Abstract
PICT1 (also known as GLTSCR2) is considered a tumor suppressor because it stabilizes phosphatase and tensin homolog (PTEN), but individuals with oligodendrogliomas lacking chromosome 19q13, where PICT1 is located, have better prognoses than other oligodendroglioma patients. To clarify the function of PICT1, we generated Pict1-deficient mice and embryonic stem (ES) cells. Pict1 is a nucleolar protein essential for embryogenesis and ES cell survival. Even without DNA damage, Pict1 loss led to p53-dependent arrest of cell cycle phase G1 and apoptosis. Pict1-deficient cells accumulated p53, owing to impaired Mdm2 function. Pict1 binds Rpl11, and Rpl11 is released from nucleoli in the absence of Pict1. In Pict1-deficient cells, increased binding of Rpl11 to Mdm2 blocks Mdm2-mediated ubiquitination of p53. In human cancer, individuals whose tumors express less PICT1 have better prognoses. When PICT1 is depleted in tumor cells with intact P53 signaling, the cells grow more slowly and accumulate P53. Thus, PICT1 is a potent regulator of the MDM2-P53 pathway and promotes tumor progression by retaining RPL11 in the nucleolus
Alterations in cell cycle control genes such as the tumor suppressor TP53 (also known as P53) contribute to tumorigenesis. In response to cellular stress, P53 induces cell cycle arrest or apoptosis. More than 50% of human cancers harbor mutations in TP53. In the remainder, the P53 pathway is often inactivated owing to overproduction of MDM2 (mouse double minute 2; ref. 1), an E3 ubiquitin ligase that targets the P53 protein for proteasomal degradation2,3. Inhibition of MDM2 therefore leads to P53 protein stabilization and accumulation.
Stresses that activate the MDM2-P53 pathway also induce cascades responsible for its regulation, including the kinases ATM-CHK2 (ataxia telangiectasia mutated–checkpoint kinase 2) and ATR-CHK1 (ataxia telangiectasia and Rad3 related–checkpoint kinase 1). These kinases phosphorylate MDM2 and P53 such that their interaction is abrogated4. MDM2 can also be inhibited by P19ARF binding triggered by infection or oncogene activation5,6; P19ARF is one of the transcript products from CDKN2A (cyclin-dependent kinase inhibitor 2A) gene. Lastly, stresses that stimulate post-translational modifications such as acetylation or sumoylation of P53 or MDM2 can influence P53 activation4.
The MDM2-P53 pathway is also regulated by ribosomal proteins7. Upon nucleolar stress, ribosomal proteins RPL5, RPL11, RPL23 and RPS7 translocate from the nucleolus to the nucleoplasm and bind to MDM2 (refs. 8–16). Nucleolar stress is often caused by disruption of ribosomal biogenesis, which in turn can be caused by serum depletion and contact inhibition17, agents like low-dose actinomycin D or mycophenolic acid18,19 or malfunction of nucleolar proteins13–15,20. RPL26 increases the translation of TP53 mRNA in response to DNA damage21, whereas RPS3 protects P53 from MDM2-mediated ubiquitination in response to oxidative stress22. Thus, ribosomal proteins can drive P53-mediated responses to stress, but how ribosomal proteins translocate from the nucleolus to the nucleoplasm to exert these functions is unknown. It is also unclear whether genes encoding ribosomal proteins that regulate P53 affect the prognosis of human cancers.
The gene encoding PICT1 (protein interacting with carboxyl terminus-1; also called GLTSCR2, glioma tumor suppressor candidate region gene 2) is located at human chromosome 19q13.32, which is frequently altered in human tumors23. Low PICT1 expression in diffuse gliomas and ovarian cancers is correlated with high malignant progression24–26, whereas PICT1 overexpression enhances apoptosis of cultured glioma cells27. At the molecular level, PICT1 stabilizes PTEN via direct physical interaction28,29. On the basis of these findings, PICT1 has been deemed a tumor suppressor. Other evidence, however, suggests that PICT1 may not always dampen cancer progression. In oligodendroglial tumors, deletion of the entire chromosome 19q arm, as well as loss of heterozygosity (LOH) at chromosome 19q13 is associated with longer disease-free survival after chemotherapy30–32. Nevertheless, in astrocytic gliomas, chromosome 19q deletions are linked to malignant progression33,34. These contradictory observations indicate that one or more genes mapped to chromosome 19q may contribute to brain cancer development. We speculated that PICT1 might be an important chromosome 19q–mapped gene that regulates tumor progression.
To clarify PICT1 functions, we carried out extensive analyses of Pict1-deficient mice and ES cells. We show here that PICT1 is a key regulator of ribosomal protein–driven P53-mediated responses to nucleolar stress and that loss of PICT1 inhibits tumor growth owing to stabilization of P53.
RESULTS
Pict1-deficient cells show cell cycle arrest and apoptosis
We generated mice bearing a null mutation of Pict1 (Supplementary Fig. 1a) but obtained no viable Pict1−/− pups (Fig. 1a and Supplementary Fig. 1b). Although we observed no morphological differences between Pict1+/+ and Pict1−/− embryos up to embryonic day 2.75 (E2.75; morula), none of the 15 Pict1−/− embryos we examined formed proper blastocysts at E3.5 and all contained apoptotic cells (Fig. 1a). This developmental failure was not due to an inadequate uterine environment, because Pict1+/− embryos of the same litter developed normally. When Pict1−/− morulae were cultured in vitro, they did not mature and died within 2–3 d (Supplementary Fig. 1c). Thus, PICT1 is essential for preimplantation embryogenesis.
Figure 1.
Pict1 loss impairs survival of mouse embryos and ES cells. (a) Top, morphologies of representative Pict1+/+, Pict1+/− and Pict1−/− embryos at the E3.5 blastocyst stage and the E2.75 morula stage (after compaction). Bottom, TUNEL staining of Pict1+/+, Pict1+/− and Pict1−/− embryos at E3.5. +/+DNase, DNase-treated E3.5 Pict1+/+ embryos (positive control). DAPI, nuclear staining. Scale bar, 50 µm. (b) Top, semiquantitative RT-PCR of Pict1 mRNA in Pict1 ES cells treated for 24 h with doxycycline (Dox) as indicated. Actb, loading control. Bottom, MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay of proliferation of Pict1 ES cells treated with Dox as indicated. Results are mean cell growth (A490) of three cultures per dose per time point. (c) Left, representative FACS profiles of Pict1 ES cells treated with or without Dox (5 ng ml−1) for 1, 2 or 3 d, stained with propidium iodide (PI), and analyzed by flow cytometry. Numbers indicate percentage of cells in G0/G1, S or G2/M phase. Right, mean ± s.e.m. percentage of ES cells in G0/G1 phase (n = 5). *P < 0.01. (d) Left, representative FACS profile of Pict1 ES cells treated with or without Dox (5 ng ml−1) for 2, 3 or 4 d, stained with TUNEL and analyzed by FACS. Right, mean ± s.e.m. percentage of TUNEL+ ES cells (n = 5). *P < 0.01. Results represent four trials.
To examine PICT1 functions in vitro, we generated mutant murine ES cells (Pict1tetTg+; Pict1Δ/−; Pict1 ES cells) that lacked endogenous Pict1 owing to gene targeting (Supplementary Fig. 1d,e) but expressed exogenous Pict1 in a doxycycline-regulatable manner such that Pict1 expression was ‘on’ without doxycyline (Dox−) but ‘off’ after doxycyline treatment (Dox+) (Supplementary Methods and Supplementary Fig. 1e,f). Because Dox− Pict1 ES cells behaved like wild-type (WT) ES cells in cell growth, cell cycle and apoptosis assays (Supplementary Fig. 1g,h), we used Dox− cells as controls throughout our study. Doxycyline treatment of Pict1 ES cells inhibited both Pict1 expression and cell growth in a dose-dependent manner (Fig. 1b). This growth inhibition was due to both cell cycle arrest (Fig. 1c) and enhanced apoptosis (Fig. 1d). Thus, PICT1 is required for the survival and proliferation of ES cells.
Effects of Pict1 loss depend on p53 but not p19Arf or Pten
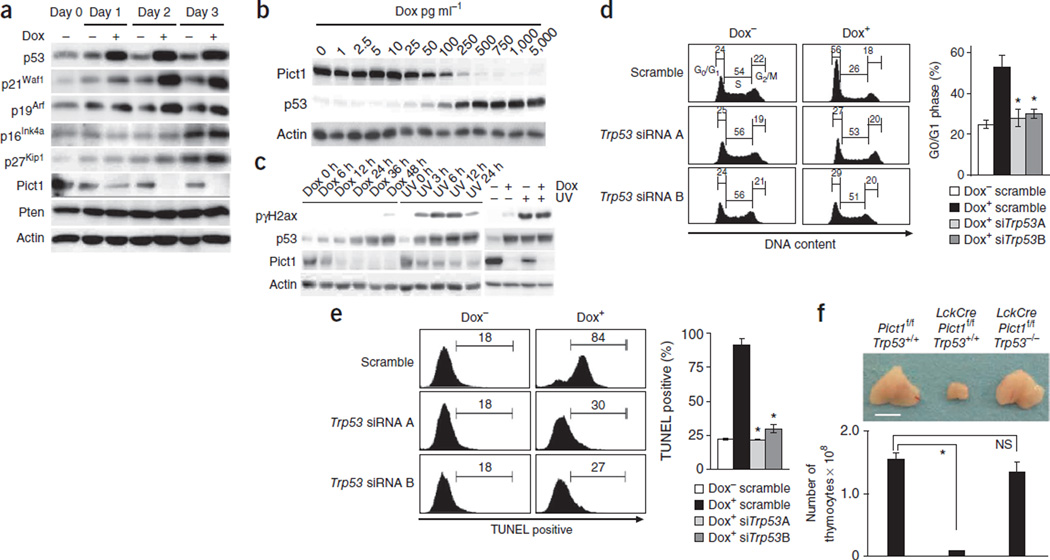
Compared with Dox− cells, Dox+ cells showed greater expression of p53, p21Waf1 and p19Arf (Fig. 2a) that, at least for p53, was correlated with the degree of doxycycline-mediated Pict1 suppression (Fig. 2b). The greater p53 expression in Dox+ cells was probably not due to DNA damage, because phospho-γ-histone family member X (pγH2ax), which is a marker of DNA damage, was much lower in Dox+ cells than in ultraviolet (UV)-treated cells with comparable p53 activation (Fig. 2c). Moreover, the elevation in pγH2ax owing to UV exposure was not further enhanced by doxycycline (Fig. 2c). Notably, siRNA-mediated p53 knockdown prevented the cell cycle arrest (Fig. 2d and Supplementary Fig. 2a) and increased apoptosis (Fig. 2e) observed in Dox+ cells. Although p19Arf expression was greater in Dox+ than in Dox− cells (Fig. 2a), siRNA-mediated p19Arf knockdown did not affect p53 protein expression, cell cycle arrest or apoptosis in Dox+ cells (Supplementary Fig. 2b–d).
Figure 2.
Effects of Pict1 deficiency are p53 dependent. (a) Immunoblot detecting the indicated proteins in Pict1 ES cells treated with or without 5 ng ml−1 doxycycline (Dox). (b) Immunoblot detecting p53 and Pict1 proteins in Pict1 ES cells treated with Dox. Actin, loading control. (c) Left, immunoblot detecting p53 protein and DNA damage (pγH2ax) in Pict1 ES cells treated with or without Dox (5 ng ml−1) or UV irradiation (80 J m−2). Right, immunoblot of Pict1 ES cells treated with Dox (5 ng ml−1, 48 h), UV irradiation (80 J m−2, 6 h) or both (final 6 h for UV). (d,e) Pict1 ES cells were transfected with scramble siRNA, Trp53 siRNA A or Trp53 siRNA B for 24 h and cultured with or without 5 ng ml−1 Dox for 2 d (d) or 4 d (e). Left, FACS profiles of percentage of cells in each cell cycle phase (d) and TUNEL+ cells (e). Right, mean ± s.e.m. (n = 5) percentage of G0–G1 phase (d) and TUNEL+ (e) ES cells. *P < 0.01. (f) Top, gross appearance of thymi from mice of the indicated genotypes (5 weeks old). Bottom, mean total thymocytes ± s.e.m. from these thymi (n = 5). *P < 0.01. Scale bar, 5 mm. Results represent three trials.
The thymus is highly sensitive to p19Arf-p53 signaling35, and our Pict1−/− mice succumbed to early embryonic lethality. We therefore generated mice deficient in Pict1 specifically in T cells by crossing LckCre transgenic mice with Pict1flox mice (Supplementary Methods). Total number of thymocytes in these mutants were <5% of those in control Pict1flox/flox mice (Fig. 2f), a defect that was markedly rescued in LckCrePict1flox/flox;Trp53−/− double knockout (DKO) mice (Fig. 2f), but not in LckCrePict1flox/flox;p19Arf−/− double-knockout mice (Supplementary Fig. 2e). Consistent with an earlier report36, p53 expression was lower in p19Arf-deficient T cells than in wild-type cells (Supplementary Fig. 2f). However, because p53 expression is still high in LckCrePict1flox/flox;p19Arf−/− double-knockout T cells (Supplementary Fig. 2f) compared with LckCrePict1flox/flox;p19Arf+/+ T cells, we do not think p19Arf mediates the higher expression of p53 associated with loss of Pict1.
Despite our observations in T cells deficient in Pict1, double-knockout mice that totally lacked Trp53 and Pict1 were still embryonic lethal (Supplementary Fig. 3a), suggesting that the phenotype of Pict1-deficient embryos involves factors in addition to p53 accumulation. To determine whether PTEN was one of these factors, we examined the stability of Pten protein without Pict1. After Pict1 ES cells with or without doxyxycline induction were treated with 100 µg ml−1 cycloheximide, immunoblotting showed that Pten degradation was faster without Pict1 (Supplementary Fig. 3b). However, expression of steady-state Pten (without cycloheximide), phospho-Pten, and phospho-Akt was comparable in Dox+ and Dox− cells (Fig. 2a and Supplementary Fig. 3c). Thus, at least in unstimulated cells, PICT1 has only a subtle stabilization effect on PTEN that occurs without obvious phosphorylation of PTEN or effector activation.
Pict1 deficiency inhibits Mdm2 function
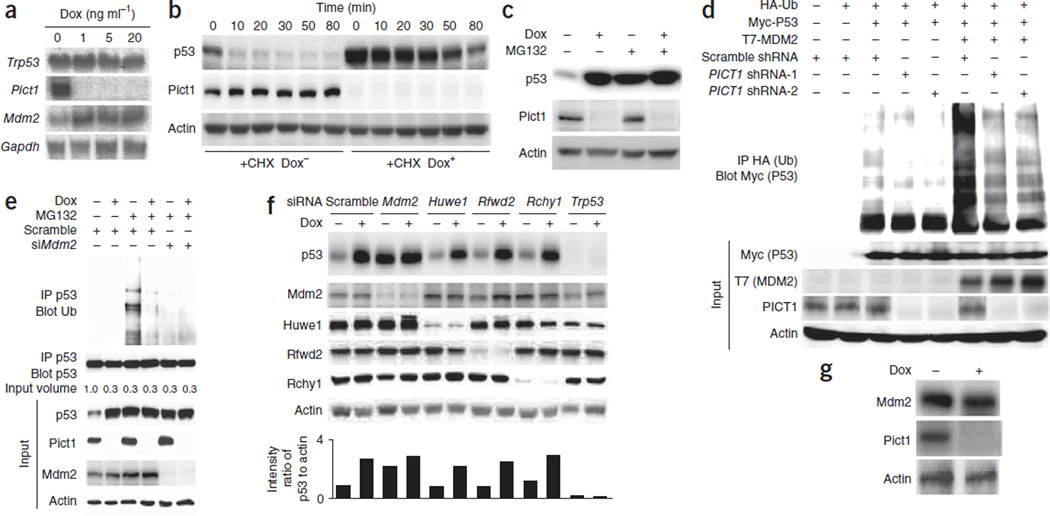
To investigate how Pict1 deficiency increases p53 expression, we first measured Trp53 mRNA levels by northern blotting. Steady-state levels of Trp53 mRNA remained constant in Dox+ cells treated with increasing doxycycline for 2 d (Fig. 3a). Through cycloheximide studies, we found that p53 protein half-life was longer in Dox+ cells compared to Dox− cells (Fig. 3b), but Pict1 deficiency had no effect on p21 protein half-life (data not shown). Studies using the proteasomal inhibitor MG132 showed that the increase in p53 protein half-life was due to protection from proteasomal degradation (Fig. 3c), suggesting that the elevated p53 abundance in Pict1-deficient cells is not due to transcriptional effects.
Figure 3.
Pict1 deficiency inhibits Mdm2 function. (a) Northern blot detecting indicated mRNAs in Pict1 ES cells treated for 48 h with Dox. (b) Immunoblot detecting p53 and Pict1 in Pict1 ES cells treated for 48 h with or without 5 ng ml−1 Dox and with or without cycloheximide (CHX; 100 µg ml−1). (c) Immunoblot detecting p53 in Pict1 ES cells treated for 36 h with or without 5 ng ml−1 Dox, with or without MG132 (20 µM). (d) Immunoblot of H1299 cells transfected with the indicated plasmids and treated with MG132 (20 µM). Lysates were immunoprecipitated and immunoblotted with antibodies to HA (ubiquitin, Ub) and Myc (ubiquitinated p53). (e) Immunoblot of Pict1 ES cells transfected with scramble siRNA or Mdm2 siRNA (siMdm2) and treated with or without 5 ng ml−1 Dox for 48 h. MG132 (20 µM) was added for 3 h before lysis. The p53 protein level in each sample was adjusted to equality before immunoprecipitation with antibody to p53 or to ubiquitin. (f) Pict1 ES cells were transfected with scramble siRNA or the indicated siRNAs and treated with or without 5 ng ml−1 Dox for 24 h. Top, immunoblot detecting indicated proteins. Bottom, quantification of ratio of p53 to actin using LAS Image Analyzer with Multi Gauge Software. (g) Immunoblot detecting indicated proteins in Pict1 ES cells treated with or without 5 ng ml−1 Dox for 48 h. Results represent three trials.
We hypothesized that PICT1 deficiency prevents P53 degradation by decreasing its MDM2-mediated ubiquitination. We transfected H1299 cells with plasmids expressing hemagglutinin-tagged ubiquitin (HA-Ub), Myc-tagged P53 and T7-tagged MDM2, and we carried out immunoprecipitation and immunoblotting to detect ubiquitinated P53 (Ub-P53). Before immunoprecipitation, we inhibited the 26S proteasome with MG132 to allow accumulation of ubiquitinated proteins. As we expected, coexpression of MDM2 and P53 produced a ladder of Ub-P53 products (Fig. 3d). However, when we also infected these MDM2- and P53-expressing H1299 cells with a lentivirus expressing PICT1 shRNA, the laddering was less intense (Fig. 3d). Notably, Pict1 deficiency also decreased the Mdm2-dependent ubiquitination of endogenous p53 (Fig. 3e and Supplementary Fig. 2a). These results show that PICT1 inhibition prevents MDM2-mediated P53 ubiquitination.
The accumulation of p53 in doxycycline-treated Pict1 deficient ES cells was dependent on Mdm2 but minimally influenced by depletion of Huwe1, Rfwd2 or Rchy1 (Fig. 3f), other E3 ligases that target p53 in cancer cell lines. Thus, Mdm2 is the major E3 ligase for p53, and other E3 ligases do not contribute to p53 degradation in mouse ES cells, consistent with earlier reports37,38. Notably, Mdm2 protein amounts were essentially equal in Dox− and Dox+ cells (Fig. 3g), in contrast to the earlier finding that Mdm2 is a direct transcriptional target of p53 (ref. 39). The reason for this discrepancy is unclear, but, considering that Mdm2 mRNA levels are elevated in the absence of Pict1 (Fig. 3a), a previously uncharacterized post-transcriptional mechanism may suppress Mdm2 protein without Pict1. The low p53 ubiquitination in Dox+ cells is not simply due to low p53-Mdm2 binding (Supplementary Fig. 4), and is probably caused by inactivation of the E3 ligase function of Mdm2 induced by loss of Pict1.
PICT1 regulates MDM2 by retaining RPL11 in the nucleolus
MDM2 function is influenced by nucleolar ribosomal proteins7,13 and other nucleolar proteins such as nucleophosmin40. When we expressed Pict1-EGFP and nucleophosmin-DsRed fusion proteins in 293T cells, these exogenous proteins colocalized in nucleoli (Supplementary Fig. 5a). Pict1 was also highly expressed in the nucleolus of Dox− cells (Fig. 4a). Nucleostemin and nucleolin are also key nucleolar proteins that regulate P53. Although deficiency in nucleophosmin, nucleostemin or nucleolin causes P53 accumulation40–42, the endogenous expression and localization of none of these proteins was impaired in Dox+ cells (Fig. 4a and Supplementary Fig. 5b,c).
Figure 4.
Pict1 regulates Mdm2 by binding to nucleolar Rpl11. (a) Immunoblot detecting indicated proteins in cytoplasmic, nuclear and nucleolar fractions (30 µg) of Pict1 ES cells treated for 1 or 2 d with or without 5 ng ml−1 Dox. Nucleostemin (Nsm), lamin and tubulin, localization controls. (b) Immunoblot detecting the indicated proteins in Pict1 ES cells transfected with vehicle (No), scramble siRNA, Trp53 siRNA (positive control) or siRNAs against the indicated ribosomal proteins and treated with or without 5 ng ml−1 Dox for 24 h. (c) Immunoblot of untreated Dox−1 ES cells immunoprecipitated and immunoblotted with antibodies to Pict1 and Rpl11, respectively. (d,e) Confocal microscopy of Pict1 ES cells transfected with plasmid encoding RPL11-DsRed and treated with or without 5 ng ml−1 Dox for 24 h or 48 h. Endogenous nucleophosmin (Npm) was detected using antibody to Npm (green). Cell fluorescence at 24 h is in d, and percentages of cells retaining RPL11 in the nucleolus at 24 h and 48 h is graphed in (e). Scale bars, 5 µm. (f) Confocal microscopy of Pict1 ES cells transfected with plasmid encoding RPL11-DsRed and treated with or without 5 ng ml−1 Dox for 48 h. Endogenous Mdm2 was detected with antibody to Mdm2 (green). Scale bars, 5 µm. (g) Immunoblot of cytoplasmic and nuclear fractions of Pict1 ES cells treated with or without 5 ng ml−1 Dox for 2 d. Rpl11 was quantified by immunoblotting and the Rpl11 protein level in each sample was adjusted to equality before immunoprecipitation with antibody to Rpl11. Lysates were immnoprecipitated with control IgG or antibody to Rpl11 followed by immunoblotting to detect Mdm2. Results represent three trials.
To identify intracellular binding partners of PICT1, we expressed Flag-PICT1 cDNA in 293T cells, immunoprecipitated the protein with antibody to Flag, and carried out nanoscale liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS; Supplementary Methods). Pict1 bound a huge complex containing many ribosomal proteins (Supplementary Fig. 5d). Among these ribosomal proteins, RPL5 and RPL11 inhibit MDM2-mediated P53 ubiquitination8–10. When we examined the binding of purified ribosomal protein fusion proteins to Pict1 in vitro, RPL5 and RPL11, which we identified in our LC-MS/MS experiments, and RPS7 and RPL23, which inhibit MDM2-mediated P53 ubiquitination11,12, physically interacted with Pict1 (Supplementary Fig. 5e). We confirmed this binding in vivo by expressing exogenous Myc-ribosomal proteins and Flag-Pict1 in 293T cells and subjecting them to immunoprecipitation (Supplementary Fig. 5f).
To determine whether any of these ribosomal proteins was responsible for the impaired Mdm2 function observed in Dox+ cells, we carried out siRNA-mediated knockdown experiments and found that only siRNA against Rpl11 blocked p53 accumulation in Dox+ cells (Fig. 4b). Furthermore, the degree of Rpl11 suppression achieved with various Rpl11 siRNAs was correlated with the degree of suppression of p53 accumulation in Dox+ cells (Supplementary Fig. 5g). We also confirmed the binding of endogenous Rpl11 to Pict1 in untreated Dox− cells by immunoprecipitation (Fig. 4c).
We next compared RPL11 localization in Dox− and Dox+ cells. Exogenous RPL11 appeared in both the nucleolus and cytoplasm in Dox− cells but was almost completely translocated out of the nucleolus in Dox+ cells (Fig. 4d,e). However, exogenous RPL5, RPL23 and RPS7 did not translocate out of the nucleolus in Dox+ cells (Supplementary Fig. 6a–c). We observed the same patterns for endogenous Rpl11, Rpl5, Rpl23 and Rps7 proteins (Fig. 4a). Immunostaining for nucleophosmin, nucleolin and nucleostemin confirmed that nucleoli were intact in our Dox+ cells (Fig. 4d and Supplementary Figs. 5c and 6a–d). Thus, PICT1 is essential for the nucleolar localization of RPL11.
Because RPL11 inhibits MDM2-mediated P53 ubiquitination by binding MDM2 (refs. 7–9,13), we analyzed RPL11-Mdm2 colocalization and binding using our Dox+ cells. Immunostaining confirmed that endogenous Mdm2 was present in the nucleoplasm in both Dox− and Dox+ cells (Supplementary Fig. 6d) and that RPL11 and Mdm2 proteins colocalized in the nucleoplasm of Dox+ cells (Fig. 4f). In addition, binding of endogenous Mdm2 to Rpl11 was much greater in the nucleus of Dox+ cells than in that of Dox− cells (Fig. 4g and Supplementary Fig. 6e). We hypothesize that, when PICT1 is not present to retain RPL11 in the nucleolus, RPL11 escapes into the nucleoplasm and binds MDM2, blocking its ubiquitination of P53. As a result, p53 accumulates in Dox+ cells. PICT1 is therefore a crucial nucleolar binding partner of RPL11 that regulates the MDM2-P53 pathway.
Suppression of PICT1 inhibits tumor cell growth
Although earlier data suggest that PICT1 is a tumor suppressor, we found that Pict1 loss led to p53 accumulation and presumed protection from oncogenesis. We therefore analyzed the onset of chemically induced skin cancers in Pict1+/− mice. Compared with Pict1+/+ controls, Pict1+/− mice were more resistant to papillomagenesis (Fig. 5a). Similarly, shRNA-mediated suppression of PICT1 in various TP53-intact cell lines derived from human gliomas or colorectal or ovarian tumors led to growth inhibition and P53 accumulation without alterations in PTEN or phospho-AKT expression (Fig. 5b). We observed neither growth inhibition nor P53 accumulation in TP53-mutated or P53-inactivated tumor cell lines that were treated with PICT1 shRNA (Supplementary Fig. 7a). Thus, PICT1 does not seem to function as a tumor suppressor when P53 signaling is intact.
Figure 5.
Reduced cancer growth and better survival with low PICT1. (a) Pict1+/+ and Pict1+/− mice treated with DMBA plus TPA were monitored for papillomagenesis for 22 weeks. Left, gross tumor appearance. Scale bars, 2 mm. Middle, number of papillomas per mouse at 22 weeks (mean ± s.e.m., *P < 0.05). Right, incidence and diameter of papillomas at the indicated number of weeks after TPA. (b) Human glioma cell lines DBTRG-05MG and D247, colorectal cancer cell lines Lovo and RKO, and ovarian cancer cell line RMG-1 (all WT TP53) were treated with scramble shRNA or shRNA against PICT1 (PICT1-1 and PICT1-2). Top, MTS assay of growth inhibition. Bottom, immunoblot detecting indicated proteins. Results represent three trials. (c) Left, Kaplan-Meier survival curves for 181 individuals with colorectal cancer and 81 individuals with esophageal cancer whose tumors showed low PICT1 mRNA (blue) or high PICT1 mRNA (orange). Middle, Kaplan-Meier curves for 67 individuals with colorectal cancer and 45 individuals with esophageal cancer whose tumors showed WT TP53 and high or low PICT1 mRNA. Right, Kaplan-Meier curves for 45 individuals with colorectal cancer and 34 individuals with esophageal cancer whose tumors showed mutated TP53 and high or low PICT1 mRNA.
We next determined whether PICT1 expression is correlated with human colorectal or esophageal cancer progression. We detected TP53 mutations in ~40% of our colorectal and esophageal cancer specimens, consistent with earlier reports43,44. Of 181 colorectal cancers examined, individuals whose tumors had a ratio of PICT1 mRNA to GAPDH mRNA of <0.58 were classified in the PICT1low group (n = 90), whereas those with ratios ≥0.58 were classified in the PICT1high group (n = 91) (Supplementary Fig. 7b). The 5-year overall survival rates for the PICT1high and PICT1low groups were 62.0% and 81.0%, respectively, but only when TP53-intact cases were considered (Fig. 5c). We found the same pattern in 81 individuals with esophageal cancer: individuals whose tumors had a ratio of PICT1 mRNA to GAPDH mRNA of <0.64 were classified as PICT1low (n = 40), and those with ratios ≥0.64 were classified as PICT1high (n = 41) (Fig. 5c and Supplementary Fig. 7b). The 5-year overall survival rates for these PICT1high and PICT1low groups (TP53-intact cases only) were 24.5% and 42.1%, respectively. We observed no difference in 5-year survival in TP53-mutated cases of either colorectal or esophageal cancer (Fig. 5c). The Cox proportional hazards model showed that PICT1 mRNA level was an independent prognostic predictor for individuals with either colorectal or esophageal cancer (Supplementary Fig. 7c). Thus, low PICT1 expression is associated with low tumor cell growth in vitro and in vivo and with a better prognosis in individuals with cancer.
DISCUSSION
Although PICT1 has been designated a tumor suppressor gene24–28,45, our work implicates PICT1 as a potentially oncogenic regulator of the MDM2-P53 pathway. Inhibition of Pict1 in ES cells and in various TP53-intact cancer cell lines led to P53 accumulation that inhibited cell growth in vitro. In vivo, Pict1+/− mice were resistant to chemically induced skin tumors; T cell–specific Pict1-deficient mice showed p53-dependent T cell developmental arrest; and individuals with colorectal or esophageal tumors with low PICT1 expression and without TP53 mutation had a better prognosis. Thus, PICT1 is a key cancer-related gene that primarily regulates P53 but is not a typical tumor suppressor. Consistent with our finding that PICT1 knockdown inhibited the growth of TP53-intact glioma cells (Fig. 5b), oligodendroglial tumors with loss of chromosome 19q13 have a better disease outcome30–32. Indeed, oligodendrogliomas show frequent loss of chromosome 19q (70%) but infrequent TP53 mutations (<15%)46. In our study, loss of PICT1 only slightly affected PTEN stability and led to a P53 accumulation in TP53-intact cells that inhibited proliferation. However, PICT1 inhibition has been linked to AKT activation in some insulin-treated cell lines29. Thus, PICT1 may act as a tumor suppressor under limited conditions that include loss of P53 function.
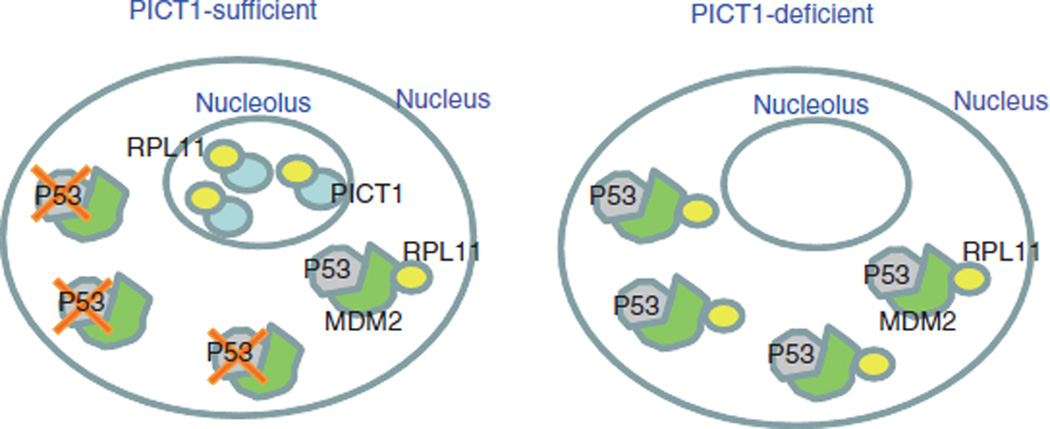
Although P53 is activated by ribosomal proteins released from the nucleolus after nucleolar stress8–12, it was unclear how these ribosomal proteins are anchored in the nucleolus. We have shown that Pict1 binds to and retains RPL11 in the nucleolus, preventing RPL11 from inhibiting Mdm2 in the nucleoplasm. Consistent with this model (Fig. 6), treatment of WT ES cells with a low dose of actinomycin D or mycophenolic acid decreased Pict1 expression and p53 accumulation (Supplementary Fig. 8). Small ribosomal subunit proteins in the nucleolus are unstable and degraded in response to stress signals47. However, we found that amounts of ribosomal proteins other than Rpl11, including Rps7, were not decreased in the nucleolus by Pict1 deficiency. In addition, the decrease in endogenous p53 ubiquitination without Pict1 was notable after the addition of MG132, and Rpl11 suppression blocked p53 accumulation in Pict1 ES cells. Thus, we think that protein degradation did not cause the observed decrease in nucleolar Rpl11, and that without retention by PICT1, RPL11 diffuses into the nucleoplasm, where it is captured by MDM2. We showed that Pict1 loss enhanced Rpl11-Mdm2 binding in the nucleoplasm and inhibited the E3 ligase activity of Mdm2, leading to p53-dependent cell cycle arrest and apoptosis in a manner consistent with earlier results8. PICT1 is therefore a major upstream regulator of P53, and the binding of PICT1 to RPL11 controls the MDM2-P53 pathway.
Figure 6.
PICT1 binding to nucleolar RPL11 regulates MDM2-P53 activity. Model of PICT1 function. Left, when PICT1 is present in the nucleolus, RPL11 is retained in the nucleolus and MDM2 is free to ubquitinate P53, promoting its degradation. Right, when PICT1 is absent, nucleolar RPL11 escapes into the nucleoplasm and binds to MDM2, blocking its ubiquitination of P53. As a result, P53 accumulates in PICT1-deficient cells.
In conclusion, our work shows that PICT1 is a key regulator that acts primarily through RPL11 and MDM2 to inhibit P53 responses to nucleolar stress. We have also identified PICT1 as a useful prognostic marker for human colorectal and esophageal cancers. Studies of factors influencing PICT1 expression or stability, or interfering with PICT1-RPL11 binding, may yield new cancer treatments, especially for individuals with TP53-intact tumors.
ONLINE METHODS
Pict1-deficient ES cells
We electroporated Pict1flox targeting vector (Supplementary Fig. 1d and Supplementary Methods) into Pict1+/− ES cells (Supplementary Fig. 1a), and used correctly targeted clones (Pict13loxP/−; Supplementary Fig. 1e) to generate Tet-regulated ES cells. We established an ES cell clone that constitutively expressed the modified Tet-regulated transactivator driven by the CAG promoter48 and co-transfected these cells with Tet-regulatable pUHD10-3Pict1.IRES.EGFP plasmid48 plus pcDNA3.1Zeo. We selected transfected cells for 10 d in medium containing 20 µg ml−1 zeocin (Invitrogen) and studied clones whose EGFP expression was tightly regulated by tetracycline (Supplementary Fig. 1f). We examined expression of the modified Pict1 genes by immunoblotting to confirm that exogenous Pict1 was driven by the Tet response promoter (tetO-CMV) and suppressed by Tet addition (Tet-off system)49. We transiently transfected Tet-regulatable Pict1-deficient ES cells (Pict1tetTg+; Pict13loxP/−; Supplementary Fig. 1e) with CAGGS-Cre DNA to eliminate the endogenous Pict1 allele. The desired mutant Pict1 ES cell clone (Pict1tetTg+; Pict1Δ/−) was maintained without Dox after Cre DNA transfection (Dox− cells). We added Dox (5 ng ml−1) to Pict1 ES cells to delete Pict1 and generate Dox+ cells (Supplementary Fig. 1e).
Immunoblotting
We carried out immunoblotting using a standard protocol and primary antibodies to PTEN (Cascade), phospho-PTEN, phospho-Akt, Akt, phospho-γ-H2ax (all from Cell Signaling), mouse p53 (Novocastra), human P53 (Santa Cruz), p21Waf1 (Calbiochem), p19Arf (Abcam), p16Ink4a (Santa Cruz), p27Kip1 (BD Biosciences), human ubiquitin (Santa Cruz), RPL5 (Abcam), nucleostemin (Millipore), nucleophosmin, nucleolin and actin (all from Sigma). Antibodies to PICT1, RPL11, RPL23 and RPS7 were affinity-purified from antisera as previously described9,11,12,50. We detected primary antibodies using horseradish peroxidase–conjugated secondary antibodies (Cell Signaling).
Transfection of siRNA and shRNA
We transfected siRNA oligonucleotides (10 nM) in OPTI-MEM (Invitrogen) into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. At 24 h after transfection, Pict1 was deleted by Dox treatment at concentrations and for the times indicated in Figures 2d,e, 3e,f and 4b and Supplementary Figures 2a–d and 5g. We assayed protein levels as described below. Target sequences for siRNAs are in Supplementary Methods.
For shRNA studies, we produced lentiviruses containing PICT1 shRNA or scrambled shRNA (lenti-shRNA) and used them to infect tumor cells as previously described51. We estimated lentivirus titers by measuring HIVp24 gag antigen concentration using ELISA (PerkinElmer Life Science). For Figure 5b and Supplementary Figure 7a, we incubated tumor cells in six-well plates (5 × 104 per well) with lenti-shRNA (1.5 × 104 transduction units) for 6 d. Gene transfection efficiency was 80–97%. Proliferation of infected cells was determined by MTS assay (Promega). Target sequences for shRNAs are in Supplementary Methods.
Ubiquitin ligase activity
We assayed ubiquitin ligase activity as described52,53 with minor modifications outlined in Supplementary Methods.
Immunoprecipitation
To detect the binding of exogenous ribosomal proteins to Pict1, we transfected the appropriate pcDNA3.1-Myc-RP cDNA plus CAG-Flag-Pict1 cDNA into 293T cells (RIKEN BioResource Center) by lipofection. At 48 h after transfection, cells were lysed and analyzed by immunoprecipitation (Supplementary Fig. 5f). To detect the binding of endogenous Pict1 to Rpl11 (Fig. 4c), Mdm2 to Rpl11 (Fig. 4g and Supplementary Fig. 6e) and Mdm2 to p53 (Supplementary Fig. 4), we lysed Pict1 ES cells at various times after addition of 5 ng ml−1 doxycycline and incubated them with antibodies to Pict1, Mdm2 (Santa Cruz), Rpl11 or p53 (Novocastra) or control IgG (Santa Cruz). We adsorbed immune complexes to protein G–Sepharose beads (GE). When used, we added MG132 (20 µM) 3 h before cell collection. After washing extensively, we analyzed samples by immunoblotting with antibodies to Rpl11, Pict1, Mdm2 or p53.
Chemical tumor induction
To induce tumors, we topically treated shaved dorsal skin of Pict1+/+ (n = 17) and Pict1+/− (n = 19) mice (6–7 weeks old) with 0.5 mg 7,12-dimethylbenz[a]anthracene (Sigma) in acetone. Two weeks later, the same area was topically treated with 5 µg phorbol-12-myristate-13-acetate (Sigma) in acetone twice weekly for 22 weeks as described51. Control mice were treated with acetone only. We measured tumor numbers and sizes weekly. The Institutional Review Board of Kyushu University approved the design of all animal studies.
Clinical samples
We acquired surgically obtained colorectal (181) and esophageal (81) cancer samples from the Department of Surgical Oncology, Kyushu University. The Institutional Research Ethics Committee of Kyushu University approved the study design, and all study participants gave written informed consent. We immediately froze resected cancer tissues in liquid nitrogen. We determined mutation of TP53 by sequencing a DNA region spanning exons 5–8, the area where most TP53 mutations occur43. We analyzed PICT1 and control GAPDH mRNA levels using quantitative real-time RT-PCR and JMP 5 for Windows software (SAS Institute). We established survival curves using Kaplan-Meier methodology and examined differences using the log-rank test. We calculated relative risk using the Cox proportional hazard model. P < 0.05 indicates statistical significance.
Additional methods
Detailed methodology is described in the Supplementary Methods.
Supplementary Material
Acknowledgments
We are grateful to T. Noda, S. Kuroda, H. Kishimoto, M. Natsui, H. Takahashi, H. Tashiro, N. Yasui, M. Suzuki, S. Suzuki, T. Shono and T. Sasaki for expert technical support and helpful discussions. We also thank H. Miyoshi (RIKEN BioResource Center, Tsukuba, Japan) for providing lentivirus vector plasmid DNA. This work was supported by grants from the Ministry of Education, Culture, Sports and Technology of Japan (MEXT), Takeda Medical Foundation, Naito Foundation, Ono Medical Research Foundation, Yasuda Medical Foundation, and Astellas Foundation for Research on Metabolic Disorders. K.M. and M.M. are supported by the Core Research for Evolutionary Science and Technology program of the Japanese Science and Technology Agency. T.Y., T.S., M.M. and A.S. are supported by the Global Centers of Excellence Program of MEXT.
Footnotes
Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
M.S. carried out the initial generation and analyses of Pict1flox mice and Pict1 ES cells. K.K. carried out subsequent major biochemical and biological experiments, and M.N. carried out mouse work. K.M., R.K. and M.M. carried out the human cancer tissue analyses. K.H. generated Pict1−/− mice. B.I. assisted with confocal microscopy. J.W., Y.K. and Y.R.Y. assisted with the introduction of shRNA into human cancer cell lines. H.H. assisted with the protein binding assays. Y.H. carried out mouse analyses. T.Y., T.K., Y. Zhang, Y. Zhu, C.P. and T.W.M. provided key materials. T.M., K.M. and A.S. conceived of the project, and M.S., K.K., K.M, T.M., M.M. and A.S. designed the experiments. M.S., K.K., M.N., K.M., R.K., T.Y., T.K., Y. Zhang, C.P., T.N., T.W.M., T.S., T.M., M.M. and A.S. discussed the hypothesis and interpreted the data. A.S. coordinated and directed the project and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 2.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 3.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 4.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 6.Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2′s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 7.Lindström MS, Deisenroth C, Zhang Y. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle. 2007;6:434–437. doi: 10.4161/cc.6.4.3861. [DOI] [PubMed] [Google Scholar]
- 8.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 11.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fumagalli S, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D, et al. Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 17.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J. Biol. Chem. 2008;283:12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulic S, et al. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19:3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 22.Yadavilli S, et al. Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst.) 2009;8:1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith JS, et al. A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics. 2000;64:44–50. doi: 10.1006/geno.1999.6101. [DOI] [PubMed] [Google Scholar]
- 24.Kim YJ, et al. Suppression of putative tumour suppressor gene GLTSCR2 expression in human glioblastomas. J. Pathol. 2008;216:218–224. doi: 10.1002/path.2401. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa Y, et al. Chromosomal imbalances in malignant peripheral nerve sheath tumor detected by metaphase and microarray comparative genomic hybridization. Oncol. Rep. 2006;15:297–303. [PubMed] [Google Scholar]
- 26.Merritt MA, et al. Expression profiling identifies genes involved in neoplastic transformation of serous ovarian cancer. BMC Cancer. 2009;9:378. doi: 10.1186/1471-2407-9-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yim JH, et al. The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death. Cell Death Differ. 2007;14:1872–1879. doi: 10.1038/sj.cdd.4402204. [DOI] [PubMed] [Google Scholar]
- 28.Okahara F, Ikawa H, Kanaho Y, Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 29.Okahara F, et al. Critical role of PICT-1, a tumor suppressor candidate, in phosphatidylinositol 3,4,5-trisphosphate signals and tumorigenic transformation. Mol. Biol. Cell. 2006;17:4888–4895. doi: 10.1091/mbc.E06-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairncross JG, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 31.Smith JS, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 32.Mariani L, et al. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J. Clin. Oncol. 2006;24:4758–4763. doi: 10.1200/JCO.2006.05.9238. [DOI] [PubMed] [Google Scholar]
- 33.von Deimling A, et al. Loci associated with malignant progression in astrocytomas: a candidate on chromosome 19q. Cancer Res. 1994;54:1397–1401. [PubMed] [Google Scholar]
- 34.Smith JS, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki M, et al. Thymocyte proliferation induced by pre–T cell receptor signaling is maintained through polycomb gene product Bmi-1–mediated Cdkn2a repression. Immunity. 2008;28:231–245. doi: 10.1016/j.immuni.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Kamijo T, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solozobova V, Blattner C. Regulation of p53 in embryonic stem cells. Exp. Cell Res. 2010;316:2434–2446. doi: 10.1016/j.yexcr.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurki S, et al. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 41.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- 43.Russo A, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005;23:7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 44.Egashira A, et al. p53 gene mutations in esophageal squamous cell carcinoma and their relevance to etiology and pathogenesis: results in Japan and comparisons with other countries. Cancer Sci. 2007;98:1152–1156. doi: 10.1111/j.1349-7006.2007.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JY, Kim HS, Lee S, Park JH. The expression of GLTSCR2, a candidate tumor suppressor, is reduced in seborrheic keratosis compared to normal skin. Pathol. Res. Pract. 2010;206:295–299. doi: 10.1016/j.prp.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto Y, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 47.Andersen JS, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 48.Era T, Witte ON. Regulated expression of P210 Bcr-Abl during embryonic stem cell differentiation stimulates multipotential progenitor expansion and myeloid cell fate. Proc. Natl. Acad. Sci. USA. 2000;97:1737–1742. doi: 10.1073/pnas.97.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitajima K, Masuhara M, Era T, Enver T, Nakano T. GATA-2 and GATA-2/ER display opposing activities in the development and differentiation of blood progenitors. EMBO J. 2002;21:3060–3069. doi: 10.1093/emboj/cdf301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okahara F, et al. Production of research-grade antibody by in vivo electroporation of DNA-encoding target protein. Anal. Biochem. 2005;336:138–140. doi: 10.1016/j.ab.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 51.Inoue-Narita T, et al. Pten deficiency in melanocytes results in resistance to hair graying and susceptibility to carcinogen-induced melanomagenesis. Cancer Res. 2008;68:5760–5768. doi: 10.1158/0008-5472.CAN-08-0889. [DOI] [PubMed] [Google Scholar]
- 52.Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.