Abstract
Colorectal cancer (CRC), the third most common cancer worldwide, also has the highest rate of cancer-related morbidity and mortality. WNT signaling is initiated by binding of WNT to various receptors, including frizzleds (FZDs), and plays a critical role in CRC and other tumor development by regulating proliferation, differentiation, migration, apoptosis, and polarity. Among the members of the FZD family, FZD6 is broadly expressed in various tissues, and its overexpression has been reported in several cancers, suggesting an important role in cancer development. In this study, we investigated the expression of FZD6 in patients with CRC and found it to be increased in tumors, as compared to paired adjacent non-tumor tissues. Additionally, we found that FZD6 expression was negatively regulated by miR199a5p in CRC cells. These results suggest that overexpression of FZD6, mediated by reduced expression of miR-199a-5p, may play an important role in the development of CRC. [BMB Reports 2015; 48(6): 360-366]
Keywords: Fzd6, MiR-199a-5p, CRC, Wnt4, miRNA
INTRODUCTION
Colorectal cancer (CRC), the third most common cancer worldwide, has the highest rate of cancer-related morbidity and mortality. There are more than 1 million cases of CRC diagnosed each year, as well as 600,000 CRC-related deaths (1). Although previous studies have characterized numerous molecules as risk factors in the oncogenesis and development of CRC, the molecular pathogenesis of CRC has yet to be fully established. Thus, many studies are currently being conducted to identify CRC-specific carcinogenesis-associated molecules, as well as to develop effective prognostic and treatment strategies.
MicroRNAs (miRNAs) are a class of endogenous small RNA molecules that consist of 2022 nucleotides. miRNAs regulate gene expression at the post-transcriptional level by binding to the 3’-UTR of specific target mRNAs (2). Aberrant miRNA expression has been associated with tumor initiation and progression. Recently, dysregulation of miRNAs and their target genes have been established in CRC (3).
The WNT/frizzled (FZD) signaling pathways can be further distinguished into canonical and non-canonical pathways (4). Nineteen members of the WNT family can bind to ten members of the FZD family, thereby activating various downstream pathways, such as the WNT/β-catenin (canonical), WNT/planar cell polarity, and WNT/Ca2+(non-canonical) pathways (4-6). Aberrant constitutive activation of the WNT/FZD pathway and subsequent uncontrolled activation of these processes in normal cells has been implicated as one of the main causes of cancer development (7).
The FZD6 gene localized on human chromosome 8q22.3-q23.1 is broadly expressed in various tissues, including the heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, thymus, prostate, testis, ovary, small intestine, and colon (8). Furthermore, increased expression of FZD6 reported in several cancers suggests that FZD6 may play an important role in cancer development (9-11).
Although members of the FZD family have been widely studied with regard to their roles and underlying molecular mechanisms in the development of CRC, the role of FZD6 in CRC has not been characterized (12-14). In the present study, we investigated the expression of FZD6 in CRC patients and found it to be increased in tumors, compared with paired adjacent nontumor tissues. Moreover, FZD6 expression was negatively regulated by miR-199a-5p in CRC cells. These results suggested that the overexpression of FZD6, mediated by reduced expression of miR-199a-5p, in CRC may play an important role in the development of CRC.
RESULTS
FZD6 expression is increased in CRC patients
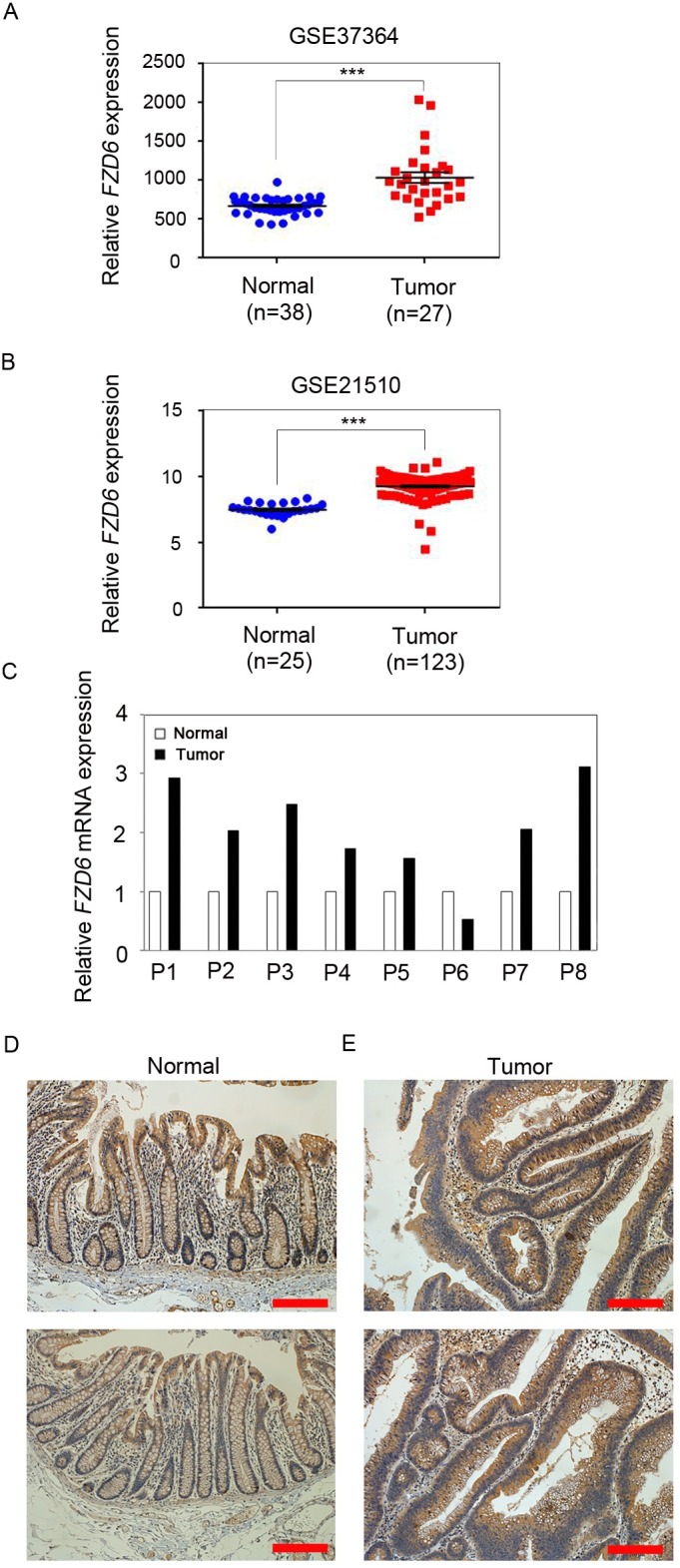
To examine whether the expression levels of FZD6 were changed in CRC, we first used the Gene Expression Omnibus (GEO) database (accession numbers: GSE37364 and GSE21510). We found that FZD6 expression was significantly upregulated in both CRC cohorts (Fig. 1A, B). To determine the expression levels of FZD6 in CRC, we performed a quantitative reverse transcription-polymerase chain reaction (qRT-PCR) in eight pairs of CRC samples and adjacent non-tumor samples. Consistent with the GEO data, the expression levels of FZD6 were found to be increased significantly in the CRC samples, compared with those in the matching non-tumor tissues (Fig. 1C). To further investigate the increased expression of FZD6 in CRC tissues, we performed immunohistochemical staining for FZD6, and detected that FZD6 protein expression was highly increased in CRC samples, compared with matching non-tumor samples (Fig. 1D and E). These results indicated that upregulation of FZD6 correlated with the development of CRC.
Fig. 1. Upregulation of FZD6 in CRC patients. (A, B) Analysis of the Gene Expression Omnibus (GEO) database (accession numbers, GSE37364 and GSE21510) showed that FZD6 was significantly increased in CRC patients, versus the control group. P values were calculated using the by Mann-Whitney test. ***P < 0.001. (C) The relative expression of FZD6 in the CRC tissues and adjacent non-tumor tissues of eight CRC patients. QRT-PCR analysis revealed that expression of FZD6 mRNA was increased in CRC tissues. The data was normalized against GAPDH mRNA expression. (D, E) Overexpression of FZD6 detected by immunohistochemistry in CRC patients compared to matching non-tumor samples. Scale bar = 200 μm.
WNT4 expression is not changed in patients with CRC
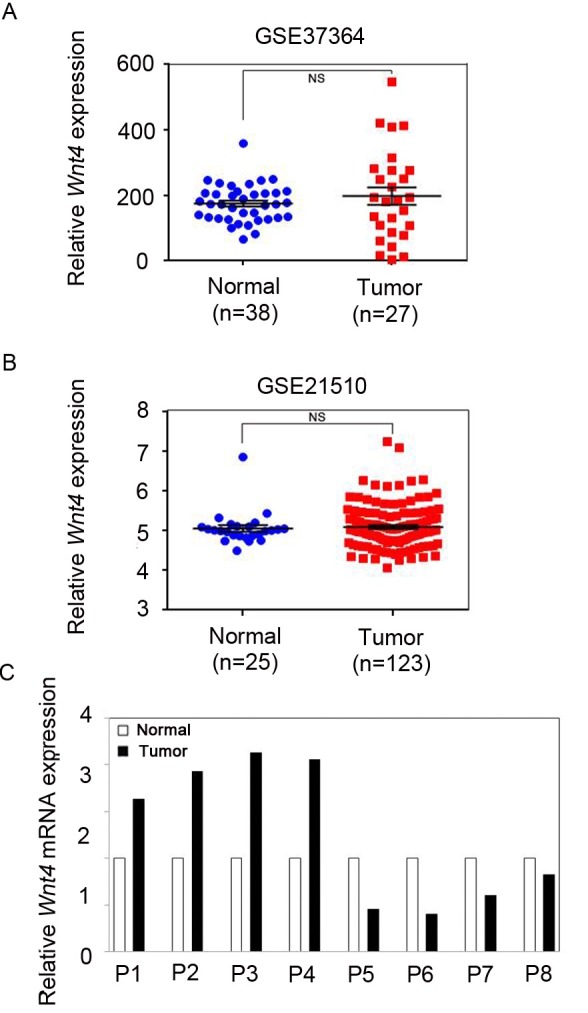
"WNT4, a member of the Wnt family, is a ligand for FZD6 that has been shown to interact with the cysteine-rich domain of FZD6 in kidney epithelial cells (15). Additionally, WNT4 has been implicated in leukemia oncogenesis by regulating cell growth (16). Because concurrent upregulation of ligands and receptors has been shown in other cancers (17, 18), we tested whether FZD6 overexpression occurred concurrently with WNT4 expression to activate WNT4/FZD6 signaling in CRC. To explore whether WNT4 expression was also increased in CRC patients, we analyzed GEO data, as well as CRC tissues. In contrast to FZD6 expression, WNT4 expression was not significantly changed between CRC and nontumor samples according to the GEO data (Fig. 2A, B). QRT-PCR analyses also showed no difference in WNT4 mRNA levels between CRC and matching adjacent non-tumor tissues (Fig. 2C). Thus, WNT4 expression was not consistently changed in CRC tissues, compared with non-tumor tissues, suggesting that WNT4 itself was not directly involved in CRC development."
Fig. 2. WNT4 expression was not altered in CRC patients. (A, B) Analysis of the Gene Expression Omnibus (GEO) database (accession numbers, GSE37364 and GSE21510) indicated that WNT4 expression showed no significant difference between CRC patients and control groups. P values were calculated using the by MannWhitney test. NS = not significant. (C) Relative expression of WNT4 in eight pairs CRC patients and their adjacent non-tumor tissues. QRT-PCR analysis also revealed that CRC tissues had no consistent expression pattern of WNT4 versus the control groups. Data were normalized against GAPDH mRNA expression.
FZD6 is a target of miR-199a-5p in CRC
Because WNT4 expression did not correlate with Fzd6 expression and CRC development, we explored an additional mechanism for the regulation of FZD6 expression. Recent studies have indicated that the expression levels of several FZDs were regulated by specific microRNAs (miRNAs), and associated with the development of various types of cancers (10, 19-22). Moreover, FZD6 expression was also reported to be inhibited by miR-194 in hepatocellular carcinoma (HCC) (10). Thus, we investigated whether the overexpression of FZD6 in CRC was caused by downregulation of a specific miRNAs. We searched for miRNAs that targeted the 3’-untranslated region (UTR) of Fzd6 mRNA and regulated its expression using computer-based algorithms such as miRBase Targets, TargetScan (release 5.2), and Microrna.org databases. Among the candidates, we selected miR-199a-5p as a putative regulator of FZD6 expression, based on the results of a recent study that showed a reduced expression of miR-199a-5p expression in CRC patients, compared with normal individuals (23). As shown in Fig. 3A, the FZD6 3’-UTR contains a putative binding sequence for miR-199a-5p that is evolutionarily conserved among various species. We thus considered that a loss or a reduction of miR-199a-5p expression may have resulted in upregulated expression of FZD6 in CRC, and that this regulation may be associated with CRC development.
Fig. 3. FZD6 is a direct target of miR-199a-5p in CRC cells. (A) The putative binding sequence of miR-199a-5p in FZD6 3’-UTR is well conserved in various species (upper panel). Diagram shows the miR-199a-5p and a putative binding sequence in 3’-UTR of FZD6 mRNA predicted by Target scan algorithm (bottom panel). (B) qRT-PCR showed that endogenous expression of FZD6 mRNA in DLD1 cells was significantly downregulated by overexpression of miR-199a-5p. Data were normalized against GAPDH mRNA expression. Results are the average of three independent experiments conducted in duplicate. **P < 0.01 (C) Western blot also revealed that overexpression of miR-199a-5p resulted in reduced expression of FZD6 at protein level. (D) Luciferase activity was significantly decreased in miR-199a-5p overexpressed DLD1 cells. Results are the average of three independent experiments conducted in duplicate. ***P < 0.001.
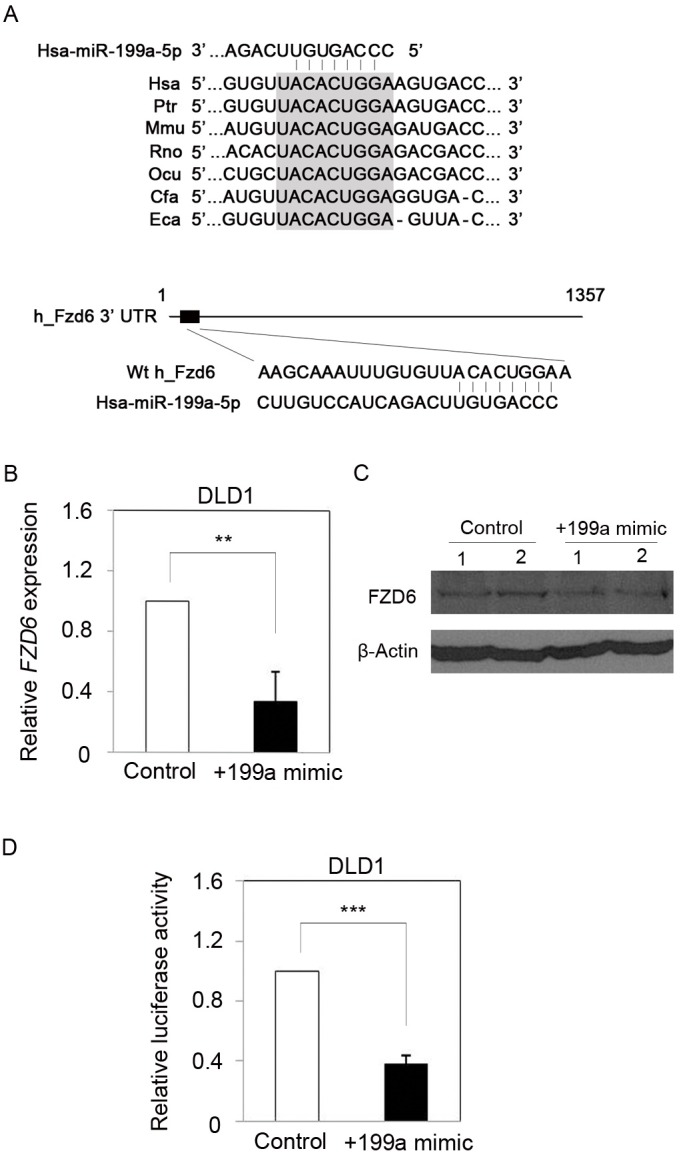
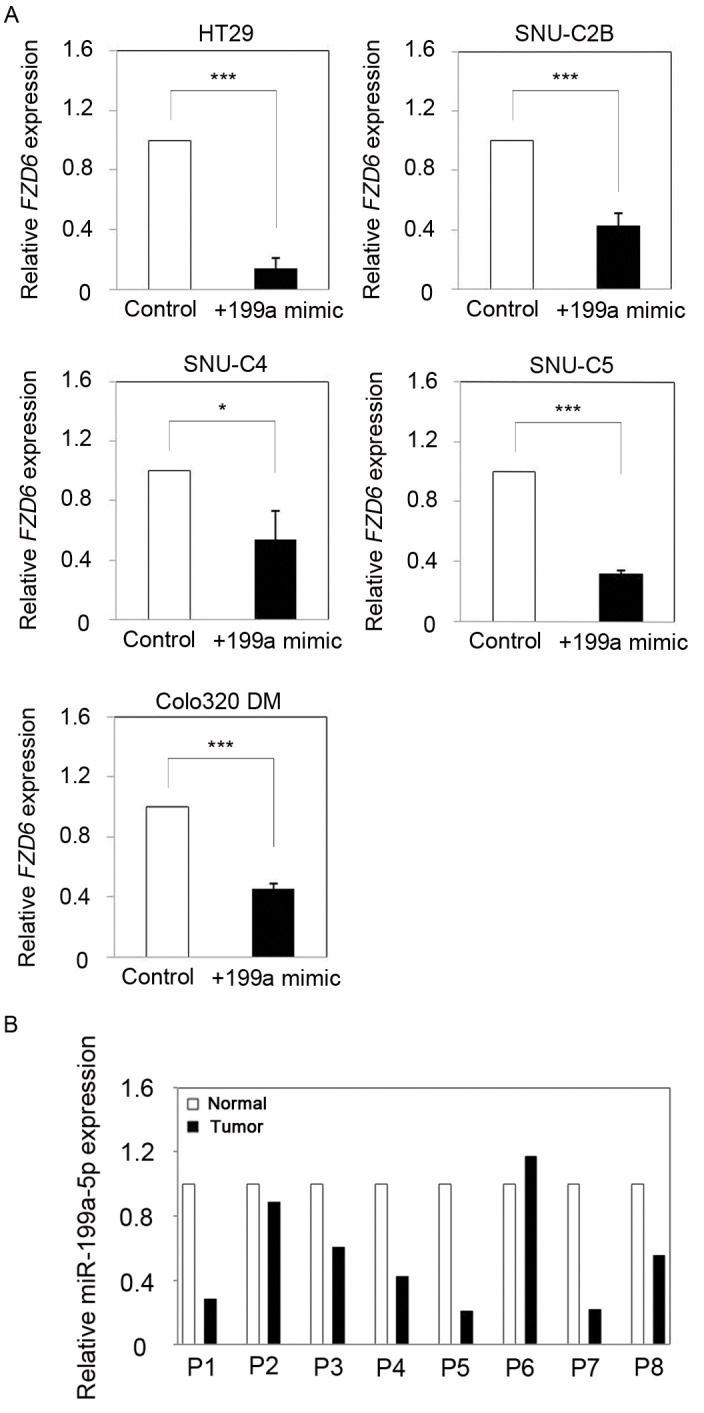
To determine whether miR-199a-5p inhibited the endogenous expression of FZD6 in CRC cells, relative expression levels of Fzd6 mRNA were assessed in DLD1 cells transfected with a miR-199a-5p mimic. QRT-PCR analysis showed that FZD6 mRNA expression was significantly downregulated in the miR-199a-5p mimic-treated DLD1 cells, compared with control transfected cells (Fig. 3B). Compared with the negative mimic-transfected cells, decreased FZD6 protein levels were also detected by Western blot analysis (Fig. 3C). To determine whether FZD6 expression was regulated directly by miR-199a-5p, we performed a luciferase reporter assay in DLD1 cells co-transfected with luciferase gene constructs containing either the full-length 3’-UTR of FZD6, a negative control mimic, or a miR-199a-5p mimic. This results showed significantly inhibited luciferase activity in the cell lysates transfected with miR-199a-5p, compared with those transfected with the negative control mimic (Fig. 3D). Additionally, we further investigated whether other CRC cell lines also showed similar regulation of FZD expression by miR-199a-5p. Using qRT-PCR, FZD6 mRNA expression levels were determined in five CRC cell lines (HT29, SNU-C2B, SNU-C4, SNU-C5, Colo320DM) transfected with a miR-199a-5p mimic. As expected, the expression of FZD6 was significantly reduced in all five cell lines transfected with a miR-199a-5p mimic, compared with those transfected with a negative control mimic (Fig. 4A). Additionally, we found that miR-199a expression was frequently decreased in CRC tissues, as assessed in CRC tumors and matching adjacent non-tumors from eight patients (Fig. 4B). As expected, this downregulation pattern was inversely related to the level of FZD6 expression (Fig. 1C). These observations suggest that miR-199a-5p targeted the 3’-UTR of FZD6 mRNA directly, resulting in inhibition of its expression in CRC.
Fig. 4. Inverse correlation between miR-199a-5p and FZD6 in CRC cells and tissues. (A) Relative expression of FZD6 mRNA in various miR-199a-5p-transfected CRC cells. All CRC cells showed reduced FZD6 expression with overexpression of miR-199a-5p. Data were normalized against GAPDH mRNA expression. Results are the average of three independent experiments conducted in duplicate. *P < 0.05, **P < 0.01, ***P < 0.001. NS = not significant. (B) The relative expression of miR-199a-5p in eight pairs CRC patients and their adjacent samples. qRT-PCR analysis revealed that expression of miR-199a-5p was frequently decreased in CRC tissues. Data were normalized against GAPDH mRNA expression.
DISCUSSION
Recently, the results of multiple studies have shown that the expression levels of several members of the FZD family, acting in both canonical and non-canonical Wnt pathways, were upregulated in various cancers. For example, FZD1 was shown to be upregulated in colon cancer, ovarian cancer, and breast cancer (24-26). Overexpression of FZD3 has been detected in lung can- cer, leukemia, and myeloma (27, 28). FZD7 expression has also been shown to be increased in gastric, esophageal, and colon cancers, as well as in HCC (14, 29-31). FZD10 is also highly expressed in colon and lung cancers (13, 32). The results from these reports suggest that the abnormal expression of FZDs induced aberrant activation of the WNT signaling pathway, which may underlie their roles in cancer development.
In the present study, we examined the expression of FZD6 mRNA and protein in patients with CRC and found it to be upregulated in tumors, compared with non-tumor tissues. The expression of FZDs is required for the activation of genes associated with the non-canonical Wnt signaling pathway. Similar to the canonical Wnt/β-catenin signaling pathway, aberrant activation of non-canonical Wnt signaling pathway has also been shown to directly promote the invasiveness and malignant progression of various types of cancers (7). A previous study demonstrated that the FZD6 expression level was increased in squamous cell carcinoma, compared with site-matched normal skin (9). Its expression was also increased in primary HCC (10). Moreover, the results of a recent study revealed that high expression levels of FZD6 correlated significantly with poor survival in human neuroblastoma (11). Based on these results, as well as the results of the current study, it seems reasonable to suggest that FZD6 is involved in WNT-mediated signaling in CRC development.
In the present study, we found that miR-199a-5p inhibited FZD6 expression directly in various CRC cell lines by targeting the 3’-UTR of its mRNA. miR-199a-5p plays an important role in tumor development through the regulation of the expression of oncogenes or tumor suppressor genes in various cancers, including ovarian, hepatocellular, gastric, and small cell carcinoma of the cervix (33-38). Recently, Hu and colleagues demonstrated that miR-199a-5p was significantly downregulated in CRC tissues and cells (23). Additionally, they found that upregulated miR-199a-5p suppressed DDR1 expression and resulted in decreased migration and invasion of CRC cells, compared with controls. These findings are comparable to the results of the current study, which demonstrated the downregulation of miR-199a-5p in CRC patients and the inverse correlation of its expression with FZD6 expression. Moreover, the results of several studies have shown that non-canonical WNT signaling controls and regulates tumor development by influencing levels of migration-associated proteins, including cytoskeletal proteins and integrin (4, 39). Although further studies are required to determine the precise role(s) of FZD6, current knowledge suggests that miR-199a-5p acts as a tumor suppressor via inhibition of DDR1 and FZD6 expression, resulting in reduced invasiveness. Additionally, downregulation of miR-199a-5p and overexpression of FZD6 have been reported in HCC tissues in two different studies (10, 35). Considering the inverse relationship between miR-199a-5p and FZD6 expression, the regulation of FZD6 by miR-199a-5p may also occur in HCC. Because there are several oncogenes known to be targets of miR-199a-5p in other cancers, further studies are required to fully determine the relationship between these target genes and miR-199a-5p in patients with CRC.
In conclusion, data from the current study indicated that FZD6 expression was increased and regulated by miR-199a-5p in CRC tissues and cells. These results provide evidence of a new role for FZD6 and miR-199a-5p in CRC, which may help in developing potential target-based therapies for patients with CRC.
MATERIALS AND METHODS
Tissue samples
All CRC and non-tumor colorectal tissue samples were obtained from the Department of Pathology in the College of Medicine at the Catholic University of Korea. All samples were approved for analysis by the Institutional Review of Board of the College of Medicine at the Catholic University of Korea.
Cell culture and transfection experiments
All colorectal cancer cells (DLD-1, HT29, ANU-C2B, SNU-C4, SNU-C5, Colo320 DM) were purchased from the Korean Cell Line Bank and maintained in RPMI-1640 Medium (Invitrogen) containing 10% fetal bovine serum with 5% CO2 in a 37℃ incubator. miR199a5p mimics and negative mimics were purchased from Dharmacon. Cells were transfected with a miR-199a-5p mimic using the DharmaFECT 1 transfection reagent (Dharmacon) according to the manufacturer’s protocol. The negative mimic was used for control purposes. After 72 h of incubation, cells were harvested and used for extraction of total RNA or protein.
miR-199a-5p target gene prediction
To predict miR-199a-5p target genes, the miRBase (http://www.mirbase.org/), TargetScan version 5.2 (http://www.targetscan.org/), and microRNA.org (http://www.microrna.org/microrna/home.do/) databases were used.
miRNA-specific qRT-PCR
Total RNA was extracted from cells using the QIAzol reagent (Qiagen) according to the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized using the Mir-X miRNA First-strand synthesis kit (Clontech) following the manufacturer’s protocol. The following primers were used to amplify miR-199a and U6, respectively: 5’CCAGTGTTCAGACTACCTGTTC3’ and 5’TGGCCCCTGCGCAAGGATG3’. The relative expression of miR-199a-5p was determined against the expression of U6 small nuclear RNA using the comparative ΔΔCt method (40).
RT-PCR and qRT-PCR
Total RNAs were isolated from cells using the QIAzol reagent (Qiagen) and reverse-transcribed into cDNA using a PrimeScript 1st strand cDNA Synthesis kit (Takara) according to the manufacturer’s protocol. RT-PCR and qRT-PCR were performed using a Thermal Cycler-100 (MJ Research) and a CFX96 (Bio-Rad Laboratories), respectively. The primer sequences and cycling conditions used are listed in Supplement Table 1. All expression levels were normalized against glyceraldehyde-3-phosphatedehydrogenase gene expression using the comparative ΔΔCt method. Results represent the average of three independent experiments measured in duplicate.
Western blot analysis
Cells were harvested from plates 72 h post-transfection. Protein extracts were prepared using radioimmunoprecipitation assay buffer (150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris-HCl, pH 8.0) according to a standard method. Cell lysates were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (41). The membrane was then incubated with a rabbit polyclonal FZD6 antibody (1:2,500, Abcam) or a mouse polyclonal β-actin antibody (1:5,000, Santa Cruz) following a standard protocol. Protein bands were visualized using an enhanced chemiluminescence system (Amersham Bioscience).
Plasmid Construction
The full length 3’-UTR cDNA of FZD6 was amplified from cDNAs generated from the total RNAs of DLD1 cells by PCR using PrimeSTAR DNA Polymerase (Takara) prior to cloning into pGEMT-easy vectors and subcloning into psiCHECK-2 vector DNA using the Not I cloning sites (Promega). The gene- specific primers are listed in Supplementary Table 1.
Luciferase reporter assay
DLD1 cells (5×105/dish) were seeded in 60 mm dishes at 70% confluency. After 24 h, cells were co-transfected with 50 nM miR-199a-5p mimic and 1 μg of a reporter construct containing the 3’-UTR of FZD6 using the Lipofectamine 2000 reagent. Luciferase activity was determined after 48 h post- transfection using the Dual-Luciferase Reporter Assay reagent (Promega) (42).
Immunohistochemistry
CRC paraffin sections were obtained from the Department of Pathology in the College of Medicine at the Catholic University of Korea. Immunohistochemistry was performed as described previously (43). Briefly, CRC paraffin wax sections were fixed prior to antigen retrieval. Slides were then incubated with the antibody against FZD6 (1:100, Santa Cruz) overnight at room temperature. Signal detection was achieved using a horseradish peroxidase-conjugated antibody (Dako, Glostrup, Denmark) incubated for 1 h at room temperature. The 3,3’-diaminobenzidine chromogen was used for the color reaction (Dako) according to the manufacturer’s instructions.
Statistical analysis
P values were determined using Student’s t-tests. A value of P < 0.05 was considered to indicate statistical significance.
Acknowledgments
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (1220180), and the author(s) also wish to acknowledge the financial support of the Catholic Medical Center Research Foundation made in the program year of 2012.
References
- 1.Atkin WS, Edwards R, Kralj-Hans I, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. (2010);375:1624–1633. doi: 10.1016/S0140-6736(10)60551-X. [DOI] [PubMed] [Google Scholar]
- 2.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. (2010);79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 3.Dassow H, Aigner A. MicroRNAs (miRNAs) in colorectal cancer: from aberrant expression towards therapy. Curr Pharm Des. (2013);19:1242–1252. doi: 10.2174/138161213804805739. [DOI] [PubMed] [Google Scholar]
- 4.Lai SL, Chien AJ, Moon RT. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. (2009);19:532–545. doi: 10.1038/cr.2009.41. [DOI] [PubMed] [Google Scholar]
- 5.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. (2004);20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 6.Huang HC, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. (2004);5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueno K, Hirata H, Hinoda Y, Dahiya R. Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int J Cancer. (2013);132:1731–1740. doi: 10.1002/ijc.27746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokuhara M, Hirai M, Atomi Y, Terada M, Katoh M. Molecular cloning of human Frizzled-6. Biochem Biophys Res Commun. (1998);243:622–627. doi: 10.1006/bbrc.1998.8143. [DOI] [PubMed] [Google Scholar]
- 9.Haider AS, Peters SB, Kaporis H, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. (2006);126:869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- 10.Krutzfeldt J, Rosch N, Hausser J, Manoharan M, Zavolan M, Stoffel M. MicroRNA-194 is a target of transcription factor 1 (Tcf1, HNF1alpha) in adult liver and controls expression of frizzled-6. Hepatology. (2012);55:98–107. doi: 10.1002/hep.24658. [DOI] [PubMed] [Google Scholar]
- 11.Cantilena S, Pastorino F, Pezzolo A, et al. Frizzled receptor 6 marks rare, highly tumourigenic stem-like cells in mouse and human neuroblastomas. Oncotarget. (2011);2:976–983. doi: 10.18632/oncotarget.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong SC, He CW, Chan CM, et al. Clinical significance of frizzled homolog 3 protein in colorectal cancer patients. PLoS One. (2013);8:e79481. doi: 10.1371/journal.pone.0079481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terasaki H, Saitoh T, Shiokawa K, Katoh M. Frizzled-10, up-regulated in primary colorectal cancer, is a positive regulator of the WNT - beta-catenin - TCF signaling pathway. Int J Mol Med. (2002);9:107–112. [PubMed] [Google Scholar]
- 14.Ueno K, Hiura M, Suehiro Y, et al. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia. (2008);10:697–705. doi: 10.1593/neo.08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyons JP, Mueller UW, Ji H, et al. Wnt-4 activates the canonical beta-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/beta-catenin activity in kidney epithelial cells. Exp Cell Res. (2004);298:369–387. doi: 10.1016/j.yexcr.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Castro B, Alvarez-Zavala M, Riveros-Magana AR, et al. Restoration of WNT4 inhibits cell growth in leukemia-derived cell lines. BMC Cancer. (2013);13:557. doi: 10.1186/1471-2407-13-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romanowska M, Evans A, Kellock D, et al. Wnt5a exhibits layer-specific expression in adult skin, is upregulated in psoriasis, and synergizes with type 1 interferon. PLoS One. (2009);4:e5354. doi: 10.1371/journal.pone.0005354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bengochea A, de Souza MM, Lefrancois L, et al. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer. (2008);99:143–150. doi: 10.1038/sj.bjc.6604422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Ma T, Huang C, et al. MiR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/ beta-catenin pathway in hepatocellular carcinoma cells. Cell Signal. (2013);25:2693–2701. doi: 10.1016/j.cellsig.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 20.Song J, Gao L, Yang G, et al. MiR-199a regulates cell proliferation and survival by targeting FZD7. PLoS One. (2014);9:e110074. doi: 10.1371/journal.pone.0110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Formosa A, Markert EK, Lena AM, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. (2014);33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 22.Ueno K, Hirata H, Majid S, et al. Tumor suppressor microRNA-493 decreases cell motility and migration ability in human bladder cancer cells by downregulating RhoC and FZD4. Mol Cancer Ther. (2012);11:244–253. doi: 10.1158/1535-7163.MCT-11-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Liu J, Jiang B, et al. MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig Dis Sci. (2014);59:2163–2172. doi: 10.1007/s10620-014-3136-0. [DOI] [PubMed] [Google Scholar]
- 24.Holcombe RF, Marsh JL, Waterman ML, Lin F, Milovanovic T, Truong T. Expression of Wnt ligands and Frizzled receptors in colonic mucosa and in colon carcinoma. Mol Pathol. (2002);55:220–226. doi: 10.1136/mp.55.4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badiglian Filho L, Oshima CT, De Oliveira Lima F, et al. Canonical and noncanonical Wnt pathway: a comparison among normal ovary, benign ovarian tumor and ovarian cancer. Oncol Rep. (2009);21:313–320. [PubMed] [Google Scholar]
- 26.Milovanovic T, Planutis K, Nguyen A, et al. Expression of Wnt genes and frizzled 1 and 2 receptors in normal breast epithelium and infiltrating breast carcinoma. Int J Oncol. (2004);25:1337–1342. [PubMed] [Google Scholar]
- 27.Lee EH, Chari R, Lam A, et al. Disruption of the non-canonical WNT pathway in lung squamous cell carcinoma. Clin Med Oncol. (2008);2008:169–179. [PMC free article] [PubMed] [Google Scholar]
- 28.Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. (2007);138:338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka S, Akiyoshi T, Mori M, Wands JR, Sugimachi K. A novel frizzled gene identified in human esophageal carcinoma mediates APC/beta-catenin signals. Proc Natl Acad Sci U S A. (1998);95:10164–10169. doi: 10.1073/pnas.95.17.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirikoshi H, Sekihara H, Katoh M. Up-regulation of Frizzled-7 (FZD7) in human gastric cancer. Int J Oncol. (2001);19:111–115. [PubMed] [Google Scholar]
- 31.Merle P, de la Monte S, Kim M, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. (2004);127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Gugger M, White R, Song S, et al. GPR87 is an overexpressed G-protein coupled receptor in squamous cell carcinoma of the lung. Dis Markers. (2008);24:41–50. doi: 10.1155/2008/857474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. (2008);14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 34.Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. (2012);279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 35.Shen Q, Cicinnati VR, Zhang X, et al. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. (2010);9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. (2006);25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. (2008);14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He XJ, Ma YY, Yu S, et al. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. (2014);14:218. doi: 10.1186/1471-2407-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Kikuchi A, Yamamoto H. Tumor formation due to abnormalities in the beta-catenin-independent pathway of Wnt signaling. Cancer Sci. (2008);99:202–208. doi: 10.1111/j.1349-7006.2007.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. (2001);25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Xiao H, Wang ZH, et al. miR-29a suppresses growth and invasion of gastric cancer cells in vitro by targeting VEGF-A. BMB Rep. (2014);47:39–44. doi: 10.5483/BMBRep.2014.47.1.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong Z, Feng Z, Sai Z, Tao S. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. (2014);47:500–505. doi: 10.5483/BMBRep.2014.47.9.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim BK, Lee HY, Kim I, Choi K, Park J, Yoon SK. Increased expression of Dkk1 by HR is associated with alteration of hair cycle in hairpoor mice. J Dermatol Sci. (2014);74:81–87. doi: 10.1016/j.jdermsci.2013.12.007. [DOI] [PubMed] [Google Scholar]


