Abstract
Over the last decade, comprehensive genome-wide sequencing studies have enabled us to find out unexpected genetic alterations of metabolism in cancer. An example is the identification of arginine missense mutations of isocitrate dehydrogenases-1 and -2 (IDH1/2) in glioma, acute myeloid leukemia (AML), chondrosarcomas, and cholangiocarcinoma. These alterations are closely associated with the production of a new stereospecific metabolite, (R)-2-hydroxyglutarate (R-2HG). A large number of follow-up studies have been performed to address the molecular mechanisms of IDH1/2 mutations underlying how these events contribute to malignant transformation. In the meanwhile, the development of selective mutant IDH1/2 chemical inhibitors is being actively pursued in the scientific community and pharmaceutical industry. The present review article briefly discusses the important findings that highlight the molecular mechanisms of IDH1/2 mutations in cancer and provides a current status for development of selective mutant IDH1/2 chemical inhibitors. [BMB Reports 2015; 48(5): 266-270]
Keywords: Cancer Metabolism, Isocitrate dehydrogenases (IDHs), Isocitrate (ICT), α-ketoglutarate (α-KG), (R)-2-hydroxyglutarate (R-2HG)
ELUCIDATION OF ARGININE MISSENSE MUTATIONS OF ISOCITRATE DEHYDROGENASE ISOFORMS IN CANCER
The belief that cellular metabolism plays an important role in cancer has been held since the observation by Otto Warburg that cancer cells prefer to produce ATP by metabolizing glucose into lactate via glycolysis instead of utilizing the efficient oxidative phosphorylation even in the presence of oxygen, a phenomenon commonly referred to as “the Warburg effect” or “the aerobic glycolysis” (1). Although it is still debated whether this metabolic switch is merely an adaptive response or a physiological event that facilitates malignant transformation, a large number of studies have demonstrated that the expression and/or catalytic activity of some glycolytic or tricarboxylic acid (TCA) cycle enzymes are significantly altered in cancer (2). For example, it is known that the expression of a pyruvate kinase isoform, pyruvate kinase M2 (PKM2), is highly elevated in cancer and it contributes to an increase in the rate of glycolysis (3). Loss of function mutations in succinate dehydrogenases (SDHs) and fumarate hydratase (FHs) that catalyzes the conversion of succinate into fumarate (for SDHs) and of fumarate into malate (for FHs) in the tricarboxylic acid (TCA) cycle, respectively were identified in particular types of tumors, such as renal carcinoma and paragangliomas (4). Most notably, a great deal of interests in cancer metabolism recently arose due to an unexpected finding of genome-wide sequencing studies: the existence of arginine missense mutations of isocitrate dehydrogenases (IDHs) in selected types of cancers (5).
Glioblastoma multiforme (GBM) is a deadly brain tumor that is refractory to chemotherapy and radiotherapy. In order to identify possible therapeutic target(s) for GBM, Parsons et al. undertook a seminal genome-wide sequencing study of 22 adult primary and secondary GBM tumors, and found a recurrent missense mutation of IDH1 in 5 of 6 secondary GBMs; however, none of the 16 primary GBMs (6). Follow-up genome-wide sequencing studies have confirmed the following findings: IDH1 missense mutation occurs in adult low-grade (grade II and III) glioma (>70%) and secondary GBM (>80%) at a high frequency (7-9). More importantly, mutations were confined to a single residue of IDH1 at arginine-132, which is mostly substituted into histidine (IDH1-R132H). When these tumors lacked IDH1 mutations, some of them harbored alternative missense mutations in functionally analogous arginine residues of IDH2 at arginine-140 or arginine-172 with the alterations predominantly substituted into glutamine or lysine (IDH2-R140Q or IDH2-R172K): both IDH1-R132 and IDH2-R140 or IDH2-R172 are located in the catalytic triad of enzymes and serve as critical residues that form a hydrogen bond with α- and β-carboxylic groups of their substrate, e.g. isocitrate (ICT) (10). In addition, the genome-wide sequencing studies revealed additional interesting features about IDH1/2 mutations: (1) IDH1/2 mutations occur in a mutually-exclusive manner, (2) these mutations are somatic mutations, and (3) they precede most critical genetic alterations (for example, TP53 mutations) that might be critical for tumor development (11, 12). The identical IDH1 or IDH2 mutations were subsequently identified in other types of cancers as well, including acute myeloid leukemia (AML), chondrosarcomas, and cholangiocarcinoma (13). However, the occurrence of IDH1/2 mutations seems to be selective, depending on the age of patients, tumor type and tissue origin: pediatric glioma and adult primary GBM possess few IDH1 or IDH2 mutations (<10%) and no IDH1/2 mutations have been identified in brain tumors with non-glial origins. Interestingly, Schwartzentruber et al. have identified somatic mutations of the gene that encodes a replication-independent histone H3 variant (histone H3.3) in pediatric GBMs with predominant substitutions occurring at two positions in the N-terminal histone H3.3 tail (H3.3-K27M or H3.3-G34R/G34V) (14).
IDH1/2 MUTATIONS ARE ASSOCIATED WITH THE PRODUCTION OF A STEREOSELECTIVE METABOLITE, (R)-2-HYDROXYGLUTARATE
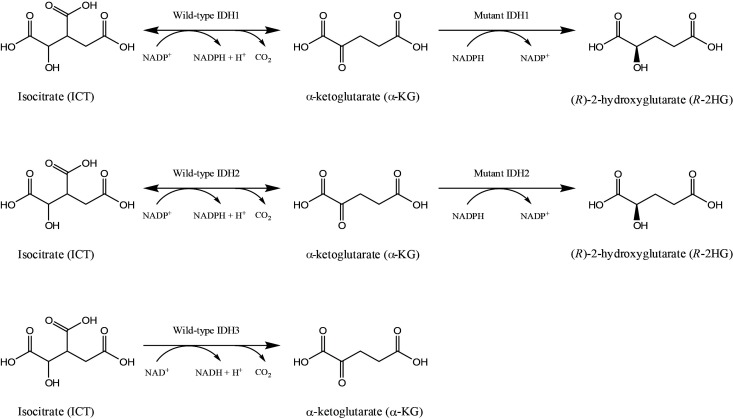
Eukaryotic cells express three different isoforms of IDHs: IDH1, IDH2 and IDH3. IDH1 and IDH2 form homodimers and catalyze the NADP+-dependent oxidative decarboxylation of ICT into α-ketoglutarate (α-KG) in the cytosol (for IDH1) and mitochondria (for IDH2), respectively (Fig. 1). On the other hand, IDH3 is known to form a heteroctamer (α2β2γ4), derived by three IDH3 gene products, e.g. IDH3A (α), IDH3B (β), and IDH3G (γ), and catalyzes NAD+-dependent decarboxylation of ICT into α-KG in the mitochondria (Fig. 1). In contrast to IDH1/2 mutations, IDH3 mutations were not identified in tumors. Because SDHs and FHs exist in the tricarboxylic acid (TCA) cycle and are bona fide tumor suppressors that generally exhibit a loss of function mutation (15), IDH1/2 mutations were initially thought to exhibit a dominant-negative activity. In line with this idea, Zhao et al. have demonstrated that introducing a missense mutation in recombinant IDH1 protein (IDH1-R132H) resulted in a reduced affinity for ICT and lowered production of α-KG in vitro (16). They observed that the overexpression of IDH1-R132H in cultured cells reduced the formation of cellular α-KG and caused an increased expression of hypoxia-inducible factor-1α (HIF-1α), whose protein stability is negatively regulated by α-KG. Finally, they observed that the HIF-1α level was higher in human gliomas, bearing an IDH1 mutation, compared with those that do not. Since, (1) IDH1/2 mutations exhibit a heterozygous pattern: only a single chromosome is mutated, and (2) they are confined to a particular residue in the enzyme’s active site, both of which are unusual features for tumor suppressor genes, the alternative speculation that IDH1/2 mutations might be oncogenic arose. Complying with this notion, Dang et al. have demonstrated that mutant IDH1/2 proteins possess a new catalytic function that can convert α-KG into a new stereospecific metabolite, (R)-2-hydroxyglutarate (R-2HG) (Fig. 1) (17). By measuring the amount of metabolites in human brain tissues, they observed that the amount of R-2HG in glioma patients harboring IDH1/2 mutations was significantly higher than that of normal people. Subsequent studies have demonstrated that R-2HG acts as an antagonist against a variety of cellular enzymes that utilize α-KG as a cofactor, such as ten eleven translocases (TETs), JmjC histone demethylases, and prolyl-hydroxylases (18). In addition, non-invasive diagnoses of glioma patients bearing IDH1/2 mutations were also attempted, based on the speculation that R-2HG could be used as a surrogate biomarker for brain IDH1/2 mutations. Indeed, magnetic resonance spectroscopy (MRS) revealed significantly higher R-2HG levels in the brain of patients bearing IDH1 mutations, while it was not detectable in normal people (19).
Fig. 1. Metabolic reactions catalyzed by wild-type and mutant isocitrate dehydrogenases (IDHs). IDH1/2/3 catalyzes the conversion of isocitrate (ICT) into α-ketoglutarate (α-KG). While, IDH1/2 utilizes NADP+ as a cofactor, IDH3 uses NAD+ instead. In particular, mutant IDH1/2 enzymes gain a new catalytic function that irreversibly converts α-KG into (R)-2-hydroxyglutarate (R-2HG), in which NADPH acts as a hydrogen donor and is oxidized into NADP+ .
To address whether IDH1 mutation can contribute to tumor formation in vivo, Mak and colleagues generated conditional heterozygous knock-in mice, in which the IDH1-R132H was inserted in the endogenous locus and the mutant IDH1 was selectively expressed either in brains (20) or in hematopoietic systems (21) by the lox-stop-lox (LSL) system. As a result, they observed that a brain-specific heterozygous IDH1-R132H knock-in expression resulted in an immediate perinatal death of mice. A massive hemorrhage was observed within the cerebral hemispheres and cerebellum at autopsy. In addition, elevated accumulation of R-2HG, stabilization of HIF-1α and impairment of collagen maturation were also observed in the brain of these mice. Due to a short lifespan, however, it could not be assessed whether heterozygous IDH1-R132H expression in brain would contribute towards glioma development in vivo. On the other hand, they observed that the hematopoietic-specific heterozygous IDH1-R132H knock-in mice were fertile and had a normal life span. In addition, these mice exhibited a decreased bone marrow cellularity and splenomegaly. Contrary to the initial expectation, however, the hematopoietic-specific heterozygous IDH1-R132H knock-in mice failed to develop leukemia, although they exhibited a number of interesting phenotypical features, such as anemia, an increased population of early hematopoietic progenitors in bone marrows, a significant R-2HG accumulation, and hypermethylated histone and DNA methylation patterns in the serum, all of which are similar to the symptoms observed in IDH1-and/or IDH2-mutant AML patients.
Based on these results, it seems possible now to raise a relevant question: Is R-2HG a genuine oncometabolite? To address this issue, Kaelin and colleagues have used TF-1 human erythroleukemia cells, whose growth is dependent on granulocytemacrophage colony stimulating factor (GM-CSF) and which retains the ability to differentiate in response to erythropoietin (EPO) (22). They observed that TF-1 cells stably infected with IDH1-R132H, but not wild-type IDH1, accumulated cellular R-2HG, spontaneously underwent leukemogenesis and failed to differentiate in response to EPO (23). Likewise, they observed that an exposure of membrane-permeable R-2HG, but not of cell-permeable S-2HG, promoted TF-1 cell leukemogenesis and its withdrawal reversed this process, suggesting a possibility that R-2HG might be an oncometabolite. On the other aspect, it is also noteworthy to take a look at a rare, inherited, and neurometabolic disorder termed as 2-hydroxyglutaric aciduria (2-HG aciduria), which is characterized by an elevated level of R-2HG or S-2HG in the central nervous system (CNS), serum, and urine of affected patients (24). Whereas 50% of D-2-hydroglutaric aciduria patients harbor homozygous mutations in the D-2HG dehydrogenase that is responsible for converting D-2HG back into α-KG (please note that D-2HG is the same enantiomer of R-2HG) (25), the genetic defects of disease in the remaining patients were unknown. Subsequent to the discovery of D-2HG producing IDH1/2 mutations, Kranendijk et al. have found that the other 50% patients with D-2HG aciduria possess a germline IDH2 mutation at arginine-140, mostly substituted into glutamine (Q) or glycine (G) (26). The most notable finding of this study is that no signs of brain or hematological malignancy were observed in patients with D-2HG aciduria, contradicting a possibility that D-2HG (or R-2HG) is an oncometabolite. Therefore, it seems ambiguous yet to answer whether R-2HG is a physiological oncometabolite and this question needs to be thoroughly addressed through more sophisticated experimental setups or clinical settings.
DEVELOPMENT OF SELECTIVE MUTANT IDH1/2 CHEMICAL INHIBITORS
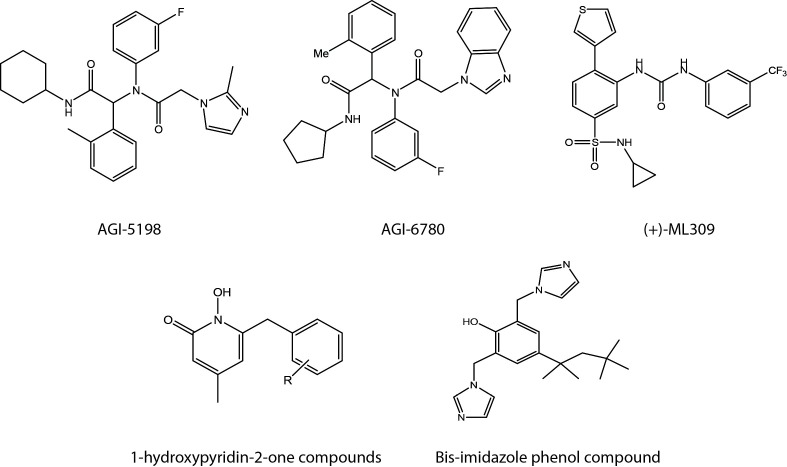
A great deal of interest for development of selective mutant IDH1/2 inhibitors recently arose due to high abundance and unique specificity of IDH1/2 mutations. In fact, IDH1/2 mutations represent by far the most frequently mutated metabolic genes in human cancer. In 2013, scientists in Agios pharmaceuticals reported AGI-5198 (27) and AGI-6780 (28) as the first selective chemical inhibitors of mutant IDH1 and mutant IDH2, respectively. Both AGI-5198 (Fig. 2), possessing a phenyl-glycine scaffold as a core pharmacophore (29) and AGI-6780 (Fig. 2), possessing a heterocyclic urea sulfonamide as a key structural element were identified from a high-throughput screening campaign, utilizing a small molecule library against purified recombinant mutant IDH1 (IDH1-R132H) or mutant IDH2 (IDH2-R140Q) proteins. They have demonstrated that AG-5198 is a competitive inhibitor of IDH1-R132H homodimers and that its treatment significantly reduced the endogenous R-2HG level and promoted astroglial differentiation of TS603 cells harboring a heterozygous genetic IDH1-R132H mutation. In addition, they observed that oral administration of AG-5198 suppressed the growth of T603 tumor xenografts in mice. On the other hand, AGI-6780 was found to be a noncompetitive inhibitor since it binds to the dimerization interface, but not to the substrate binding site of mutant IDH2 homodimers, as revealed by kinetic and X-ray crystallography analyses. In addition, treatment of AGI-6780 significantly reduced endogenous R-2HG level, reversed IDH2-R140Q-induced differentiation block in TF-1 cells and efficiently induced differentiation of primary human AML cells bearing IDH2-R140Q mutation ex vivo. Identification of selective mutant IDH1/2 inhibitors by scientists in Agios pharmaceuticals was soon followed by discoveries of additional selective chemical inhibitors of mutant IDH1. Davis et al. have identified a stereospecific mutant IDH1 selective chemical inhibitor, also referred to as (+)-ML309 (Fig. 2), which harbors a phenyl-glycine scaffold like AG-5198 and observed that treatment of (+)-ML309 significantly suppressed the production of R-2HG in U87MG glioma cells, transfected with IDH1-R132H (30). Liu et al. have identified a series of 1-hydroxypyridin-2-one compounds (Fig. 2) as new selective chemical inhibitors of mutant IDH1 dimers (IDH1-R132H or IDH1-R132C) (31). After a careful structure-activity relationship (SAR)-guided optimization, they have developed selective inhibitors for IDH1-R132H with Ki values as low as 140 nM. Most recently, Deng et al. have reported a new selective mutant IDH1 chemical inhibitor, bearing a bis-imidazole phenol structure (Fig. 2), which binds to the dimer surface of IDH1-R132H and affects direct contact of enzyme with a catalytically essential divalent cation, such as Mg2++(32). With reference to rapid pace of progress, it is expected that additional selective mutant IDH1/2 chemical inhibitors with a greater selectivity will be developed in the near future.
Fig. 2. Structures of selective mutant isocitrate dehydrogenase-1/2 (IDH1/2) chemical inhibitors. Note that all chemicals are selective mutant IDH1 chemical inhibitors except for AGI-6780.
CONCLUDING REMARKS
After the initial elucidation of IDH1/2 missense mutations in 2008, a great deal of progress has been made in understanding the molecular mechanisms of IDH1/2 mutations. However, we are still confronted with another important issue to address, i.e. IDH1/2 mutations and/or the production of R-2HG are oncogenic in vivo? This is a critical question because it can be assumed that, if not, a targeted therapy of cancer patients harboring IDH1/2 mutations with selective mutant IDH1/2 chemical inhibitors will eventually fail. Although a number of cell-based studies have illustrated that these events are oncogenic, studies with conditional IDH-R132H knock-in mice and observations from D-2-hydroglutaric aciduria patients illustrate that it is not necessarily so. Currently, phase I/II clinical trials for targeted therapy against cancer patients, who harbor IDH1/2 mutations are currently undergoing with selective mutant IDH1 (AG-120) and IDH2 (AG-221) chemical inhibitors (please refer to the following website for more information, http://www.agios.com/pipeline-idh.php) and the final outcomes of these clinical trials will reveal the feasibility of targeting metabolic alterations for mechanism-based cancer therapies.
Acknowledgments
This work was supported by Bio-Theme Cluster program of Korea Industrial Complex Corp. (KICOX) (THMSD02R02 for Y.-S. Keum and THMCC01R1 for B.-Y. Choi). We would like to thank Mr. Hyo-Joon Kim (College of Pharmacy, Dongguk University, Republic of Korea) for preparation of figures in the manuscript.
References
- 1.Warburg O. On the origin of cancer cells. Science. (1956);123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. (2013);339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. (2012);23:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teicher BA, Linehan WM, Helman LJ. Targeting cancer metabolism. Clin Cancer Res. (2012);18:5537–5545. doi: 10.1158/1078-0432.CCR-12-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. (2010);16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. (2008);321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. (2009);27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. (2009);360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. (2009);30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, Zhao J, Xu Z, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. (2004);279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 11.Oermann EK, Wu J, Guan KL, Xiong Y. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol. (2012);23:370–380. doi: 10.1016/j.semcdb.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. (2009);174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Losman JA, Kaelin WG., Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. (2013);27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. (2012);482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 15.Pollard PJ, Ratcliffe PJ. Cancer. Puzzling patterns of predisposition. Science. (2009);324:192–194. doi: 10.1126/science.1173362. [DOI] [PubMed] [Google Scholar]
- 16.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. (2009);324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. (2009);462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. (2013);3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 19.Andronesi OC, Rapalino O, Gerstner E, et al. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest. (2013);123:3659–3663. doi: 10.1172/JCI67229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki M, Knobbe CB, Itsumi M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. (2012);26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki M, Knobbe CB, Munger JC, et al. mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. (2012);488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura T, Tange T, Terasawa T, et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. (1989);140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 23.Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. (2013);339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kranendijk M, Struys EA, Gibson KM, et al. Evidence for genetic heterogeneity in D-2-hydroxyglutaric aciduria. Hum Mutat. (2010);31:279–283. doi: 10.1002/humu.21186. [DOI] [PubMed] [Google Scholar]
- 25.Struys EA, Salomons GS, Achouri Y, et al. Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am J Hum Genet. (2005);76:358–360. doi: 10.1086/427890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kranendijk M, Struys EA, van Schaftingen E, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science. (2010);330:336. doi: 10.1126/science.1192632. [DOI] [PubMed] [Google Scholar]
- 27.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. (2013);340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. (2013);340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 29.Popovici-Muller J, Saunders JO, Salituro FG, et al. Discovery of the First Potent Inhibitors of Mutant IDH1 That Lower Tumor 2-HG in Vivo. ACS Med Chem Lett. (2012);3:850–855. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis MI, Gross S, Shen M, et al. Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate dehydrogenase IDH1. J Biol Chem. (2014);289:13717–13725. doi: 10.1074/jbc.M113.511030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Yao Y, Kogiso M, et al. Inhibition of cancerassociated mutant isocitrate dehydrogenases: synthesis, structure-activity relationship, and selective antitumor activity. J Med Chem. (2014);57:8307–8318. doi: 10.1021/jm500660f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng G, Shen J, Yin M, et al. via Disruption of a Metal Binding Network by an Allosteric Small Molecule. J Biol Chem. (2015);290:762–774. doi: 10.1074/jbc.M114.608497. [DOI] [PMC free article] [PubMed] [Google Scholar]