Abstract
PTPRT/RPTPρ is the most recently isolated member of the type IIB receptor-type protein tyrosine phosphatase family and its expression is restricted to the nervous system. PTPRT plays a critical role in regulation of synaptic formation and neuronal development. When PTPRT was overexpressed in hippocampal neurons, synaptic formation and dendritic arborization were induced. On the other hand, knockdown of PTPRT decreased neuronal transmission and attenuated neuronal development. PTPRT strengthened neuronal synapses by forming homophilic trans dimers with each other and heterophilic cis complexes with neuronal adhesion molecules. Fyn tyrosine kinase regulated PTPRT activity through phosphorylation of tyrosine 912 within the membrane-proximal catalytic domain of PTPRT. Phosphorylation induced homophilic cis dimerization of PTPRT and resulted in the inhibition of phosphatase activity. BCR-Rac1 GAP and Syntaxin-binding protein were found as new endogenous substrates of PTPRT in rat brain. PTPRT induced polymerization of actin cytoskeleton that determined the morphologies of dendrites and spines by inhibiting BCR-Rac1 GAP activity. Additionally, PTPRT appeared to regulate neurotransmitter release through reinforcement of interactions between Syntaxin-binding protein and Syntaxin, a SNARE protein. In conclusion, PTPRT regulates synaptic function and neuronal development through interactions with neuronal adhesion molecules and the dephosphorylation of synaptic molecules. [BMB Reports 2015; 48(5): 249-255]
Keywords: PTPRT, Receptor-type protein tyrosine phosphatase, Neuronal synapse formation, Dendritic arborization, Neuronal development
INTRODUCTION
Tyrosine phosphorylation is a fundamental mechanism for numerous important aspects of eukaryotic physiology, human health and diseases (1-4). Although tyrosine phosphorylation is regulated by the equal and balanced action of protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs), the important roles of PTPs had not been recognized until recently. It is known that the human genome has a set of 107 genes that encode members of PTP families whereas is also a set of 90 genes encoding PTK families. PTPs are considered to have comparable substrate specificities and non-redundancies in cellular functions to PTKs.
Based on the amino acid sequences of their catalytic domains, the PTPs can be grouped into four separate families, each with a range of substrate specificities: the cysteine-based PTPs of classes I, II, and III, and aspartate-based PTPs. There are 38 strictly tyrosine specific ‘classical’ PTPs among the 99 class I cysteine-based family of PTPs. All receptor-type PTPs (RPTPs) belong to the classical PTPs and they are represented in the human genome by 21 genes (5). They usually possess two intracellular phosphatase domains, and their extracellular domains are highly variable. RPTPs can be classified into several subtypes, based on the structure of their extracellular segments: types I, IIA, IIB, III, IV, and V (6). The structure of RPTPs suggests that they function as an interface between the extracellular environment of a cell and its intracellular signaling pathways (7-9). The type II class of RPTPs can be defined as a combination of immunoglobulin-like (Ig) domains and fibronectin type III (FN-III) repeats in the ectodomain. There is a potential proteolytic cleavage site within the membrane-proximal FN-III repeat of type II RPTPs, and extracellular N-terminal segment and an intracellular membrane bound C-terminal segment could be generated (10-12). The extracellular segment is generally thought to remain non-covalently associated with the membrane-bound intracellular segment. These transmembrane molecules are known to be connected with signal transduction, cell adhesion, and neurite outgrowth (6, 13-15).
PTPRT LOCALIZED TO NEURONAL SYNAPSES
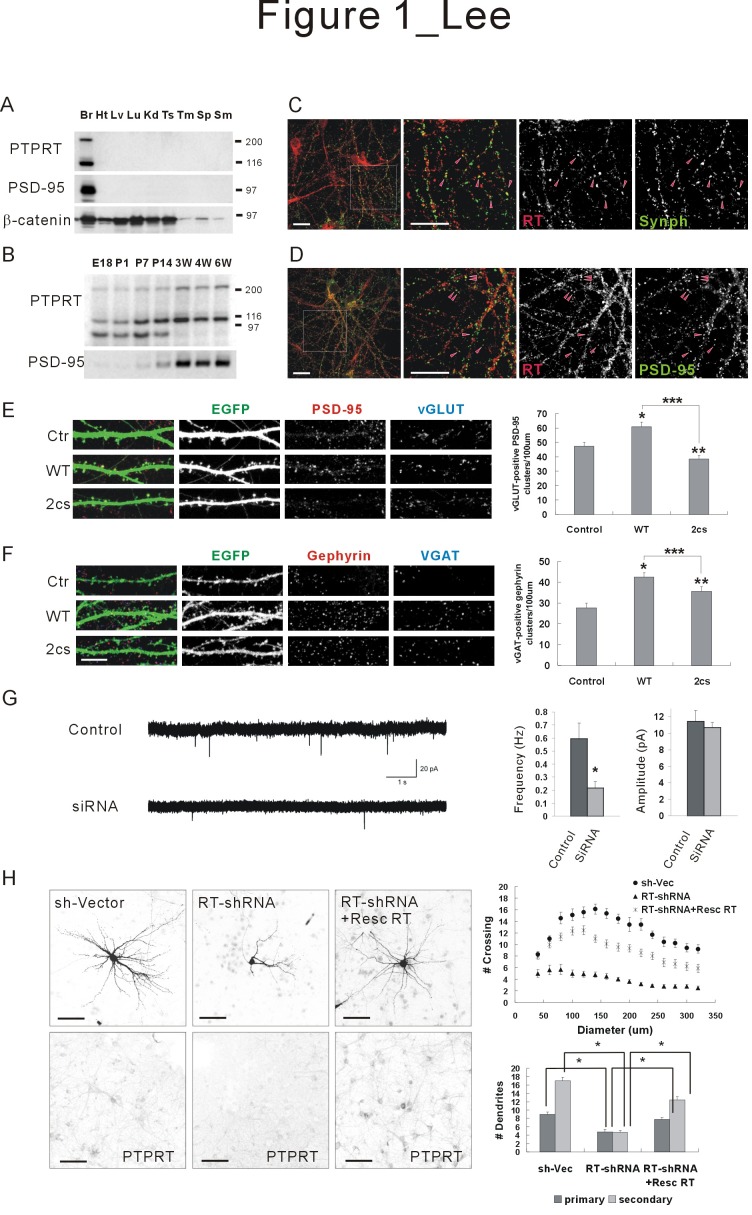
PTPRT (protein tyrosine phosphatase receptor T; receptor protein tyrosine phosphatase rho, RPTPρ) is a member of the type IIB RPTP family. Among the type II class, type IIB RPTPs are characterized by the presence of a N-terminal meprin/A5/PTPμ (MAM) domain. To date, four type IIB phosphatases have been reported: PTPμ, PTPκ, PCP-2, and PTPRT. PTPRT is the most recently isolated member of the IIB family, and the role of PTPRT had not been well understood (16, 17). The expression of PTPRT has been known to be largely restricted to the central nervous system (CNS) through Northern blot and in situ hybridization studies (18). When a Western blot of rat organs was carried out with a monoclonal antibody specific to the PTPRT catalytic domain, PTPRT proteins were expressed only in the brain and expression of PTPRT was shown to be developmentally regulated (12) (Fig. 1A, B). A 90-kDa cleavage fragment of PTPRT was evident in the early development stages of the rat brain, but its expression became undetectable from 3 weeks. On the other hand, the expression level of the full-length (210 kDa) and another fragmented (110 kDa) proteins, assumed to be a membrane-bound intracellular segment, increased gradually up to 3 weeks of age.
Fig. 1. PTPRT regulates synaptic formation and dendritic arborization in hippocampal neurons. (A) Tissue distribution patterns of PTPRT revealed by Western blot analysis. Monoclonal antibodies specific for PTPRT were used. Br, brain; Ht, heart; Lv, liver; Lu, lung; Kd, kidney; Ts, testis; Tm, thymus; Sp, spleen; Sm, skeletal muscle. PSD-95 and β-catenin were used as controls. (B) Developmental pattern of PTPRT in the rat brain. E, embryonic; P, postnatal days. PSD-95 was used as a control. (C, D) PTPRT co-localized to neuronal synapses stained with synaptophysin and PSD-95. Scale bar, 20 μm. (E) The number of excitatory synapses (vGLUT-positive PSD-95 clusters). Mean ± SEM. n = 30 dendrites for control, 31 for WT-RT (wild-type), and 30 for 2cs-RT (inactive). Scale bar, 10 μm. (F) The number of inhibitory synapses (vGAT-positive gephyrin clusters). Mean ± SEM. n = 27 dendrites for control, 28 for WT-RT, and 28 for 2cs-RT. Scale bar, 10 μm. (G) PTPRT knock-down of PTPRT in cultured neurons decreased the frequency, but not amplitude, of mEPSCs (n = 12 for control and n = 10 for shRNA). (H) Attenuated dendritic arborization upon knockdown of PTPRT. When rat hippocampal neurons were transfected with PTPRT-shRNA, the numbers of dendrites and arborization were decreased significantly and rescued by the addition of Resc PTPRT in this context. Mean ± SEM of data from 9 control neurons, 10 PTPRTshRNA neurons, and 9 PTPRT-shRNA + Resc PTPRT neurons are shown. Scale bar, 100 μm.
In cultured hippocampal neurons, PTPRT was localized to both excitatory and inhibitory synapses (12) (Fig. 1C, D). Overexpression of PTPRT increased the number of dendritic spines, and excitatory and inhibitory synapses (Fig. 1E, F). In contrast, knockdown of PTPRT inhibited synaptic formation, decreased synaptic transmission, and attenuated dendritic arborization of hippocampal neurons (12, 19) (Fig. 1G, H). From these results, PTPRT newly appeared as a potentially important player in synaptic function and neuronal development.
REGULATION OF SYNAPTIC FORMATION BY PTPRT
There are several suggestions about the ways in which PTPRT regulates synaptic formation. First, PTPRT induces synaptic formation by acting as a cell adhesion molecule (Fig. 2A). Although many RPTPs are orphan receptors and their functional ligands remain largely unknown, they have been shown to be involved in the regulation of neuronal adhesion, axon growth and guidance (7). Most of the type IIB RPTP forms a homophilic trans (antiparallel) dimer (20). The crystal structure of the PTPRM (RPTPμ) ectodomain has been described to form a homophilic trans (antiparallel) dimer with an extended and rigid architecture (21). PTPRT also has been shown to form a homophilic trans dimer through its extracellular ectodomain when its recombinant ectodomain was tested as a pre-bound form to protein A-agarose beads (12). In addition, the possibility of forming a homophilic trans dimer was checked in hippocampal neurons; dendritic spines and synaptic puncta were decreased when hippocampal neurons were incubated with PTPRT ectodomain. Recombinant PTPRT ectodomain decreased synaptic formation by inhibiting the interactions between ectodomains of PTPRT protruded from pre- and post-synapses.
Fig. 2. cis-homophilic interactions of PTPRT are reinforced through tyrosine phosphorylation by Fyn tyrosine kinase. (A, B) PTPRT appears to induce synaptic formation through trans-homophilic interactions with each other, and cis-heterophilic interaction with neuroligin and neurexin. (C) Synaptic molecules could be recruited to PTPRT by neuroligin and neurexin. Activated PTPRT dephosphorylates many synaptic substrates and regulates synaptic functions. (D) When Fyn tyrosine kinase phophorylates tyrosine 912 of PTPRT, cis-homophilic interaction is induced, PTPRT activity is inhibited, and synaptic formation is attenuated. (F) In the crystal structure of PTPRK/RPTPκ (pdb code: 2c7s), tyrosine 892 is equivalent to tyrosine 912 fin PTPRT. Left, overall view of PTPRK D1 catalytic domain. The wedge moiety is shown as cyan, while the rest of the molecule is shown as yellow. The catalytic cysteine and the tyrosine residue are colored green. Right, close-up view of PTPRK near tyrosine 892. The residues involving the close contacts with tyrosine 892 are shown as sticks.
Secondly, PTPRT induces synaptic formation through interactions with neuroligin and neurexin (12) (Fig. 2B). Neuroligin and neurexin have been known to play important roles for synaptic formation through their rigorous interactions, and each of them recruits synaptic components in pre- and post-synapses, respectively (22, 23). Neuroligin was found as an interaction partner of PTPRT from rat synaptosomes using a novel method of sequential immunoprecipitation with an anti-PTPRT monoclonal antibody and then identification with tandem mass spectrometry. In an experiment using recombinant proteins, PTPRT appeared to cis-interact with neuroligin and neurexin (12). PTPRT functionally interacts with neuroligin and neurexin in synaptic formation; overexpression of PTPRT increased the puncta of neuroligin, and knockdown of PTPRT inhibited the induction of synaptic formation by neuroligin in hippocampal neurons.
In cultured hippocampal neurons, PTPRT knockdown decreased the frequency of mEPSCs; this result indicates that PTPRT is involved in pre-synaptic functions like neurotransmitter release (12). Through sequential immunoprecipitation and tandem mass spectrometry, Syntaxin-binding protein (Sec/Munc18-like protein) also was found as a candidate interaction partner of PTPRT (24). Syntaxin-binding protein is not a member of the SNARE proteins (soluble N-ethylmaleimidesensitive factor attachment protein receptor protein), but it is linked to synaptic vesicle fusion by virtue of its tight binding to Syntaxin (25, 26). The tyrosine 145 residue, located around the linker between Domain 1 and 2 of Syntaxin-binding protein, was shown to be dephosphorylated by PTPRT, and the interaction of Syntaxin-binding protein with Syntaxin was enhanced by dephosphorylation of tyrosine 145 (24). Previously, PTPRT was suggested to localize to synaptic vesicles when sub-cellular fractionation was carried out with rat brains (12). Thus, PTPRT appears to regulate synaptic vesicle fusion machinery and to control neurotransmitter release in presynaptic vesicles through the dephosphorylation of Syntaxin-binding protein. As a result, PTPRT induces neuronal synaptic formation by acting as a neuronal adhesion molecule, and by controlling neurotransmitter release.
REGULATION OF DENDRITIC ARBORIZATION BY PTPRT
PTPRT knockdown elicited the attenuation of dendritic arborization, as well as synaptic formation in hippocampal neurons (12, 19). Regulation of dendritic arborization is very important for neuronal development and effective neural connections because the dendritic branching pattern determines the numbers and type of inputs that a neuron can receive (27). Actin cytoskeletal formation is fundamental component for morphogenetic processes in neurons, and Rho GTPases mediate morphological changes during neuronal development by serving as critical regulators of actin cytoskeletal formation (28-30). Among Rho GTPases, Rac1 is one of the most extensively studied members in the field of dendritic arborization and spine morphology (31).
Breakpoint cluster region (BCR) protein is a Rac1 GTPase-activating protein that is expressed abundantly in the nervous system (32-34). BCR was found to be a new neuronal substrate of PTPRT, and PTPRT is known to regulate dendritic arborization through BCR (19). The activity of the BCR GTPase-activating domain was modulated by means of conversions between the intra- and inter-molecular interactions, which are finely regulated through the dephosphorylation of a specific tyrosine residue by PTPRT. When tyrosine 177, located on the BCR N-terminus, was dephosphorylated by PTPRT, the interaction between the BCR N- and C-termini was enhanced. As a result, multimerization of BCR molecules was attenuated and the GTPase activity of BCR was inhibited. Paxiliin has been found to be dephosphorylated by the intracellular part of PTPRT expressed in HEK cells, and identified as a substrate of PTPRT (35). Paxiliin is a multi-domain adaptor found at the interface between the plasma membrane and the actin cytoskeleton, and PTPRT appears to control the linkage of the actin cytoskeleton to focal adhesions (36). In conclusion, PTPRT dephosphorylated BCR, decreased GTPase activity, increased polymerization of the actin cytoskeleton, and enhanced dendritic arborization as well as spine formation in hippocampal neurons.
REGULATION MECHANISM OF PTPRT ACTIVATION
What mechanism could control the activity of PTPRT catalytic domains? Fyn tyrosine kinase was found to interact with PTPRT and it phosphorylated the tyrosine 912 residue of PTPRT (12) (Fig. 2C). Fyn tyrosine kinase inhibited PTPRT activation and attenuated synaptic formation when co-expressed with PTPRT in hippocampal neurons (Fig. 2D). Tyrosine 912 is located in the interface between the wedge and the body of the membrane-proximal catalytic domain of PTPRT. Phosphorylation of tyrosine 912 appeared to induce the detachment of the wedge from the body of the catalytic domain and allowed the flexible wedge to enter the catalytic pocket of the other monomer deeply (37, 38) (Fig. 2E). In an experiment using PTPRT tagged differentially, multimerization of PTPRT was enhanced when Fyn kinase was coexpressed. Phosphorylation of PTPRT strengthened the dimerization of the cis-form, and eventually inhibited the catalytic activity of PTPRT. A PTPRT mutant mimicking phosphorylation of tyrosine 912 lost its phosphatase activity in an in vitro PTP assay carried out with recombinant PTPRT catalytic domain protein. Inhibition of PTPRT activation elicited dysfunction of synaptic molecules and disorder in neuronal development. Dimerization of PTPRT also appeared to prevent heterophilic interactions between PTPRT and neuroligin, and then attenuated synaptic formation (12).
MUTATIONS IN THE PTPRT GENE IN NEURONAL DISEASES
The human PTPRT gene is located on chromosome 20q12-13.1, while the mouse gene is located in a syntenic region of chromosome 2 (5, 18). Although the expression of PTPRT has been shown to be restricted to the murine brain and spinal cord, many missense and nonsense mutations were identified in PTPRT gene from human colorectal, lung, breast cancers (16, 18, 39). Among them, five missense mutations were found to reduce phosphatase activity, and expression of wild-type, but not a mutant, PTPRT in human cancer cells inhibited cell growth. In fact, colon tumors were easily induced in PTPRT knockout mice by carcinogens, and thus PTPRT could be a tumor suppressor ordinarily (36).
Recently, exome sequencing in small families with intellectually disabled siblings identified potentially pathogenic variants in the PTPRT gene (40). Although PTPRT mutation has any chance of becoming polymorphism, the combination of a heterozygous missense variant, p.Thr1365Met, and a heterozygous intronic deletion of 150 kb was reported to elicit severe intellectual disability. From this result, PTPRT was suggested to regulate synaptic functions and neuronal development of human brains. Previously, it was reported that chromosome 20q11.21-q13.12, containing the PTPRT gene, was found as a significant region for autism spectrum disorder when a high-density, single-nucleotide polymorphism (SNP), genome-wide scan was performed on a six-generation pedigree (41). Knock-in mice carrying the PTPRT D1046A trapping mutation have been found to have higher social approach scores than wild-type animals (42). Mutation of aspartate to alanine (D1046A) in the PTPRT catalytic domain was known to inactivate phosphatase activity but to retain substrate binding. The behavioral changes suggest that the function of PTPRT phosphatase is indispensable in modulating neural pathways involved in mouse social behaviors, possibly relevant to the symptoms in human autism spectrum disorder patients.
CONCLUSIONS
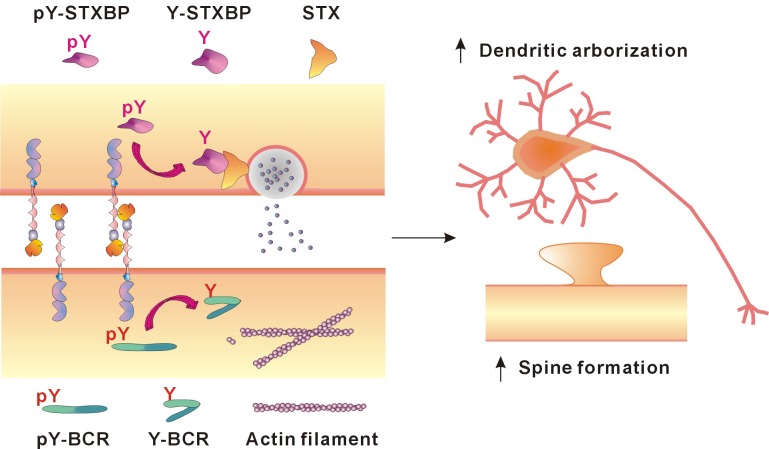
PTPRT performs its role in the extracellular environment through its ectodomain and concurrently carries out intracellular functions with its phosphatase domain. The ectodomain of PTPRT resembles a cell-adhesion molecule and PTPRT forms a homophilic dimer. PTPRT is activated when it forms a homophilic trans (antiparallel) dimer, and is inactivated when it forms a homophilic cis (parallel) dimer. The PTPRT trans dimer dephosphorylates synaptic molecules, and interacts with neuronal adhesion molecules. PTPRT phosphorylation of tyrosine 912 induces cis dimerization of PTPRT by letting the flexible wedge obstruct the catalytic pocket of the other monomer. PTPRT could control Rac1 GTPase activity of BCR, enhance the polymerization of the actin cytoskeleton, and regulate formation of dendritic spines and arborization of hippocampal neurons (Fig. 3). PTPRT also controls the interaction of Syntaxin binding protein with SNARE proteins and the release of neurotransmitter.
Fig. 3. Dendritic arborization and spine formation regulated by PTPRT activation. In pre-synapses the interaction between Syntaxin-binding protein and SNARE proteins is strengthened by PTPRT. PTPRT dephosphorylates tyrosine 145 located around the linker between Domains 1 and 2 of Syntaxin-binding protein, and the structure of Syntaxin-binding protein is changed for enhancement of interaction between Syntaxin-binding protein and Syntaxin that elicits neuronal transmission through increased neurotransmitter release. Actin cytoskeletal formation is a fundamental component for morphogenetic processes in neurons, and Rac1 GTPases mediate morphological changes during neuronal development. The activity of the BCR Rac1 GTPase-activating domain is modulated through the dephosphorylation of a specific tyrosine residue by PTPRT. When tyrosine 177 located on BCR N-terminus is dephosphorylated by PTPRT, the interaction between the BCR N- and C-termini is enhanced. As a result, GTPase activity of BCR is inhibited and polymerization of the actin cytoskeleton is increased. pY, phosphorylated tyrosine residue; Y, dephosphorylated tyrosine residue.
Recently, many mutations of the PTPRT gene have been found in human patients with psychiatric disorders, like autism and intellectual disability, and PTPRT becomes noticed for its role in the central nervous system. Because PTPRT is localized in neuronal synapses and regulates synaptic formation, the deficit of PTPRT activity appears to elicit delayed brain development and neuronal diseases. Thus, PTPRT and its signal pathway in the brain could be possible targets for brain disease treatment.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation (NRF) (2012R1A2A2A02014520 to JRL), the Postgenomic Research Foundation of Korea (NRF-2014M3C9A2064619 to JRL), and a grant from KRIBB Research Initiative Program (KGM1141413 & KGM4701511 to JRL).
References
- 1.Alonso A, Sasin J, Bottini N, et al. Protein tyrosine phosphatases in the human genome. Cell. (2004);117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Mustelin T, Vang T, Bottini N. Protein tyrosine phosphatases and the immune response. Nat Rev Immunol. (2005);5:43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- 3.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. (2006);7:833–346. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 4.Lemmon MA, Schlessinger J. Cell signaling by receptor-tyrosine kinases. Cell. (2010);141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen JN, Jansen PG, Echwald SM, et al. A genomic perspective on protein tyrosine phosphatases: gene structure, pseudogenes, and genetic disease linkage. FASEB J. (2004);18:8–13. doi: 10.1096/fj.02-1212rev. [DOI] [PubMed] [Google Scholar]
- 6.Besco JA, Frostholm A, Popesco MC, et al. Genomic organization and alternative splicing of the human and mouse RPTPρ genes. BMC Genomics. (2001);2:1. doi: 10.1186/1471-2164-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul S, Lombroso PJ. Receptor and non-receptor protein tyrosine phosphatases in the nervous system. Cell Mol Life Sci. (2003);60:2465–2482. doi: 10.1007/s00018-003-3123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunah AW, Hueske E, Wyszynski M, et al. LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat Neurosci. (2005);8:458–467. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- 9.Woo J, Kwon SK, Choi S, et al. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci. (2009);12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 10.Streuli M, Krueger NX, Ariniello PD, et al. Expression of the receptor-linked protein tyrosine phosphatase LAR: proteolytic cleavage and shedding of the CAM-like extracellular region. EMBO J. (1990);9:2399–2407. doi: 10.1002/j.1460-2075.1992.tb05128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders L, Mertins P, Lammich S, et al. Furin-, ADAM 10-, and γ-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of β-catenin’s transcriptional activity. Mol Cell Biol. (2006);26:3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim SH, Kwon SK, Lee MK, et al. Synapse formation regulated by protein tyrosine phosphatase receptor T through interaction with cell adhesion molecules and Fyn. EMBO J. (2009);28:3564–3578. doi: 10.1038/emboj.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burden-Gulley SM, Brady-Kalnay SM. PTPμ regulates N-cadherin-dependent neurite outgrowth. J Cell Biol. (1999);144:1323–1336. doi: 10.1083/jcb.144.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burden-Gulley SM, Ensslen SE, Brady-Kalnay SM. Protein tyrosine phophatase-μ differentially regulates neurite outgrowth of nasal and temporal neurons in the retina. J Neurosci. (2002);22:3615–3627. doi: 10.1523/JNEUROSCI.22-09-03615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson KG, Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol Rev. (2003);83:1–24. doi: 10.1152/physrev.00016.2002. [DOI] [PubMed] [Google Scholar]
- 16.McAndrew PE, Frostholm A, White R, et al. Identification and characterization of RPTPρ, a novel μ/κ -like receptor protein tyrosine phosphatase whose expression is restricted to the central nervous system. Mol Brain Res. (1998);56:9–21. doi: 10.1016/S0169-328X(98)00014-X. [DOI] [PubMed] [Google Scholar]
- 17.McAndrew PE, Frostholm A, Evans JE, et al. Novel receptor protein tyrosine phosphatase (RPTPρ) and acidic fibroblast growth factor (FGF-1) transcripts delineate a rostrocaudal boundary in the granule cell of the murine cerebellar cortex. J Comp Neurol. (1998);391:444–455. doi: 10.1002/(SICI)1096-9861(19980222)391:4<444::AID-CNE3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Besco J, Popesco MC, Davuluri RV, et al. Genomic structure and alternative splicing of murine R2B receptor protein tyrosine phosphatases (PTPκ, μ, ρ and PCP-2). BMC Genomics. (2004);5:14. doi: 10.1186/1471-2164-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park AR, Oh D, Lim SH, et al. Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a substrate of PTPRT. J Cell Sci. (2012);125:4518–4531. doi: 10.1242/jcs.105502. [DOI] [PubMed] [Google Scholar]
- 20.Zondag GCM, Koningstein GM, Jiang YP, et al. Homophilic interactions mediated by receptor tyrosine phosphatase μ and κ. J Biol Chem. (1995);270:14247–14250. doi: 10.1074/jbc.270.24.14247. [DOI] [PubMed] [Google Scholar]
- 21.Aricescu AR, Siebold C, Choudhuri K, et al. Structure of a Tyrosine Phosphatase Adhesive Interaction Reveals a Spacer-Clamp Mechanism. Science. (2007);317:1217–1220. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- 22.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. (2005);307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 23.Boucard AA, Chubykin AA, Comoletti D, et al. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α-and β-neurexins. Neuron. (2005);48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 24.Lim SH, Moon J, Lee M, et al. PTPRT regulates the interaction of Syntaxin-binding protein 1 with Syntaxin 1 through dephosphorylation of specific tyrosine residue. Biochem Biophys Res Comm. (2013);439:40–46. doi: 10.1016/j.bbrc.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 25.Rizo J, Sudhof TC. SNAREs and Munc 18 in synaptic vesicle fusion. Nat Rev Neurosci. (2002);3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 26.Deak F, Xu Y, Chang WP, et al. Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming. J Cell Biol. (2009);184:751–764. doi: 10.1083/jcb.200812026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Chen G, Zhou B, et al. Actin filament assembly by myristoylated, alanine-rich C kinase substratephosphatidylinositil-4,5-diphosphate signaling is critical for dendrite branching. Mol Biol Cell. (2008);19:4804–4813. doi: 10.1091/mbc.E08-03-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. (2002);18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Firestein BL. RhoA regulates dendritic branching in hippocampal neurons by decreasing cypin protein levels. J Neurosci. (2007);27:8378–8386. doi: 10.1523/JNEUROSCI.0872-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama AY, Harms MB, Luo L. Small GTPase Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. (2000);20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. (2000);1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 32.Chuang TH, Xu X, Kaartinen V, et al. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein gamily. Proc Natl Acad Sci U S A. (1995);92:10282–10286. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kweon SM, Cho YJ, Minoo P, et al. Activity of the Bcr GTPase-activating domain is regulated through direct protein/protein interaction with the Rho guanine nucleotide dissociation inhibitor. J Biol Chem. (2008);283:3023–3030. doi: 10.1074/jbc.M705513200. [DOI] [PubMed] [Google Scholar]
- 34.Oh D, Han S, Seo J, et al. Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J Neurosci. (2010);30:14134–14144. doi: 10.1523/JNEUROSCI.1711-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner CE. Paxillin and focal adhesion signaling. Nat Cell Biol. (2000);2:E231–E236. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Zhang X, Gudac K, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci. (2010);107:2592–2597. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilwes AM, Hertog J, Hunter T, et al. Novel Structural basis for inhibition of receptor protein-tyrosine phosphatase-α by dimerization. Nature. (1996);382:555–559. doi: 10.1038/382555a0. [DOI] [PubMed] [Google Scholar]
- 38.Xie Y, Massa SM, Ensslen-Craig SE, et al. Wedge Domain Peptides: a novel approach for inhibition of PTP function and augmentation of protein-tyrosine kinase function. J Biol Chem. (2006);281:16482–16492. doi: 10.1074/jbc.M603131200. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. (2004);304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 40.Schuurs-Hoeijmakers JH, Vulto-van Silfhout AT, Vissers LEL, et al. Identification of pathogenic gene variants in small families with intellectually disabled siblings by exome sequencing. J Med Genet. (2013);50:802–811. doi: 10.1136/jmedgenet-2013-101644. [DOI] [PubMed] [Google Scholar]
- 41.Allen-Brady K, Miller J, Matsunami N, et al. A highdensity SNP genome-wide linkage scan in a large autism extended pedigree. Mol Psych. (2009);14:590–600. doi: 10.1038/mp.2008.14. [DOI] [PubMed] [Google Scholar]
- 42.Rajamani KT, O’Neill B, Han DD, et al. Inactivation of the catalytic phosphatase domain of PTPRT/RPTPρ increases social interaction in mice. Autism Res. (2014) doi: 10.1002/aur.1390. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]