Abstract
Human epidermal growth factor receptor (HER) family plays an important role in various types of cancers. As a result, antibodies against HER and the mechanism of antigen-antibody binding action are under active investigation. We previously constructed a single-chain variable fragment (ScFv) against HER2, i.e. anti-Her2 ScFv, for expressing in the Escherichia coli. In the present study, we report the optimization of anti-Her2 ScFv expression in an E. coli host of BL21 (DE3) pLysS using response surface methodology based on tuning of three cultivation variables, including isopropyl-beta-D-thiogalactopyranoside (IPTG) concentration, temperature and post-induction time. A model for protein expression according to the Box-Behnken design predicted a maximal anti-Her2 ScFv expression at 37 °C, a post-induction time of 10.45 h and 0.75 mM IPTG. In addition, strategies based on inclusion body isolation and affinity chromatography were applied to purify anti-Her2 ScFv. The purity of the final product for inclusion bodies isolation and purification by Ni-NTA resin were 70 % and 95 %, respectively. The solubilization of the inclusion bodies was carried out using two denaturant agents, guanidine hydrochloride and urea. The present study showed that guanidine hydrochloride was more effective than urea in solubilizing the inclusion bodies.
Keywords: Response surface methodology, Escherichia coli, Single-chain variable fragment (ScFv), Antibody, Human epidermal growth factor receptor (HER)
INTRODUCTION
Single-chain variable fragments (ScFvs) are small antibody fragments consisting of variable regions of light chain and heavy chain linked together by a flexible peptide linker of 10–25 amino acids. These molecules often retain the original antigen-binding site of their parental antibodies, but have rapid biodistribution and better penetration into tumor tissue (1). Furthermore, ScFvs are easy to be genetically engineered for targeted delivery of drugs, toxins, or radioisotopes (2,3,5). ScFvs represent an alternative to intact antibodies in many therapeutic and diagnostic applications and a number of ScFv therapeutics have been under clinical trials (6).
ScFvs have been expressed in various expression systems, among which bacterial expression systems are popular for high-level production of non-glycosylated antibody fragments (7). Being the mostly used bacterial host for the overexpression of recombinant proteins (8), Escherichia coli can also be a suitable host for production of antibody fragments with high concentrations up to 2 g/L (9). Due to these advantages, production of ScFv in E. coli can represent an effective bioprocess for large-scale manufacturing of antibody fragments (10).
High-level expression of heterologous proteins in E. coli usually results in the formation of insoluble aggregates known as inclusion bodies (11). Inclusion bodies are resistant to proteolysis by E. coli proteases and can be easily harvested in a cost-effective downstream processing scheme (12). However, the yield of recombinant protein expression is dependent on a variety of biological and genetic parameters such as expression vector design, promoter strength, expression host strain, codon usage, as well as cultivation conditions (13).
Optimization of cultivation conditions for overexpression of recombinant proteins are traditionally achieved by changing one variable at a time (14,15). This method is not only time-consuming, but also results in misinterpretation of the results when the interaction between different variables is present (16). There are many alternative approaches to simultaneously analyze several variables such as full factorial and fractional factorial designs, response surface designs, and Taguchi method. Taguchi method and response surface methodology (RSM) are the most common multivariate analysis methods (17). Taguchi method can handle discrete variables however, it ignores parameter interactions (18). RSM can be applied to determine the optimum culture conditions for protein expression by simultaneously changing several variables based on a minimum number of experiments. Furthermore RSM can identify potential interactions among experimental variables, based on a reasonable prediction of culture conditions for optimal protein expression (19,20,21,22).
Recently, we have developed a new ScFv against HER2 (23). In the present study, we used RSM to optimize culture conditions for expression of this ScFv in E. coli by selecting three variables including isopropyl-beta-D-thiogalactopyranoside (IPTG) concentration, temperature and post-induction time. In addition, the interaction between these variables on ScFv expression was investigated. The present work also evaluated alternative methodologies for ScFv harvesting and purification.
MATERIALS AND METHODS
Bacterial strains and plasmids
The synthesized anti-Her2-ScFv gene was used as a template for polymerase chain reaction (PCR) to append NdeI and XhoI restriction sites. The amplified fragment was cloned into pTZ57R/T (InsTAcloneTM PCR cloning kit, Thermo Fisher Scientific co, USA) cloning vector. The resulting construct was subsequently digested with NdeI and XhoI, and the obtained fragment containing anti-Her2-ScFv gene was sub-cloned into pET-22b (Novagen, Madison, WI, USA) expression vector using the corresponding restriction sites, in frame with histidine (6xHis) tags. This recombinant vector was transformed into competent E. coli BL21(DE3) pLysS (Novagen, Madison, WI, USA) using the CaCl2 method. A positive clone was sequenced to ensure no mutations and to confirm the correct reading frame for expression.
Expression
For expression of anti-Her2-ScFv protein, a positive recombinant clone was inoculated into 5 ml Luria-Bertani (LB) broth containing 100 μg/ml ampicillin and incubated at 37 °C for 16 h at 180 rpm. This overnight culture was used to inoculate (1 % v⁄v) 50 ml of fresh LB medium in 250 ml Erlenmeyer flasks. Cells were incubated at 37 °C until they reached the exponential phase (an OD600 nm of 0.4-0.6) then expression of protein was induced by addition of IPTG. Initial determination of the expression of anti-Her2-ScFv protein was performed with 1 mM IPTG at 37 °C for 2 h. For the optimization of protein expression, each experiment was performed under various conditions of induction (IPTG concentration, post-induction time and temperature) as described in experimental design. These variables have shown greatest effects on induction conditions in previous studies (24,25). At the end of expression time, cell density of each sample (OD600) was measured and the cells were harvested by centrifugation at 7,500 g for 10 min.
Experimental design and optimization of cultivation by response surface methodology
To systematically evaluate the effects of three independent variables including IPTG concentration (factor A), post-induction time (factor B) and temperature (factor C), on the production of anti-Her2-ScFv in E. coli BL21(DE3) pLysS, an experimental design was developed using Box-Behnken factorial design scheme (26). Each variable was investigated at levels of -1 (the lower value of the variable), +1 (the higher value of the variable) and 0 (the central point of the variable) (Table 1). As a result, a total of 15 experiments were carried out including the one in triplicate at the center point (Table 2). Data analysis of experimental design and surface response methodology was performed using Design Expert software (version 8.0.7.1, Stat-Ease Inc., Minneapolis, USA).
Table 1.
Variables and levels used in the experimental design.
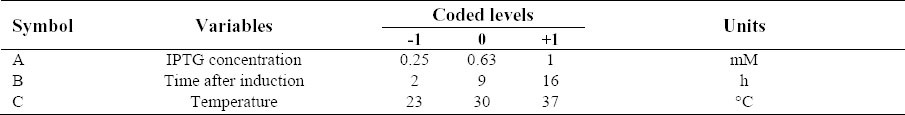
Table 2.
Box–Behnken experimental design of 3 factors and 3 levels.
Isolation of inclusion bodies and purification of protein
To isolate inclusion bodies, the cells were harvested by centrifugation at 7,500 g for 10 min. The pellet was resuspended in 500 μl of TE buffer (20 mM ethylenediaminetetraacetic acid (EDTA), 0.1 M Tris-HCl, pH 8.0). Then 1.5 g/ml of 0.1 mm glass bead was added to the suspension and cell disruption was performed using a Micro Smash MS-100 (Tomy, Japan).
Lysozyme was added to the cell suspension at final concentration of 0.25 mg/ml. After 30 min incubation at room temperature, 500 μl of buffer containing 0.5 mM NaCl and 5% Triton X-100 was added to the mixture and bead mill cell disruption was repeated. The cell suspension was centrifuged at 16,000 g for 20 min at 4 °C and the pellet was washed four times with buffer containing 20 mM EDTA, 0.1 M Tris-HCl and Triton X-100 (The concentration of Triton X-100 was decreased gradually from 2.5 to 0 %) and recovered through centrifugation at 4 °C and 16,000 g for 20 min. The guanidine hydrochloride (GdnHCl) buffer (6 M GdnHCl, 20 mM sodium phosphate, 500 mM NaCl, pH 7.8) was added to the pellet and incubated at room temperature for 1 h to solubilize inclusion bodies.
To purified protein, after cell harvesting, lysozyme was added to the cell suspension. The suspension was sonicated for 1 min and then incubated on ice for 1 additional min. Total sonication time was 5 min. After cell disruption, insoluble fraction was collected and solubilized with the GdnHCl buffer. The mixture was transferred to a purification column containing Ni-NTA agarose resin (Invitrogen, Carlsbad, CA, USA) and protein was purified as described by the manufacturer.
Electrophoretic analysis and quantitation of protein expression by densitometry analysis
The cells were equally diluted to normalize the amount of expressed protein as compared to total cell protein (100 × OD600 μl of PBS was used for suspending cell pellets). The protein samples were analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie Blue R-250. Protein quantification was performed by densitometry analysis of acrylamide gels using TL120 software (Nonlinear Inc, Durham NC, USA). The band intensities of expressed anti-Her2-ScFv were analyzed and normalized to protein molecular weight marker (SM0441, Thermo Fisher Scientific co, USA). The purity of protein was determined by densitometry analysis of polyacrylamide gels using TL120 software (Nonlinear Inc, Durham NC, USA). Purity was calculated as a percentage of total protein (27).
RESULTS
Experimental design and modeling for single-chain variable fragments production
Cultivation for the production of anti-Her2-ScFv in E. coli BL21(DE3) pLysS by changing three variables (i.e. IPTG concentration, post-induction time, and temperature) was performed in shaking flasks so that their individual and synergistic effects on anti-Her2 ScFv expression can be evaluated using a Box-Behnken factorial design. The measured levels of protein expression for the 15 cultures are summarized in Table 2.
A wide range of anti-Her2-ScFv expression from 75 to 233 mg/l was observed under these investigated culture conditions, indicating the necessity of identifying an optimal condition. To evaluate experimental error and reproducibility of the model, cultivation was performed in triplicate under the central point conditions (i.e. 0.63 mM IPTG, 9 h of induction and 30 °C). The anti-Her2-ScFv concentration was 203 ± 5 (mg/l) with the coefficient of variation (defined as the ratio of the standard deviation to the mean) being less than 2.4%.
Correlation of anti-Her2-ScFv concent-ration and the investigated variables was developed using the Design Expert software and was represented by the following quadratic equation.
Protein expression (mg/l)=203.45 + 10.17 A + 50.32 B + 24.41 C + 6.40 AB - 0.85 AC + 2.35 BC - 5.90 A2 - 57.18 B2 - 4.31 C2 (1)
Model analysis and validation
The statistical significance of the above equation was confirmed by an F-test and the analysis of variance (ANOVA) for response surface quadratic model is summarized in Table 3.
Table 3.
ANOVA for Response Surface Quadratic Model.
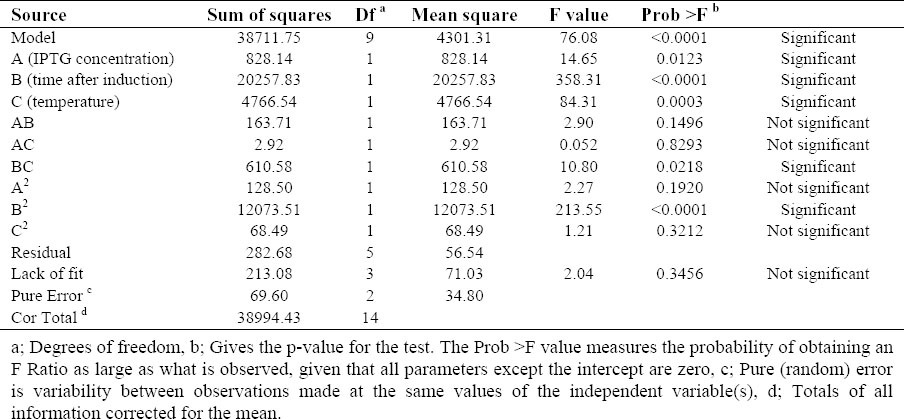
The model correlation coefficient (R-squared) value is high (0.9928) and the “Pred R-Squared” of 0.9086 is in reasonable agreement with the “Adj R-Squared” of 0.9797, implying a good correlation based on the developed model. “Adeq Precision” was used to measure the signal (meaningful information) to noise (unwanted signal) ratio, for which a value greater than 4 is normally desired (28).
The high value of adequate precision (24.345) indicates that this model was of an adequate signal and could be applicable and used to navigate the design space. The “lack of fit F-value” of 2.04 indicates the lack of fit was not significant (0.3456) and there was a 34.56% chance that a “Lack of Fit F-value” could occur because of noise (Table 3). As shown in Table 3, the “Model F-value” of 76.08 with a very low probability value [(Prob > F) < 0.0001] implied that the model is significant and there was only a 0.01% chance that a “Model F-Value” could occur due to the noise.
In this case, A, B, C, BC and B2 are significant model terms (P<0.05). The linear and square terms of post-induction time, the linear terms of temperature and IPTG concentration, and the term of interaction between post-induction time and temperature are highly significant. The other interaction terms AB and AC, and the square terms A2 and C2 are not significant in terms of anti-Her2-ScFv production (P>0.05).
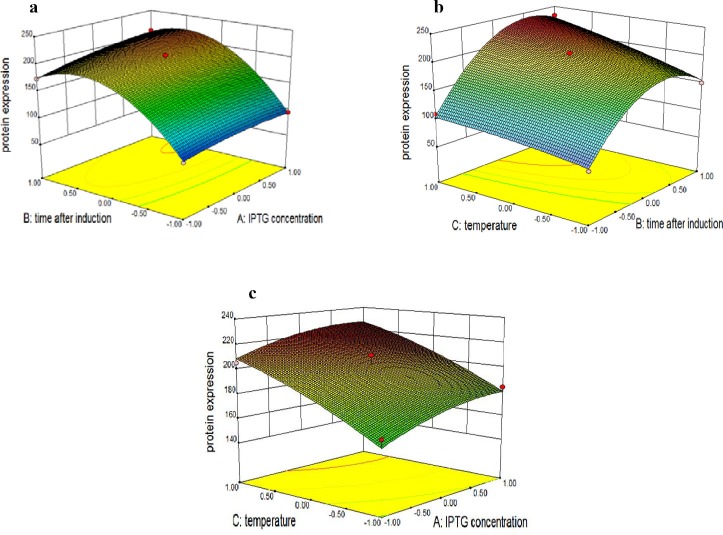
Fig. 1a shows the response surface plot of the dependency of IPTG concentration and post-induction time on anti-Her2-ScFv production when the temperature remains at its zero level (30 °C). Based on the graph, anti-Her2-ScFv expression increases significantly with post-induction time, especially when IPTG concentration is high.
Fig. 1.
Response surface of ScFv expression represents the interaction between two factors in the expression of ScFv (mg/l) by keeping other factor constant. a; IPTG concentration (mM) and time after induction (h), b; time after induction (h) and temperature (°C), c; IPTG concentration (mM) and temperature (°C).
Higher influence of post-induction time is confirmed by the low P value of the coefficient B (<0.0001), as presented in Table 3. On the other hand, the dependency of the post-induction time and temperature on anti-Her2-ScFv production when IPTG concentration is fixed at its zero level (0.63 mM) is presented in Fig. 1.b. The interaction between these two variables is significant as indicated by the low p value of the coefficient BC of 0.0218. (Table 3). As shown in Fig. 1.b, anti-Her2-ScFv production increases significantly with both post-induction time and temperature. However, the increasing trend with post-induction time ceases at a certain point, indicating the minimum dependency of the post-induction time on anti-Her2-ScFv production. Fig. 1.c shows the response surface plot of the dependency of IPTG concentration and temperature on anti-Her2-ScFv production when the post-induction time is kept at its zero level (9 h). According to Table 3, the interaction between IPTG concentration and temperature was not significant due to the high p value of the coefficient AC (P>0.05). As shown in Fig. 1.c, temperature appears to be highly influential on anti-Her2-ScFv production in comparison with IPTG concentration.
The studentized residuals measure the number of standard deviations separating the actual and predicted values. The normal probability plot of the studentized residuals is one of the effective diagnostic methods to evaluate the accuracy and predictability of a model. As Fig. 2 revealed, the normal probability plot of anti-Her2-ScFv production model shows a normal distribution and the linear trend of the data points. The results suggest an excellent adequacy and significance of the quadratic regression model.
Fig. 2.
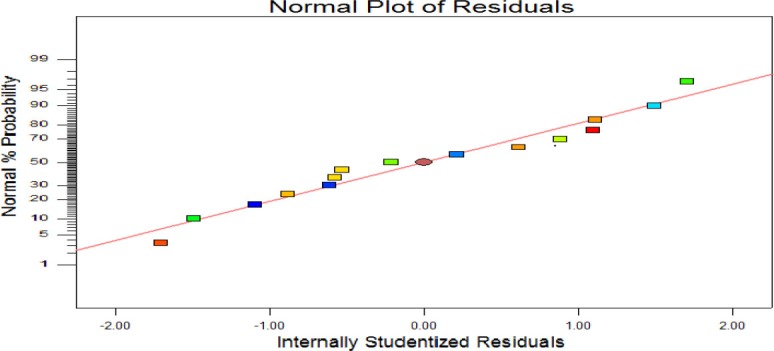
Normal (%) probability plot of the ‘studentized’ residuals for the model of ScFv expression.
Based on the developed model, the optimal culture conditions for ScFv production in E. coli were predicted to be as follows: the temperature of 37 °C, post-induction time of 10.45 h, and 0.75 mM IPTG. These optimal levels led to a ScFv concentration of 236 mg/l. The predicted local optimal culture conditions were used for large scale cultivation with an anti-Her2-ScFv concentration of 240 mg/l, which is close to the predicted optimum.
Purification of anti-Her2-ScFv
We observed that GdnHCl was more effective than urea in solubilizing the inclusion bodies (Fig. 3). For affinity chromatography, the post-sonication cell lysate was subjected to purification by Ni-NTA resin. The fractions obtained after each experiment were analyzed by SDS-PAGE and the purity of anti-Her2-ScFv was quantified using TL120 software. As shown in Fig. 3, a large amount of anti-Her2-ScFv was expressed, representing approximately 30% of total cell protein. The purity of anti-Her2-ScFv in the inclusion body fraction was 35% and the purity increased to 55% after Triton X-100 washing. The purity of the final product for inclusion bodies isolation and affinity chromatography were 70% and 95%, respectively.
Fig. 3.
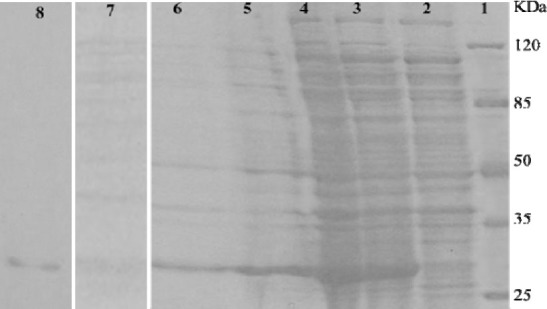
SDS–PAGE of primary isolation of ScFv expressed in E. coli strain BL21(DE3) pLysS in the form of inclusion bodies. Lane 1; molecular weight marker, lane 2; bacterial culture before induction, lane 3; bacterial culture after induction, lane 4; inclusion bodies fraction after centrifugation, lane 5; inclusion bodies fraction after Triton X-100 wash, lane 6; inclusion bodies fraction solubilized with GndHCl, lane 7; inclusion bodies fraction solubilized with Urea, lane 8; purified ScFv with Ni-NTA column.
DISCUSSION
E. coli BL21(DE3) pLysS is a widely used host cell deficient in two protease genes of ompT and lon. This strain also contains pLysS plasmid expressing T7 lysozyme, which is a natural inhibitor of T7 RNA polymerase for tight regulation of the leaky expression of recombinant proteins (29).
The aim of this study was to optimize induction conditions for recombinant anti-Her2-ScFv production in E. coli BL21(DE3) pLysS. We selected three major variables at three levels for evaluation of their effects on anti-Her2-ScFv production. Box-Behnken design was used to formulate our cultivation experiments. This method is effective due to the requirement of fewer experimental runs compared to other RSM designs (30). Jafari and coworkers (31) reported the optimization of culture conditions for anti-keratin ScFv TS1-218 production in the yeast expression system of Pichia pastoris. By evaluating the effects of temperature, pH, and methanol concentration (serving as both an inducer and a carbon source) and formulate the experimental runs using the Box-Behnken design, the anti-keratin ScFv TS1-218 yield was increased by 21 fold compared to that prior to the optimization (31). The present study shows that using optimization a 3 fold increase in expression of ScFv was obtained. The optimum culture conditions were found to be the temperature at 37 °C, a post-induction time of 10.45 h and 0.75 mM IPTG, leading to the anti-Her2-ScFv concentration of 240 mg/l, which is in good agreement with the theoretical value of 236 mg/l predicted based on our developed model.
The IPTG concentration for induction of gene expression ranges widely from 0.005 to 5 mM (32). High IPTG concentration may lead to a significantly reduced growth rate and production of bacterial proteases degrading heterologous proteins. Considering the high cost and potential toxicity of IPTG, determination of optimum concentration of this inducer is important. Larentis and colleagues reported that the optimization led to the improved expression of mature rPsaA in E. coli BL21(DE3) star at a concentration of IPTG 10 fold lower than usually used (25).
Another critical factor influencing the expression of recombinant proteins is cultivation temperature. Santala and coworkers (33) reported that optimal expression of anti-thyroid stimulating hormone (TSH) ScFv in E. coli Origami B occurred at a low temperature of 24 °C. Volontθ and colleagues (34) observed that expression of recombinant protein was optimal at a growth temperature of 37 °C with IPTG addition at the middle-exponential growth phase. On the other hand, formation of inclusion bodies occurs frequently at a high temperature due to the temperature dependency of hydrophobic interactions (35). Zhang and coworkers presented the optimum condition for extracellular human anti-HBsAg ScFv expression in E. coli with a post-induction time of 8 h and 0.5 mM IPTG (36). These studies suggest the significant variance of culture conditions for different host/vector systems.
In the present study, the majority of anti-Her2-ScFv was expressed as inclusion bodies. Therefore, we evaluated various strategies for harvesting and purification of anti-Her2-ScFv. In particular, lysozyme-EDTA treatment facilitated cell disruption. Triton X-100 as a nonionic detergent increased the efficacy of washing for removal of nonspecifically adsorbed proteins. The isolated inclusion bodies were solubilized using both GdnHCl and urea, but GdnHCl appeared to be a more effective chaotropic agent than urea for solubilization of anti-Her2-ScFv inclusion body. The pH of buffer should also be optimized once urea is used for more efficient solubilization. Using this method, inclusion bodies were isolated with 70% purity. These results are in agreement with another study on isolation of inclusion bodies, which reported the isolation of hygromycin B phosphotransferase inclusion bodies with 73% purity (37). However, after cell disruption using sonication and isolation of inclusion bodies following four steps of washing, they used 0.3% sarkosyl to solubilize inclusion bodies.
CONCLUSION
The RSM was proved useful for the optimization of anti-Her2-ScFv production in E. coli. The optimization was performed by varying culture conditions based on only 15 cultivation runs. The utilization of Box-Behnken design simplified the experimental setup and data analysis. In addition, potential interactions between the investigated variables were identified. The optimal culture conditions were temperature of 37 °C, a post-induction time of 10.45 h, and 0.75 mM IPTG, leading to the anti-Her2-ScFv concentration of 240 mg/l. Therefore, these methodologies and results could be exploited for subsequent studies for bioreactor scale-up.
ACKNOWLEDGMENTS
We thank Mrs. Fatemeh Moazen for her excellent technical assistance. The content of this paper is extracted from the Ph.D thesis NO. 189113 submitted by V. Akbari which was financially supported by the Research Department of Isfahan University of Medical Sciences, Isfahan, I.R. Iran.
REFERENCES
- 1.Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res. 1992;52:3402–3408. [PubMed] [Google Scholar]
- 2.Weisser NE, Hall JC. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol Adv. 2009;27:502–520. doi: 10.1016/j.biotechadv.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Adams GP, Schier R, McCall AM, Simmons HH, Horak EM, Alpaugh RK, et al. High affinity restricts the localization and tumor penetration of single-chain fv antibody molecules. Cancer Res. 2001;61:4750–4755. [PubMed] [Google Scholar]
- 4.Cao Y, Marks J, Liu Z, Cheung L, Hittelman W, Rosenblum M. Design optimization and characterization of Her2/neu-targeted immunotoxins: comparative in vitro and in vivo efficacy studies. Oncogene. 2014;33:429–439. doi: 10.1038/onc.2012.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monnier PP, Vigouroux RJ, Tassew NG. In Vivo Applications of Single Chain Fv (Variable Domain)(scFv) Fragments. Antibodies. 2013;2:193–208. [Google Scholar]
- 6.Nelson AL. Antibody fragments. Hope and hype MAbs. 2010;2:77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma R, Boleti E, George A. Antibody engineering: comparison of bacterial, yeast, insect and mammalian expression systems. J Immunol Methods. 1998;216:165–181. doi: 10.1016/s0022-1759(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen R. Bacterial expression systems for recombinant protein production E. coli and beyond. Biotechnol Adv. 2012;30:1102–1107. doi: 10.1016/j.biotechadv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Snedecor B, Nishihara J, Joly J, McFarland N, Andersen D, et al. High-level accumulation of a recombinant antibody fragment in the periplasm of Escherichia coli requires a triple-mutant (degP prc spr) host strain. Biotechnol Bioeng. 2004;85:463–474. doi: 10.1002/bit.20014. [DOI] [PubMed] [Google Scholar]
- 10.Miethe S, Meyer T, Wöhl-Bruhn S, Frenzel A, Schirrmann T, Dübel S, et al. Production of Single Chain Fragment Variable (scFv) Antibodies in Escherichia coli Using the LEX™ Bioreactor. J Biotechnol. 2013;163:105–111. doi: 10.1016/j.jbiotec.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 11.García-Fruitós E. Inclusion bodies: a new concept. Microb Cell Fact. 2010;9:80–85. doi: 10.1186/1475-2859-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatti‐Lafranconi P, Natalello A, Ami D, Doglia SM, Lotti M. Concepts and tools to exploit the potential of bacterial inclusion bodies in protein science and biotechnology. FEBS J. 2011;278:2408–2418. doi: 10.1111/j.1742-4658.2011.08163.x. [DOI] [PubMed] [Google Scholar]
- 13.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011;10:32–37. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czitrom V. One-factor-at-a-time versus designed experiments. Am Stat. 1999;53:126–131. [Google Scholar]
- 15.Frey DD, Engelhardt F, Greitzer EM. A role for” one-factor-at-a-time” experimentation in parameter design. Res Eng Des. 2003;14:65–74. [Google Scholar]
- 16.Araujo PW, Brereton RG. Experimental design II. Optimization. TrAC Trends Anal Chem. 1996;15:63–70. [Google Scholar]
- 17.Myers RH, Khuri AI, Carter WH. Response surface methodology: 1966-l988. Technometrics. 1989;31:137–157. [Google Scholar]
- 18.Antony J. Taguchi or classical design of experiments: a perspective from a practitioner. Sensor Rev. 2006;26:227–230. [Google Scholar]
- 19.Bezerra MA, Santelli RE, Oliveira EP, Villar LS, Escaleira LA. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta. 2008;76:965–977. doi: 10.1016/j.talanta.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Mandenius CF, Brundin A. Bioprocess optimization using design-of-experiments methodology. Biotechnol Prog. 2008;24:1191–1203. doi: 10.1002/btpr.67. [DOI] [PubMed] [Google Scholar]
- 21.Myers RH, Anderson-Cook CM. New Jersey: John Wiley & Sons; 2009. Response surface methodology: process and product optimization using designed experiments. [Google Scholar]
- 22.Suhail AH, Ismail N, Wong S, Ialil NA. Response Surface Methodology. J Appl Sci. 2011;11:308–315. [Google Scholar]
- 23.Akbari V, Mir Mohammad Sadeghi H, Jafrian-Dehkordi A, Abedi D, Chou CP. Functional expression of a single-chain antibody fragment against human epidermal growth factor receptor 2 (HER2) in Escherichia coli. J Ind Microbiol Biotechnol. 2014;20(41) 6:947–956. doi: 10.1007/s10295-014-1437-0. [DOI] [PubMed] [Google Scholar]
- 24.Sadeghi H, Rabbani M, Rismani E, Moazen F, Khodabakhsh F, Dormiani K, et al. Optimization of the expression of reteplase in Escherichia coli. Res Pharm Sci. 2011;6:87–92. [PMC free article] [PubMed] [Google Scholar]
- 25.Larentis AL, Argondizzo APC, Esteves GdS, Jessouron E, Galler R, Medeiros MA. Cloning and optimization of induction conditions for mature PsaA (pneumococcal surface adhesin A) expression in Escherichia coli and recombinant protein stability during long-term storage. Protein Expr Purif. 2011;78:38–47. doi: 10.1016/j.pep.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Box GE, Behnken D. Some new three level designs for the study of quantitative variables. Technometrics. 1960;2:455–475. [Google Scholar]
- 27.Wu X, Zhang L, Zhang J, Zhang C, Zhu L, Shi Y. Recombinant early secreted antigen target 6 protein as a skin test antigen for the specific detection of Mycobacterium tuberculosis infection. Clin Exp Immunol. 2008;152:81–87. doi: 10.1111/j.1365-2249.2008.03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong C, Haiyun W. Optimization of volatile fatty acid production with co-substrate of food wastes and dewatered excess sludge using response surface methodology. Bioresource Technol. 2010;101:5487–593. doi: 10.1016/j.biortech.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira SC, Bruns R, Ferreira H, Matos G, David J, Brandão G, et al. Box-Behnken design: An alternative for the optimization of analytical methods. Anal Chim Acta. 2007;597:179–186. doi: 10.1016/j.aca.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Jafari R, Sundström BE, Holm P. Optimization of production of the anti-keratin 8 single-chain Fv TS1-218 in Pichia pastoris using design of experiments. Microb Cell Fact. 2011;10:34–41. doi: 10.1186/1475-2859-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie L, Hall D, Eiteman M, Altman E. Optimization of recombinant aminolevulinate synthase production in Escherichia coli using factorial design. Appl Microbiol Biotechnol. 2003;63:267–273. doi: 10.1007/s00253-003-1388-2. [DOI] [PubMed] [Google Scholar]
- 33.Santala V, Lamminmäki U. Production of a biotinylated single-chain antibody fragment in the cytoplasm of Escherichia coli. J Immunol methods. 2004;284(1):165–75. doi: 10.1016/j.jim.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Volontè F, Piubelli L, Pollegioni L. Optimizing HIV-1 protease production in Escherichia coli as fusion protein. Microb Cell Fact. 2011;10:53–59. doi: 10.1186/1475-2859-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahdev S, Khattar SK, Saini KS. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Wang J, Zhong R, Niu B. Optimization of human anti-HBsAg scFv secretary expression in Escherichia coli. Chin J Exp Clin Virol. 2009;23(1):50–5. [PubMed] [Google Scholar]
- 37.Zhuo Q, Piao J-h, Wang R, Yang X-G. Refolding and purification of non-fusion HPT protein expressed in Escherichia coli as inclusion bodies. Protein Expr Purif. 2005;41:53–60. doi: 10.1016/j.pep.2004.12.026. [DOI] [PubMed] [Google Scholar]