI] [PubMed] [Google Scholar]
- 14.Rabien A, SanchezRuderisch H, Schulz P, et al. Tumor suppressor p16 INK4a controls oncogenic K-Ras function in human pancreatic cancer cells. Cancer Sci. 2012;103:169–175. doi: 10.1111/j.1349-7006.2011.02140.x. [DOI] [PubMed] [Google Scholar]
- 15.Kamb A, ShattuckEidens D, Eeles R, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Pei R, Lu Z, Rao X, Liu B. Methylation of p16 CpG islands correlated with metastasis and aggressiveness in papillary thyroid carcinoma. J Chin Med Assoc. 2013;76:135–139. doi: 10.1016/j.jcma.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Tian Y, Hou Y, Zhou X, Cheng H, Zhou R. Tumor suppressor RASSF1A promoter: p53 binding and methylation. PLoS One. 2011;6:e17017. doi: 10.1371/journal.pone.0017017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CunhaDda S, Simões MI, Viviani DN, et al. Carotid artery rupture following radioiodine therapy for differentiated thyroid carcinoma. Arq Bras Endocrinol Metabol. 2011;55:419–425. doi: 10.1590/S0004-27302011000600009. [DOI] [PubMed] [Google Scholar]
- 19.Gao T, Wang S, He B, et al. The association of RAS association domain family Protein1A (RASSF1A) methylation states and bladder cancer risk: a systematic review and meta-analysis. PLoS One. 2012;7:e48300. doi: 10.1371/journal.pone.0048300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreijerink K, Braga E, Kuzmin I, et al. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci USA. 2001;98:7504–7509. doi: 10.1073/pnas.131216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yee KS, Grochola L, Hamilton G, et al. A RASSF1A polymorphism restricts p53/p73 activation and associates with poor survival and accelerated age of onset of soft tissue sarcoma. Cancer Res. 2012;72:2206–2217. doi: 10.1158/0008-5472.CAN-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura N, Carney JA, Jin L, et al. RASSF1A and NORE1A methylation and BRAFV600E mutations in thyroid tumors. Lab Invest. 2005;85:1065–1075. doi: 10.1038/labinvest.3700306. [DOI] [PubMed] [Google Scholar]
- 23.Dammann R, Schagdarsurengin U, Seidel C, et al. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645–663. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- 24.vanVlodrop IJ, Niessen HE, Derks S, et al. Analysis of promoter CpG island hypermethylation in cancer: location, location, location! Clin Cancer Res. 2011;17:4225–4231. doi: 10.1158/1078-0432.CCR-10-3394. [DOI] [PubMed] [Google Scholar]
- 25.Santoro A, Pannone G, Carosi MA, et al. BRAF mutation and RASSF1A expression in thyroid carcinoma of southern Italy. J Cell Biochem. 2013;114:1174–1182. doi: 10.1002/jcb.24460. [DOI] [PubMed] [Google Scholar]
- 26.Huang P, Li DX, Zhang Y. Methylation of p16 gene and expression of p16 protein in human thyroid neoplasms. J Practical Oncol. 2006;21:49–51. (In Chinese) [Google Scholar]
- 27.Schagdarsurengin U, Gimm O, HoangVu C, et al. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002;62:3698–3701. [PubMed] [Google Scholar]
- 28.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 29.Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 30.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 31.Dai YL, Zhang F, Jiang ZR, et al. Association the methylation of p16 and RASSF1A with papillary thyroid carcinoma. Chin J Int Med. 2011;50:428–429. [Google Scholar]
- 32.Li XF, Jin YL, He ZL, Gu DK. Detecting the abnormal methylation of plasma p16 promoter in patients with thyroid carcinoma by nested-methylation-specific polymerase chain reaction. Chin J Prev Contr Chronic Dis. 2013;21:34–36. (In Chinese) [Google Scholar]
- 33.Peng ZL, Cao RX, Wen GB, Liu JH, Wen F. The methylation of p16 gene in papillary thyroid carcinoma. J Mod Oncol. 2006;14:1501–1503. (In Chinese) [Google Scholar]
- 34.Qu F, Xue WJ. RASSF1A methylation and its clinical roles in papillary thyroid carcinoma. J Nantong Univ (Med Sci) 2012;32:490–492. (In Chinese) [Google Scholar]
- 35.Tang JD, Su XL. Research of CpG island methylation status of NIS and RASSF1A gene promoters in papillary thyroid carcinomas. China J Mod Med. 2010;20:3282–3285. (In Chinese) [Google Scholar]
- 36.Wang C, Shi JH, Wang WY. Expressions of p53 and p16 and their clinical significance in thyroid papillary carcinoma. J Bengbu Med Col. 2009;34:871–873. (In Chinese) [Google Scholar]
- 37.Wang XH, Zhang GC, Liu YL, et al. Detecting the abnormal methylation of RASSF1A in patients with thyroid carcinoma. Shaanxi Med J. 2009;38:790–792. (In Chinese) [Google Scholar]
- 38.Barroeta JE, Baloch ZW, Lal P, et al. Diagnostic value of differential expression of CK19, Galectin-3, HBME-1, ERK, RET, and p16 in benign and malignant follicular-derived lesions of the thyroid: an immunohistochemical tissue microarray analysis. Endocr Pathol. 2006;17:225–234. doi: 10.1385/EP:17:3:225. [DOI] [PubMed] [Google Scholar]
- 39.Lam AK, Lo CY, Leung P, et al. Clinicopathological roles of alterations of tumor suppressor gene p16 in papillary thyroid carcinoma. Ann Surg Oncol. 2007;14:1772–1779. doi: 10.1245/s10434-006-9280-9. [DOI] [PubMed] [Google Scholar]
- 40.Boltze C, Zack S, Quednow C, et al. Hypermethylation of the CDKN2/p16INK4A promotor in thyroid carcinogenesis. Pathol Res Pract. 2003;199:399–404. doi: 10.1078/0344-0338-00436. [DOI] [PubMed] [Google Scholar]
- 41.Xing M, Cohen Y, Mambo E, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64:1664–1668. doi: 10.1158/0008-5472.CAN-03-3242. [DOI] [PubMed] [Google Scholar]
- 42.Fernandes MS, Carneiro F, Oliveira C, Seruca R. Colorectal cancer and RASSF family - a special emphasis on RASSF1A. Int J Cancer. 2013;132:251–258. doi: 10.1002/ijc.27696. [DOI] [PubMed] [Google Scholar]