SUMMARY
In budding yeast, the actin-binding protein Bud6 cooperates with formins Bni1 and Bnr1 to catalyze the assembly of actin filaments. The nucleation-enhancing activity of Bud6 requires both a “core” domain that binds to the formin and a “flank” domain that binds monomeric actin. Here we describe the structure of the Bud6 flank domain in complex with actin. Two helices in Bud6flank interact with actin; one binds in a groove at the barbed-end of the actin monomer in a manner closely resembling the helix of WH2 domains, a motif found in many actin nucleation factors. The second helix rises along the face of actin. Mutational analysis verifies the importance of these Bud6-actin contacts for nucleation-enhancing activity. The Bud6 binding site on actin overlaps with that of the formin FH2 domain and is also incompatible with inter-subunit contacts in F-actin, suggesting that Bud6 interacts only transiently with actin monomers during filament nucleation.
INTRODUCTION
Controlled assembly of actin filaments underlies diverse cellular processes, including adhesion, migration, myosin-based intracellular transport, and cytokinesis (Pollard and Cooper, 2009). Assembly of most actin-based structures requires the activity of one or both of two major classes of actin nucleating proteins, the Arp2/3 complex and formins (Goode and Eck, 2007; Pollard, 2007). The Arp2/3 complex assembles the branched networks of actin filaments found at the leading edge of migrating cells and sites of endocytosis. The multi-subunit assembly of the Arp2/3 complex includes a substructure that binds the side of existing actin filaments, as well as two actin-like subunits that form a “seed” for nucleating a new “daughter” filament that elongates as a branch from the anchoring “mother” filament. Filament nucleation by the Arp2/3 complex is controlled by binding of WASP/WAVE-family proteins, which serve as nucleation promoting factors (NPFs) by bringing the actin-like subunits of the complex into proper register for nucleation and by recruiting actin monomers via short actin-binding motifs termed WASP homology-2 (WH2) domains (Campellone and Welch, 2011).
Formins employ a structurally distinct mechanism to nucleate linear, unbranched filaments that give rise to diverse actin structures, including stress fibers, filopodia, cytokinetic rings, and polarized cables. In formins, the FH2 domain (formin homology-2 domain) is required and sufficient for actin filament nucleation and elongation in vitro (Pring et al., 2003;Zigmond et al., 2003;Moseley et al., 2004). The FH2 domain is a dimer consisting of two rod-shaped domains connected by flexible linkers at either end to form a closed ring (Otomo et al., 2005b; Xu et al., 2004). Each of these rod-shaped domains can bridge between two actin subunits, and the dimer is thought to seed a nascent filament by capturing or organizing two or three actin subunits into a filament-like structure. After a filament is nucleated, the dimeric FH2 domain remains attached to the growing barbed end of the filament as additional subunits are incorporated. This stair-stepping behavior, termed processive capping, is a hallmark of formin function.
Regions flanking the FH2 domain can aid in actin nucleation and elongation. The proline-rich FH1 domain binds profilin and thereby recruits profilin-bound actin monomers to the growing filament end (Paul and Pollard, 2009; Kovar et al., 2006). More recently it has been shown that additional ‘tail’ segments just C-terminal to the FH2 domain can bind monomeric actin and are important for efficient nucleation and elongation (Gould et al., 2011; Heimsath and Higgs, 2012; Vizcarra et al., 2014). The actin assembly activity of formins is controlled in part by regulatory domains; in diaphanous-family formins, binding of GTP-loaded Rho GTPases to N-terminal domains releases autoinhibitory interactions with the C-terminal diaphanous autoregulatory domain (DAD) (Nezami et al., 2010; Nezami et al., 2006; Otomo et al., 2005a; Otomo et al., 2010; Rose et al., 2005; Maiti et al., 2012; Li and Higgs, 2003).
Formins are ubiquitously expressed in eukaryotes, and constitute a large gene/protein family including 15 distinct formins in humans (Higgs and Peterson, 2005). Like the Arp2/3 complex, some formins directly interact with actin-monomer binding proteins that act as NPFs in promoting formin-mediated nucleation. The Drosophila formin Cappuccino, as well as its mammalian orthologs FMN1 and FMN2, bind to Spire, a protein with actin nucleation activity conferred by a tandem array of four WH2 domains (Bosch et al., 2007; Quinlan et al., 2005). Spire binds to the C-terminal tail of the formin, apparently blocking its contribution to filament nucleation (Pechlivanis et al., 2009; Quinlan et al., 2007; Rasson et al., 2014; Vizcarra et al., 2011). However, Spire associates with the barbed end of filaments (Ito et al., 2011) and interacts with the C-terminal tail of the formin FMN2 to recruit it to the barbed end, promoting processive elongation in vitro (Montaville et al., 2014). In vivo, both are required for assembly of an actin mesh in the course of Drosophila oogenesis, and disruption of the gene encoding either protein yields a similar phenotype (Pfender et al., 2011). The mammalian diaphanous-family formin mDia1 acts in concert with the adenomatous polyposis coli protein (APC), which aids nucleation by recruiting actin monomers despite the fact that it lacks recognizable WH2 domains (Breitsprecher et al., 2012; Okada et al., 2010). Upon filament polymerization, APC dissociates from mDia1, remaining at the nucleation site, while the formin tracks the barbed end of the growing filament. In budding yeast, the actin monomer-interacting protein Bud6 (also called Aip3) binds to the DAD-containing tail regions of both yeast formins, Bni1 and Bnr1, to stimulate actin nucleation in vitro and promote actin cable formation in vivo (Graziano et al., 2011; Graziano et al., 2013; Moseley et al., 2004).
The domain structure of Bud6 contains an N-terminal region of unknown structure that is required for its localization to the bud tip and neck, and for cortical capture of astral microtubules, perhaps via direct binding to microtubules and/or EB1 (Delgehyr et al., 2008; Ten Hoopen et al., 2012). The C-terminal portion of the protein (residues 550-788, C-Bud6) directly binds to formins Bni1 and Bnr1 and functions as an NPF; it stimulates actin nucleation by the formin, but has little if any effect on the rate of elongation (Graziano et al., 2011; Graziano et al., 2013). The C-terminal region can be further subdivided into a “core” region (Bud6core, residues 550-688) that is sufficient to bind formins, and a “flank” (Bud6flank, residues 699-788) that binds monomeric actin (Figure 1A), and crystal structures reveal that Bud6core forms a rod-shaped dimer ~120 Å in length (Tu et al., 2012). While no crystal structure of Bud6 in complex with formins is available, biochemical studies indicate that it binds the formin with a 2:1 stoichiometry; that is, two Bud6 dimers bind to a single formin dimer, with one Bud6 dimer engaging each tail of the formin. Our studies of Bud6flank revealed that it binds monomeric actin with a 1:1 stoichiometry, and thus Bud6 could coordinate as many as four actin subunits in association with the formin dimer (Tu et al., 2012).
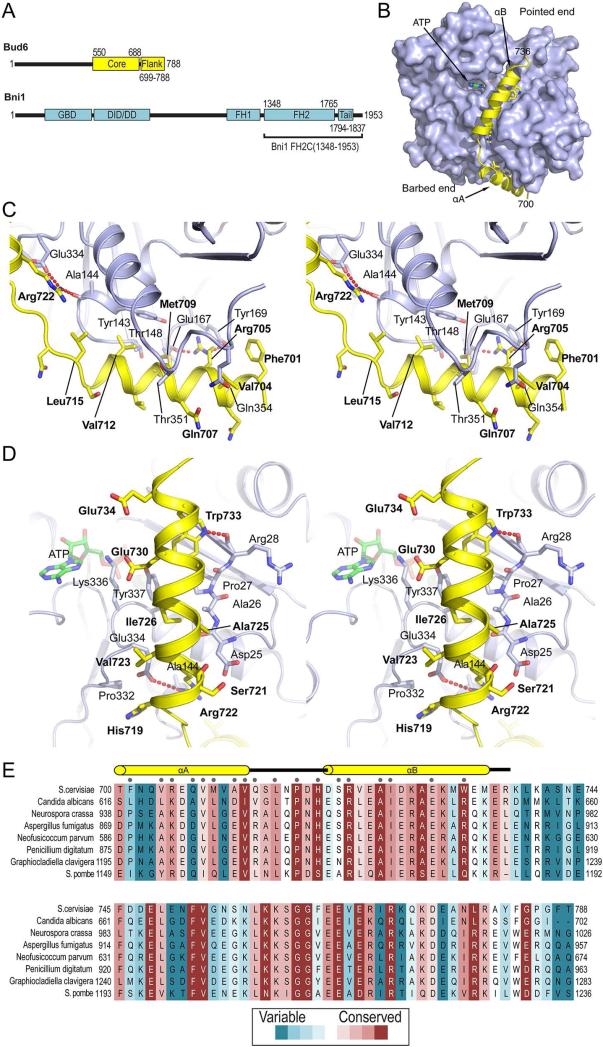
Figure 1. Crystal structure of Bud6flank in complex with actin.
A, Domain structures of Bud6 and the formin Bni1. B, Overview of the complex. Actin is shown in a surface representation, and Bud6 as a yellow ribbon. ATP is present in the nucleotide-binding cleft of actin. C and D, Stereo views of the interactions of helices αA and αB, respectively. Actin is shown as a blue ribbon, Bud6 as a yellow ribbon. Selected side chains are shown in stick form, Bud6 residues are labeled in bold text and actin residues in plain text. Electron density in the region of helix αA is shown in Figure S1. E, Evolutionary conservation of Bud6flank. Selected Bud6 sequences are aligned and shaded according to rate of evolutionary variation based on analysis of sequences of Bud6 from 46 fungal species as previously described (Tu et al., 2012). Analysis was carried out with the CONSURF server (Ashkenazy et al., 2010); shading ranges from dark magenta to teal (most conserved to most variable, respectively). Secondary structure elements are indicated above the alignment, and residues in contact with actin (as determined by a 4.0 Å distance cutoff) are indicated by grey dots.
Developing a mechanistic understanding of how formins and their associated NPFs promote nucleation and actin assembly is a major goal in the field, and will require detailed structural information about the interactions of each NPF with its formin partner and with actin. Until now, such information has been limited to Spire, for which crystal structures of its KIND domain in complex with the FMN1 tail (Vizcarra et al., 2011; Zeth et al., 2011) and its WH2 domains in complex with actin are available (Chen et al., 2012; Ducka et al., 2010). Here we investigated Bud6-actin interactions and determined the crystal structure of Bud6flank in complex with G-actin. We find that a ~40-residue segment of Bud6flank binds to G-actin, forming two helices that interact extensively with the actin monomer. Helix A packs in the groove between subdomains 1 and 3 in a manner reminiscent of the helical portion of the WH2 motif. Bud6flank lacks the “LKKT” sequence that is characteristic of the WH2 domain, and contains instead Helix B, which packs across the front face of the actin monomer and also contacts both subdomains 1 and 3. Point mutations in key interacting arginine residues in Bud6 ablate its NPF activity. The Bud6-binding site on actin overlaps with the binding sites for profilin and formin, and Bud6 interactions also overlap with longitudinal contacts in F-actin. Together these findings suggest that Bud6 may function by interacting transiently with actin monomers, before handing them off to the formin or directly to a nascent filament nucleated by the formin.
RESULTS
Crystal Structure of Bud6flank in Complex with Actin
We expressed Bud6flank (residues 699-788 of S. cerevisiae Bud6) as a GST-fusion in E. Coli as previously described (Tu et al., 2012) and crystallized it in complex with rabbit skeletal muscle actin in 3.5M sodium formate, 0.1M CaCl2, and 5 mM TCEP. The crystals were of space group P32, with 12 Bud6/actin complexes in the asymmetric unit. The structure was determined by molecular replacement using ATP-bound G-actin as a search model (PDB ID: 3MN7). Examination of the crystal lattice revealed that the 12 actin/Bud6flank complexes in the asymmetric unit are arranged in two columns of six, with each column formed form a stack of three dimers related by a local two-fold axis. We do not ascribe any biological significance to these assemblies. Non-crystallographic symmetry averaging revealed continuous, readily interpretable density for the Bud6 portion of the structure, despite the modest resolution of the diffraction data (Figure S1). The structure was ultimately refined to an R-value of 0.21 (Rfree=0.25) using data to 3.5 Å resolution (Table 1).
Table 1.
Crystallographic data collection and refinement statistics.
| PDB ID Code | 4WYB |
| Data collection | |
| Space group | P32 |
| Cell dimensions | |
| a, b, c (Å) | 138.753, 138.753, 356.650 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution* (Å) | 44.11- 3.5 (3.56-3.5) |
| Rmerge* | 0.10 (0.48) |
| I/σ* | 9.6 (1.8) |
| Completeness* (%) | 98.8 (99.4) |
| Redundancy* | 2.3 (2.2) |
| Refinement | |
| Resolution (Å) | 44.11-3.5 |
| No. of Reflections | 91200 |
| Rwork/ Rfree | 0.213/0.253 |
| No. of Atoms | |
| Protein | 37129 |
| Ligand/ion (ATP, Ca++) | 384/12 |
| B-factors | |
| Protein | 98.06 |
| Ligand/ion (ATP, Ca++) | 89.59 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.483 |
| Ramachandran Plot | |
| Most favored | 4423 (94.1%) |
| Allowed | 260 (5.6%) |
| Outliers | 16 (0.3%) |
Numbers in parentheses are for the highest resolution shell.
Residues 700-736 of Bud6flank form two α-helices connected by a short linker. The two helices interact extensively with actin, but not with each other (Figure 1B). Helix αA packs at the base of the actin monomer (barbed end), in the groove between subdomains 1 and 3, while helix αB extends along the “front” of subdomains 1 and 3 toward the pointed end. A number of highly conserved residues in both helices make specific interactions with actin. In helix αA, Arg705 hydrogen bonds with the backbone carbonyl of Glu167 and its guanidinium group stacks with the phenyl ring of Tyr169 in actin (Figure 1C). Residues Val708, Met709 and Val712 and Leu715 of Bud6flank interact along the length of the relatively hydrophobic binding groove. Helix αB of Bud6flank is anchored on one end by Arg722, which forms a salt bridge with Glu334 in actin subdomain 3 and also hydrogen bonds with the backbone carbonyl of Ala144 in actin (Figure 1C, D). Near the C-terminal end of αB, Trp733 hydrogen bonds with the carbonyl of Arg28 and is in van der Waals contact with Pro27 and Val30 in subdomain 1 as well as Tyr337 in subdomain 3 of actin. Glu730 hydrogen bonds with the side chains of actin residues Lys336 and Tyr337 (Figure 1D). Intervening hydrophobic residues Ala725, Ile726 and Ala729 complete the interface of helix αB with actin. Beyond Trp733, helix αB is no longer in contact with actin.
Evolutionary conservation in Bud6flank is shown in Figure 1E. For the most part, highly conserved residues are in direct contact with actin, including Pro717 and His719 in the linker connecting helices αA and αB. The entire flank region of Bud6 was included in the crystallized construct, but no density was observed for residues 740-788.
Comparison with the WH2 Domain
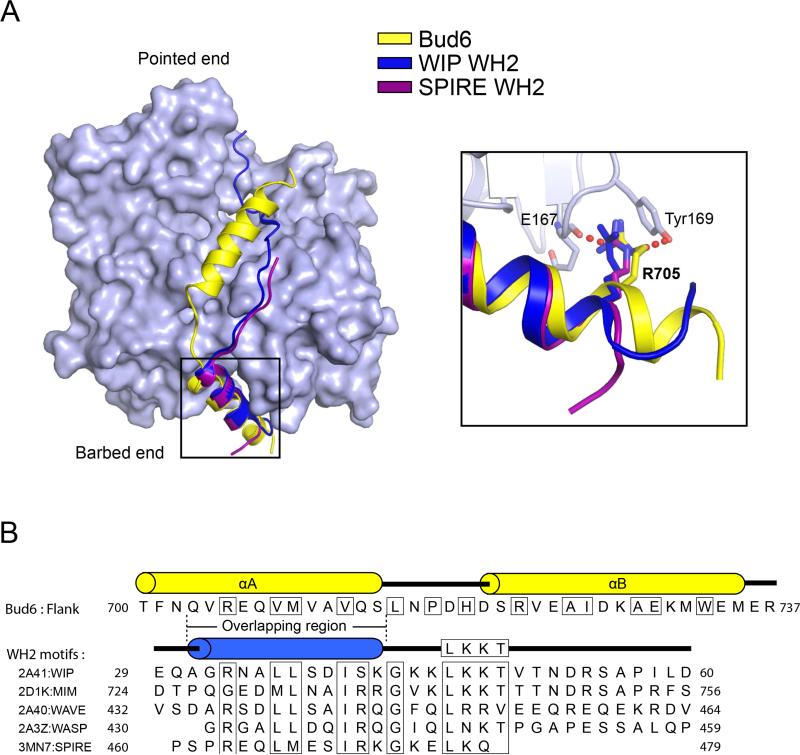
Many actin-binding proteins target the barbed-end groove occupied by helix αA of Bud6flank (Dominguez, 2009). In particular, WH2 domains contain a helix that binds this site in a manner quite similar to Bud6 (Chereau et al., 2005). The actin monomer-sequestering protein β-thymosin also contains the WH2 motif and forms a similar interaction with actin (Dominguez, 2007). The structures of two representative WH2 domains (those of WASP-interacting protein and Spire) are superimposed on the Bud6flank structure in Figure 2A, and Bud6 and WH2 motif sequences are compared in Figure 2B. The WH2 domain and Bud6flank interact with actin in a highly similar manner in the region of helix αA, and several interacting residues are the same or similar. WH2 domains contain an arginine residue that is equivalent to Arg705 in Bud6flank, and identical or conservatively altered hydrophobic residues equivalent to Val 708, Met709 and Val712 in Bud6flank, respectively. Outside this core region of helix A (residues Val704 to Ser714 of Bud6), the structures diverge. The Bud6 helix is approximately one turn longer at its N-terminal end, and the WH2 fold has no equivalent to helix αB. Instead, WH2 domains include an “LKKT” sequence that binds a distinct site on the front face of actin in an extended conformation. Thymosin β4 and other β-thymosins also contain the LKKT motif (Figure S2). It is unclear whether WH2 domains and the Bud6flank actin-binding region arose from a common ancestral domain, or whether their similar mechanisms of binding in the barbed-end groove reflect convergent evolution.
Figure 2. Comparison of the Bud6 actin-binding domain with WH2 domains.
A, Superposition of actin-bound Bud6flank (yellow) and the WH2 domains of WIP (WASP-interacting protein, dark blue, PDB ID 2A41) and Spire (purple, PDB ID 3MN7). Structures were superimposed using the actin-binding portion of each of the three structures; the light blue surface corresponds to the actin-binding portion of the present structure. The WH2 domain helix overlaps closely with helix αA of Bud6flank, but there is no equivalent of helix αB in the WH2 domains. Both Bud6flank and these two WH2 domains position an arginine residue between Glu167 and Tyr169 in actin (inset). Note also that the Bud6flank helix is one turn longer than that of the WH2 domains. See also Figure S2. B, Comparison of Bud6flank and WH2 domain sequences. Sequences of Bud6flank and selected WH2 domains are shown, with conserved actin-binding residues boxed. Respective secondary structures are shown above the sequences, and the structurally overlapping region of Bud6flank and the WH2 motif is indicated. Note that there is no equivalent of the “LKKT” WH2 sequence motif in Bud6flank.
Structure-Function Analysis of Bud6flank
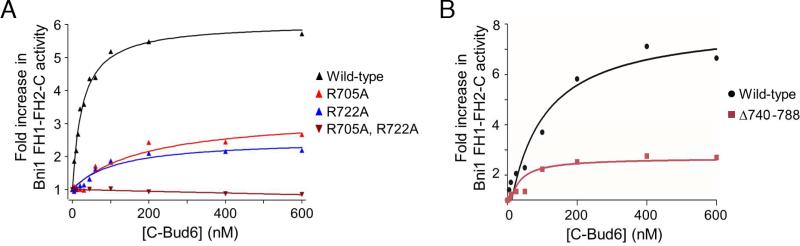
Prior functional studies of Bud6 have established that its stimulation of actin assembly requires interactions of Bud6core with the formin C-terminal tail in addition to interactions of Bud6flank with actin monomers (Graziano et al., 2011; Tu et al., 2013). A triple alanine mutant of conserved residues in the flank region (Arg705Ala, Glu706Ala, Val708Ala) was found to be defective in actin monomer binding and nucleation enhancement (Graziano et al., 2011). In vivo, this mutant yielded defects in actin cable formation and cell growth, even more severe than a complete deletion of the BUD6 gene. As noted above, the present structure reveals that these residues are part of helix αA, and that both Arg705 and Val708 are in direct contact with actin. To further probe the role of the Bud6 flank region in actin assembly, we tested the effects of mutating two arginine residues that are highly conserved and directly interact with actin, Arg705 in helix αA and Arg722 in helix αB (Figures 3A and S3, A-D). Wild type C-Bud6 potently stimulated Bni1-mediated actin assembly, as previously reported. Single alanine substitutions in each of these residues decreased, but did not eliminate the ability of C-Bud6 to enhance Bni1-mediated actin assembly, and the R705A, R722A double mutant was completely inactive. At higher concentrations, the double mutant modestly inhibited Bni1-mediated actin assembly (Figure S3D). A similar effect was observed with Bud6core, which lacks the entire flank region and is defective in actin binding (Tu et al., 2013).
Figure 3. Structure-function analysis of the Bud6-actin interaction.
A, Concentration-dependent effects of wild type and mutant C-Bud6 polypeptides. 2 μM monomeric actin was polymerized in the presence of 10 nM Bni1 FH1-FH2-C and indicated concentrations of wild type or mutant C-Bud6 (550-788). Fold increase in actin assembly activity (relative to Bni1 FH1-FH2-C alone) is plotted as a function of C-Bud6 concentration. B, As in A, but the activity of intact C-Bud6 (residues 550-788) is compared with C-Bud6Δ740-788 (residues 550-739). See Figure S3 for raw actin assembly curves.
As noted above, only the N-terminal half of Bud6flank is observed in our structure. Thus we asked whether the remaining C-terminal portion of the flank is important for Bud6 function. A C-terminal truncation mutant spanning Bud6 residues 550-739 (CBud6Δ740-788) retained the ability to stimulate Bni1-mediated actin assembly, but was impaired relative to the intact C-Bud6 construct (Figures 3B and S3, E and F). This finding is not surprising, given that the C-terminal half of Bud6flank includes regions with a high degree of evolutionary sequence conservation (Figure 1E). While the truncated construct clearly retains the ability to bind actin, we cannot exclude the possibility that residues in the disordered, c-terminal portion of the flank also contribute to actin binding.
Discussion
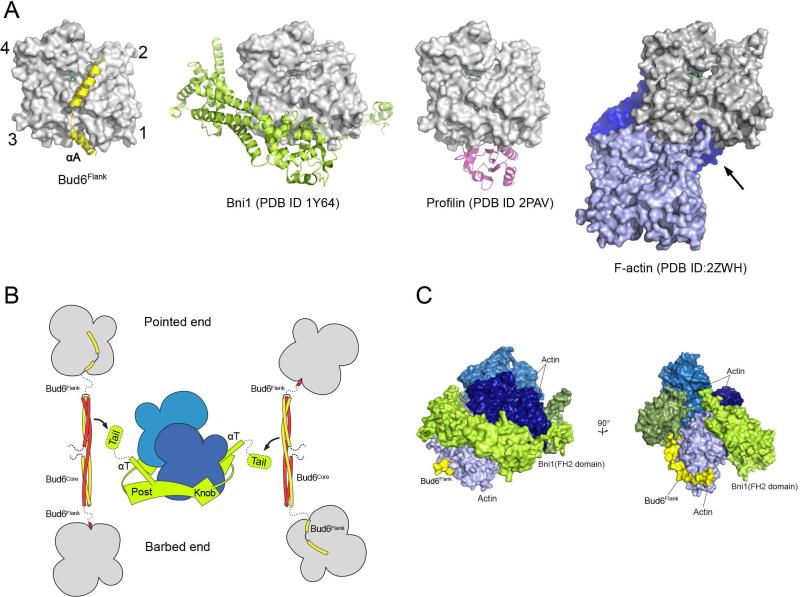
Our discovery and characterization of a WH2-like element in Bud6flank highlights a related structural and mechanistic basis between Bud6 and other NPFs that cooperate with either formins or the Arp2/3 complex. The WH2 domain and its variations have been widely adapted for actin monomer recruitment in actin nucleating proteins or protein complexes. In diverse NPFs, the small WH2 motif is fused, alone or in multiples, to domains that bind the nucleation partner. While some WH2 proteins (e.g., Spire and Cobl) have “autonomous” actin assembly activity, Bud6 requires partnership with one of the yeast formins, Bni1 or Bnr1. Our emerging structural understanding of the C-terminal half of Bud6 explains its lack of independent nucleation activity; the rod-shaped dimeric Bud6core domain positions its two actin binding flank elements 116 Å apart, which likely disfavors filament-like interactions between the two actin monomers bound by a single Bud6 dimer. In addition, the interactions of Bud6flank with actin are sterically incompatible with longitudinal contacts in F-actin (Figure 4A). Interestingly, the site occupied by Bud6flank at the barbed end of the actin monomer also directly overlaps with the binding site of the “knob” region of the formin FH2 domain, and with the binding site of profilin (Figure 4A). Because Bud6 binds with high affinity and stimulates actin assembly in both the presence and absence of profilin (Graziano et al., 2011; Moseley et al., 2004; Tu et al., 2012), the Bud6flank interaction appears to effectively compete with profilin from actin.
Figure 4. Mechanistic implications of the Bud6 structure and mode of actin binding.
A, The Bud6flank binding site on actin overlaps with that of other actin-assembly factors and is partially blocked in F-actin. Crystal structures of Bud6flank, profilin, and the Bni1 FH2 domain in complex with actin are shown in the same orientation. The barbed-end groove occupied by helix αA in Bud6 is also part of the binding surface for profilin and the FH2 domain. The groove is also blocked in F-actin by the DNAse I binding loop (arrow) of a longitudinally apposed subunit (medium blue) in the helical filament. Three actin subunits of a filament are drawn based on the X-ray fiber diffraction structure of F-actin (PDB ID 2ZWH); examination of a cryo-electron microscopy reconstruction leads to the same conclusion (PDB ID 3MFP, not shown). B, Schematic summary of available structural information for the Bni1 FH2 domain and C-Bud6 and their interactions with actin. Components are drawn approximately to scale, and the illustration is based on structures of the Bni1 FH2 domain (green) bound to actin (blue), the Bud6core domain (yellow and red), and Bud6flank in complex with actin (yellow or red, with actin in gray). No structure is available for Bud6core in complex with Bni1, but biochemical studies map the binding interaction to the “tail” of Bni1, which lies just C-terminal to the long “αT” helix of the FH2 domain. The Bni1 tail and the ~20 residue linker that connects Bud6 flank and core domains are shown as dotted lines, because they are not present in available crystal structures. The Bni1 FH2 dimer is thought to promote nucleation by bridging between two or more actin subunits in a filament-like orientation, via contacts of its “knob” and “post” elements. C, Superposition of Bud6flank on a Bni1/actin complex. Three actin subunits and a Bni1 FH2 domain dimer from the crystal structure of the complex (PDB ID 1Y64) are shown in shades of blue and green, respectively. The interaction with the formin arranges the actin subunits in a filament-like orientation that is proposed to lead to formation of a nascent filament (Otomo et al., 2005b). Bud6flank (yellow) is docked based on superposition of the present structure with the light-blue actin subunit. The Bud6 binding site on the medium and dark blue actin subunits is blocked by contact with the FH2 domain, but it is accessible on the light blue subunit, which is in contact with only the post-site of the FH2 domain. We speculate that this mode of interaction could allow Bud6 to contribute to filament nucleation by the FH2 domain (see text). There is a modest steric clash between the end of Helix αB in Bud6 and the opposite subunit in the FH2 dimer (rotated view), but the precise orientation of the two FH2 subunits that leads to this clash arises from crystallographic symmetry and is not thought to be directly relevant to Bni1-mediated nucleation. Note that one of the flexible linkers connecting the two halves of the FH2 dimer is not illustrated; due to an artifact in the crystal structure, it connects to an adjacent FH2 subunit in the lattice rather than closing the FH2 dimer.
A key role of the Bni1/Bud6 complex is to overcome the kinetic barrier that prevents spontaneous assembly of actin (or profilin-bound actin) into F-actin via formation of a stable nucleus of actin subunits. The formin FH2 domain has low affinity for actin monomers on its own, thus Bud6 is hypothesized to facilitate monomer recruitment (Moseley et al., 2004). Consistent with this, Bud6 function requires binding to both actin and the formin (Graziano et al., 2011; Tu et al., 2012). Current structural and biochemical understanding of Bni1 and C-Bud6 and their interactions with actin is summarized in Figure 4B. Precisely how Bud6 binds Bni1 is not yet known, but the Bni1 dimer can bind two Bud6 dimers and the interaction requires the C-terminal tail region of Bni1 and conserved surfaces on the Bud6core domain (Tu et al., 2012). Two Bud6 dimers, in association with the formin dimer, could in principle bind as many as four actin subunits, but how Bud6-bound actin subunits might be incorporated into a stable actin nucleus or how Bud6flank might otherwise participate in nucleation remains unclear. Nonetheless, examination of the structure of the Bni1 FH2 domain in complex with actin suggests one possibility in which Bni1 and Bud6 might simultaneously engage the same actin subunit. Each half of the formin dimer has two actin-binding sites, termed the “knob” and “post” sites (Otomo et al., 2005b; Xu et al., 2004). A formin dimer with two actin subunits bound in a filament-like orientation is expected to have one post-site unoccupied (Otomo et al., 2005b). The Bud6flank binding site does not overlap with the post site on actin (Figure 4C), thus Bud6 could promote binding of a third actin subunit in a nucleus by delivering an actin subunit to this free post site, or stabilizing it there once it was bound. Such a mode of interaction would have to be transient; stepping of the opposite subunit of the Bni1 dimer to allow elongation would require displacement of Bud6flank so that the formin knob could engage the barbed-end groove of this newly recruited actin subunit.
Although structurally plausible, this speculative “barbed-end” model is not particularly satisfying; it requires participation of only one of the two flank domains of the Bud6 dimer, and only one of the two Bud6 dimers that may be associated with the formin dimer. Furthermore, it does not explain how Bud6 contributes to nucleation without affecting the rate of elongation. Unless elongation promotes Bud6 dissociation, Bud6flank could continue to interact with incoming actin subunits in the course of elongation. Alternative models in which Bud6 recruits or stabilizes subunits to the pointed-end to create a stable nucleus avoid this conundrum; one or two steps of elongation would be expected to take the Bni1/Bud6 complex out of reach of the pointed end. However, steric considerations argue against such models; as noted above, the Bud6flank binding site interferes with the longitudinal contact in F-actin. Release of the Bud6flank interaction prior to incorporation of the actin subunit into a stable nucleus would presumably allow the actin to diffuse away. Partial dissociation of Bud6flank involving release of helix αA while Helix αB remained associated could allow such a pointed-end contribution, but it is entirely unclear whether these two binding elements are independent and whether such subsite-dissociation can occur. Clearly, further study is required. The structure described here and the considerations discussed above will guide our ongoing structural and mechanistic studies of this actin assembly device.
Materials and Methods
Protein Preparation
Rabbit skeletal muscle actin (RMA) was purified as previously described (Spudich and Watt, 1971). Bud6core (550-788) and Bud6flank (699-788) were expressed as N-terminal GST-TEV-tagged fusion proteins using a modified pET-30 vector. C-Bud6 (550-788) and C-Bud6Δ740-788 were expressed similarly, but with the addition of a C-terminal His8-tag. Plasmids were transformed into BL21(DE3) cells (Novagen) and grown at 37°C to an optical density of 0.5. The temperature of the culture was then shifted to 30°C, and cells were induced with 0.5 mM IPTG for 6 h. Cells were lysed by sonication in lysis buffer (1X PBS, 5 mM DTT, 2 mM EDTA, 1 mM PMSF) and cleared by high-speed centrifugation. The supernatant was incubated with glutathione sepharose resin (GE healthcare) for 3 h at 4°C, washed, and the protein eluted with elution buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM DTT, 10 mM glutathione). The protein was incubated with TEV protease at 4°C overnight to remove the GST tag , then purified further by anion-exchange followed by size-exclusion chromatography in 10 mM Tris (pH 8.0), 0.2 mM ATP, 0.2 mM CaCl2, and 0.2 mM DTT (G-buffer for actin). For the C-Bud6 (550-788) and C-Bud6Δ740-788 proteins, a Nickel-NTA agarose (Qiagen) affinity purification step was added after TEV cleavage.
Crystallization and Structure Determination
Bud6flank (residues 699–788) was mixed with G-actin at a molar ratio 3:1 and concentrated to 7 mg/ml. Crystals of the complex were grown using hanging-drop vapor diffusion at 20°C by mixing 2 μl of the complex wit h an equal volume of a well solution containing 3.5 M sodium formate, 0.1 M CaCl2, and 5 mM TCEP. Crystals were frozen in liquid nitrogen after addition of 20% glycerol to the mother liquor as a cryoprotectant. Diffraction data were collected on the NE-CAT beamlines ID24-C and E at Argonne National Laboratory at 100K, and were processed and merged with HKL2000 (Otwinowski et al., 2003). The structure was determined by molecular replacement using monomeric actin as a search model (PDB ID 3MN7). Iterative twelve-fold non-crystallographic symmetry averaging was carried out using PHENIX (Adams et al., 2010) and the Bud6flank portion of the structure was built into the averaged map and included in subsequent refinement cycles. Repeated rounds of manual refitting and crystallographic refinement were performed using COOT (Emsley et al., 2010) and PHENIX (Adams et al., 2010). Crystallographic data are presented in Table 1.
In vitro Actin Assembly Assays
Gel-filtered monomeric actin (2 μM final; 5% pyrene-labeled) in G-buffer (10 mM Tris (pH 8.0), 0.2 mM ATP, 0.2 mM CaCl2, and 0.2 mM DTT) was converted to Mg-ATP-actin 2 min prior to use in reactions. A total of 42 μl of G-actin was added to 15 μl control buffer or proteins in the same buffer and 3 μl 20x initiation mix (40 mM MgCl2, 10 mM ATP, 1 M KCl). Pyrene fluorescence was monitored at excitation 365 nm and emission 407 nm at 25°C in an Infinite M200 plate reader (Tecan, Männe dorf, Switzerland). Rates of assembly were calculated from slopes of the curves at 20-40% polymerization.
Supplementary Material
Highlights.
Crystal structure of the actin-binding domain of Bud6 in complex with actin
Bud6 binds actin using a novel WH2-like motif
Mutations in Bud6 actin-binding residues impair stimulation of actin assembly
Structural considerations inform potential models for collaboration with formin Bni1
ACKNOWLEDGMENTS
This work was supported by NIH grants GM071834 (MJE) and GM083137 (BLG). We thank the staff of the Northeastern Collaborative Access Team (NE-CAT) beamlines at the Advanced Photon Source. NE-CAT is supported by a grant from the National Institutes of Health (P41 GM103403).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Suppl):W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Le KH, Bugyi B, Correia JJ, Renault L, Carlier MF. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Sawaya MR, Phillips ML, Reisler E, Quinlan ME. Multiple forms of Spire-actin complexes and their functional consequences. J Biol Chem. 2012;287:10684–10692. doi: 10.1074/jbc.M111.317792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Lopes CS, Moir CA, Huisman SM, Segal M. Dissecting the involvement of formins in Bud6p-mediated cortical capture of microtubules in S. cerevisiae. J Cell Sci. 2008;121:3803–3814. doi: 10.1242/jcs.036269. [DOI] [PubMed] [Google Scholar]
- Dominguez R. The beta-thymosin/WH2 fold: multifunctionality and structure. Ann N Y Acad Sci. 2007;1112:86–94. doi: 10.1196/annals.1415.011. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducka AM, Joel P, Popowicz GM, Trybus KM, Schleicher M, Noegel AA, Huber R, Holak TA, Sitar T. Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation. Proc Natl Acad Sci U S A. 2010;107:11757–11762. doi: 10.1073/pnas.1005347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol. 2013;201:595–611. doi: 10.1083/jcb.201212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimsath EG, Jr., Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem. 2012;287:3087–3098. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Narita A, Hirayama T, Taki M, Iyoshi S, Yamamoto Y, Maeda Y, Oda T. Human spire interacts with the barbed end of the actin filament. J Mol Biol. 2011;408:18–25. doi: 10.1016/j.jmb.2010.12.045. [DOI] [PubMed] [Google Scholar]
- Montaville P, Jegou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biology. 2014;12:e1001795. doi: 10.1371/journal.pbio.1001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami A, Poy F, Toms A, Zheng W, Eck MJ. Crystal structure of a complex between amino and carboxy terminal fragments of mDia1: insights into autoinhibition of diaphanous-related formins. PLoS One. 2010;5(9):e12992. doi: 10.1371/journal.pone.0012992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami AG, Poy F, Eck MJ. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005a;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Machius M, Rosen MK. Crystal structure of the Formin mDia1 in autoinhibited conformation. PLoS One. 2010;5(9):e12896. doi: 10.1371/journal.pone.0012896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005b;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta crystallographica Section A, Foundations of crystallography. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlivanis M, Samol A, Kerkhoff E. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem. 2009;284:25324–25333. doi: 10.1074/jbc.M109.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr Biol. 2011;21:955–960. doi: 10.1016/j.cub.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasson AS, Bois JS, Pham DS, Yoo H, Quinlan ME. Filament Assembly by Spire: Key Residues and Concerted Actin Binding. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.09.002. doi: 10.1016/j.jmb.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Ten Hoopen R, Cepeda-Garcia C, Fernandez-Arruti R, Juanes MA, Delgehyr N, Segal M. Mechanism for astral microtubule capture by cortical Bud6p priming spindle polarity in S. cerevisiae. Curr Biol. 2012;22:1075–1083. doi: 10.1016/j.cub.2012.04.059. [DOI] [PubMed] [Google Scholar]
- Tu D, Graziano BR, Park E, Zheng W, Li Y, Goode BL, Eck MJ. Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc Natl Acad Sci U S A. 2012;109:E3424–3433. doi: 10.1073/pnas.1203035109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Bor B, Quinlan ME. The Role of Formin Tails in Actin Nucleation, Processive Elongation, and Filament Bundling. J Biol Chem. 2014 doi: 10.1074/jbc.M114.588368. doi: 10.1074/jbc.M114.588368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci U S A. 2011;108:11884–11889. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Zeth K, Pechlivanis M, Samol A, Pleiser S, Vonrhein C, Kerkhoff E. Molecular basis of actin nucleation factor cooperativity: crystal structure of the Spir-1 kinase non-catalytic C-lobe domain (KIND)*formin-2 formin SPIR interaction motif (FSI) complex. J Biol Chem. 2011;286:30732–30739. doi: 10.1074/jbc.M111.257782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.