Abstract
Phytoremediation to clean up arsenic-contaminated environments has been widely hailed as environmentally friendly and cost effective, and genetic engineering is believed to improve the efficiency and versatility of phytoremediation. Successful genetic engineering requires the thorough understanding of the mechanisms involved in arsenic tolerance and accumulation by natural plant species. Key mechanisms include arsenate reduction, arsenic sequestration in vacuoles of root or shoot, arsenic loading to the xylem, and volatilization through the leaves. Key advances include the identification of arsenic (As) translocation from root to shoot in the As hyperaccumulator, Pteris vittata, and the characterization of related key genes from hyperaccumulator and nonaccumulators. In this paper we have proposed three pathways for genetic engineering: arsenic sequestration in the root, hyperaccumulation of arsenic in aboveground tissues, and phytovolatilization.
Introduction
Arsenic is introduced into the environment through both geological and anthropogenic processes and is considered as a global contaminant. Arsenic is among the top carcinogens, and arsenic elevation in soil (and thus food) and drinking water has been reported to affect millions of people around the globe [1,2]. Long-term exposure to arsenic is associated with a variety of diseases, including cancer and diabetes [3–5]. Worldwide, substantial effort has been directed to the removal of arsenic from water and soil through chemical and physical remediation processes, but these are expensive and therefore have limited applicability in many areas where arsenic contamination and poverty coexist, as a result, the consumption of arsenic-tainted water and food remains the major route of arsenic exposure in humans. Alternative and environmental-friendly technologies are thus urgently needed to combat global arsenic contamination.
Among various technologies available so far or under development currently, phytoremediation is considered to be the most environmentally friendly and cost effective. The phytoremediation of arsenic-contaminated environments includes the following types: firstly, phytostabilization, which refers to the use of plants (or vegetation) to minimize arsenic dispersion from soil to water or air; secondly, phytoextraction (or phytofiltration), which refers to the use of plants to remove arsenic from soil or water; thirdly, phytovolatilization is a newly conceived and specialized form of phytoremediation and involves the production of volatile arsenic compounds and their emission from plants. However, a common feature of most types of phytoremediation is arsenic tolerance and accumulation, and both tolerance and accumulation involve arsenic compartmentation and translocation. This paper will thus review some recent progress in genetic engineering related to arsenic compartmentation and translocation in plants, and will discuss some of the key issues of how to make phytoremediation work in reality.
Brief overview of plant uptake and metabolism of arsenic
Plant uptake and metabolism of arsenic has recently been reviewed by Tripathi et al. [6] and Zhao et al. [7••], so here we will only provide a brief overview. Arsenic in the environment mainly exists in two inorganic oxidation states, arsenate (As(V)) and arsenite (As(III)). As(V) and As(III) enter plant cells via phosphate transporters and aquaglyceroporins, respectively, as reviewed in [8]. Once taken up, As(V) is reduced to As(III), catalyzed largely by arsenate reductases, members of the superfamily of protein tyrosine phosphatase (PTPase) [9]. As(III) can then be complexed with glutathione (GSH) or phytochelatins (PCs), Raab et al. [10] identified up to 14 different species of arsenic complexes in sunflower plants. As(III) or complexed As(III) is then transported across the tonoplast and sequestered in the vacuole. Most data support the idea that arsenic is translocated from the roots to the tissues above ground, mostly in the form of As(III) [11,12••]. As(III) can be methylated to form monomethylarsenate (MMAs(V)), dimethylarsenate (DMAs(V)), and trimethylarsine oxide (TMAO(V)) in planta [7••,13].
Genetic engineering for arsenic sequestration in vacuole
Complexation of As(III) with PCs or GSH is an efficient way to detoxify arsenic, probably because the complexes are pumped and sequestered in the vacuole catalyzed by the homologs of multidrug resistance proteins (MRPs), members of the ABC superfamily [14,15]. Targeting increased accumulation or synthesis of PCs and/or GSH may be one way to develop arsenic phytoremediation. Increased expression of phytochelatin synthase (PCS), the rate-limiting step in PC biosynthesis, has been attempted to increase plant tolerance to and accumulation of arsenic. Gasic and Korban [16] found that the overexpression of PCS in Indian mustard increased its tolerance to arsenic but did not enhance arsenic accumulation significantly. The lack of response in accumulation could be due to the fact that PC synthesis is also limited by the production of GSH. A more recent study by Guo et al. [17] showed that overexpressing AtPCS1 and GSH1, which encodes γ-glutamylcysteine synthetase (γ-ECS), the rate-limiting step in GSH biosynthesis, individually in Arabidopsis thaliana increased arsenic tolerance and accumulation. Although these studies indicated the feasibility of overexpressing PCS and/or γ-ECS for increasing arsenic accumulation and concomitantly tolerance, there are no direct data on the site of arsenic storage in these transgenic lines; thus it remains unclear whether the complexed As(III) is primarily vacuolar or remains in the cytoplasm. It is possible that transport of complexed As(III) or even free As(III) across the tonoplast membrane is potentially the rate-limiting step in overall arsenic tolerance and accumulation. Yet, to date, there are no reports of genetic engineering of tonoplast transport.
Even if arsenic is sequestered in the vacuole, its overall effect on tolerance will depend on the spatial expression of the relevant genes in planta. If these genes are overexpressed in root, this could result in reduced arsenic translocation from root to shoot, which is of practical relevance for phytostabilization. On the contrary, if these genes were specifically overexpressed in above ground tissues, then the genetically modified plant might be useful for phytoextraction. Of course, the ultimate efficiency of spatial distribution may depend on the machinery of arsenic translocation from root to shoot, as discussed below.
Genetic engineering for arsenate reduction
It is clear that both arsenic sequestration and translocation are closely linked to the forms of arsenic that exist within plant tissues. Trivalent arsenite is most easily trapped in the root through vacuole sequestration of thiol conjugates, but, under aerobic conditions, much of the arsenic coming into the cells is as pentavalent arsenate. Therefore the reduction of arsenate to arsenite is a key step in arsenic metabolism. To express the genes for arsenate reductases, genetically engineering plants have been applied for phytoremediation. Dhankher et al. [18] found that overexpressing the gene for the Escherichia coli arsenate reductase gene, arsC, in A. thaliana under the control of a light-responsive transcription factor (so that it would be expressed only in above ground tissues) led to hypersensitivity to arsenic. Although this seemed at first a counter-intuitive result, the authors considered the possibility that the product, arsenite, was more toxic than the substrate, arsenate, without sufficient thiols to form the As(GS)3 conjugate. For that reason the gene for the E. coli γ-ECS, which would increase the rate of GSH biosynthesis, was coexpressed with arsC, resulting in higher tolerance to and accumulation of arsenic. This is consistent with tolerance being related to the sequestration of As(III)–thiol conjugates in the vacuole.
Although phytoremediation might be facilitated by the expression of heterologous arsenate reductases in above ground tissues, it is now clear that plants have their own such enzymes that are expressed constitutively [19•,20]. One reason for the slow translocation of arsenic from root to shoot could be due to endogenous arsenate reductase activity in the root, so that the arsenate entering root cells is partly reduced to arsenite, conjugated with thiols, and sequestered in the root vacuole. Therefore, if expression of the native arsenate reductase gene(s) could be reduced in the root, more arsenate might be available for movement to aboveground tissues. In support of this possibility, Dhankher et al. [21] reported that silencing the arsenate reductase gene AtACR2 in the root of A. thalina resulted in the hyperaccumulation of arsenic in the shoot, although the authors did not present direct evidence on the speciation of arsenic in different plant tissues and in the xylem.
Genetic engineering for increased translocation from root to shoot
One of the key properties of arsenic hyperaccumulators such as Pteris vittata is a highly efficient system of arsenic translocation from root to shoot [11,22], while most non-hyperaccumulators usually have a low mobility rate compared to P. vittata. Arsenic mobility from root to shoot varies considerably among different plant species, suggesting that it is under genetic control. A key step in arsenic translocation from root to shoot is arsenic loading to the xylem, a process that is not well understood. Recently, Ma et al. [23,24] identified a gene encoding an efflux protein, Lsi2, which is responsible for loading arsenite into the xylem, as arsenite is the dominant arsenic species in the xylem. An Lsi2 mutation resulted in a nearly 50% reduction in arsenic accumulation in the shoot. Lsi2 is a homolog of the E. coli ArsB, which is an As(III)/H+ exchanger that confers bacterial arsenite resistance [25]. The plant efflux protein apparently transports both metalloids As(III) and Si(IV). It will be interesting to examine whether overexpression of genes such as Lsi2 will enhance arsenic translocation from root to shoot.
Genetic engineering for volatilization
Many organisms, including bacteria, fungi, and animals, methylate arsenic. Methylated arsenic species have been detected in several plant species, including rice grain [26,27], and recent data suggest that this is the result of endogenous methylation by the plants themselves [13]. The final product of the methylation pathway is the gas trimethylarsine (TMAs(III)), which can be volatilized from the plant. Recently, Qin et al. [28••] cloned a gene encoding an As(III)-S-adenosylmethionine methyltransferase (arsM) from the soil bacterium Rhodopseudomonas palustris. Expression of the arsM gene in an arsenic-sensitive strain of E. coli that had been genetically engineered to remove all arsenic detoxification genes resulted in the biosynthesis of several methylated forms of arsenic, including volatile TMAs(III) and concomitant arsenic tolerance. These results indicate that the expression of the single methyltransferase gene is sufficient to produce both volatilization of and tolerance to arsenic. More recently Rosen and coworkers have identified the gene for an ArsM homolog in a primitive plant, the eukaryotic alga Cyanidioschyzon merolae [29••]. Cells expressing CmArsM similarly methylate As(III), as does the purified enzyme. Whether similar processes are also present in higher plants remains unclear, but, in a rice microarray study, a putative gene annotated as a methyltransferase was upregulated by exposure to arsenate in the growth solution [30•]. These results point to the possibility of engineering arsenic volatilization for the phytoremediation of arsenic-contaminated water and soil and also to improve the safety of the food supply by reducing the arsenic content of the rice grain.
Conclusions
Successful phytoremediation depends largely on our understanding of the bioavailability of arsenic in soil, and of plant tolerance to and accumulation of arsenic. We have discussed the complexity of plant tolerance and accumulation of arsenic; it is clear that arsenic accumulation in plants is modulated by a network of functional genes, their expression patterns, and temporal/spatial coordination. Here we summarize the key mechanisms of arsenic tolerance and accumulation and point out key pathways that can be manipulated for phytoremediation (Figure 1). To date, there have been a few studies that indicate the feasibility of manipulating one or more genes for the phytoremediation of arsenic-contaminated environments. However, successful phytoremediation may require a more complex system-wide approach that combines many of these single methodologies. Finally, whether transgenic plants designed for the remediation of arsenic-contaminated soil or water can work under field conditions is a major question. Practical applications of phytoremediation will require the analysis of the following topics:
Further elucidation of the molecular mechanisms of arsenic tolerance and accumulation, particularly where and how arsenic is translocated and stored in plants.
Genetic modification of fast-growing and high-biomass crop plants to accumulate substantial arsenic in above ground tissues for phytoextraction and/or volatilization, or to sequester arsenic in roots for phytotstabilization.
Integration (molecular design) of multiple pathways of arsenic uptake and metabolism for the optimization of phytoremediation using genetically modified ‘smart’ plants.
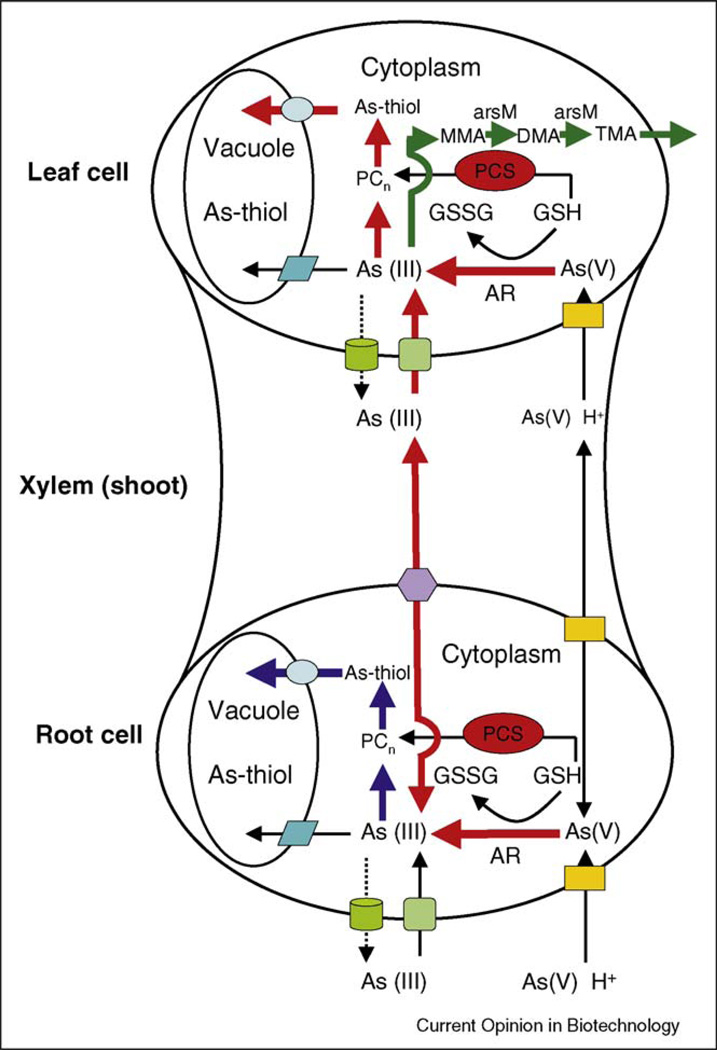
Figure 1.
Schematic diagram of arsenic uptake and metabolism in plants and possible genetic manipulations that can be designed for efficient phytoremediation. The pathway in red is for phytoextraction, in dark blue for phytostabilization, and in dark green for phytovolatilization. AR, arsenate reductase; PCS, phytochelatin synthase; arsM, As(III)-S-adenosylmethionine methyltransferase.  Phosphate/arsenate transporter;
Phosphate/arsenate transporter;  plasma membrane aquaporin channel;
plasma membrane aquaporin channel;  unidentified arsenite efflux transporter;
unidentified arsenite efflux transporter;  As-thiol transporter;
As-thiol transporter;  tonoplast aquaporin channel for As(III) transporter to vacuole;
tonoplast aquaporin channel for As(III) transporter to vacuole;  arsenite efflux carrier Lsi2.
arsenite efflux carrier Lsi2.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Abernathy CO, Liu YP, Longfellow D, Aposhian HV, Beck B, Fowler B, Goyer R, Menzer R, Rossman T, Thompson C, Waalkes M. Arsenic: health effects, mechanisms of actions, and research issues. Environ Health Perspect. 1999;107:593–597. doi: 10.1289/ehp.99107593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abernathy CO, Thomas DJ, Calderon RL. Health effects and risk assessment of arsenic. J Nutr. 2003;133:1536S–1538S. doi: 10.1093/jn/133.5.1536S. [DOI] [PubMed] [Google Scholar]
- 3.Tchounwou PB, Centeno JA, Patlolla AK. Arsenic toxicity, mutagenesis, and carcinogenesis — a health risk assessment and management approach. Mol Cell Biochem. 2004;255:47–55. doi: 10.1023/b:mcbi.0000007260.32981.b9. [DOI] [PubMed] [Google Scholar]
- 4.Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure — a critical review. Toxicol Pathol. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- 5.Tseng CH, Tseng CP, Chiou HY, Hsueh YM, Chong CK, Chen CJ. Epidemiologic evidence of diabetogenic effect of arsenic. Toxicol Lett. 2002;133:69–76. doi: 10.1016/s0378-4274(02)00085-1. [DOI] [PubMed] [Google Scholar]
- 6.Tripathi RD, Srivastava S, Mishra S, Singh N, Tuli R, Gupta DK, Maathuis FJ. Arsenic hazards: strategies for tolerance and remediation by plants. Trends Biotechnol. 2007;25:158–165. doi: 10.1016/j.tibtech.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 7. Zhao FJ, Ma JF, Meharg AA, McGrath SP. Arsenic uptake and metabolism in plants. New Phytol. 2009 doi: 10.1111/j.1469-8137.2008.02716.x. This paper provides a thorough review of the recent progress in research on the plant uptake and metabolism of arsenic, particularly on arsenite uptake, arsenate reduction, arsenic xylem loading, translocation from root to shoot, and potential volatilization.
- 8.Bhattacharjee H, Mukhopadhyay R, Thiyagarajan S, Rosen BP. Aquaglyceroporins: ancient channels for metalloids. J Biol. 2008;7:33. doi: 10.1186/jbiol91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect. 2002;110(Suppl 5):745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raab A, Schat H, Meharg AA, Feldmann J. Uptake, translocation and transformation of arsenate and arsenite in sunflower (Heliannthus annuus): formation of arsenic–phytochelatin complexes during exposure to high arsenic concentrations. New Phytol. 2005;168:551–558. doi: 10.1111/j.1469-8137.2005.01519.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu XY, McGrath SP, Zhao FJ. Rapid reduction of arsenate in the medium mediated by plant roots. New Phytol. 2007;176:590–599. doi: 10.1111/j.1469-8137.2007.02195.x. [DOI] [PubMed] [Google Scholar]
- 12. Su YH, McGrath SP, Zhu YG, Zhao FJ. Highly efficient xylem transport of arsenite in the arsenic hyperaccumulator Pteris vittata. New Phytol. 2008;180:434–441. doi: 10.1111/j.1469-8137.2008.02584.x. This paper, together with several early publications from the group demonstrates that the key mechanism of As hyperaccumulation is rapid arsenate reduction to arsenite in the root, and efficient loading of arsenite into xylem, and then transported to the fronds for storage, thus accumulation in above ground tissues.
- 13.Wu JH, Zhang R, Lilley RM. Methylation of arsenic in vitro by cell extracts from the bentgrass (Agrostis tenuis): effect of acute exposure of plants to arsenate. Funct Plant Biol. 2002;29:73–80. doi: 10.1071/PP01022. [DOI] [PubMed] [Google Scholar]
- 14.Lu YP, Li ZS, Rea PA. AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc Natl Acad Sci U S A. 1997;94:8243–8248. doi: 10.1073/pnas.94.15.8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tommasini R, Vogt E, Fromenteau M, Hortensteiner S, Matile P, Amrhein N, Martinoia E. An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 1998;13:773–780. doi: 10.1046/j.1365-313x.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- 16.Gasic K, Korban SS. Transgenic Indian mustard (Brassica juncea) plants expressing an Arabidopsis phytochelatin synthase (AtPCS1) exhibit enhanced As and Cd tolerance. Plant Mol Biol. 2007;64:361–369. doi: 10.1007/s11103-007-9158-7. [DOI] [PubMed] [Google Scholar]
- 17.Guo JB, Dai XJ, Xu WZ, Ma M. Overexpression of GSH1 and AsPCS1 simultaneously increase the tolerance and accumulation of cadmium ad arsenic in Arabidopsis thaliana. Chemosphere. 2008;72:1020–1026. doi: 10.1016/j.chemosphere.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Dhankher OP, Li Y, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA, Meagher RB. Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat Biotechnol. 2002;20:1140–1145. doi: 10.1038/nbt747. [DOI] [PubMed] [Google Scholar]
- 19. Duan GL, Zhou Y, Tong YP, Mukhopadhyay R, Rosen BP, Zhu YG. A CDC25 homologue from rice functions as an arsenate reductase. New Phytol. 2007;174:311–321. doi: 10.1111/j.1469-8137.2007.02009.x. This paper reports the cloning and characterization of arsenate reductase genes from an important crop plant — rice. The gene products were purified and shown to reduce arsenate into arsenite. Mutagenesis of cysteine residues in the putative active site HC(X)5R motif led to nearly complete loss of both phosphatase and arsenate reductase activities.
- 20.Ellis DR, Gumaelius L, Indriolo E, Pickering IJ, Banks JA, Salt DE. A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiol. 2006;141:1544–1554. doi: 10.1104/pp.106.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhankher OP, Rosen BP, McKinney EC, Meagher RB. Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2) Proc Natl Acad Sci U S A. 2006;103:5413–5418. doi: 10.1073/pnas.0509770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan GL, Zhu YG, Tong YP, Cai C, Kneer R. Characterization of arsenate reductase in the extract of roots and fronds of Chinese brake fern, an arsenic hyperaccumulator. Plant Physiol. 2005;138:461–469. doi: 10.1104/pp.104.057422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma JF, Tamai K, Ichii M, Wu GF. A rice mutant defective in Si uptake. Plant Physiol. 2002;130:2111–2117. doi: 10.1104/pp.010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma JF, Tamai K, Yamaji N, Mitani N, Konishi K, Katsuhara M, Ishiguro M, Murata Y, Yano M. A silicon transporter in rice. Nature. 2006;440:688–691. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- 25.Meng YL, Liu Z, Rosen BP. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J Biol Chem. 2004;279:18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- 26.Williams PN, Price AH, Raab A, Hossain SA, Feldmann J, Meharg AA. Variation in arsenic speciation and concentration in paddy rice related to dietary exposure. Environ Sci Technol. 2005;39:5531–5540. doi: 10.1021/es0502324. [DOI] [PubMed] [Google Scholar]
- 27.Zhu YG, Sun GX, Lei M, Teng M, Liu YX, Chen NC, Wang LH, Carey AM, Deacon C, Raab A, et al. High percentage inorganic arsenic content of mining impacted and nonimpacted Chinese rice. Environ Sci Technol. 2008;42:5008–50013. doi: 10.1021/es8001103. [DOI] [PubMed] [Google Scholar]
- 28. Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci U S A. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. This is the first paper reporting the cloning and characterization of arsM, the study shows that when arsM is expressed heterologously in an Assensitive strain of E. coli, it conferred As tolerance by catalyzing the formation of methylated As species.
- 29. Qin J, Lehrb CR, Yuan C, Le XC, McDermott TR, Rosen BP. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0900238106. This study reports the cloning of two arsenic methyltransferase genes, CmarsM7 and CmarsM8, from Cyanidioschyzon sp. isolate 5508, a primitive plant. These two genes were demonstrated to confer resistance to As(III) in an arsenite hypersensitive strain of Escherichia coli. The recombinant CmArsMs were purified and shown to transform As(III) into monomethylarsenite, DMAs(V), TMAO, and trimethylarsine gas, with a Topt of 60–70 °C.
- 30. Norton GJ, Lou-Hing DE, Meharg AA, Price AH. Rice–arsenate interactions in hydropinics: whole genome transcriptional analysis. J Exp Bot. 2008;59:2267–2276. doi: 10.1093/jxb/ern097. In this study, RNA extracted from the roots of rice plants grown for one week in phosphate-free nutrient solution with or without arsenate was used to challenge the Affymetrix (52K) GeneChip Rice Genome array. A large number of responses from genes involved in glutathione synthesis, metabolism, and transport suggest that glutathione conjugation and arsenate methylation may be important biochemical responses to arsenate challenge.