Abstract
Introduction
Elucidating the microbial ecology of endodontic infections (EI) is a necessary step in developing effective intra-canal antimicrobials. The aim of the present study was to investigate the bacterial composition of symptomatic and asymptomatic primary and persistent infections in a Greek population, using high throughput sequencing methods.
Methods
16S amplicon pyrosequencing of 48 root canal bacterial samples was conducted and sequencing data were analyzed using an oral microbiome-specific (HOMD) and a generic (Greengenes; GG) database. Bacterial abundance and diversity were examined by EI type (primary or persistent) and statistical analysis was performed by using non-parametric and parametric tests accounting for clustered data.
Results
Bacteroidetes was the most abundant phylum in both infection groups. Significant, albeit weak associations of bacterial diversity were found, as measured by UniFrac distances with infection type (ANOSIM R=0.087, P=0.005) and symptoms (ANOSIM R=0.055, P=0.047). Persistent infections were significantly enriched for Proteobacteria and Tenericutes as compared to primary ones; at the genus level, significant differences were noted for 14 taxa, including increased enrichment of persistent infections for Lactobacillus, Streptococcus, and Sphingomonas. More but less-abundant phyla were identified using the GG database; among those, Cyanobacteria (0.018%) and Acidobacteria (0.007%) were significantly enriched among persistent infections. Persistent infections showed higher Phylogenetic Diversity (asymptomatic: PD=9.2, [standard error (se)=1.3]; symptomatic: PD=8.2, se=0.7) compared to primary infections (asymptomatic: PD=5.9, se=0.8; symptomatic: PD=7.4 se=1.0).
Conclusions
The present study revealed a high bacterial diversity of EI and suggests that persistent infections may have more diverse bacterial communities than primary infections.
Keywords: Primary endodontic infection, persistent infection, bacterial diversity, pyrosequencing, oral microbiome
Endodontic infections have been linked to the commensal oral microbiota, which colonize and proliferate in the root canal system as a consequence of pulp necrosis secondary to caries, tooth trauma, defective restorations (1), or due to a failed endodontic treatment (2). A thorough understanding of the microbial etiology and characteristics of endodontic infections is a necessary step in developing effective intra-canal antimicrobial protocols. Nevertheless, the exploration and identification of endodontic pathogens remains one of the most challenging aspects in endodontic microbiology, with the majority of bacteria still unknown or uncultivated (3). Broad-range PCR followed by cloning and Sanger sequencing as well as molecular fingerprinting techniques such as denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism analysis (T-RFLP) have offered initial insights into the bacterial diversity of the infected root canal system (4,5). Nevertheless, despite their high sensitivity, these methods can detect only the most prevalent bacterial community members.
The development and application of molecular biology methods has facilitated the identification and linkage of specific bacterial species with periradicular disease and thus have led to the discovery of novel endodontic pathogens (3). Next-generation sequencing is now part of the toolbox available for 16S rRNA-based bacterial diversity analyses (6). The technology enables a large number of reads in a single run, providing increased sampling depth compared to other techniques (7) and has the major advantage of enabling the detection of low-abundant genera (7,8). So far, only eight studies have used this approach to investigate different types of endodontic infections (7,9–15). From those, only two investigations have examined the endodontic microbiome in teeth with failed endodontic treatment (14,15). Even though these persistent infections present an important clinical problem, there is a knowledge gap in their microbial etiology, especially regarding the low-abundant bacteria. Accumulating evidence indicates substantial heterogeneity in the microbiology of endodontic infections among geographically diverse populations (16–18). It is unclear whether this heterogeneity is a manifestation of random variation or a reflection of genetic or environmental differences between different populations; nevertheless, it reinforces the importance of examining the endodontic infection microbiome among diverse populations, as this may open the door for possible optimization of intracanal antimicrobial protocols at a population- or individual level. The aim of the present study was to investigate the composition and diversity of bacterial population inhabiting both symptomatic and asymptomatic primary and persistent endodontic infections in a Greek population by using 16S amplicon pyrosequencing.
Materials and Methods
Participants’ recruitment and tooth selection
Participants were recruited from a private endodontic clinic in Athens, Greece, between January and June 2013. The study protocol was approved by the Ethics Committee of Athens University School of Dentistry, and a written informed consent was obtained from all study participants. Forty four adult patients aged 23 to 65 years comprised the study sample. A complete medical and dental history was obtained at the initial study visit. None of them had severe systemic illnesses, need for antimicrobial prophylaxis prior to treatment, or received antibiotic treatment during the three months preceding the initial examination. Teeth were excluded if they were cracked, the pulp chamber was exposed to the oral cavity, had periodontal pockets >4 mm and/ or had prosthodontic restorations.
The selected teeth had either a non-vital pulp or were endodontically treated at least 4 years previously. Radiographically, a periapical lesion was always present. Clinical signs and symptoms such as spontaneous pain or pain during mastication, tenderness to percussion, pain to palpation, mobility, presence of a sinus tract, and presence of localized or diffuse swelling were recorded.
A total of 48 teeth comprised the final sample and were classified in 4 groups according to their primary or secondary EI status and the presence of symptoms. The first 2 groups (primary EI) included 24 single or multi-rooted teeth, all with necrotic pulps (confirmed by cold and electric pulp sensibility tests) and radiographic evidence of apical periodontitis, characterized by bone destruction around the root apex. Thirteen teeth were diagnosed with acute apical periodontitis or acute apical abscess and thus were classified as symptomatic, whereas the remaining 11 teeth were diagnosed with chronic apical periodontitis and were classified as asymptomatic. Five of the 11 asymptomatic teeth had a preoperative sinus tract. Symptomatic patients were defined as those with spontaneous pain or moderate to severe pain to percussion or palpation of the involved tooth and/or had swelling. The remaining 2 groups (persistent EI) also included 24 single or multi-rooted endodontically-treated teeth with radiographic evidence of apical periodontitis. Thirteen teeth were diagnosed with clinical symptoms whereas the remaining 11 teeth were asymptomatic. Radiographic appearance of most endodontic treatments among the persistent infection group (79%) was of poor quality. In the majority of cases, termini of root canal fillings were 3–6mm short of the radiographic apex. In these cases, the root fillings were poorly compacted with no enlargement of the apical third of the canal. Nevertheless, all teeth showed intact coronal restorations with no direct exposure of the filling material to the oral cavity.
Microbiome sample collection and DNA isolation
Root canal microbial samples were obtained from each tooth by the first author (GT), an experienced endodontist. Strict aseptic conditions were maintained throughout the endodontic sampling procedure according to a protocol previously described by Siqueira and colleagues (19). Briefly, each tooth was initially cleansed with pumice and isolated with a rubber dam. The tooth and the surrounding field was then cleansed with 3% hydrogen peroxide and decontaminated with a 2.5% sodium hypochlorite (NaOCl) solution. Endodontic access was completed with a sterile high-speed carbide bur. After access completion and caries removal, the tooth, clamp and adjacent rubber dam were again disinfected with 2.5% NaOCl; 5% sodium thiosulfate was used for NaOCl inactivation. A small amount of sterile saline solution was introduced into the root canal by a 27G syringe (27G) (Ultradent, South Jordan, UT, USA), and the canal walls were filed as follows: initially, a K file No 10 (Dentsply Maillefer, Balaigues, Switzerland) was used to ensure apical patency with the aid of an electronic apex locator (Root-ZX, Morita, USA). Coronal pre-flaring was performed with SX ProTaper instrument (DentsplyMaillefer, Balaigues, Switzerland). A K-file No 15 was then introduced to the working length determined by using again electronic apex locator and a gentle filing motion was applied with files No 20, No 25 and No 30. Root canal irrigation with sterile saline solution was performed before sample collection. Subsequently, the root canal contents were absorbed into a minimum of 4 paper points. Each paper point was kept into the canal for at least 30 seconds. In multi-rooted teeth with more than one periradicular lesion, samples were taken from all root canals associated with apical periodontitis. The endodontic files with the handle cut off and the paper points were transferred to cryotubes containing TE buffer (10 mM Tris-HCl, 0.1mM EDTA, pH 7.6) and immediately frozen at −20°C.
In teeth with persistent EI, the coronal gutta-percha was removed using sterile Gates-Glidden burs (Dentsply Maillefer, Ballaigues, Switzerland) and the apical filling material was retrieved with K-type or Hedstrom files without use of chemical solvents. Instrumentation and sample collection was made as already described. When possible, filling material retrieved from the root canals was transferred to the TE buffer-containing cryotubes.
Total genomic DNA was extracted from root canal samples using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. A step of pre-incubation with lysozyme for 30 min was introduced to the protocol to ensure optimal DNA yield from Gram-positive bacteria. Before microbiome analysis, total DNA samples, including control samples obtained to verify the sterility of the working field, were quantified using the NanoDrop 2000 UV-Vis spectrophotometer at 260nm (Thermo Fisher Scientific Inc. Waltham, MA, USA).
16S amplicon pyrosequencing
Amplification of the hypervariable V1–V2 region of the bacterial 16S rRNA was performed on total DNA from 48 collected samples as previously described (20). Master mixes for PCR reactions contained the Qiagen Hotstar Hi-Fidelity Polymerase Kit (Qiagen, Valencia CA) with a forward primer composed of the Roche Titanium Fusion Primer A (5’-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3’), a 10bp Multiplex Identifier (MID) sequence (Roche, Indianapolis, IN), unique to each sample, and the universal bacterial primer 8F (5'-AGAGTTTGATCCTGGCTCAG-3') (21). The reverse primer was composed of the Roche Titanium Primer B (5’-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG -3’), the identical 10bp MID sequence as the forward primer, and the reverse bacterial primer 338R (5’-GCTGCCTCCCGTAGGAGT-3’) (22). The barcoded 16S rDNA amplicons (330nt) were pooled and sequenced on a 454 Genome Sequencer FLX Titanium instrument (Roche, Indianapolis, IN) in the Microbiome Core Facility (University of North Carolina, Chapel Hill, NC), using the GS FLX Titanium XLR70 sequencing reagents and protocols indicated by the manufacturer. Initial data analysis, base pair calling, and sequence trimming were performed by Research Computing at the University of North Carolina at Chapel Hill.
Sequencing data analysis
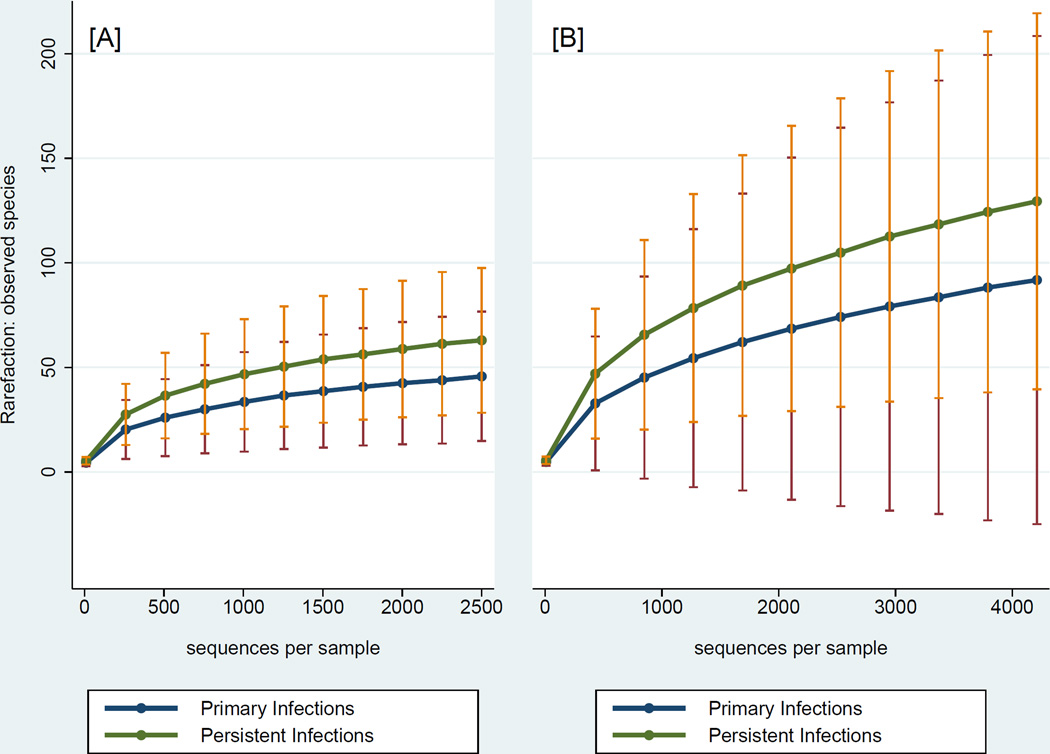
Bioinformatics analysis of bacterial 16S amplicon pyrosequencing data was carried out using the Quantitative Insights into Microbial Ecology (QIIME) software pipeline (23). Generated sequencing data plus metadata were de-multiplexed, filtered for quality control (sequences shorter than 150nt were discarded), and de-noised using Denoiser in QIIME (24). Sequences were aligned and clustered into Operational Taxonomic Units (OTUs) using UCLUST (25), and the Human Oral Microbiome Database (HOMD; http://database.oxfordjournals.org/cgi/content/full/2010/0/baq013) was used for taxonomy assignment of OTUs. After taxonomic assignment, sequences were aligned and phylogenetic trees were built (26). Rarefaction analyses were performed using a random selection of 2,500 sequences from each sample to ensure an even sampling depth (Fig. 1A). Alpha diversity estimates were calculated on rarefied OTU tables to determine Species richness (S), Shannon, Chao1, and Phylogenetic Diversity (PD) metrics. Beta diversity estimates were calculated within QIIME using weighted and unweighted UniFrac distances (27) between samples. To identify bacteria potentially not covered by HOMD, we used a second, generic database (Greengenes) (28) and a 4,218 sampling depth (Fig. 1B) and compared our findings using the 2 databases.
Figure 1.
Rarefaction curves illustrating the number of observed species-level OTUs and 95% confidence limits according to database and sampling depth. Panel A: Human Oral Microbiome Database (HOMD) and panel B: Greengenes database
Statistical analysis
Summary and descriptive statistics [mean, median, range, standard error, and 95% confidence intervals (CI)] were generated for all samples and according to the 4 groups of interest (i.e., combinations of primary vs. secondary infections and symptomatic vs. asymptomatic teeth) and presented using tabular and graphical means. Bacterial abundance (proportion of microbiome) and detection (proportion of samples positive) of phyla and genera overall, and across groups was examined using Wilcoxon’s rank-sum and Fisher’s Exact tests, using a conventional P<0.05 statistical significance threshold. Differences in bacterial diversity [Phylogenetic diversity (PD), observed species, Chao1, and Shannon indices] between the 4 EI groups were tested using a mixed-effects linear regression model, accounting for clustering of observations within samples and individuals, and applied a Bonferroni multiple-testing correction to account for multiple pairwise comparisons. To formally test between-group differences in microbial communities Analyses of Similarity (ANOSIM, n=10,000 permutations) were employed to calculate R and P values using the phylogeny-based unweighted UniFrac distance metric. Differences in bacterial community structures is reflected by high (closer to 1) R and low (less than 0.05) P values. Stata 13.1 (StataCorp LP, College Station, TX) was used for statistical analyses and generation of figures.
Results
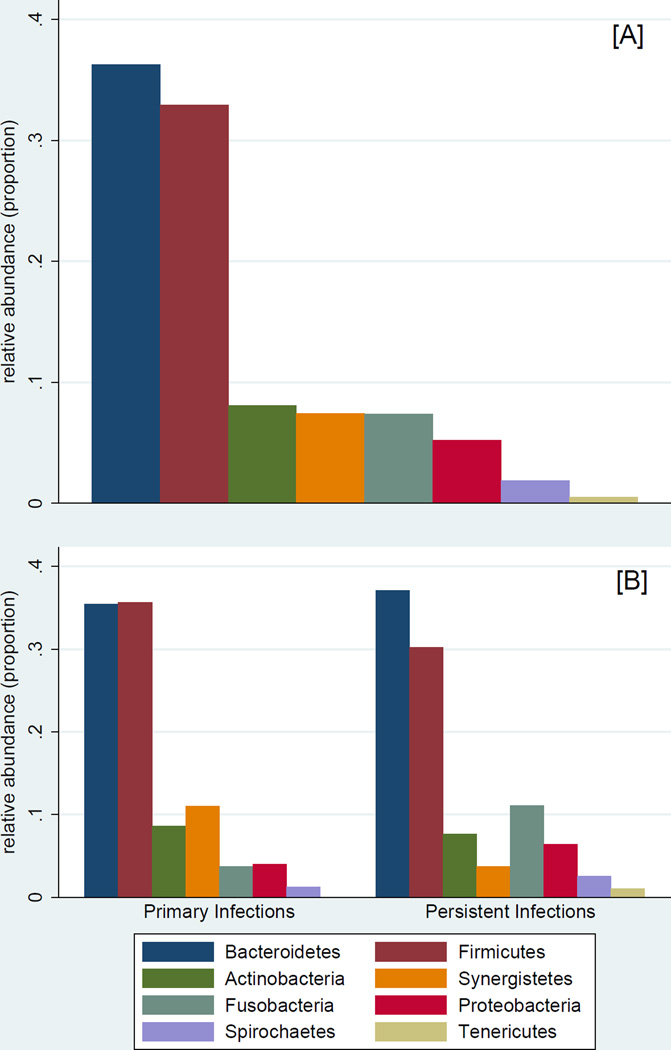
The study sample comprised 44 participants (mean age=43 years; 50% female) and 48 teeth with EI (Table 1). A total of 406,070 sequences were obtained from the 48 samples after quality filtering and de-noising corresponding to 8,460 reads per sample (range: 2,500–19,024). A total of 339 Operational Taxonomic Units (OTUs) were assigned to 11 phyla, 60 families, and 109 genera. Phyla with a representation of 0.5% or higher (relative abundance) are presented in Figure 2: Bacteroidetes (36.2%), Firmicutes (32.9%), Actinobacteria (8.1%), Synergistetes (7.4%), Fusobacteria (7.4%), Proteobacteria (5.2%), Spirochaetes (1.9%), and Tenericutes (0.5%). Identified phyla are presented in Table 2. Persistent infections were significantly enriched for Proteobacteria (6.4% vs. 4.0%; P=0.02) and Tenericutes (1.0% vs. <0.05%; P=0.03) compared to primary ones. Tenericutes were detected in 42% of persistent infections versus 12% of primary infections (P<0.05). Using the GG database, 18 additional less-abundant phyla were identified, all at less than 0.2% abundance. Among those, Cyanobacteria (0.018%) and Acidobacteria (0.007%) were the most abundant, and were significantly enriched among persistent infections: Cyanobacteria were detected in 67% of samples with an abundance of 0.3%; Acidobacteria were detected in 42% of samples with an abundance of 0.1%.
TABLE 1.
Clinical and History Information of the 48 Teeth Included in the Analytical Sample
| Entire sample | Initial treatment | Retreatment | |||
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | P* | ||
| 48 (100) | 24 (50) | 24 (50) | |||
| Edema | 0.60 | ||||
| no | 40 (83) | 21 (88) | 19 (79) | ||
| yes | 8 (17) | 3 (12) | 5 (21) | ||
| Spontaneous pain | 0.13 | ||||
| no | 31 (65) | 18 (75) | 13 (54) | ||
| yes | 17 (35) | 6 (25) | 11 (46) | ||
| Clinical symptoms | 1.00 | ||||
| no | 22 (46) | 11 (46) | 11 (46) | ||
| yes | 26 (54) | 13 (54) | 13 (54) | ||
| Previous treatment quality | . | ||||
| good | 2 (8) | . | 2 (8) | ||
| moderate | 3 (13) | . | 3 (13) | ||
| poor | 19 (79) | . | 19 (79) | ||
| Sinus tract | 0.08 | ||||
| no | 42 (88) | 19 (79) | 23 (96) | ||
| yes | 6 (12) | 5 (21) | 1 (4) | ||
| Tooth type | 0.54 | ||||
| Incisor/canine | 9 (19) | 6 (25) | 3 (12) | ||
| Premolar | 28 (58) | 13 (54) | 15 (63) | ||
| Molar | 11 (23) | 5 (21) | 6 (25) | ||
corresponding to X2 tests for categorical variables and t test for continuous variables
Figure 2.
Abundance of observed phyla with relative abundance of ≥0.5% in the entire sample (panel A) and according to endodontic infection type (panel B)
TABLE 2.
Abundance of phyla identified in the entire sample, and by endodontic infection type.
| Entire sample | Primary Infections | Persistent Infections | P | |||||
|---|---|---|---|---|---|---|---|---|
| Phyla | abundance | detected | abundance | detected | abundance | detected | A1 | D2 |
| Bacteroidetes | 0.362 | 1.00 | 0.354 | 1.00 | 0.371 | 1.00 | 0.8 | 1.0 |
| Firmicutes | 0.329 | 1.00 | 0.356 | 1.00 | 0.302 | 1.00 | 0.4 | 1.0 |
| Actinobacteria | 0.081 | 0.98 | 0.086 | 1.00 | 0.076 | 0.96 | 0.7 | 1.0 |
| Synergistetes | 0.074 | 0.81 | 0.110 | 0.92 | 0.037 | 0.71 | 0.4 | 0.1 |
| Fusobacteria | 0.074 | 0.88 | 0.037 | 0.83 | 0.111 | 0.92 | 0.09 | 0.7 |
| Proteobacteria | 0.052 | 0.88 | 0.040 | 0.79 | 0.064 | 0.96 | 0.02 | 0.2 |
| Spirochaetes | 0.019 | 0.83 | 0.012 | 0.83 | 0.025 | 0.83 | 0.1 | 1.0 |
| Tenericutes | 0.005 | 0.27 | <0.0005 | 0.12 | 0.010 | 0.42 | 0.03 | 0.05 |
| TM7 | 0.001 | 0.27 | 0.001 | 0.17 | 0.001 | 0.38 | 0.2 | 0.2 |
| Chloroflexi | <0.0005 | 0.19 | <0.0005 | 0.08 | 0.001 | 0.29 | 0.07 | 0.1 |
| SR1 | <0.0005 | 0.08 | <0.0005 | 0.08 | <0.0005 | 0.08 | 0.9 | 1.0 |
| other | 0.002 | 0.94 | 0.002 | 0.96 | 0.002 | 0.92 | 0.9 | 1.0 |
Comparison of abundance between primary and persistent infections, derived from Wilcoxon rank-sum tests
Comparison of number of samples where phylum was identified, derived from Fisher’s Exact tests
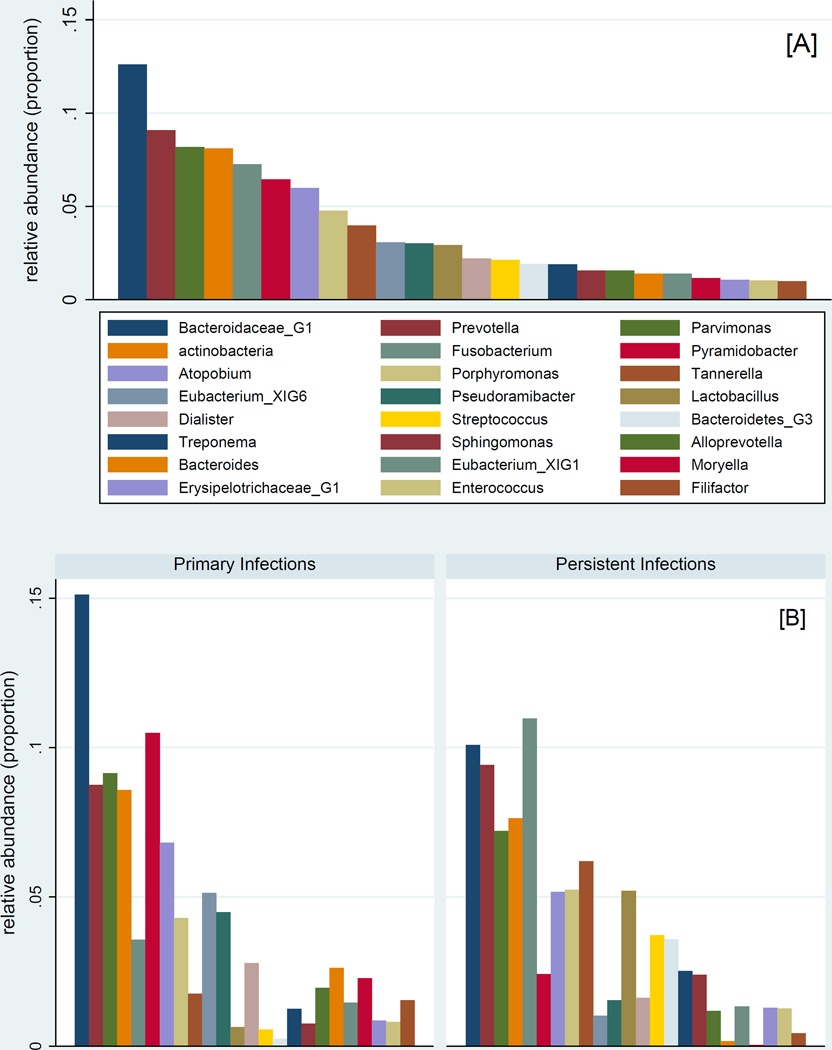
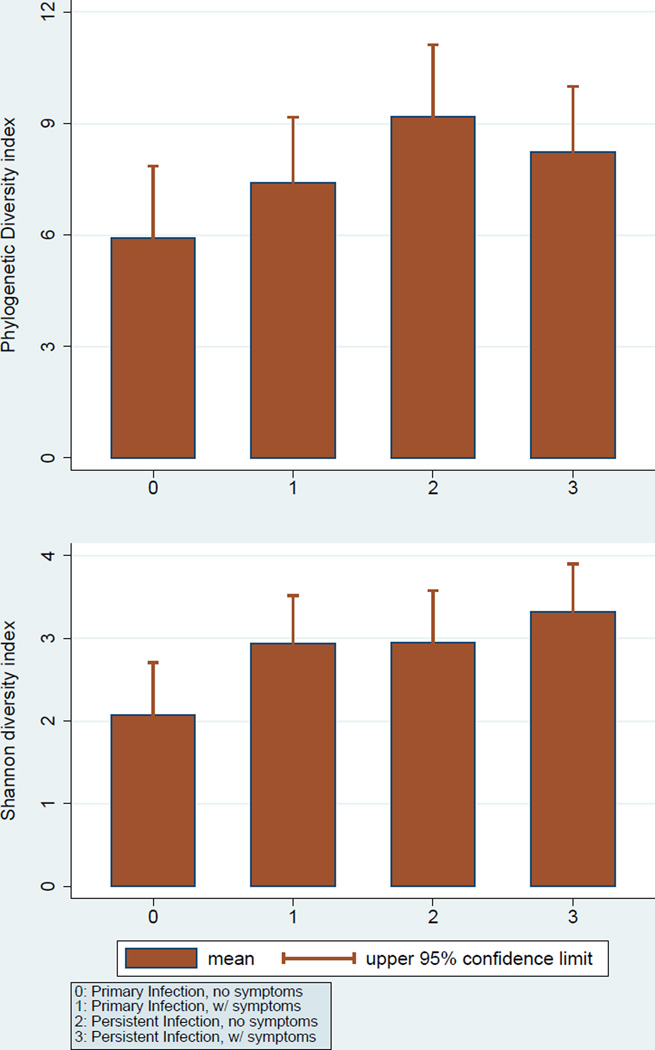
At the genus level, Bacteroidaceae_unclassified, Pyramidobacter, and Parvimonas were the most abundant in primary infections whereas Fusobacterium, Bacteroidaceae_unclassified, and Prevotella were the most abundant in teeth with persistent infections (Table 3). Significant differences were observed for 14 taxa (Figure 3), including increased enrichment of persistent infections for Lactobacillus, Streptococcus, Sphingomonas and Ralstonia (Table 3). In primary infections, symptomatic ones were more diverse that the asymptomatic ones; in persistent infections the opposite was found. Persistent infections showed higher Phylogenetic Diversity compared to primary infections (Table 4 and Figure 4).
TABLE 3.
Abundance of selected(abundant at >0.4%and those with statistically significant differences) genera(of 109 identified) in the entire sample, and by endodontic infection type.
| Genera | Entire sample |
Primary Infections | Persistent Infections | P* |
|---|---|---|---|---|
| Bacteroidaceae_G1 | 0.126 | 0.151 | 0.101 | 0.5 |
| Prevotella | 0.091 | 0.087 | 0.094 | 0.9 |
| Parvimonas | 0.082 | 0.091 | 0.072 | 0.3 |
| Actinobacteria | 0.081 | 0.086 | 0.076 | 0.7 |
| Fusobacterium | 0.073 | 0.036 | 0.110 | 0.08 |
| Pyramidobacter | 0.065 | 0.105 | 0.024 | 0.2 |
| Atopobium | 0.060 | 0.068 | 0.052 | 0.9 |
| Porphyromonas | 0.048 | 0.043 | 0.052 | 0.8 |
| Tannerella | 0.040 | 0.018 | 0.062 | 0.3 |
| Eubacterium_XIG6 | 0.031 | 0.051 | 0.010 | 0.6 |
| Pseudoramibacter | 0.030 | 0.045 | 0.015 | 0.3 |
| Lactobacillus | 0.029 | 0.006 | 0.052 | 0.005 |
| Dialister | 0.022 | 0.028 | 0.016 | 0.8 |
| Streptococcus | 0.021 | 0.006 | 0.037 | 0.003 |
| Bacteroidetes_G3 | 0.019 | 0.002 | 0.036 | 0.4 |
| Treponema | 0.019 | 0.012 | 0.025 | 0.1 |
| Sphingomonas | 0.016 | 0.007 | 0.024 | 0.002 |
| Alloprevotella | 0.016 | 0.020 | 0.012 | 0.01 |
| Bacteroides | 0.014 | 0.026 | 0.002 | 0.9 |
| Eubacterium_XIG1 | 0.014 | 0.015 | 0.013 | 0.7 |
| Moryella | 0.012 | 0.023 | <0.0005 | 0.002 |
| Erysipelotrichaceae_G1 | 0.011 | 0.009 | 0.013 | 0.4 |
| Enterococcus | 0.010 | 0.008 | 0.013 | 0.1 |
| Filifactor | 0.010 | 0.015 | 0.004 | 0.9 |
| Oribacterium | 0.009 | 0.013 | 0.005 | 0.5 |
| Fretibacterium | 0.008 | 0.005 | 0.011 | 0.8 |
| Olsenella | 0.008 | 0.012 | 0.003 | 0.3 |
| Solobacterium | 0.008 | 0.008 | 0.008 | 0.2 |
| Veillonella | 0.007 | 0.008 | 0.007 | 0.3 |
| Ralstonia | 0.007 | 0.001 | 0.013 | 0.001 |
| Rothia | 0.005 | <0.0005 | 0.009 | 0.2 |
| Mycoplasma | 0.005 | <0.0005 | 0.010 | 0.03 |
| Xanthomonadaceae>Other | 0.005 | 0.011 | <0.0005 | 0.4 |
| Neisseria | 0.004 | 0.003 | 0.005 | 0.1 |
| Propionibacterium | 0.004 | 0.002 | 0.007 | 0.08 |
| Caulobacter | 0.004 | 0.001 | 0.007 | 0.004 |
| Peptostreptococcaceae_XIG1 | 0.004 | 0.004 | 0.003 | 0.6 |
| Afipia | 0.003 | 0.002 | 0.004 | 0.05 |
| Pasteurellaceae>Other | 0.0005 | <0.0005 | 0.001 | 0.04 |
| Finegoldia | <0.0005 | 0 | .0001 | 0.02 |
| Catonella | <0.0005 | <0.0005 | 0.0006 | 0.02 |
| Enterobacter | <0.0005 | <0.0005 | 0.0005 | 0.03 |
| Campylobacter | <0.0005 | <0.0005 | 0.001 | 0.05 |
| Unassigned | 0.002 | 0.002 | 0.002 | 0.9 |
derived from Wilcoxon rank-sum tests
Figure 3.
Abundance of most abundant observed genera (≥1%) among the entire sample (panel A) and according to endodontic infection type (panel B)
TABLE 4.
Measures of alpha diversity (phylogenetic diversity, identified species, Chao1 and Shannon) in strata of primary and persistent endodontic infections, with and without symptoms, using an oral health-specific (HOMD) and a generic (Greengenes) database.
| Primary infections | Persistent infections | ||||
|---|---|---|---|---|---|
| Group A | Group B | Group C | Group D | Comp* | |
| Asymptomatic | Symptomatic | Asymptomatic | Symptomatic | ||
| HOMD | |||||
| Phylogenetic Diversity, mean (95% CI) | 5.9 (4.1–7.8) | 7.4 (5.2–9.6) | 9.2 (6.2–12.2) | 8.2 (6.6–9.9) | C>A1 |
| Species, median (range) | 28.0 (7–103) | 42.5 (16–152) | 64.5 (12–172) | 53.0 (21–97) | C>A1 |
| Chao1, mean (95% CI) | 54.5 (31.3–77.7) | 72.5 (46.7–98.3) | 95.1 (58.3–132) | 75.6 (58.3–92.9) | C>A1 |
| Shannon, mean (95% CI) | 2.08 (1.34–2.81) | 2.94 (2.28–3.59) | 2.95 (1.96–3.94) | 3.32 (2.85–3.78) | B>A1 D>A 2 |
| Greengenes database | |||||
| Phylogenetic Diversity, mean (95% CI) | 10.4 (7.2–13.5) | 14.5 (7.4–21.7) | 18.9 (10.5–27.3) | 15.4 (11.7–19.0) | C>A1 |
| Species, median (range) | 53 (13–171) | 69.5 (37–647) | 103 (16–378) | 99.5 (31–246) | |
| Chao1, mean (95% CI) | 103 (63.8–142) | 198 (5.4–390) | 214 (109–319) | 176 (122–230) | |
| Shannon, mean (95% CI) | 2.33 (1.55–3.12) | 3.38 (2.47–4.29) | 3.46 (2.21–4.71) | 3.44 (2.94–3.93) | C>A1 D>A1 |
Pairwise comparisons were based on a mixed-effects linear regression model accounting for clustering of observations within samples, and multiple comparison (Bonferroni)-corrected contrast of predicted marginal effects
Nominally significant difference (unadjusted P<0.05)
Bonferroni-corrected significant difference (adjusted P<0.05)
Figure 4.
Measures of bacterial diversity (Phylogenetic Diversity index, top panel; Shannon index, bottom panel) according to endodontic infection type and presence of symptoms
ANOSIM indicated statistically significant, albeit weak associations of infection type (R=0.087, P=0.005), symptoms (R=0.055, P=0.047), and combined strata (R=0.093, P=0.007) with UniFrac-assessed bacterial community composition.
Using the GG database, in primary infections, a total of 24 phyla and 280 genera were identified, whereas these numbers were 28 and 347, respectively in persistent infections. In primary infections, we identified on average 10 phyla, 50 genera, and 112 species-level phylotypes per sample, whereas these numbers were higher (12 phyla, 80 genera, and 162 species-level phylotypes) in teeth with persistent infections. ANOSIM indicated statistically significant but small-in-magnitude association (R=0.115, P=0.004) of bacterial composition and infection type according to GG database. Finally, bacteria classified as Elusimicrobia, OP3, OP8, Planctomycetes and WS5 were not detected in primarily infected canals whereas Gemmatimonadetes was the only phylum that was not found in endodontically-treated teeth with persistent infections. Several additional genera were detected using the GG database. Candidatus solibacter, Sharpea, Methylobacterium, Novosphingobium, and Jathinobacterium were the most abundant in this group.
Discussion
The present study investigated the bacterial diversity of primary and persistent endodontic infections in teeth with and without symptoms. To our knowledge, only three previous studies have explored the bacterial diversity of persistent infections using high throughput sequencing methods (14,15,29). The first was restricted to primary and persistent chronic asymptomatic cases (14), the second examined only persistent infections in asymptomatic and symptomatic teeth (15) whereas the third included a small number of samples with a secondary/persistent infection (29). Thus, this study is the first investigation of primary and persistent infections in both symptomatic and asymptomatic teeth. This is also the first pyrosequencing study contrasting results from two databases, HOMD and Greengenes.
In the present study, the paper point sampling technique was used because the examined teeth were to be retained in the oral cavity. Of the 8 pyrosequencing studies performed so far, 5 used the same technique for root canal sampling (7,10,13–15). The other three obtained samples either from periapical lesions after apical surgery (12), or cryo-pulverized root segments after teeth extraction (9,11). Obtaining cryo-pulverized root segments to study endodontic microbiota may offer advantages over the paper point technique; (30,31) however, this procedure necessitates tooth extraction and unless prosthodontic or orthodontic reasons warrant the latter, it is not suitable for the study of teeth that either have favorable prognosis or that could be retained after endodontic treatment.
In our study, Bacteroidetes was the most abundant phylum without significant differences between the two infection groups. In primary infections, Bacteroidetes and Firmicutes were found in equal abundance whereas Bacteroidetes were more abundant in persistent infections. These results are in agreement with results of other pyrosequencing studies performed in the US and Korea (7,13,14) but are in contrast with the findings of other Brazilian, Dutch and Sudanese studies, which found that the most abundant phyla were Firmicutes (10,15) and Proteobacteria (9,11). However, studies which have found Proteobacteria as the most abundant phylum used a different sampling methodology since they obtained their samples after tooth extraction or apical surgery. It is also likely that a possible geographic-related bacterial pattern may play a role for the observed differences. In the present study, Proteobacteria were found in lower abundance compared with results of earlier studies (9,11,12). However, our analysis revealed significant differences in the abundance of Proteobacteria and Tenericutes between persistent and primary infections. Root canals with persistent infections harbored significantly more Proteobacteria and Tenericutes than primarily infected canals. This is in contrast to a previous report (14) and, to the best of our knowledge, a novel finding based on pyrosequencing analyses. In addition, a tendency was detected for more Fusobacteria in persistent infections and more Synergistetes in primary infections. These differences however, were not statistically significant.
Our findings, especially from GG database, among the examined group of Greek patients coupled with previously reported evidence from pyrosequencing (7,9–15) and molecular broad-range studies (19,32,33) suggest a high bacterial diversity of endodontic infections. Also, our finding of higher bacterial diversity among persistent versus primary endodontic infections is a novel one, demonstrating that persistent endodontic infections are polymicrobial infections and not caused by a single or few pathogens. Because this finding could be a reflection of the poor quality previous endodontic treatments, these results require further validation and replication in future studies, among larger and more diverse patient populations.
Our results also showed that primary symptomatic infections tended to be more diverse than primary asymptomatic infections; in contrast persistent symptomatic infections were less diverse than persistent asymptomatic ones. Regarding primary infections, our results are compatible with the results of a previous similar study which has reported significant differences between asymptomatic and symptomatic cases (10). With regard to persistent infections, our results are consistent with findings of a recent pyrosequencing study of asymptomatic and symptomatic persistent infections where similar diversity was found, albeit it was not statistically significant except for Proteobacteria (15). Several plausible mechanistic explanations for the observed differences in diversity between symptom groups within infection type groups exist, including presence of keystone pathogens, virulent clonal types, bacterial interactions (34), and clinical/environmental conditions (i.e., restoration quality). It is also possible that symptoms are not linked to bacterial diversity. While no definitive answer is possible with current knowledge, this is an important area for future studies. The complex interplay between clinical/environmental conditions and the endodontic microbiome may be the key to understand the transition from asymptomatic to symptomatic states, and vice versa.
In the present study, Dialister, Erysipelotrichaceae_G1 and Peptostreptococcaceae_X1G4 were found in symptomatic persistent infections in a statistically higher relative abundance in relation to the asymptomatic state. The latter two genera are as-yet uncultivated phylotypes. With regard to persistent infections, it is well-known that environmental conditions are adversely modified for microbes in root canal-treated teeth. Under these conditions (pH change, substrate availability and nutritional supply, bacterial resistance), it can be argued that some resistant and fast-growing microorganisms proliferate in symptomatic infections against others which are slow-growing or their growth is inhibited by metabolic byproducts of faster-growing microorganisms, thereby decreasing the relative diversity related to the presence or absence of taxa.
At the genus level, Bacteroidaceae_unclassified, Prevotella, Parvimonas, Atopobium and Porphyromonas, were all found in relatively high abundance both in primary and persistent infections, in agreement with previous studies (7,10–15). Pyramidobacter was found in high abundance only in primary infections whereas Fusobacterium, Tannerella, and Lactobacillus were detected in high abundance in persistent infections which are consistent with the findings of previous similar studies (9,14,15). The difference for Lactobacillus was statistically significant. The above findings for Fusobacterium possibly suggest that its role on the development, maintenance and relapse of periapical disease may have been underestimated. Interestingly also, the abundance of Tannerella was relatively higher in persistent compared to primary infections. This is a notable finding, considering that Tannerella is a Gram (−) obligate anaerobe which has been associated with symptomatic cases of primary infections (35). Enterococcus which has been found to be the most frequently isolated microorganism in root-filled teeth with periapical lesions was detected in a notably low abundance. This finding is in agreement with the results of previous studies using pyrosequencing and gene clone library analysis (14,15,32,33) suggesting probably a previous overestimation of its role in treatment failure.
In conclusion, the present pyrosequencing study offers a novel, detailed characterization of the endodontic microbiome both in primary and persistent infections. These results suggest a high bacterial diversity of endodontic infections and a more diverse bacterial community profile in persistent versus primary infections. Using the GG database, a substantial number of microorganisms was not possible to be taxonomically classified and may be associated with the development of apical periodontitis. Further endodontic microbiome studies are warranted to identify and characterize these microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
Financial support was provided by the University of North Carolina-Chapel Hill School of Dentistry start-up funds. The Microbiome Core Facility is supported in part by the NIH/National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK34987.
I affirm that We have no financial affiliation (e.g., employment, direct payment, stock holdings, retainers, consultantships, patent licensing arrangements or honoraria), or involvement with any commercial organization with direct financial interest in the subject or materials discussed in this manuscript, nor have any such arrangements existed in the past three years.
Footnotes
The authors deny any conflicts of interest related to this study.
No other potential conflict of interest is disclosed.
References
- 1.Kirkevang LL, Vaeth M, Hörsted-Bindslev P, et al. Risk factors for developing apical periodontitis in a general population. Int Endod J. 2007;40:290–299. doi: 10.1111/j.1365-2591.2007.01224.x. [DOI] [PubMed] [Google Scholar]
- 2.Moreno JO, Alves FR, Goncalves LS, et al. Periradicular status and quality of root canal fillings and coronal restorations in an urban Colombian population. J Endod. 2013;39:600–604. doi: 10.1016/j.joen.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Siqueira JF, Jr, Rocas IN. Diversity of endodontic microbiota revisited. J Dent Res. 2009;88:969–981. doi: 10.1177/0022034509346549. [DOI] [PubMed] [Google Scholar]
- 4.Munson MA, Pitt-Ford T, Chong B, et al. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res. 2002;81:761–766. doi: 10.1177/0810761. [DOI] [PubMed] [Google Scholar]
- 5.Siqueira JF, Jr, Rocas IN, Rosado AS. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of endodontic infections. J Endod. 2005;31:775–782. doi: 10.1097/01.don.0000155221.33667.bb. [DOI] [PubMed] [Google Scholar]
- 6.Siqueira JF, Jr, Fouad AF, Rocas IN. Pyrosequencing as a tool for better understanding of human microbiomes. J Oral Microbiol. 2012;4:1–15. doi: 10.3402/jom.v4i0.10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Hsiao WW, Nandakumar R, et al. Analyzing endodontic infections by deep coverage pyrosequencing. J Dent Res. 2010;89:980–984. doi: 10.1177/0022034510370026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored rare biosphere. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siqueira JF, Jr, Alves FR, Rocas IN. Pyrosequencing analysis of the apical root canal microbiota. J Endod. 2011;37:1499–1503. doi: 10.1016/j.joen.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Santos LA, Siqueira JF, Jr, Rocas IN, et al. Comparing the Bacterial Diversity of Acute and Chronic Dental Root Canal Infections. PLoS ONE. 2011;6:e28088. doi: 10.1371/journal.pone.0028088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozok AR, Persoon IF, Huse SM, et al. Ecology of the microbiome of the infected root canal system: a comparison between apical and coronal root segments. Int Endod J. 2012;45:530–541. doi: 10.1111/j.1365-2591.2011.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saber MH, Schwarzberg K, Alonaizan FA, et al. Bacterial flora of dental periradicular lesions analyzed by the 454-pyrosequencing technology. J Endod. 2012;38:1484–1488. doi: 10.1016/j.joen.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 13.Hsiao WW, Li KL, Liu Z, et al. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genomics. 2012;13:345. doi: 10.1186/1471-2164-13-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong BY, Lee TK, Lim SM, et al. Microbial analysis in primary and persistent endodontic infections by using pyrosequencing. J Endod. 2013;39:1136–1140. doi: 10.1016/j.joen.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Anderson AC, Al-Ahmad A, Elamin F, et al. Comparison of the bacterial composition and structure in symptomatic and asymptomatic endodontic infections associated with root-filled teeth using pyrosequencing. PLoS One. 2013;8:e84960. doi: 10.1371/journal.pone.0084960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumgartner JC, Siqueira JF, Jr, Xia T, et al. Geographical differences in bacteria detected in endodontic infections using polymerase chain reaction. J Endod. 2004;30:141–144. doi: 10.1097/00004770-200403000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Rocas IN, Baumgartner JC, Xia T, et al. Prevalence of selected bacterial named species and uncultivated phylotypes in endodontic abscesses from two geographic locations. J Endod. 2006;32:1135–1138. doi: 10.1016/j.joen.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Machado de Oliveira JC, Siqueira JF, Jr, et al. Bacterial community profiles of endodontic abscesses from Brazilian and USA subjects as compared by denaturing gradient gel electrophoresis analysis. Oral Microbiol Immunol. 2007;22:14–18. doi: 10.1111/j.1399-302X.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- 19.Siqueira JF, Jr, Rocas IN, Rosado AS. Investigation of bacterial communities associated with asymptomatic and symptomatic endodontic infections by denaturing gradient gel electrophoresis fingerprinting approach. Oral Microbiol Immunol. 2004;19:363–370. doi: 10.1111/j.1399-302x.2004.00170.x. [DOI] [PubMed] [Google Scholar]
- 20.Devine AA, Gonzalez A, Speck KE, et al. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS One. 2013;8:e73140. doi: 10.1371/journal.pone.0073140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards U, Rogall T, Blocker H, et al. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierer N, Hamady M, Lauber CL, et al. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vengerfeldt V, Spilka K, Saag M, et al. Highly diverse microbiota in dental root canals in cases of apical periodontitis (Data of Illumina Sequencing) J Endod. 2014;40:1778–1783. doi: 10.1016/j.joen.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Alves FR, Siqueira JF, Jr, Carmo FL, et al. Bacterial community profiling of cryogenically ground samples from the apical and coronal root segments of teeth with apical periodontitis. J Endod. 2009;35:486–492. doi: 10.1016/j.joen.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 31.Rocas IN, Alves FR, Santos AL, et al. Apical root canal microbiota as determined by reverse-capture checkerboard analysis of cryogenically ground root samples from teeth with apical periodontitis. J Endod. 2010;36:1617–1621. doi: 10.1016/j.joen.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Zakaria MN, Takeshita T, Shibata Y, et al. Microbial community in persistent apical periodontitis: a 16S rRNA gene clone library analysis. Int Endod J. 2014 doi: 10.1111/iej.12361. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 33.Sakamoto M, Siqueira JF, Jr, Rocas IN, et al. Molecular analysis of the root canal microbiota associated with endodontic treatment failures. Oral Microbiol Immunol. 2008;23:275–281. doi: 10.1111/j.1399-302X.2007.00423.x. [DOI] [PubMed] [Google Scholar]
- 34.Siqueira JF, Jr, Rôças IN. Microbiology and treatment of acute apical abscesses. Clin Microbiol Rev. 2013;26:255–273. doi: 10.1128/CMR.00082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassone LM, Fidel RA, Faveri M, et al. A microbiological profile of symptomatic teeth with primary endodontic infections. J Endod. 2008;34:541–545. doi: 10.1016/j.joen.2008.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.