Abstract
Background
Malignant high-grade gliomas (HGGs), including the most aggressive form, glioblastoma multiforme, show significant clinical and genomic heterogeneity. Despite recent advances, the overall survival of HGGs and their response to treatment remain poor. In order to gain further insight into disease pathophysiology by correlating genomic landscape with clinical behavior, thereby identifying distinct HGG molecular subgroups associated with improved prognosis, we performed a comprehensive genomic analysis.
Methods
We analyzed and compared 720 exome-sequenced gliomas (136 from Yale, 584 from The Cancer Genome Atlas) based on their genomic, histological, and clinical features.
Results
We identified a subgroup of HGGs (6 total, 4 adults and 2 children) that harbored a statistically significantly increased number of somatic mutations (mean = 9257.3 vs 76.2, P = .002). All of these “ultramutated” tumors harbored somatic mutations in the exonuclease domain of the polymerase epsilon gene (POLE), displaying a distinctive genetic profile, characterized by genomic stability and increased C-to-A transversions. Histologically, they all harbored multinucleated giant or bizarre cells, some with predominant infiltrating immune cells. One adult and both pediatric patients carried homozygous germline mutations in the mutS homolog 6 (MSH6) gene. In adults, POLE mutations were observed in patients younger than 40 years and were associated with a longer progression-free survival.
Conclusions
We identified a genomically, histologically, and clinically distinct subgroup of HGGs that harbored somatic POLE mutations and carried an improved prognosis. Identification of distinctive molecular and pathological HGG phenotypes has implications not only for improved classification but also for potential targeted treatments.
Keywords: better prognosis, glioblastoma, polymerase epsilon, germline MSH6 mutation, ultramutated tumor
Gliomas are tumors of the neuroepithelial tissue with an incidence of 6.02 per 100 000 and account for ∼28% of all brain and central nervous system tumors. Among these, high-grade gliomas (HGGs), including anaplastic astrocytomas and glioblastoma multiforme (GBM), account for 80% of all malignant brain tumors and carry a poor prognosis, with 5-year survival rates of 26.5% and 4.7%, respectively.1,2 It has been well established that HGGs can form from the malignant transformation of low-grade gliomas, the so-called “secondary” HGGs, or they can arise de novo as “primary” malignant tumors. The genomic architecture and prognosis of primary versus secondary HGGs differ such that the latter typically harbor the recurrent neomorphic isocitrate dehydrogenase 1 (IDH1) R132 mutation and confer a better prognosis.3 Primary HGGs, on the other hand, are frequently characterized by loss of loci of cyclin-dependent kinase inhibitor 2A and B (CDKN2A/B) and phosphatase and tensin homolog (PTEN) on chromosomes 9 and 10, respectively, and amplification of the epidermal growth factor receptor gene (EGFR) on chromosome 7.
While certain well-known clinical features, such as age, Ki-67 values, and Karnofsky performance scores, can be used to predict prognosis in GBM,4 few molecular markers have offered such value. In the gene expression–based clustering of The Cancer Genome Atlas (TCGA), the proneural GBM subgroup was found to show a trend toward improved survival.5 In general, however, none of the genomic variants, including somatic driver mutations in genes such as PTEN, TP53, EGFR, CDKN2A, and MDM2, have prognostic value.6
Herein we report the identification of a specific subgroup of primary HGGs with distinct genomic architecture identifiable with a potential marker and favorable clinical prognosis based on exome sequencing, mutational signature, clonality, and copy number variation (CNV) analyses.
Materials and Methods
Clinical Materials
Institutional review board approvals for genetic studies, along with written consent from all study subjects, were obtained at the participating institutions.
Whole-Exome Sequencing
We performed whole-exome capture and next-generation sequencing of 136 adult gliomas. These samples were classified, based on IDH1-R132 mutation status, as primary versus secondary gliomas.3 Ninety-one of these samples had matching blood samples, 53 of which were IDH1 wild-type primary gliomas. Mean coverages of 194.3 for tumor and 121.3 for matching blood were achieved. The average percentages of reads with at least 20× coverage were 91.0% and 88.4% for tumor and blood, respectively (Supplementary Table S1). We performed a quality control step on the raw reads before alignment for filtering out low-quality reads and adapter contamination as detailed previously.7 The alignment is performed to the human genome reference sequence (version GRCh37) with a Burrows–Wheeler Aligner (v0.5.9-r16)8 followed by Stampy (v1.0.16).9 For the tumor–blood matched samples, we determined the somatic mutations using Haplotyper caller implemented in Genome Analysis Toolkit (GATK v2.5).10 Variant annotation was performed after variant calling using the Ensembl database (v69) with the help of the Variant Effect Predictor v2.7 tool (http://useast.ensembl.org/info/docs/variation/vep/vep_script.html, last accessed date January 20, 2015). Missense variants were annotated to be deleterious, based on the predictions using either SIFT11 or Polyphen2.12 We selected the most deleterious consequence out of all annotated transcripts for each variant site. We applied further filtering on the raw somatic calls based on the sequencing quality metrics and the frequency of variants reported in control databases such as the National Heart, Lung, and Blood Institute's Exome Variant Server (http://evs.gs.washington.edu/EVS/, last accessed date January 20, 2015) and 1000 Genomes13 (see Supplementary material).
We calculated the signatures of somatic mutations of individual tumors by considering 6 major mutation classes—G:C > T:A, G:C > A:T, G:C > C:G, A:T > G:C, A:T > C:G, A:T > T:A (Supplementary Table S2).
Copy Number Variation and Admixture Rate Calculation From Exome Data
Somatic CNVs were calculated using the ExomeCNV14 tool. False positive CNV events were corrected by calculating minor B-allele frequencies in each CNV segment. We also estimated the admixture rate in tumor samples using the copy number loss regions identified (see Supplementary material).
Clonality Analysis
Clonality rate is defined as the percent of tumor cells harboring the identified somatic mutation and correlates with the temporal evolution of the tumor.15,16 Clonality rate of each somatic mutation was calculated based on the variant allele frequency and ploidy at that site, taking into account the admixture rate of each tumor as described previously (see Supplementary material).17 We also analyzed the distribution of variant allele frequency of all somatic mutations for each sample and compared the distributions among samples.
Clinical and Histological Features
Clinical features for all tumors in the Yale glioma cohort (n = 136), such as age at diagnosis and progression-free survival (PFS) time, were analyzed, whenever available (Supplementary Table S3). Histological features of the samples (n = 136) were analyzed by 2 independent neuropathologists using hematoxylin and eosin, glial fibrillary acidic protein (GFAP), and nuclear p53 stainings, whenever available. We have also performed survival analysis on the time-to-recurrence metrics for the patients, when the information is available.
Results
Genomic Profile of Primary Glioblastoma Multiforme
Of the 136 gliomas that were exome sequenced at Yale, 91 had matching blood samples. Initial analysis of the number of somatic coding mutations of these 91 gliomas identified 2 outlier samples. Both of these gliomas were among the 53 of the 91 samples that were IDH1 wild-type primary HGGs. Excluding these 2 tumors (GBM-10468 and GBM-60001), the average numbers of total coding and protein altering somatic mutations in the remaining adult primary HGGs (n = 51) were 76.18 (range, 1–1023) and 53.51 (range, 1–659), respectively. In marked contrast, the 2 outlier samples had a total of 6174 and 10 020 coding somatic mutations (4652 and 7527 protein altering variants, respectively) (Fig. 1A). Previous studies in colon and endometrial cancer have correlated the ultramutated phenotype with mutations within the exonuclease domain of the polymerase epsilon gene (POLE), affecting its proofreading function.18–25 Based on this established relationship between DNA polymerases and the ultramutated cancer phenotype, we searched for POLE or POLD1 mutations in these 2 glioma samples and identified POLE-P286R and POLE-V411L mutations, respectively. Both of these mutations, which were located within the exonuclease domain of POLE, were predicted to be deleterious and have previously been reported to be recurrent in the ultramutated endometrial and colorectal cancer samples18–20,22,26 (Fig. 2A). We did not identify any other somatic mutations affecting the exonuclease domain of POLE or POLD1 in the remaining primary HGG dataset (n = 51). Detailed analysis of this dataset also did not reveal any of the germline POLE or POLD1 mutations previously reported in familial colorectal cancer cases.24,27
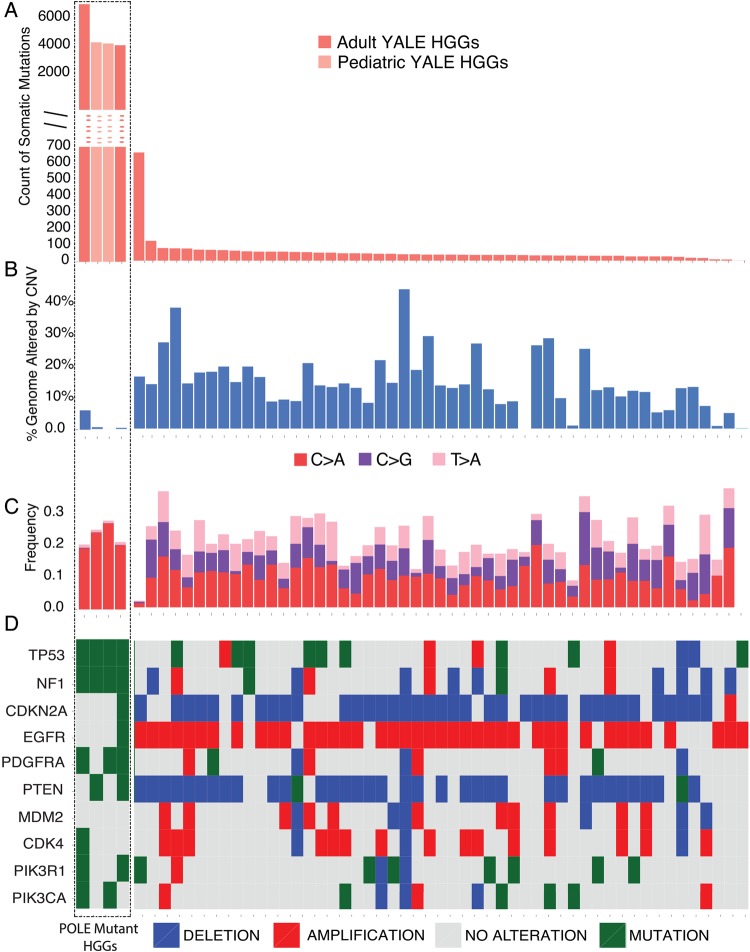
Fig. 1.
Mutation spectrum for 55 primary HGGs. Each of the 55 tumor samples is plotted along the horizontal axis. The vertical axis shows (A) the number of protein altering somatic mutations or (B) percent genome altered. The 4 HGG cases (marked by the dotted-border line, from left to right, GBM-60001, GBM-60003, GBM-60004, and GBM-10468) are ultramutated. These samples show a significantly increased number of somatic mutations but are genomically stable based on the frequency of large-scale copy number alterations (shown as percent genome altered) (B). The sample with increased number of somatic mutations in the non-POLE mutant group had a somatic MSH6 mutation, characterized by increased C > T transitions (Supplementary Fig. S1). (C) Analysis of mutation signatures as determined by the relative percentages of all 6 possible types of single nucleotide mutation types. Only the signatures with significant changes between the ultramutated and non-ultramutated cases are presented. The ultramutated samples, which cluster to the left side of the panel and are marked by the box, are characterized by an increased percentage of C > A transversions and decreased C > G and T > G mutations relative to the other samples. See Supplementary Fig. S1 for the relative distribution of all mutational signature subtypes. (D) Oncoprint representing the CNV and single nucleotide polymorphism (SNP)/insertion-deletion (Indel) status for the frequently altered genes in GBM. SNPs and Indels are depicted as “Mutations.” For the ultramutated cases, there are no CNV events affecting genes that are frequently amplified or deleted in GBM, such as EGFR, PTEN, PDGFRA, CDKN2A, CDK4, and MDM2 (Supplementary Table S4).
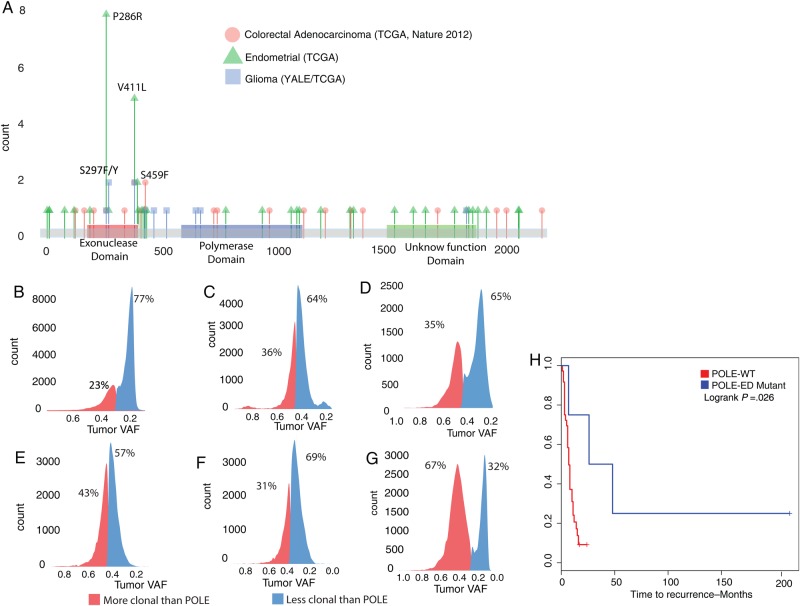
Fig. 2.
(A) Distribution of somatic POLE mutations in various cancers. Exonuclease domain mutations in glial tumors with ultramutated phenotype have previously been reported in ultramutated endometrial and colon cancer cases. (B–G) Clonality analysis showing the distribution of the variant allele frequencies (VAFs) for all somatic mutations with respect to the POLE mutations in 6 HGG cases (B: GBM-60001, C: GBM-10468, D: TCGA-06-5416, E: GBM-60003, F: GBM-60004, G: TCGA-DU-6392). In each tumor, somatic mutations with clonality less than that of POLE are shown in blue. In all cases, POLE mutations occur early, as evidenced by the fact that the majority of the observed mutations carry a lower variant allele frequency compared with the POLE mutations. (H) Kaplan–Meier plot for PFS of ultramutated POLE exonuclease domain mutant (POLE-ED Mutant) adult HGGs (n = 4) show statistically significantly better prognosis compared with POLE wild-type (POLE-WT) HGGs. The adult cases presented in PFS analysis are 2 adult cases each from the Yale (GBM-60001, GBM-10468) and TCGA (TCGA-DU-6392, TCGA-06-5416) cohorts compared with the 51 adult POLE-WT HGGs.
Consistent with the ultramutated phenotype in other cancers, the 2 ultramutated HGG cases were genomically stable without any large-scale chromosome abnormalities. Indeed, the percents of genome altered by CNVs for these 2 samples were 0.34% and 6.08%, respectively, compared with the mean value of 14.61% (range, 0%–43.89%) for the remainder of the primary HGG dataset (n = 51; Fig. 1B, Supplementary Fig. S3). The majority of the CNV alterations on the ultramutated samples were deletions and did not involve loci known to be commonly altered in primary HGGs, including chromosomes 7 (EGFR amplification), 9 (CDKN2A deletion), and 10 (PTEN deletion) (Fig. 1D, Supplementary Table S4). In these ultramutated cases, we also did not identify any potential activating mutations in known GBM driver genes.
The analysis of mutational signature as characterized by the relative proportions of 6 classes of transition and transversion mutations revealed a significant enrichment on the C > T transitions (mean, 64.78%; range, 37.50%–100%) (Supplementary Figs. S1 and S2 and Table S3) in the discovery cohort of 53 primary adult HGGs. Interestingly, the ultramutated HGG samples revealed a unique mutation signature profile. Compared with the rest of the primary HGGs (n = 51), these samples showed an increased rate of C > A transversion (20% and 19% vs an average of 9.3% for the rest; range, 0%−19.61%; P = 3.8e-06) and decreased T > A and C > G transversions (P = 1.9e-14 and 4.1e-16, respectively) (Fig. 1C, Supplementary Figs. S1 and S2). This is consistent with the published literature, which showed the exact same mutation signature in other ultramutated cancers with POLE exonuclease domain mutations.18,19,22
We next compared the clonality rates of somatic mutations for the ultramutated cases to assess the temporal evolution of the increased number of somatic mutations (see Supplementary material). For POLE exonuclease domain mutations to be the cause of the ultramutated phenotype, they should exist in the majority of the cancer cells and be clonal (ie, they should occur early in tumorigenesis, prior to or at the same time as the majority of other somatic mutations). In the 2 ultramutated gliomas, we calculated both POLE mutations to have a clonality rate of >90%, suggesting that POLE mutations indeed occurred early during tumor formation. Consistent with this observation, in both tumors, the variant allele frequency for the majority of the remaining somatic mutations (64% and 77%, respectively) was less than that of POLE mutations (Fig. 2B and C). These findings are in line with the previous reports of POLE exonuclease domain mutations leading to the increased number of somatic mutations.
Additional POLE Mutated Adult High-Grade Gliomas
In an attempt to replicate our results by identifying additional ultramutated HGG samples, we next accessed the glioma28,29 database of TCGA through cBioPortal.30,31 We identified 2 additional samples from this dataset: a primary GBM (TCGA-06-5416) and an anaplastic astrocytoma (TCGA-DU-6392). Similar to our results with the discovery cohort of 53 primary adult HGGs, both of these tumors lacked the recurrent R132H IDH1 mutation and harbored 14 074 and 15 545 total coding somatic mutations (10 292 and 11 290 protein altering, respectively). As predicted, they both carried POLE exonuclease domain mutations: V411L and S297Y, respectively (Fig. 2A, Table 1). Both samples showed a genomic profile similar to POLE mutant HGGs identified from the Yale cohort: they were also genomically stable (3.3% and 13.1% genome alteration by CNVs) and had an increased percentage of C > A transversion mutations (both 25%). Distribution of variant allele frequency with respect to the POLE exonuclease domain mutation showed a similar pattern for TCGA-06-5416 (Fig. 2D). However, the distribution for TCGA-DU-6392 did not show a similar enrichment for the variants that are less clonal than POLE mutation (Fig. 2G).
Table 1.
Ultramutated HGG cases
| Patient ID | Age, y, at Diagnosis | % Genome Altered | Total Mutation Count | Somatic POLE Mutation | Germline MSH6 Mutation |
|---|---|---|---|---|---|
| GBM-60001 | 42 | 6.09 | 10 020 | V411L | V379A, homozygous |
| GBM-60003 | 8 | 0.55 | 4918 | S297F | Q160*, homozygous |
| GBM-60004 | 2 | 0.01 | 4813 | S459F | Q160*, homozygous |
| GBM-10468 | 42 | 0.34 | 6174 | P286R | WT |
| TCGA-05-5416 | 23 | 3.3 | 14 074 | V411L | WT |
| TCGA-DU-6392 | 35 | 13.1 | 15 545 | S297Y | WT |
Abbreviation: WT, wild-type for deleterious mutation. Overall, POLE exonuclease domain mutant HGGs are observed in younger patients and somatically reveal genomic stability but significantly increased number of coding mutations.
Clinical Characteristics
These observations, based on the analyses of number of coding somatic mutations, clonality, percent genome altered, and mutational signature, suggest that the ultramutated POLE exonuclease domain mutant primary HGGs define a distinct molecular subgroup. We next investigated whether these patients (GBM-10468, GBM-60001, TCGA-DU-6392, and TCGA-06-5416) who harbored ultramutated gliomas also differed with regard to clinical characteristics compared with the rest of the patients with primary HGGs (n = 51). Interestingly, patients with the ultramutated HGGs were statistically significantly younger at the time of diagnosis compared with the rest of the primary HGG patients (average age, 35.5 vs 58 y; range, 23–42 vs 22–83; P = .005) (see Supplementary material).1
In addition, ultramutated glioma patients showed a statistically significantly longer PFS (mean, 26.93 vs 6.93 mo; range, 6.8–214 vs 2–16; log rank P = .03 with Kaplan–Meier analysis) (Fig. 2H, Supplementary Figs. S6 and S7). Even though the sample size for POLE mutant samples is small for a comprehensive survival analysis, these findings replicate the findings in endometrial cancer, suggesting that exonuclease domain mutations on POLE in adults predict a longer PFS.20,22
Pediatric GBM Cases With the Ultramutated Phenotype
We next searched the Yale Brain Tumor Genomics dataset for cases with an increased number of somatic mutations to potentially identify additional POLE mutant tumors. This database contains exome sequencing results of 464 brain tumor samples (285 blood matched), including low- and high-grade gliomas, meningiomas, and other brain tumors. We identified only 2 other outlier samples (GBM-60003 and GBM-60004) that harbored a >3 standard deviation increase in the number of somatic mutations.
The 2 identified samples were found to be pediatric GBM cases obtained from 2 siblings, diagnosed at the ages of 2 and 8. Both samples were primary gliomas, lacking the R132H IDH1 mutation and carrying a total of 4918 and 4813 coding somatic mutations (4861 and 4780 of them, respectively, being protein altering) (Fig. 1A). Similar to the cases of adult ultramutated HGGs presented above, these cases also failed to show large-scale genomic alterations due to copy number changes (Fig. 1B). As predicted, we identified somatic POLE exonuclease domain mutations in both tumors. Interestingly, the tumors harbored different deleterious somatic missense POLE exonuclease domain variants: the GBM resected from the younger sibling (GBM-60004) carried the S459F mutation, whereas the tumor from the older sibling (GBM-60003) harbored the S297F mutation. Both of these variants were previously found to be recurrent in the ultramutated phenotype in colorectal and endometrial cancer, respectively18,27 (Fig. 2A). We next performed the clonality analysis on these pediatric cases, and similar to the adult cases, we identified the POLE mutations to be clonal, with the majority of the remaining mutations less frequent (Fig. 2E and F).
Given the rarity of GBM in the pediatric population (incidence rate of 0.14 in 100 000 in the 0–14 age group),1 we next analyzed the exome sequencing results of their germline DNA and identified a homozygous loss-of-function mutation in the mutS homolog 6 gene (MSH6) (Q160X) that was shared by both siblings. The mutation segregated in the family as expected, with both parents being heterozygous for the mutation (Supplementary Fig. S4). Homozygous germline mutations in the DNA mismatch genes, including MSH6, have previously been reported in a pediatric case with multiple tumors.32 Another study reported a germline homozygous MSH6 mutation to cause Turcot syndrome in a family with childhood onset brain tumors.33 Heterozygous germline mutations in MSH6 and other DNA mismatch repair genes, including MLH1 and MSH2, have also been reported to cause an increased risk for colorectal cancer in Lynch syndrome.34 To differentiate whether a defect in DNA mismatch repair due to germline MSH6 mutations or loss of proofreading function due to somatic POLE exonuclease domain mutations was responsible for the increased number of somatic mutations in these 2 GBM cases, we analyzed their mutational signatures. As detailed above, while defects in DNA mismatch result in an increased burden of C > T transitions,29,35 POLE exonuclease domain mutations lead to an increased number of C > A transversions.18,22,26 Similar to the adult cases, analysis of mutational signatures of both tumors showed an increased percentage of C > A transversions (24% and 27%, respectively, vs an average of 9.2% in HGGs; P = 3.8e-06) (Fig. 1C), suggesting that although the inherited MSH6 mutation was most likely responsible for the initiation of tumorigenesis, both tumors later acquired POLE exonuclease domain mutations, leading to tumor progression through the ultramutated somatic profile.36
Germline Mismatch Deficiency in POLE Mutant High-Grade Gliomas
Based on the germline mismatch repair deficiency found in the pediatric cases, we also analyzed the germline alterations in the discovery cohort of 53 primary GBM samples with matching blood. Interestingly, one of the ultramutated adult Yale cases (GBM-60001) harbored a germline homozygous predicted deleterious (by SIFT and probably damaging by Polyphen2) missense mutation in the MSH6 gene (V379A). This variant has been reported in the Single Nucleotide Polymorphism Database with a population frequency <1%, all due to heterozygous mutations. DNA from other family members was not available to study the family segregation. There were no other samples in the discovery cohort with germline homozygous deleterious mutations on MSH6, or other mismatch repair genes.
Ultramutated GBM Defines a Distinct Histology
We next investigated whether the distinctive ultramutated HGG molecular profile correlated with a particular histological phenotype. Review of the available pathological slides revealed a unique histology (Fig. 3 for selected representative cases, and Supplementary Fig. S5 for all cases). Specifically, the tumors were characterized by the predominance of numerous large and bizarre multinucleated giant cells and smaller cells with rounded or fusiform appearance. The giant cells displayed a variety of highly pleomorphic and grotesque shapes and frequently contained numerous eccentrically placed nuclei (Fig. 3A, B, D, E, G, and H; and Supplementary Fig. S5, black arrowheads). The number and appearance of nuclei ranged from a few large bilobed or clumped nuclei to the presence of more than 10 smaller nuclei positioned in a peripheral location. Nuclei also often contained multiple prominent nucleoli. Mitoses were frequent and tended to display a variety of atypical mitotic figures with clumped, fragmented, or dispersed chromatin patterns. Numerous tumor cells, including bizarre forms, showed variable immunohistochemical staining with GFAP (Fig. 3C), and nuclear p53 expression was prominent in the majority of cells (Fig. 3F).
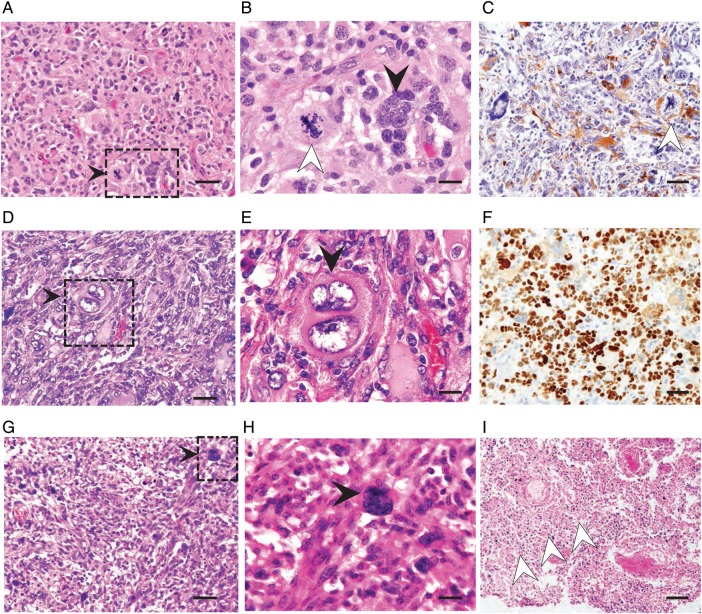
Fig. 3.
(A–C) GBM-60001: Hematoxylin and eosin (H&E) stained (A) low and (B) high magnification histological images are shown (scale bars = 50 μm and 20 μm, respectively). These reveal numerous large multinucleated giant cells with clumped nuclei, and cells with many smaller, eccentrically placed nuclei (black arrowhead). (C) GFAP immunohistochemistry demonstrates astrocytic differentiation of large cells (scale bar = 50 μm). There are frequent atypical mitotic figures (white arrowheads in B and C). (D–F) GBM-60004: H&E stained sections with multinucleated giant cells, some showing bilobed nuclei. These cells appear very large and show bizarre appearance (black arrowheads in D and E). There is strong expression of nuclear p53 in the majority of tumor cells (F). Scale bars = 50 μm, 20 μm, and 50 μm for (D), (E), and (F), respectively. (G–I) GBM-60003: H&E stained section shows numerous smaller cells with rounded appearance, consistent with infiltrating immune cells. Black arrowheads point to multi bizarre-shaped nucleated cells (G and H). Noted is extensive geographic necrosis (white arrow heads in I). Scale bars = 50 μm, 20 μm, and 100 μm for (G), (H), and (I), respectively.
Necrosis, as characterized by areas of geographic—and only occasionally as pseudopalisading—type, was present but not as prominent as classic GBM (Fig. 3I). In addition, microvascular proliferation was rather subtle. Interestingly, we also noted rather prominent lymphocytic infiltrates within the perivascular cuffs as well as the tumor parenchyma. TCGA cases also showed similar histological attributes37 (Supplementary Fig. S5E and F).
Discussion
We have identified a subgroup of primary HGGs with somatic POLE exonuclease domain mutations displaying a longer PFS (Table 1). POLE mutations led to an ultramutated genomic landscape, characterized by more than an average of 7000 protein altering somatic mutations with a stable genome without large-scale chromosomal abnormalities and a distinct mutational signature. Indeed, DNA polymerases encoded by the POLD1 and POLE genes manage the proofreading of spontaneous errors during DNA replication in the lagging and leading strands, respectively.38 The strong mutator phenotype of mutant DNA polymerases was first shown in T4 bacteriophage.39,40 More recent studies showed mutations in the exonuclease domain of POLE to cause an ultramutated phenotype, in organisms ranging from yeast to mice.41–43 Finally, exonuclease domain POLE mutations have been described in subgroups of endometrial and colon cancers (∼10% vs ∼4%, respectively).21,22
Pathological examination of these tumors revealed a characteristic histological subtype, remarkable for the presence of multinucleated giant tumor cells as well as abundant infiltrating immune cells. Consistent with this observation, previous clinical studies have identified the giant cell GBM histological subtype to carry a better prognosis. It has been previously reported that giant cell GBM represents a rare population of all GBM, ranging from 1% to 5%, with a higher prevalence in pediatric cases.44 Therefore we believe that the somatic POLE altered ultramutated GBM represents a subset of all giant cell GBM, and the exact percentage of somatic POLE mutations in the giant cell histological HGG subgroup remains to be determined.
Clinically, the adult patients with the somatic POLE exonuclease mutated HGGs were younger (average age 35.5 vs 58 y in the Yale series, P = .005) and experienced longer PFS (mean 26.93 vs 6.93 mo, log rank P = .03). Even though our sample size for the POLE mutant HGGs is small, we believe that further studies will be able to confirm somatic POLE mutant HGGs to be more prevalent among younger GBM patients. Overall, our results define an important clinical subtype of long-term survivors of primary HGGs with distinctive molecular and histological features, characterized by somatic POLE exonuclease domain mutations.
In 1 adult and 2 pediatric patients (who were siblings), we identified inherited homozygous mutations in the DNA mismatch repair gene MSH6. Heterozygous mutations in MSH6 and other DNA mismatch repair genes, including MLH1 and MSH2, have previously been reported in Lynch syndrome, characterized by an increased risk for colorectal cancer, among others.34 In addition, germline homozygous MSH6 mutations have been previously documented to cause multiple pediatric cancers, including brain tumors.32,33 Previous studies also emphasized the strong synergistic functionalities of mismatch repair and proofreading deficiencies and their impact on increased risk for tumorigenesis and mutation burden.38,41 These observations suggest that mutations in DNA mismatch genes might be responsible for tumor initiation, followed by acquisition of somatic POLE exonuclease mutations, resulting in the ultramutated phenotype. The POLE-induced ultramutated somatic phenotype is distinguishable from the hypermutated tumors that are caused by somatic mutations in mismatch repair genes (such as MSH6) based on the number of mutations as well as the mutation signatures.35,45 POLE exonuclease domain mutant tumors harbor increased C > A transversion mutations as opposed to the increased number of C > T transitions prevalent in MSH6 mutant tumors.29 Based on the extensive literature linking POLE exonuclease domain mutations with a specific mutation signature as well as the clonality analysis presented in this study, we were able to conclude that the ultramutated somatic phenotype observed in the HGG cases was due to somatic POLE mutations.
Consistent with findings in other cancers, the adult ultramutated glioma cases carried a better overall prognosis with prolonged PFS. Interestingly, this paradoxical relationship with ultramutated profile and better prognosis has been also observed in other cancer types, such as endometrial cancer.20,22
Overall, we identified a genomically, histologically, and clinically distinct subgroup of HGGs, which harbored an ultra-large number of somatic alterations due to POLE mutations. This ultramutated primary GBM was observed at a younger age in adults (typical age of diagnosis <40 y) and carried an improved clinical prognosis. Somatic POLE exonuclease domain mutations can serve as a new prognostic molecular marker for better prognosis in HGGs. Identification of distinctive molecular genetic and pathological HGG phenotypes has implications not only for improved classification but also for potential future targeted treatments.
Supplementary Material
Funding
This study was supported by the Gregory M. Kiez and Mehmet Kutman Foundation.
Supplementary Material
Acknowledgments
We are grateful to the patients and their families who have contributed to this study.
Conflict of interest statement. None declared.
References
- 1.Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(suppl 2):ii1–ii56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 3.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. [DOI] [PubMed] [Google Scholar]
- 4.Scott JN, Rewcastle NB, Brasher PM, et al. Which glioblastoma multiforme patient will become a long-term survivor? A population-based study. Ann Neurol. 1999;46(2):183–188. [PubMed] [Google Scholar]
- 5.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraus J, Glesmann N, Beck M, et al. Molecular analysis of the PTEN, TP53 and CDKN2A tumor suppressor genes in long-term survivors of glioblastoma multiforme. J Neurooncol. 2000;48(2):89–94. [DOI] [PubMed] [Google Scholar]
- 7.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunter G, Goodson L. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;216:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.1000 Genomes Project Consortium, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sathirapongsasuti JF, Lee H, Horst BAJ, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics. 2011;2719:2648–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13(11):795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Church DN, Briggs SEW, Palles C, et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;2214:2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74(7):1895–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng B, Hoang LN, McIntyre JB, et al. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol. 2014;134(1):15–19. [DOI] [PubMed] [Google Scholar]
- 21.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida R, Miyashita K, Inoue M, et al. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet. 2011;19(3):320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valle L, Hernández-Illán E, Bellido F, et al. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet. 2014;23(13):3506–3512. [DOI] [PubMed] [Google Scholar]
- 25.Zou Y, Liu F-Y, Liu H, et al. Frequent POLE1 p.S297F mutation in Chinese patients with ovarian endometrioid carcinoma. Mutat Res Fundam Mol Mech Mutagen. 2014;761:49–52. [DOI] [PubMed] [Google Scholar]
- 26.Shinbrot E, Henninger EE, Weinhold N, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;2411:1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palles C, Cazier J-B, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Cancer Genome Atlas Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menko F, Kaspers G, Meijer G, et al. A homozygous MSH6 mutation in a child with café-au-lait spots, oligodendroglioma and rectal cancer. Fam Cancer. 2004;3(2):123–127. [DOI] [PubMed] [Google Scholar]
- 33.Hegde MR, Chong B, Blazo ME, et al. A homozygous mutation in MSH6 causes Turcot syndrome. Clin Cancer Res. 2005;11(13):4689–4693. [DOI] [PubMed] [Google Scholar]
- 34.Baglietto L, Lindor NM, Dowty JG, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102(3):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66(8):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutman DA, Cobb J, Somanna D, et al. Cancer Digital Slide Archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J Am Med Inform Assoc. 2013;20(6):1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20(5):281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speyer JF. Mutagenic DNA polymerase. Biochem Biophys Res Commun. 1965;21(1):6–8. [DOI] [PubMed] [Google Scholar]
- 40.Speyer JF, Karam JD, Lenny AB. On the role of DNA polymerase in base selection. Cold Spring Harb Symp Quant Biol. 1966;31:693–697. [DOI] [PubMed] [Google Scholar]
- 41.Albertson TM, Ogawa M, Bugni JM, et al. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;10640:17101–17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lujan SA, Clausen AR, Clark AB, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;2411:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804(5):1049–1063. [DOI] [PubMed] [Google Scholar]
- 44.Kozak KR, Moody JS. Giant cell glioblastoma: a glioblastoma subtype with distinct epidemiology and superior -prognosis. Neuro Oncol. 2009;11(6):833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15(14):4622–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.