Abstract
Alzheimer's disease is the most common form of dementia. Diffusion imaging provides information on white matter integrity not available with standard MRI, revealing additional information on how Alzheimer’s disease affects the brain. Here we implemented and tested a multi-atlas labeling algorithm to segment the fornix and a point-correspondence tract matching scheme to assess fiber integrity in the fornix in diffusion MRI from 210 participants scanned as part of the Alzheimer’s Disease Neuroimaging Initiative. Various diffusion-derived measures were used to relate fornix degeneration to cognitive decline. On 3D parametric tract models, mean diffusivity (MD) was more sen-sitive to group differences than fractional anisotropy (FA). Compared to previous studies, we mapped diffusion information along the fornix, yielding 3-D maps of degenerative changes along the tract in people with different stages of Alzheimer’s disease.
Keywords: Alzheimer’s disease, DTI, fornix, clustering, label fusion
1. INTRODUCTION
Alzheimer’s disease (AD) is a progressive, degenerative disease that affects the brain’s nerve cells, or neurons, resulting in behavioral changes, loss of memory, language function, and general cognitive decline. An estimated 26.6 million people worldwide have AD in 2013. Therefore, there is a worldwide effort to identify cheaper and less invasive biomarkers of disease and its progression. More recently, diffusion tensor imaging (DTI) [1] has become a popular method to reconstruct the local profile of water diffusion in tissues, yielding information on white matter (WM) integrity and connectivity that is not available from standard anatomical MRI. Region-of-interest (ROI)-based voxel analyses have been used to identify AD-related abnormalities in the corpus callosum, fronto-occipital and inferior longitudinal fasciculi, cingulum, and forceps major based on DTI-derived parameters, such as fractional anisotropy (FA) and measures of mean (MD), radial, or axial diffusivity [2].
Among the WM fiber tracts, the fornix is critical for normal cognitive functioning. It is the major output tract of the hippocampus, arching around the thalamus and connecting the medial temporal lobes to the hypothalamus. Hippocampal atrophy is one of the most widely used MRI biomarkers of AD [3], and the fornix is the main white matter pathway to and from the hippocampus, but the fornix is less well-studied as a predictor of cognitive impairment as AD progresses. This is mainly because the fornix is very hard to segment due to its small volume relative to the typical voxel size of MRI or DTI. Even so, AD typically begins in the hippocampus and medial temporal lobes, so it makes sense to evaluate the tracts innervating this crucial region for learning and memory. Prior studies either manually delineated ROIs for the fornix [4] or automatically deformed a template onto each subject in a population [5]. The 1-D mean FA of the main fornix body or the 2-D “crest line” of locally maximal FA intensities across voxels have been used in statistical analyses, such as tract-based spatial statistics, usually in small cohorts of subjects.
To overcome some of these limitations, here we take a different approach to automatically segment the fornix from 3-D whole-brain tractography. Based on the fornix ROI from a publicly available WM atlas [6], we first manually construct five fornix atlases (from different individuals) as prior anatomical information. Then, we transfer the tract label to new subjects by selecting only fibers that are similar to the corresponding fornix atlases, based on a mathematically defined similarity measure. Multiple atlases are used, to help adapt to the variability of tract shapes in new subjects. Next, we use a label fusion scheme to fuse the clustered results obtained from individual atlases. Many fiber clustering methods have been proposed, but it is not always clear how to apply them to large-scale group studies. Here, we implement a novel point-wise matching scheme to match fiber points across the population. To test the robustness of our algorithm, we applied it to a study of a cohort of 210 participants scanned with DTI. We studied the 3-D profile of the fornix in the cohort, at sub-voxel resolution. Our goal was to compare a variety of DTI-derived measures across different diagnostic groups in the cohort to demonstrate the value of the algorithm for clinical research, and to define tract-based measures sensitive to disease progression.
2. METHODS
2.1. Subjects and Data Acquisition
The DTI data of our subjects were downloaded from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) data-base. We analyzed baseline DWI data from 210 participants (Age: 55–90; mean: 72.2+/−7.4 SD; 124 males/86 females). 52 of them were cognitively normal elderly people, 113 had mild cognitive impairment (MCI) – which is associated with increased risk of progressing to AD – and 45 were AD patients. Each subject underwent cognitive and clinical evaluations, including the Mini-Mental State Examination (MMSE) and the Clinical Dementia Rating Scale Sum of Boxes (CDR-SOB) tests.
Diffusion scans were acquired with 3-Tesla GE Medical Systems scanners at 14 sites across North America. Each 3D volume consisted of 59 axial slices with isotropic voxel size 2.7 mm with a 256 × 256 acquisition matrix. 46 volumes were acquired per subject: 5 with T2-weighted b0 images and 41 diffusion-weighted volumes (b = 1000 s/mm2).
2.2. Probabilistic Tractography
We performed whole-brain tractography with Camino (http://cmic.cs.ucl.ac.uk/camino/). We used the Probabilistic Index of Connectivity method (PICo) [7] to generate probabilistic tractography. Seed points were chosen at those voxels whose FA values were greater than 0.3. Monte Carlo simulation was used to generate fibers proceeding from the seed points throughout the entire brain with 4th-order Runge-Kutta interpolation. The maximum fiber turning angle was set to 40°/voxel, and tracing stopped at any voxel whose FA was less than 0.2.
2.3. Fornix Atlas Construction
We manually constructed five fornix atlases from a healthy twins’ data set. A single-subject template called the “Type II Eve Atlas” (a 32-year old healthy female) [6] was registered to the FA images of each atlas. The “Eve” fornix ROI was re-assigned to the five atlases with the resulting deformation fields by ANTs. Fibers that traversed the ROI were extracted and manually edited to form our fornix atlases. We opted to use data from healthy adults (twins) instead of elderly individuals from ADNI because their fornix tracts are intact and can be more completely extracted.
2.4. Fiber Clustering
For each subject in our data set, the same registration registered the subject’s FA image to each of the five fornix atlas-es’ FA images and the “Eve” FA image, respectively. Each fornix atlas and the “Eve” fornix ROI were then warped to the subject space with the corresponding deformation fields. Fiber alignment is improved significantly with this type of registration [8].
We first chose the fibers that traversed the warped Eve fornix ROI. This reduced the number of fibers from millions to only a few hundreds. To further refine the result, we defined a fiber distance metric to select the fibers whose distances were close to one of the warped atlas fibers, based on a validated empirical threshold (15 mm) in [9]. For any pair of fibers γi and γj, the symmetric Hausdorff distance is:
where dH′(γi,γj) = maxx∈γiminy∈γj‖x − y‖. ‖·‖ is the Euclidean norm and the ordered pair (γi,γj) indicates an asymmetric distance from γi to γj,. The x’s and y’s are the coordinate points along fibres γi and γj, respectively [10]. The Hausdorff distance is conservative and keeps only the fibers whose shapes and locations are similar to the atlases. This is intended to facilitate large population studies.
2.5. Label Fusion
Due to the variability of individual atlases, different atlases may “nominate” different candidate fibers in new subjects’ data as belonging to a specific tract (here the fornix). We implemented a label fusion scheme to combine the results from individual atlases. A mean fiber distance was defined to rank the fibers nominated by individual atlases:
where di is the Hausdorff distance between an unlabeled subject’s fiber and the i-th atlas, dcutoff is the empirical cutoff threshold chosen in Section 2.4, dsup is the upper bound Hausdorff distance within which a subject fiber can be possibly considered a candidate for a given tract, and n is the number of atlases. We ranked all the candidate fibers from different atlases based on their dmean’s. The smaller its dmean, the higher its rank. A fusion percentage was defined to include fibers whose dmean’s were among the top specified percentage. Here we set the fusion percentage to be 95% because no other confounding fibers are adjacent to the fornix, and the number of false positive fibers is relatively low.
2.6. Fiber Matching
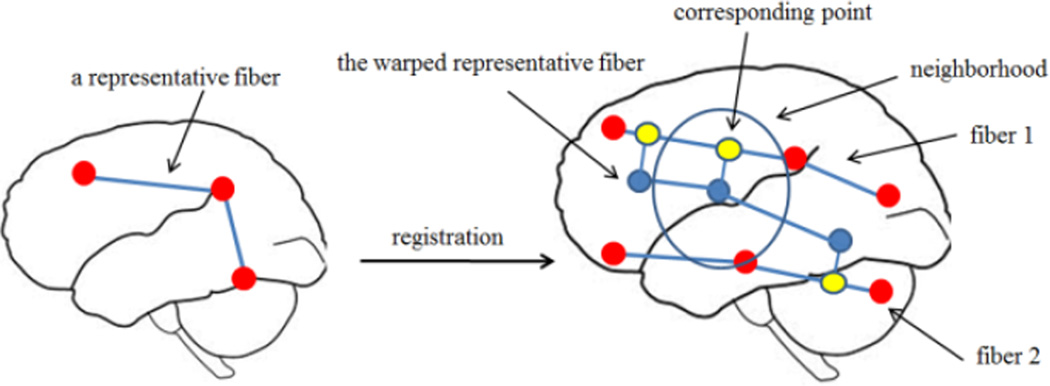
To perform group studies, we need to establish a computed correspondence between fibers of the segmented fornix tracts across the cohort. First, we chose a representative sample fornix tract from our ADNI population. Each point on that representative tract was mapped to the rest of the population. The point on each fornix tract in the cohort with the closest Euclidean distance to that sample point was considered the corresponding point. More specifically, the point on the representative tract (the representative point) was warped to each subject’s space. It was then projected onto the fibers that intersect with the neighborhood of the representative point. The projection point with the shortest distance to the representative point was taken as the corresponding point for that subject. If there were no fibers crossing the neighborhood (a sphere with the 10 mm radius), the warped representative point location is used as the correspondence point. Figure 1 illustrates our fiber matching method.
Figure 1.
An illustration of our fiber matching scheme.
2.7. Statistical Analysis
Statistical analysis was performed comparing groups with a variety of metrics, such the mean FA/MD of the volume that those fibers cross, and the 3-D FA/MD profile, to demonstrate the utility of the technique.
3. RESULTS
3.1 3-D FA and MD Profiles
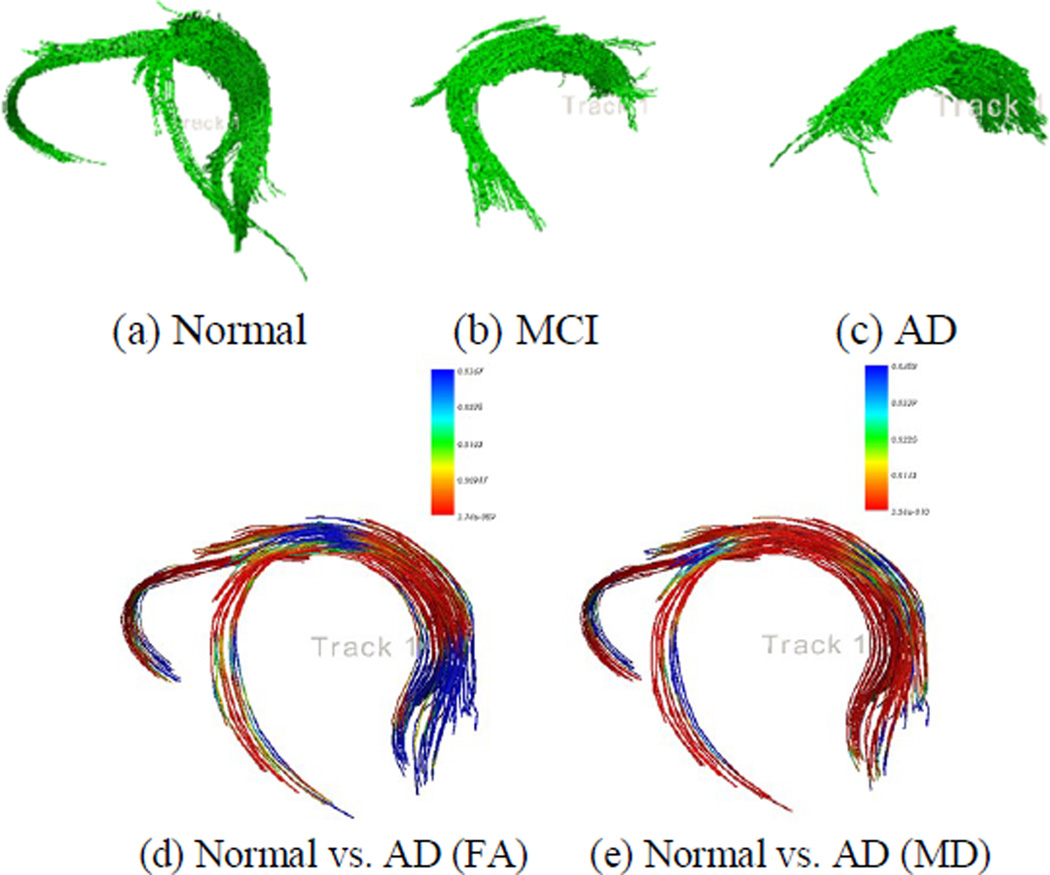
Figure 2 shows the three individual representative fornix tracts for the three groups and the 3-D sub-voxel profiles (a point-wise t-test between different diagnostic groups) of the differences in FA and MD values between the normal group and the AD group, respectively. Results are corrected for multiple comparisons using the false discovery rate method [11]. MD shows more sensitivity in detecting the differences, at least in this sample.
Figure. 2.
A representative fornix tract is shown for each group. 3D color maps reveal differences in FA and MD values between normal elderly and AD groups, respectively. Redder colors show greater group differences in FA or MD values at those points.
3.2 Quantitative Validation
Ten subjects (4 normal; 3 MCI; and 3 AD) were randomly selected from the dataset. A trained neuroscientist performed manual segmentation on the fornix tracts from the whole-brain tractography. The results were taken as ground truth to validate our automated clustering results. The Dice’s coefficient is defined as , where N(a) and N(b)are the sets of fibers from ground truth and our clustering results. The average Dice’s coefficient over the ten subjects was 0.97 (standard deviation: 0.01). This suggests that our algorithm can reliably segment the fornix tract.
3.3 Mean FA and MD
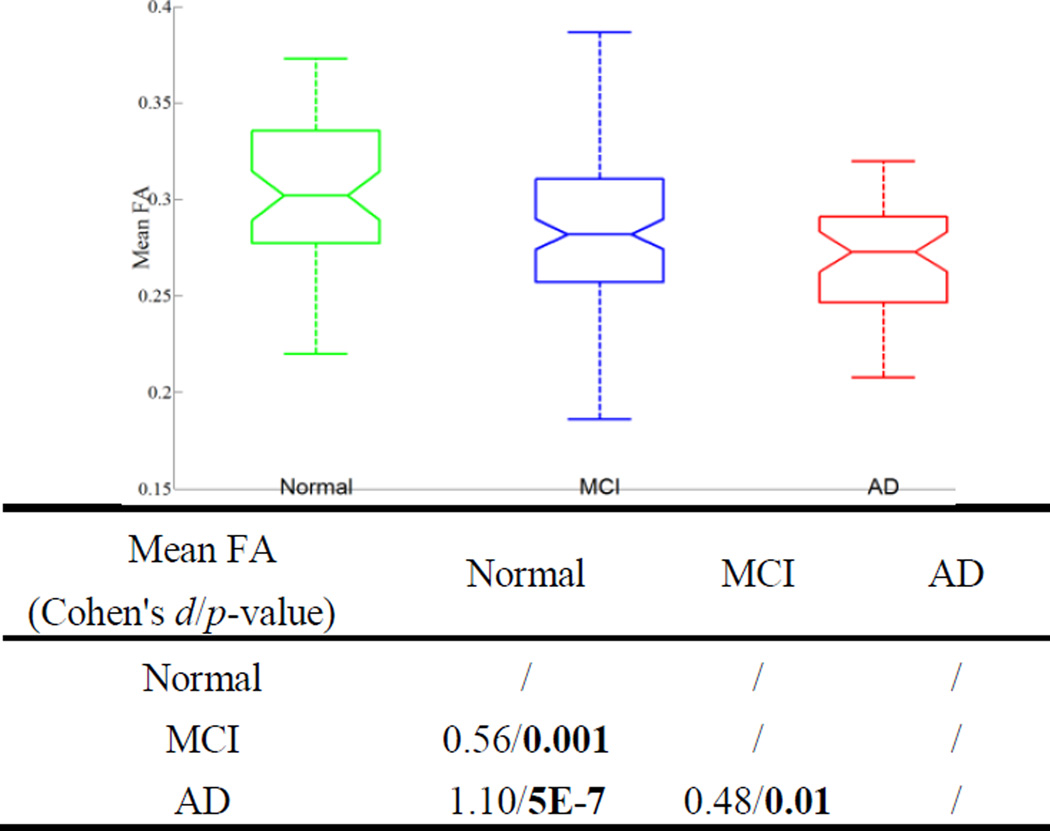
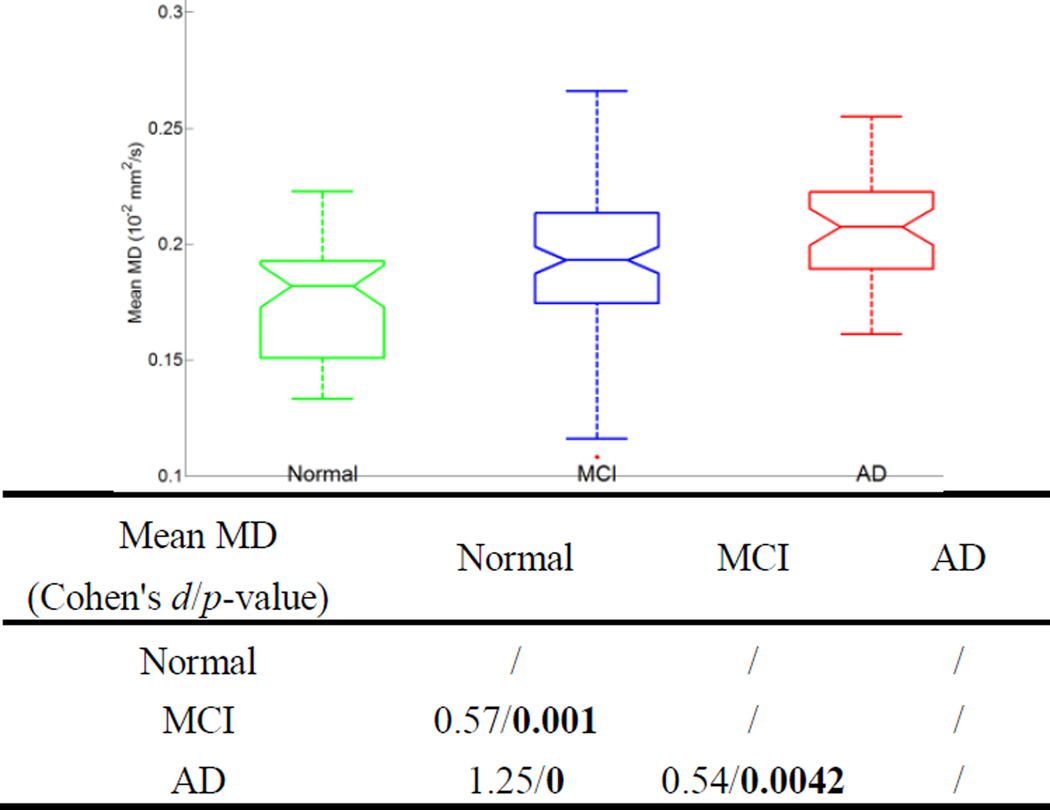
Figure 3 and 4 show box plots of the mean FA and MD values of the entire fornix for the three diagnostic groups and the effect size (Cohen’s d) and the p-values of the t-tests comparing groups. The five horizontal lines in each group represent the minimum, lower quartile, median, upper quartile, and the maximum. Outliers are marked with a +. Notches offer a rough guide to significance of the difference in medians. The mean FA or MD value for each subject was calculated by averaging the FA or MD values of all the voxels the labeled fornix tract traversed and the voxels which the corresponding fiber points were in (in case tractography was poor in some subjects). We used 10−2 mm2/s as the unit for MD to make its values comparable to the range of FA values (which are dimensionless). The median of mean FA decreases from the normal toward the AD group, while the median of mean MD increases. The diffusivity increase may be because myelin breaks down in AD and offers less hindrance to water diffusion. The mean MD difference is more marked than that of FA between groups in terms of both effect size and the associated p-values.
Figure 3.
The box plot on the top shows the mean FA values of the fornix for three groups. The group-wise Cohen’s d’s and p-values of the t-tests are listed at the bottom.
Figure 4.
The box plot on the top shows the mean MD values of the fornix for three groups. The associated group-wise Cohen’s d’s and p-values of the t-tests are listed at the bottom.
3.4 Correlations and Clinical Scores
We performed a linear regression analysis on the MMSE and CDR-SOB scores for each subject versus the mean FA and MD values of their fornix tracts, adjusting for age and sex. The statistics are shown in Table 1. The mean FA/MD of the fornix explains about 10%/12% of the variance (R2) in MMSE scores and 10%/11% in CDR-SOB, respectively. The relationships between clinical scores and mean FA/MD are both statistically significant.
Table 1.
Regression statistics relating MMSE and CDR-SOB scores (measures of clinical decline) to mean FA and MD of the fornix, after adjusting for age and sex.
| Clinical Score |
Variable | R2 |
p- value |
Variable | R2 |
p- value |
|---|---|---|---|---|---|---|
| FA | 2E-3 | MD | 2E-4 | |||
| MMSE | age | 0.10 | 0.02 | age | 0.12 | 0.10 |
| sex | 0.69 | sex | 0.56 | |||
| FA | 2E-5 | MD | 8E-6 | |||
| CDR-SOB | age | 0.10 | 0.59 | age | 0.11 | 0.97 |
| sex | 0.84 | sex | 0.76 | |||
4. DISCUSSION AND CONCLUSION
Here we presented an automatic tract labeling technique that uses anatomical information from multiple manual atlases. We implemented it to segment the fornix in 210 subjects. We then used a point-wise fiber matching scheme to establish tract correspondence across a population and perform large-scale group studies.
We studied the fornix in detail by comparing how Alzheimer’s disease affects different DTI-derived measures as AD progresses. Our results are consistent with prior findings that fornix degeneration is associated with cognitive decline [4,5]. MD may be a better measure for detecting AD-related differences than FA [12].
The contribution of our work is to present a reliable general workflow for large population studies and provide extra information (such as 3-D sub-voxel profiles) for tract analysis. This will be extended to group studies of various neurological and psychiatric conditions, and to research in imaging genetics.
Acknowledgments
Thanks to NIH U54 EB020403 (BD2K) for funding.
REFERENCES
- 1.Basser PJ, et al. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66(1):259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teipel SJ, et al. Multivariate network analysis of fiber tract integrity in Alzheimer’s disease. NeuroImage. 2007;34(3):985–995. doi: 10.1016/j.neuroimage.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 3.Morra JH, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. NeuroImage. 2008;45(1):59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oishi K, et al. The fornix sign: a potential sign for Alzheimer’s disease based on diffusion tensor imaging. J. Neuroimaging. 2012;22(4):365–374. doi: 10.1111/j.1552-6569.2011.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fletcher E, et al. Loss of fornix white matter volume as a predictor of cognitive impairment in cognitively normal elderly individuals. JAMA Neurol. 2013;70(11):1389–1395. doi: 10.1001/jamaneurol.2013.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oishi K, et al. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping. NeuroImage. 2009;46(2):486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker GJM, et al. A framework for a streamline-based probabilistic index of connectivity (PICo) using a structural interpretation of MRI diffusion measurements. J. Magn. Reson. Imaging. 2003;18(2):242–254. doi: 10.1002/jmri.10350. [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, et al. 3D elastic registration improves HARDI-derived fibre alignment and automated tract clustering. 8th IEEE ISBI. 2011:822–826. [Google Scholar]
- 9.Jin Y, et al. Automated clustering of white matter fibers in brain diffusion MRI with an application to genetics. NeuroImage. 2014;100:75–90. doi: 10.1016/j.neuroimage.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerig G, et al. Analysis of brain white matter via fiber tract modeling. IEEE Eng. Med. Biol. Soc. 2004;6:4421–4424. doi: 10.1109/IEMBS.2004.1404229. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, et al. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57(1):289–300. [Google Scholar]
- 12.Nir TM, et al. Effectiveness of regional DTI measures in distinguishing Alzheimer’s disease, MCI, and normal aging. NeuroImage Clin. 2013;3:180–195. doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]