Abstract
Our understanding of network breakdown in Alzheimer’s disease (AD) is likely to be enhanced through advanced mathematical descriptors. Here, we applied spectral graph theory to provide novel metrics of structural connectivity based on 3-Tesla diffusion weighted images in 42 AD patients and 50 healthy controls. We reconstructed connectivity networks using whole-brain tractography and examined, for the first time here, cortical disconnection based on the graph energy and spectrum. We further assessed supporting metrics - link density and nodal strength - to better interpret our results. Metrics were analyzed in relation to the well-known APOE-4 genetic risk factor for late-onset AD. The number of disconnected cortical regions increased with the number of copies of the APOE-4 risk gene in people with AD. Each additional copy of the APOE-4 risk gene may lead to more dysfunctional networks with weakened or abnormal connections, providing evidence for the previously hypothesized “disconnection syndrome”.
Index Terms: graph spectrum, energy, Alzheimer’s disease, APOE-4, disconnection syndrome
1. INTRODUCTION
Within the nervous system, the brain’s white matter consists of highly myelinated axons that allow transfer of information among cortical, subcortical and other brain regions. Unlike standard MRI, diffusion weighted imaging (DWI) captures subtle changes in white matter integrity through measures that are sensitive to tissue microstructure. In addition, tractography methods can be used to infer connectivity patterns of neural pathways leading to complex mathematical metrics describing fiber networks.
Studies of the human connectome are increasingly popular [3–5, 11], and there is growing interest in analyzing the brain using network theory. Brain networks may be represented as matrices or graphs - these contain nodes, typically defined as anatomical regions segmented from MRI, and edges that connect these regions. This line of work is still in its formative stages, and we are in constant search for metrics that might differentiate disease from normal variation and disease progression.
Here, we implemented ideas from spectral graph theory using measures computed on the Laplacian matrix to describe the spectrum of a graph. The spectrum can reveal information about the disconnected components (i.e., nodes) of a network. The graph energy is a distinct concept, from organic chemistry, normally used to quantify the stability of molecular orbitals associated with π-electrons [9]. We applied this concept to study the stability of connections in the brain’s networks, in addition to more commonly computed metrics – link density and nodal strength.
We analyzed the brain spectrum of 50 healthy elderly participants and 42 patients with Alzheimer’s disease (AD) in relation to the genetic variant – apolipoprotein E epsilon 4 allele (APOE-4), a known genetic risk factor for developing AD [3, 11]. Individuals who carry one or two copies of the APOE-4 risk gene have an increased chance of developing AD and tend to have a lower mean age at onset than noncarriers [3]. Therefore, neuroimaging studies involving APOE-4 carriers aged 60 or older are critically important for understanding genetic contributions to AD risk and cognitive decline [3].
AD is characterized by severe neuronal atrophy that typically starts in the temporal, parietal and limbic cortices and spreads – over time – to the frontal cortex [3]. This may lead to the previously hypothesized ‘disconnection syndrome’ between cortical regions of the brain, resulting in episodic memory loss, among other deficits [6]. This hypothesis was assessed here using spectral graph theory metrics to quantify disconnection. We found that disconnections between cognitive areas of the brain in AD participants were located in the entorhinal, frontal and temporal lobes bilaterally. We also found that the energy of the brain network was reduced in AD carriers of more copies of the APOE-4 risk gene – suggesting instability of network components especially in the temporal lobe, followed by the parietal and frontal lobes.
2. METHODS
2.1. Participants and diffusion-weighted imaging
We analyzed diffusion-weighted images (DWI) from 92 participants scanned as part of the Alzheimer’s Disease Neuroimaging Initiative (ADNI; http://adni.ini.usc.edu). Table 1 shows the demographics of the participants we studied including their age, sex and APOE-4 status (0, 1 and 2 indicate the number of alleles). All 92 participants underwent whole-brain MRI on 3-Tesla GE Medical Systems scanners, at 16 sites across North America. Standard anatomical T1-weighted IR-FSPGR (inverse recovery fast spoiled gradient recalled echo) sequences were collected (256×256 matrix; voxel size = 1.2×1.0×1.0 mm3; TI = 400 ms; TR = 6.984 ms; TE = 2.848 ms; flip angle = 11°) in the same session as the DWI (128×128 matrix; voxel size: 2.7×2.7×2.7 mm3; scan time = 9 min). 46 separate images were acquired for each scan: 5 T2-weighted images with no diffusion sensitization (b0 images) and 41 diffusion-weighted images (b = 1000 s/mm2).
Table 1.
Demographic information for 50 controls (CTL) and 42 AD subjects scanned with diffusion MRI as part of the ADNI project. 6 AD participants did not have APOE-4 information.
| CTL | AD | Total | |
|---|---|---|---|
| N | 50 | 42 | 92 |
| Age | 72.6 ± 6.1 SD | 75.5 ± 8.9 SD | 73.9 ± 7.6 SD |
| APOE-4 | 35(0)/15(1) | 16(0)/15(1)/5(2) | 51(0)/30(1)/5(2) |
| Sex | 22M/28F | 28M/14F | 50M/42F |
2.2 NxN Connectivity Matrix Creation
Whole-brain tractography was performed using the Hough transform [1] to recover fiber tracts based on a constant solid angle orientation distribution function (CSA-ODF) to model the local diffusion propagator. We computed ~10,000 for each subject (3D curves) and extracted 34 cortical labels per hemisphere (68 per whole-brain) from aligned T1-weighted structural MRI scans with FreeSurfer (http://surfer.nmr.mgh.harvard.edu/).
We created a 68×68 connectivity matrix for each subject with each element depicting the number of fibers that passes through a pair of ROIs, normalized by the total number of fibers detected across the whole brain. We removed the “weak” connections through a k-core decomposition method, by computing the nodal degree – the number of connections (edges) linked to an ROI (node): ki = ∑j∈n cij, where cij is a connection status between components of the network (1 if connected, 0 otherwise). For a graph G = (n, e) with |n| nodes and |e| edges, a k-core is computed by assigning a subgraph, H = (B, e|B) where set B ⊆ n is a k-core of order k iff ∀ υ ∈ B: degreeH ≥ k, and H is the maximum subgraph satisfying this property [5]. We set the threshold to k=10 so that each node of the network has a nodal degree of at least 10 as computed on the original matrices. These weighted 10-core matrices, H(nk, ek), were used for all computations in this study.
2.3 Spectral graph theory
Spectral graph theory is a branch of mathematics that uses linear algebra and matrix theory to study the properties of graphs [4, 13]. The spectrum of a graph is the set of all eigenvalues computed from the Laplacian matrix of a graph (explained below). Structural networks are usually modeled as undirected, symmetric graphs, H(nk, ek), and we used these as the adjacency matrix, A(H)=wij, where wij is a weight representing the proportion of fibers in the brain linking a pair of nodes. Next, we computed the Laplacian matrix of graph G, L(H)=lij, where L(H)=D(H)-A(H). D(H) is the nxn (i.e., 68×68) diagonal degree adjacency matrix (i.e., diag(sum(H))). Then, the eigenvalues, λi, were computed from the Laplacian matrix, where 0=det(L-λI) and I is the nxn identity matrix. For each λi we computed the corresponding eigenvector, x, as (L-λI)x=0 [7]. The spectrum of each graph is a resorted 10-core matrix (same elements are kept) based on x corresponding to the second smallest eigenvalue, also known as the Fiedler value [7].
From the eigenvalues of L(H), we computed the total number of disconnected components, indicated by the number of zero eigenvalues, λi=0. A closely related measure – the energy of the graph – is defined as the sum of the absolute values of the eigenvalues, [2, 9]. The concept of graph energy originates from organic chemistry and allows the computation of the energy associated with π-electron orbitals (defined by eigenvectors) using the skeleton of hydrocarbons as the adjacency matrix [9]. When the energy is zero we have a completely disconnected graph; for each λi>0 there exist many edges (e) that make up the graph, H. Therefore, the more e, the more densely interconnected a graph is, allowing more abundant communication between nodes linked through shorter connections [2]. To quantify this, we computed the link density of H, as . Finally, a measure that is embedded in the computation of all aforementioned metrics is the strength of connections between communicating nodes: . All metrics were computed with tools adapted from the MIT Strategic Engineering website (http://strategic.mit.edu).
We used all four measures – the number of connected components, graph energy, link density and strength to describe the level of disconnection among components of the brain network in AD participants, and quantify the density and strength of connections among nodes of the brain, relative to healthy brain networks. To do this, we fitted a linear regression to each metric, covaried for age, sex, and APOE-4 status, and tested for network differences between AD and healthy controls (with AD coded as 1 and controls coded as 0). We will only report results for which the significant p-value threshold was below 0.05/4 (p<0.0125), where 4 is the number of tests we ran. In a similar analysis, we assessed the association between the three global brain metrics (number of connected components, graph energy and link density) and the APOE-4 risk factor in AD participants while covarying for age and sex. Using matrix spectral decomposition, we estimated the number of independent tests performed using a correlation matrix consisting of the three metrics across all AD subjects and adjusted the p-value significant threshold accordingly (http://gump.qimr.edu.au/general/daleN/matSpD/).
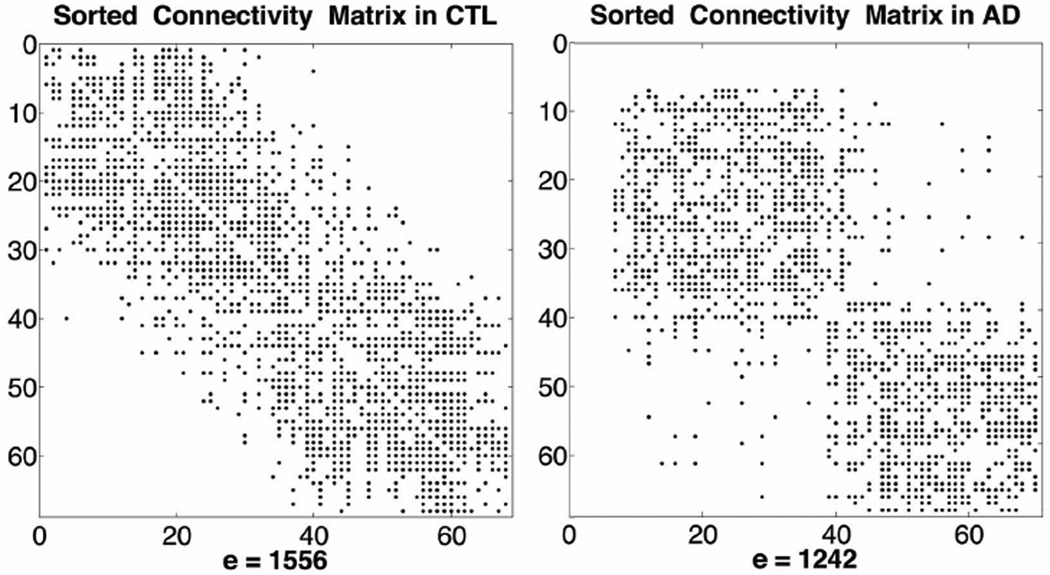
AD participants had a significantly higher number of disconnected components in their brain networks, relative to healthy controls (p-value=9.3×10−4). Most disconnected nodes were in the entorhinal, temporal and frontal poles bilaterally, sites that commonly show signs of AD pathology. We illustrate the disconnected components in Figure 1 by computing the spectrum of a graph as a sorted matrix, H, based on the eigenvector corresponding to the Fiedler value. The magnitude of the Fiedler value is proportional to the level of interconnectedness of a graph – i.e., how difficult it is to “tear a graph apart”. Figure 1 illustrates the sparsely interconnected network in one AD participant, qualitatively, compared to one healthy control.
Figure 1.
Spectrum of a graph obtained from the connectivity matrices sorted as a function of the eigenvectors and Fiedler value. Empty rows and columns indicate disconnected nodes in AD. The number of edges, e, is also listed. *CTL=healthy controls.
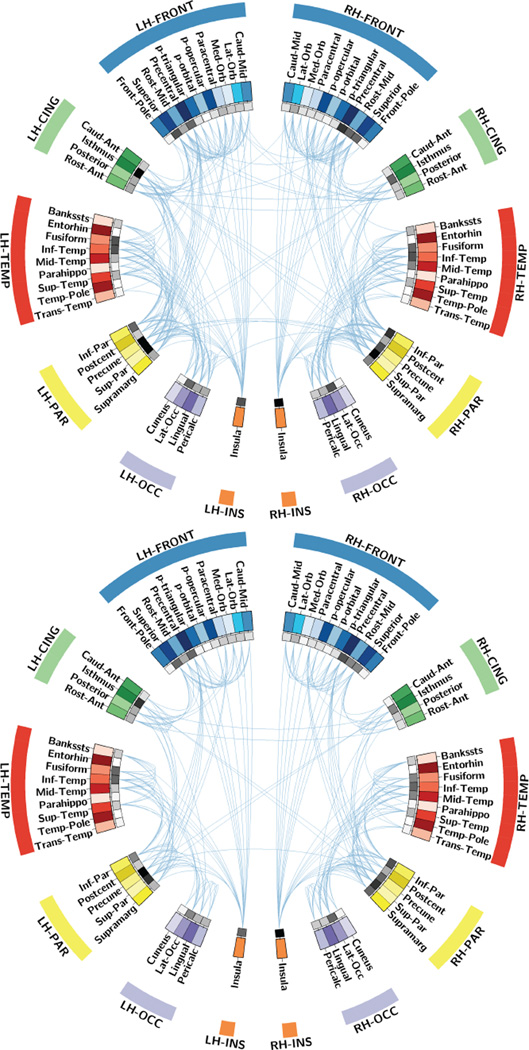
The energy of the brain networks was significantly lower in AD patients than in controls (p-value=3.7×10−4). The energy was computed across all components of the network, including those that are disconnected. As shown in organic chemistry, energy is a direct measure of the stability of a structure (i.e., for bonds between molecules) [9]. Here, the graph energy is linear, as is expected in the case of relatively small networks (i.e., 68×68 matrix) with low link densities [2]; the larger its magnitude, the more densely interconnected and stable the brain networks are. These findings are further supported by a significantly lower link density in AD participants, relative to healthy controls (p-value= 2.4×10−3). A decrease in links connecting regions of the brain may indicate reduced communication, eventually leading to disconnected nodes also shown in the connectograms [10] from Figure 2.
Figure 2.
Connectogram depicting connections between ROIs in the brain for controls (top) and AD (bottom). Inner ring (gray pallet) indicates the strength of connections passing through each ROI with darker squares indicating higher strength.
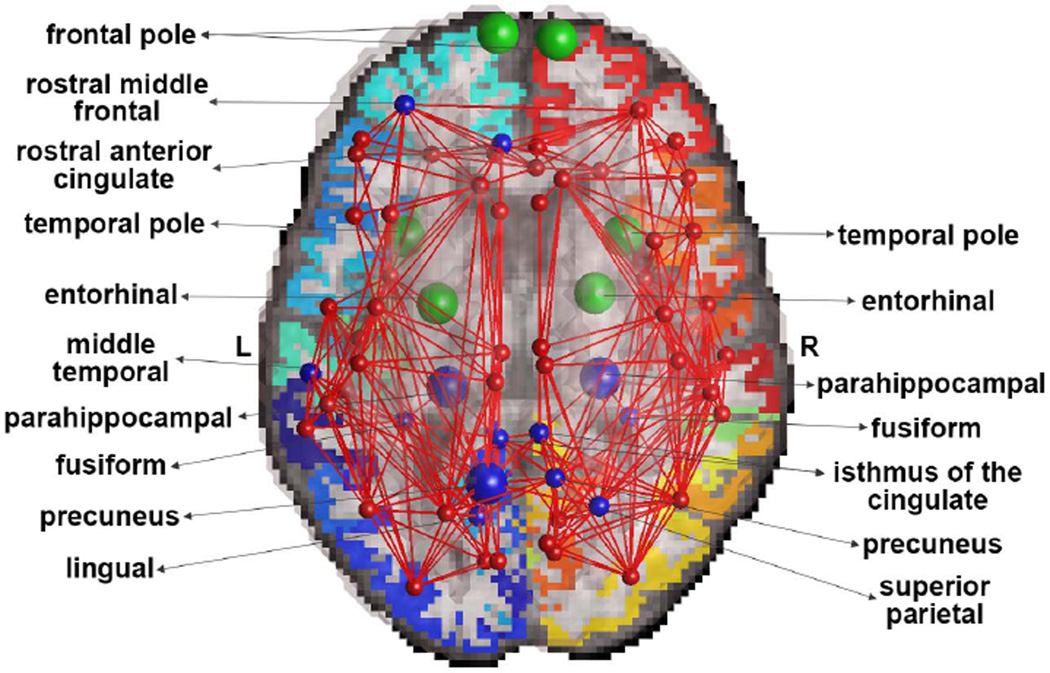
Finally, the strength of connections between network nodes was significantly lower in AD participants (FDR critical p-value=8.8×10−3), relative to controls, in 13 distinct brain regions illustrated in Figure 3. Most prominently affected regions were the left precuneus (p-value=5.0×10−5) and left and right parahippocampal regions (p-values<3.2×10−5). Although nodal strength was previously studied in AD participants [3, 11], here we computed it on thresholded matrices – unlike what was done before, to aid the interpretation of the spectral graph theory metrics that were not previously applied on AD networks in this context. We also used this measure to compute the average strength of connectivity in the frontal, temporal, parietal and occipital lobes in AD, in relation to healthy participants. We found that the lobes of the brain, in the order mentioned, were 13%, 26%, 18% and 9% less interconnected in AD.
Figure 3.
Average nodes and edges illustrating brain areas with significant alterations in AD, relative to controls. Green indicates disconnected nodes in AD, and blue indicates reduced strength in AD. Larger blue nodes denote more severe alterations.
3.2 APOE-4 associations with spectral metrics in AD
The energy of the brain networks in AD participants decreased with increasing number of copies of the APOE-4 risk gene (p-value=0.02). We also found a decrease in the related metric, link density (p-value=0.02) and we illustrate the sparsely interconnected connectome in AD participants in Figure 3. The significant p-value threshold was 0.035, as adjusted based on the number of independent variables. The association between the number of disconnected components and APOE-4 status did not pass this threshold (p-value=0.04), possibly due to the low statistical power (i.e., 42 AD participants).
We also tested for associations between brain metrics in healthy controls and their APOE-4 status but did not find significant results. Note that 70% of healthy participants had no copies of the APOE-4 risk gene and the rest had only one copy of the allele (Table 1). In contrast, there were proportionally more APOE-4 carriers among the AD participants; some had 2 copies of the allele, leading to more significant results indicative of brain network damage.
4. DISCUSSION
In this study we introduced the application of spectral graph theory to analyze the structure of brain networks in Alzheimer’s disease participants, and also related differences to the commonly studied APOE-4 genetic risk factor. To our knowledge, this may be the first study to assess the spectrum of a network and its energy (i.e., stability) as associated with APOE-4.
The notion of a “disconnection syndrome” in AD has been long hypothesized and proposes that brain dysfunction arises due to altered connectivity between regions of the brain [6]. Several studies report alterations in the brain connectome in AD patients [3–5, 11], suggesting disconnection. Here, we were able to directly quantify, using the spectrum of a graph (Fig. 1), that the entorhinal, frontal and temporal lobes bilaterally may be the most disconnected components of a network in AD, relative to controls (Figs. 2 and 3). Note that this does not indicate that, in reality, these regions have no white matter fibers passing through them; instead, the remaining communication between specific regions of the brain may be highly disrupted making these mathematical metrics useful descriptors of diseases that involve disconnection. The spectrum of a graph reveals the stability of its network components – an important property describing the topological layout of a graph that may also influence the brain’s physiology [8]. Our results suggest that carriers of more copies of the APOE-4 risk gene are more prone to instabilities in the structure of their brain networks. These subjects are also known to have more brain amyloid than those with no copies of the risk gene, which may lead to the observed alterations in brain connectivity.
Spectral graph theory metrics are most often applied to non-medical applications such as the online web [2]. Our study used these to assess disease characterization of network disruption in AD and APOE-4 carriers with multiple copies of the risk gene. We found that these metrics have a unique added value and are likely to enhance our understanding of dysfunction in the aging brain.
REFERENCES
- 1.Aganj I, Lenglet C, Sapiro G, Yacoub E, Ugurbil K, Harel N. Reconstruction of the Orientation Distribution Function in Single and Multiple Shell Q-Ball Imaging within Constant Solid Angle. Magn Reson Med. 2010;64(2):466–554. doi: 10.1002/mrm.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bounova G, de Weck OL. Overview of metrics and their correlation patterns for multiple-metric topology analysis on heterogeneous graph ensembles. Phys. Rev. E. 2012;85:016117. doi: 10.1103/PhysRevE.85.016117. [DOI] [PubMed] [Google Scholar]
- 3.Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KJ, Small GW, Bookheimer SY. Brain network local interconnectivity loss in aging APOE-4 allele carriers. PNAS. 2011;108(51):20760–20765. doi: 10.1073/pnas.1109038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daianu M, Jahanshad N, Nir TM, Leonardo CD, Jack CR, Jr, Weiner MW, Bernstein M, Thompson PM. Algebraic connectivity of brain networks shows patterns of segregation leading to reduced network robustness in Alzheimer’s disease. MICCAI’14 CDMRI Workshop. 2014 doi: 10.1007/978-3-319-11182-7_6. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daianu M, Jahanshad N, Nir TM, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Breakdown of Brain Connectivity between Normal Aging and Alzheimer’s Disease: A Structural k-core Network Analysis. Brain Connectivity. 2013;3(4):407–422. doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbeuck X, Van der Linden M, Collette F. Alzheimer's disease as a disconnection syndrome? Neuropsychol Rev. 2003;13(2):79–92. doi: 10.1023/a:1023832305702. [DOI] [PubMed] [Google Scholar]
- 7.Fiedler M. Algebraic connectivity of graphs. Czechoslovak Math. J. 1973;23:298–305. [Google Scholar]
- 8.Gray RT, Robinson PA. Stability constraints on large-scale structural brain networks. Front Comput Neurosci. 2013;12(7):31. doi: 10.3389/fncom.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutman I. The energy of a graph. Ber. Math. Statist. Sekt. Forsch-ungszentram Graz. 1978;103 [Google Scholar]
- 10.Irimia A, Chambers MC, Torgerson CM, Van Horn JD. Circular representation of human cortical networks for subject and population-level connectomic visualization. NeuroImage. 2012;60:1340–1351. doi: 10.1016/j.neuroimage.2012.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahanshad N, Nir TM, Toga AW, Jack CR, Jr, Bernstein MA, Weiner MW, Thompson PM. Seemingly unrelated regression empowers detection of network failure in dementia. Neurobiol Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.02.032. S0197-4580(14)00539-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohar B. The Laplacian spectrum of graphs. Graph Theory, Combinatorics, and Applications. 1991;2:1–28. [Google Scholar]
- 13.Norman B. Algebraic Graph Theory. 2nd ed. Cambridge: Cambridge University Press; 1993. [Google Scholar]