SUMMARY
Synaptic adhesion molecules regulate diverse aspects of synapse development and plasticity. SALM3 is a PSD-95-interacting synaptic adhesion molecule known to induce presynaptic differentiation in contacting axons, but little is known about its presynaptic receptors and in vivo functions. Here, we identify an interaction between SALM3 and LAR family receptor protein tyrosine phosphatases (LAR-RPTPs) that requires the mini-exon B splice insert in LAR-RPTPs. In addition, SALM3-dependent presynaptic differentiation requires all three types of LAR-RPTPs. SALM3 mutant (Salm3−/−) mice display markedly reduced excitatory synapse number but normal synaptic plasticity in the hippocampal CA1 region. Salm3−/− mice exhibit hypoactivity in both novel and familiar environments but perform normally in learning and memory tests administered. These results suggest that SALM3 regulates excitatory synapse development and locomotion behavior.
Graphical Abstract
In Brief
SALM3 is a postsynaptic adhesion molecule known to regulate synapse development, but the underlying mechanism remains unclear. Li et al. find that SALM3 interacts with presynaptic LAR family receptor protein tyrosine phosphatases (LAR-RPTPs) in a splicing-dependent manner. In addition, they show that SALM3-mutant mice display reduced excitatory synapse number and hypoactivity.
INTRODUCTION
Synaptic adhesion molecules regulate aspects of synapse formation, maturation, and plasticity. Recent studies have identified a large number of synaptogenic adhesion molecules, including neuroligins and neurexins (Biederer and Stagi, 2008; Dalva et al., 2007; de Wit and Ghosh, 2014; de Wit et al., 2011; Krueger et al., 2012; Missler et al., 2012; Shen and Scheiffele, 2010; Siddiqui and Craig, 2011; Song and Kim, 2013; Südhof, 2008; Takahashi and Craig, 2013; Um and Ko, 2013; Valnegri et al., 2012; Yamagata et al., 2003; Yuzaki, 2011). These molecules regulate synapse development in different spatiotemporal contexts. Although our understanding of these functions is rapidly increasing, many important questions still remain to be addressed. For instance, the types of trans-synaptic adhesions and the mechanisms underlying their functions are incompletely understood. Moreover, the available evidence is insufficient to establish the relative importance of currently identified trans-synaptic adhesion types and mechanisms in vivo.
SALM (also known as Lrfn) encompasses a family of leucinerich repeat (LRR)-containing synaptic cell adhesion molecules (Ko et al., 2006; Morimura et al., 2006; Nam et al., 2011; Wang et al., 2006). There are five known members in the family: SALM1/Lrfn2, SALM2/Lrfn1, SALM3/Lrfn4, SALM4/Lrfn3, and SALM5/Lrfn5. SALMs share a similar domain structure consisting of six LRRs flanked by NTLRR and CTLRR domains, an immunoglobulin (Ig) domain, a fibronectin III (FNIII) domain, a transmembrane domain, and a C-terminal PDZ-binding motif; the latter is present in SALMs 1–3, but not in SALMs 4 and 5. However, with the exception of C-terminal PDZ-binding motifs in SALMs 1–3, the cytoplasmic regions of SALMs differ in length and amino acid (aa) sequence, suggesting that SALMs may have distinct functions.
Indeed, a wealth of data supports this presumption. For instance, SALMs 3 and 5, but not other SALMs, induce presynaptic differentiation in contacting axons (Mah et al., 2010). In addition, SALM1 interacts with and clusters NMDA (N-methyl-D-aspartate) receptors (NMDARs), but not AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors (AMPARs) (Wang et al., 2006). In contrast, SALM2 associates with both NMDARs and AMPARs (Ko et al., 2006). Moreover, SALMs 1–3 form heteromeric complexes with each other, whereas SALMs 4 and 5 do not (Seabold et al., 2008), but SALMs 4 and 5, unlike SALMs 1–3, can form trans-cellular and homomeric complexes (Seabold et al., 2008). To date, however, none of the SALMs has been characterized in knockout mice; thus, in vivo data supporting this functional diversity are lacking. In addition, the presynaptic ligands that mediate SALM3/5-dependent presynaptic differentiation have not been identified.
The leukocyte common antigen-related (LAR) subfamily of receptor protein tyrosine phosphatases (LAR-RPTPs), consisting of the three members, LAR (PTPRF), PTPσ (PTPRS), and PTPδ (PTPRD), has been implicated in the organization of synapse development (Takahashi and Craig, 2013; Um and Ko, 2013). Early studies on LAR-RPTP homologs in Drosophila and C. elegans have implicated LAR in the regulation of axon guidance and presynaptic development (Ackley et al., 2005; Johnson and Van Vactor, 2003; Stryker and Johnson, 2007). Studies on mammalian LAR-RPTPs have demonstrated that they regulate dendrite and excitatory synapse development through postsynaptic mechanisms (Dunah et al., 2005; Hoogenraad et al., 2007) in addition to their reported regulation of axon development.
More recent studies have identified several LAR-RPTP-inter-acting postsynaptic adhesion molecules, including NGL-3, TrkC, IL1RAPL1, IL-1RAcP, and Slitrks (Slitrk1–6) (Takahashi et al., 2011, 2012; Valnegri et al., 2011; Woo et al., 2009; Yim et al., 2013; Yoshida et al., 2011, 2012). These trans-synaptic interactions regulate both excitatory and inhibitory synapse development, suggesting that LAR-RPTPs act as presynaptic organizers. Moreover, LAR-RPTPs display differential distribution patterns in various brain regions (Kwon et al., 2010), and alternative splice inserts in LAR-RPTPs differentially regulate their trans-synaptic interactions with postsynaptic adhesion molecules (Takahashi et al., 2011; Yoshida et al., 2011, 2012), adding additional layers of complexity. These results highlight the importance of comprehensively identifying and characterizing existing LAR-RPTP-related trans-synaptic interactions and the underlying mechanisms.
Defects in SALMs and LAR-RPTPs have been implicated in diverse neuropsychiatric disorders, providing support for the clinical importance of these proteins. Specifically, SALM1/Lrfn2 and SALM5/Lrfn5 are associated with autism spectrum disorders and intellectual disability (de Bruijn et al., 2010; Mikhail et al., 2011; Voineagu and Yoo, 2013; Wang et al., 2009), as well as schizophrenia (Xu et al., 2009). In addition, PTPδ has been linked to autism spectrum disorders (Pinto et al., 2010), attention deficit/hyperactivity disorder (Elia et al., 2010), bipolar disorder (Malhotra et al., 2011), and restless leg syndrome (Schormair et al., 2008; Yang et al., 2011). Most recently, LAR has been implicated in schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014).
In the present study, we demonstrated that SALM3 trans-synaptically interacts with all three types of presynaptic LAR-RPTPs in an alternative splicing-dependent manner. Salm3−/− mice show a reduced excitatory synapse number but normal synaptic plasticity in the hippocampus. Behaviorally, Salm3−/− mice show hypoactivity, but no abnormalities in the learning and memory tests administered.
RESULTS
SALM3 Interacts with LAR-RPTPs
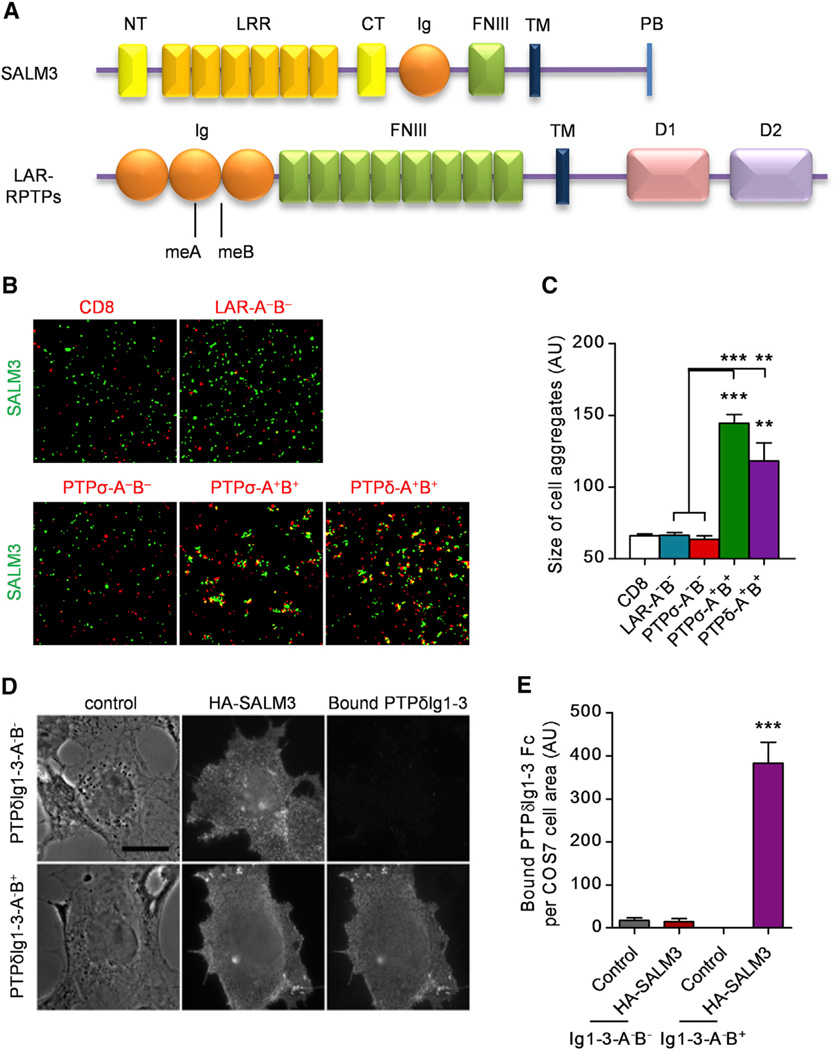
To identify presynaptic ligands for SALM3, we performed cell-aggregation assays in which one group of L cells expressing SALM3 was mixed with another expressing candidate synaptic adhesion molecules or surface membrane proteins (Figure S1). We found that SALM3-expressing cells coaggregated with cells expressing PTPσ and PTPδ, two known members of the LAR-RPTP family.
In subsequent quantitative cell aggregation experiments, SALM3 interacted with selected LAR-RPTPs (Figures 1A–1C). Specifically, PTPσ and PTPδ containing the two mini exons, meA and meB (PTPσ-A+B+ and PTPδ-A+B+), in the N-terminal Ig domain region (Ig1–3) interacted more strongly with SALM3 than the negative control CD8, but the same PTPσ construct and a similar LAR construct lacking both exons (PTPσ-A−B− and LAR-A−B−) did not. These results suggest that SALM3 may interact with LAR-RPTPs in a splice variant-dependent manner.
Figure 1. SALM3 Interacts with LAR-RPTPs.
(A) Domain structures of SALM3 and LAR-RPTPs (LAR, PTPσ, and PTPδ). LRR, leucine rich repeat; NT/CT, N/C-terminal domain; FNIII, fibronectin type-III; TM, transmembrane; PB, PDZ domain binding; D1/D2, phosphatase domain.
(B and C) SALM3 interacts with a subset of LAR-RPTPs in cell aggregation assays. A group of L cells expressing SALM3 (pDisplay) was mixed with another expressing the indicated LAR-RPTPs (pDisplay) for cell aggregation. Scale bar represents 100 µm. Means ± SEM (n = 3 for CD8 [control], LAR, PTPσ, and PTPδ, **p < 0.01, ***p < 0.001, ANOVA).
(D and E) SALM3 interacts with PTPδ in a cell-based binding assay. COS7 cells expressing HA-SALM3 were incubated with PTPδIg1-3-Fc A−B− or A−B+ and assessed for bound Fc fusion protein. Scale bar represents 10 µm (n = 20 cells each from two independent experiments, ANOVA p < 0.0001 and ***p < 0.001 by Bonferroni posthoc test compared with neighbor non-transfected cells). See also Figure S1 and Table S1.
In addition to this candidate approach, we performed an unbiased proteomics screen for PTPδ binding proteins. We used the full ectodomain of PTPδ fused to human Fc (PTPδ-Fc, A+B+ form) or just the N-terminal three Ig domains (PTPδIg1–3-Fc, a mix of A−B− and A−B+ forms) as bait in a pull-down assay with rat brain crude synaptosomal fraction. Bound material was analyzed by mass spectrometry. Proteins isolated by high salt elution with both PTPδ-Fc and PTPδIg1–3-Fc, but not with the neurexin ectodomain neurexin1 β-Fc as a negative control, are listed in Table S1. Prominent in this list of putative interacting proteins was SALM3/Lrfn4. Another protein previously shown to interact with PTPδ, IL1RAPL1 (Yoshida et al., 2011; Valnegri et al., 2011), was also isolated by subsequent low pH elution.
To confirm the interaction with SALM3 and differentiate among the PTPδIg1–3 forms, we expressed SALM3 with an extracellular HA tag, HA-SALM3, on the surface of COS7 cells and performed a cell-based binding assay. PTPδIg1–3-Fc A−B+ form, but not A−B− form, bound to cells expressing HA-SALM3 (Figures 1D and 1E). These results further support the interpretation that SALM3 interacts with LAR-RPTPs in a splice variant-dependent manner and show that the Ig1–3 domains of PTPδ are sufficient for interaction.
Mini-Exon B in LAR-RPTPs Determines SALM3 Binding
To more systematically test the possibility that meA and meB splice inserts in LAR-RPTPs regulate interaction with SALM3, we performed cell-based binding assays in the reverse orientation. We incubated the SALM3 ectodomain fusion protein SALM3-Fc with COS7 cells expressing PTPσ, PTPδ, or LAR containing or lacking meA and/or meB. All of these LAR-RPTPs show good cell surface expression patterns (Yoshida et al., 2011), and all the PTPσ isoforms bind TrkC-Fc (Takahashi et al., 2011).
We found that LAR-RPTPs containing meB alone (A−B+) exhibited significant SALM3 binding, whereas SALM3 binding was undetectable for LAR-RPTPs containing only meA (A+B−) or lacking both meA and meB (A−B−) (Figures 2A and 2B), indicating that meB alone is sufficient for SALM3 binding. However, the addition of meA to meB (A+B+) significantly increased SALM3 binding, suggesting that meA in the presence of meB substantially increases SALM3 binding. These results indicate that SALM3 binds to meB-containing forms of PTPσ, PTPδ, and LAR and that meA exerts a modulatory effect.
Figure 2. The meB Splice Insert in Ig Domains of LAR-RPTPs Is Sufficient for SALM3 Binding, whereas meA Is Modulatory.
(A and B) SALM3 Fc protein was incubated with COS7 cells expressing full-length PTPσ, PTPδ, or LAR with or without meA and meB splice inserts together with YFP to mark transfected cells (meA+, nine residue insert for PTPσ and PTPδ, six residue insert for LAR). Scale bar represents 10 µm. Means ± SEM (n = 20 cells each from two independent experiments, ANOVA p < 0.0001 and *p < 0.05, **p < 0.01, ***p < 0.001 by Bonferroni posthoc test compared with YFP alone, †p < 0.05 and †††p < 0.001 by Bonferroni post hoc test comparing A−B+ with A+B+ for individual RPTPs).
(C) Amino acid sequence alignment and relative abundance of LAR-RPTP splice variants expressed in the rat hippocampus (P12), determined by RT-PCR and DNA sequencing of 50 randomly chosen independent clones for each protein.
To explore in vivo relevance of these findings, we determined the types and relative abundance of LAR-RPTP splice variants expressed in the hippocampus, a brain region frequently used in coculture synapse formation assays (see below). When mRNA samples from rat hippocampi at postnatal day 12 (P12) were subjected to RT-PCR and DNA sequencing, we found that the hippocampus expresses LAR-RPTP splice variants in abundances that are similar to those obtained the whole mouse brain at P11 (Yoshida et al., 2011), with the exception that the PTPδ variants missing the A splice insert (PTPδA−B+ and PTPδA−B−) are more abundant in the hippocampus (58%) than in the whole brain (10%) (Figure 2C). The relative abundances of LAR-RPTP variants containing the splice B insert, a key determinant of SALM3 interaction, were 38%, 80%, and 10% for PTPσ, PTPδ, and LAR, respectively, which are largely comparable to the 42%, 96%, and 10% reported for the mouse whole brain (Yoshida et al., 2011). These data suggest that mini exon B-dependent adhesions may be more important in PTPδ than in PTPσ or LAR in rats and mice, although these are the differences at the mRNA level.
SALM3-Dependent Synaptogenesis Requires All Three LAR-RPTPs
Next, we tested which LAR-RPTPs are critical for SALM3-dependent presynaptic differentiation. To this end, we cocultured heterologous cells expressing SALM3 with hippocampal neurons expressing knockdown constructs for individual LAR-RPTPs, LAR (Figure S2) (Mander et al., 2005), PTPσ (Figure S2) (current study), and PTPδ (Takahashi et al., 2012), or all three LAR-RPTPs. These cocultured neurons were then stained for synapsin I as a measure of presynaptic differentiation, which has been associated with the positive uptake of synaptotagmin luminal domain antibodies, indicative of its functionality (Mah et al., 2010). In triple knockdown of LAR-RPTPs, SALM3-induced synapsin I clustering was decreased by 85.2% ± 3.0%, suggesting that LAR-RPTPs play a major role in SALM3-dependent presynaptic differentiation. Knockdown of individual LAR-RPTPs revealed a rank order of contribution of PTPδ > LAR > PTPσ. However, the differences were small, indicating that efficient SALM3-induced synapsin I clustering requires all three LAR-RPTPs (Figures 3A and 3C). In control experiments, presynaptic differentiation induced by neuroligin 2 was not inhibited by knockdown of LAR-RPTPs (Figures 3B and 3D).
Figure 3. SALM3-Dependent Presynaptic Differentiation Requires All Three LAR-RPTPs.
(A and C) SALM3-expressing COS7 cells were co-cultured with hippocampal neurons (DIV 10–11) nucleofected to express individual knockdown constructs of LAR-RPTPs (sh-LAR, PTPσ, PTPδ), or all three (TKD), followed by monitoring of presynaptic clustering of synapsin I (a presynaptic marker) as a measurement of presynaptic differentiation. Means ± SEM (n = 28 for sh-control, 26 for sh-LAR, 28 for sh-PTPσ, 25 for sh-PTPδ, and 26 for TKD from three independent experiments, ANOVA p < 0.0001 and **p < 0.01, ***p < 0.001 by Bonferroni post hoc test compared with the control sh-control).
(B and D) In control experiments, neuroligin 2 (NL2)-dependent presynaptic differentiation is not affected by LAR-RPTP knockdown (n = 29 for sh-control, 22 for sh-LAR, 27 for sh-PTPσ, 25 for sh-PTPδ, and 29 for TKD from three independent experiments, ANOVA p > 0.05). Scale bar represents 10 µm.
See also Figure S2.
Generation and Characterization of Salm3−/− Mice
In order to explore the in vivo functions of SALM3, we generated Salm3−/− mice in which all SALM3-coding exons (exons 2 and 3) were replaced with a β-geo cassette (β-galactosidase/neomycin fusion) (Figures 4A and 4B). SALM3 protein was undetectable in immunoblots of Salm3−/− whole-brain lysates (Figure 4C).
Figure 4. Generation and Characterization of Salm3−/− Mice.
(A) Strategy to generate Salm3−/− mice.
(B) Genotyping of Salm3−/− mice by PCR. WT, Salm3+/+; heterozygous (HT), Salm3+/−; knockout (KO), Salm3−/−.
(C) Absence of SALM3 proteins in Salm3−/− whole brain lysates, revealed by immunoblot analysis with two different SALM antibodies (1929 and 1816).
(D) SALM3 expression patterns visualized by X-gal staining of Salm3+/− brain slices. Ctx, cortex; Hp, hippocampus; Str, striatum; Ob, olfactory bulb; Cb, cerebellum. Scale bar represents 1 mm.
(E–G) Normal gross morphology of the brain (E) and neuron number in Salm3−/− mice, as revealed by staining for NeuN (a neuronal marker) (F and G; hippocampal CA1 pyramidal neurons). Scale bar represents 0. 252 mm. Means ± SEM (n = 3; ns, not significant; Student’s t test).
(H) Salm3−/− brain displays normal levels of synaptic proteins, including other SALMs, and other synaptogenic adhesion molecules (NGL-3, Slitrk1/3, IL1RAPL1, and IL-1RAcP) in the whole brain (WB) and hippocampus (Hp) (n = 3; ns, not significant; Student’s t test). See also Figure S3.
SALM3 expression, visualized by X-gal staining, was detected in multiple brain regions, including the hippocampus, cortex, striatum, olfactory bulb, and cerebellum (Figures 4D and S3). In the hippocampus, SALM3 expression was stronger in the CA1 and CA3 subfields than in the dentate gyrus region.
The Salm3−/− brain exhibited normal gross morphology and neuronal numbers, as revealed NeuN (neuronal marker) staining (Figures 4E–4G). SALM3 deletion had no effect on the expression levels of diverse synaptic proteins in the whole brain or in the hippocampus, including other SALMs (SALM1/2/4/5) and synaptogenic adhesion molecules (NGL-3, Slitrk1, Slitrk3, IL1RAPL1, and IL-1RAcP) (Figure 4H).
Decreased Excitatory Synapse Number in Salm3−/− Neurons
Given that SALM3 can induce excitatory and inhibitory presynaptic differentiation in contacting axons in fibroblast-neuron coculture assays (Mah et al., 2010), we tested whether neuronal synapse number or function was altered in Salm3−/− mice. We found a substantial decrease (~52.3%) in the frequency, but not the amplitude, of miniature excitatory postsynaptic currents (mEPSCs) in Salm3−/− CA1 pyramidal neurons compared with WT neurons (Figures 5A–5C). In contrast, neither the frequency nor the amplitude of miniature inhibitory postsynaptic currents (mIPSCs) was altered (Figures 5D–5F).
Figure 5. Reduced mEPSC Frequency and Excitatory Synapse Number but Normal Paired Pulse Ratio, AMPA-NMDA Ratio, and Synaptic Plasticity in the Salm3−/− Hippocampus.
(A–C) Reduced frequency but normal amplitude of mEPSCs in Salm3−/− hippocampal CA1 pyramidal neurons (P17–21). Means ± SEM (n = 14 for WT and KO, **p < 0.01; ns, not significant; Student’s t test).
(D–F) Normal mIPSC frequency and amplitude in Salm3−/− CA1 pyramidal neurons (P17–21) (n = 14 for WT and 10 for KO, ns, not significant, Student’s t test).
(G–K) Salm3−/− mice (P14) show reduced PSD (arrows) density (G and H), but normal PSD perforation (G and I; arrowheads), thickness (J), and length (K) in EM analysis of hippocampal CA1 region (stratum radiatum). Scale bar represents 500 nm (n = 3 for WT and KO, *p < 0.05; ns, not significant; Student’s t test).
(L–N) SALM3 expression in Salm3−/− hippocampal CA1 pyramidal neurons by lentiviral gene delivery (P9/10-23/24; an example shown on the left) normalizes the reduced mEPSC frequency, whereas a control virus (EGFP alone) has no effect (n = 16 for WT-EGFP, 13 for KO-EGFP, 12 for WT-EGFP, and 12 for KO-EGFP, **p < 0.01; ns, not significant; two-way ANOVA).
(O) Normal paired pulse facilitation at Salm3−/− SC-CA1 synapses (3–4 weeks) (n = 27 slices from 15 animals for WT and 24, 13 for KO, Student’s t test).
(P) Normal AMPA-NMDA ratio at Salm3−/− SC-CA1 synapses, based on AMPAR and NMDAR EPSCs (2–3 weeks) (n = 9, 9 for WT and KO; ns, not significant; Student’s t test).
(Q) Normal LTP induced by theta burst stimulation at Salm3−/− SC-CA1 synapses (3–4 weeks) (n = 14, 9 for WT and 10, 6 for KO).
(R) Normal LTD induced by low-frequency stimulation (1 Hz, 900 pulses) at Salm3−/− SC-CA1 synapses (3–4 weeks) (n = 10, 6 for WT and 9, 4 for KO).
(S) Normal late LTP induced by three trains of high-frequency stimulation (100 Hz) at Salm3−/− SC-CA1 synapses (P26–32) (n = 6, 5 for WT, 6, 4 for KO).
See also Figures S4 and S5.
Electron microscopic (EM) analyses of the hippocampal CA1 region revealed a significant reduction (~14.5%) in the density of excitatory synapses, defined by the sites of postsynaptic density (PSD) apposed to axon terminals (Figures 5G and 5H). However, there were no significant differences in the fraction of perforated synapses (a measure of excitatory synaptic maturity), PSD thickness, or PSD length (Figures 5I–5K). An independent set of EM experiments analyzing PSDs apposed to axon terminals immunopositive for the excitatory neurotransmitter glutamate yielded similar results (Figure S4).
In a rescue experiment, SALM3 expression in Salm3−/− hippocampal CA1 pyramidal neurons by lentiviral gene delivery (P9/10–23/24) reversed the reduced mEPSC frequency to levels comparable to those of WT neurons, whereas a control virus carrying EGFP alone had no effect (Figures 5L–5N). SALM3 viral expression did not affect mEPSC amplitude in WT or Salm3−/− neurons. In contrast to the results from the hippocampus, Salm3−/− dorsolateral striatal neurons showed normal frequency or amplitude of mEPSCs (Figure S5). These results suggest that SALM3 deletion selectively and markedly decreases excitatory synapse number in CA1 pyramidal neurons.
Normal Excitatory Transmission and Synaptic Plasticity at Salm3−/− Synapses
SALM3 deletion, which resulted in a marked reduction in excitatory synapse number, could also influence synaptic transmission or plasticity in the remaining excitatory synapses. However, we found no change in paired pulse ratio at Salm3−/− SC-CA1 synapses compared with WT synapses (Figure 5O), suggesting that presynaptic neurotransmitter release is unlikely to be changed. In addition, the ratio of evoked AMPAR- and NMDAR-mediated synaptic transmission (AMPA-NMDA ratio) was comparable between genotypes (Figure 5P).
Next, we tested whether Salm3−/− synapses display altered synaptic plasticity. However, neither long-term potentiation (LTP) induced by theta burst stimulation nor long-term depression (LTD) induced by low-frequency stimulation (1 Hz, 900 pulses) was altered at Salm3−/− SC-CA1 synapses compared with WT synapses (Figures 5Q and 5R). In addition, late LTP induced by three trains of high-frequency stimulation, known to involve dopamine modulation (Lisman et al., 2011), was unaltered at Salm3−/− SC-CA1 synapses (Figure 5S). Together, these results suggest that SALM3 deletion has no effect on excitatory synaptic transmission or synaptic plasticity tested here.
Hypoactivity of Salm3−/− Mice in Novel and Familiar Environments
Next, we explored behavioral alterations in Salm3−/− mice. We found that Salm3−/− mice displayed hypoactivity in a novel environment, compared with WT mice, as measured by the total distance moved in an open field arena (Figures 6A and 6B). Movement speed of the mice during the first 5 min in the open field arena were also comparable between genotypes (Figure 6C), indicating that this parameter does not contribute to the difference. Notably, the frequency of immobility was increased in Salm3−/− mice, whereas the duration of each immobility was similar between genotypes (Figures 6D and 6E), suggesting that frequent episodes of immobility contribute to the hypoactivity of Salm3−/− mice. Salm3−/− mice were also hypoactive in a familiar environment (home cages), especially during the first half of the light-off period, when mice are usually quite active (Figure 6F).
Figure 6. Salm3−/− Mice Display Hypoactivity but Normal Anxiety-like Behavior, Motor Coordination, and Stereotypy.
(A–E) Salm3−/− mice show reduced locomotor activity in open field assays compared with WT mice (A and B; 60 min). Note that Salm3−/− mice show normal velocity of movement (C; first 5 min), increased frequency of immobility (D; first 30 min), and normal duration of each immobility (E; first 30 min). Means ± SEM (n = 12 for WT and 10 for KO, **p < 0.01; ns, not significant; repeated-measures ANOVA and Student’s t test).
(F) Salm3−/− mice show hypoactivity in their home cages (n = 14 for WT and 15 for KO, *p < 0.05, repeated-measures ANOVA).
(G) Time spent in the center region of an open field arena.
(H and I) Elevated plus maze test measuring time spent in and number of entries into closed/open arms (n = 12 for WT and 15 for KO; ns, not significant; Student’s t test).
(J) Motor coordination and learning in the rotating rod test (n = 10 for WT and KO, repeated-measures ANOVA).
(K and L) Grooming and digging behavior (grooming, n = 8 for WT and 9 for KO; digging, n = 10 for WT and 9 for KO; ns, not significant; Student’s t test).
Salm3−/− mice did not show anxiety-like behaviors in the open field arena, spending normal amount of time in the center region (Figure 6G). In the elevated plus-maze test, both genotypes showed comparable levels of time spent in closed and open arms and similar numbers of entries into closed and open arms (Figures 6H and 6I).
Finally, motor coordination and motor learning in the rotating rod test (Figure 6J), and stereotypic behaviors, including grooming and digging (Figures 6K and 6L), were normal in Salm3−/− mice.
Normal Hippocampus-Dependent Learning and Memory in Salm3−/− Mice
Next, we subjected Salm3−/− mice to a battery of learning and memory behavioral tests, starting from the spontaneous and rewarded T-maze arm alternation tests, which are sensitive to hippocampal dysfunctions (Deacon and Rawlins, 2006). In the spontaneous alternation test, where spontaneous and explorative target arm alternations of a mouse are monitored, Salm3−/− and WT mice showed comparable levels of spontaneous arm alternations (Figure 7A). In the rewarded alternation test, where a mouse has to use working memory to avoid a recently visited target arm for food reward, WT and Salm3−/− mice showed comparable improvements over 12 test days (six blocks) (Figure 7B).
Figure 7. Salm3−/− Mice Perform Normally in T-maze, Morris Water Maze, Object Recognition, and Contextual Fear Conditioning and Extinction Tests.
(A) Spontaneous alternations in a T-maze. Means ± SEM (n = 11 for WT and 16 for KO; ns, not significant; Student’s-test).
(B) Rewarded alternations in a T-maze (n = 13 for WT and 11 for KO, repeated-measures ANOVA).
(C–F) Morris water maze learning and 24-hr spatial memory. Mice were trained for 5 days (learning curve, C) and subject to the probe test on day 6 for target quadrant occupancy (D), exact platform crossing (E), and swimming speed (F) (n = 10 for WT and 9 for KO; ns, not significant; repeated-measures ANOVA and Student’s t test).
(G and H) Morris water maze and 7-day spatial memory. Mice after the 5-day training sessions were tested on day 12 for quadrant occupancy (G) and exact platform crossing (H) (n = 10 for WT and 9 for KO; ns, not significant; Student’s t test).
(I and J) Novel and displaced object recognition. Mice were exposed to two identical objects, and one of the two objects was replaced with a new one (I), or translocated (or displaced) to a new position in the same box (novel object, n = 9 for WT and KO, novel location, n = 12 for WT and 10 for KO; ns, not significant; Student’s t test).
(K and L) Contextual fear conditioning. Mice exposed to foot shock were re-exposed to the same environment 24 hr (K) or 7 days (L) after the training and monitored of freezing responses (24 hr, n = 10 for WT and 9 for KO; 7 days, n = 10 for WT and 9 for KO; ns, not significant; Student’s t test).
(M) Contextual fear extinction. Mice exposed to foot shock were re-exposed to the same environment for 5 days starting from 24 hr after the training (n = 10 for WT and 9 for KO, repeated-measures ANOVA).
Next, we evaluated Salm3−/− in a Morris water maze test, which measures hippocampus-dependent spatial learning and memory (Morris, 1984). Salm3−/− mice performed normally during the learning phase of the maze (Figure 7C). In probe tests, performed on day 6 after 5-day learning sessions, Salm3−/− mice showed normal levels of target quadrant occupancy, exact platform crossing number, and swimming speed (Figures 7D–7F). Long-term memory, determined by performing the probe test on day 12, which is 7 days after the last training (1-week memory), was also unaltered in Salm3−/− mice (Figures 7G and 7H).
In the novel object recognition test, which measures non-spatial hippocampus-dependent memory (Ennaceur and Delacour, 1988), mice were exposed to the same two objects 24 hr before the test. On the day of test, when one of the two objects was replaced with a new object, Salm3−/− mice showed novel object preference comparable to that of WT mice (Figure 7I). In addition, when one of the two objects was translocated or displaced to a new position in the same box in a spatial version of the object preference test (Dix and Aggleton, 1999), Salm3−/− mice preferred to explore the displaced object to an extent comparable to that of WT mice (Figure 7J). These results suggest that Salm3−/− mice have normal recognition memory.
In a contextual fear conditioning test, which measures hippocampus- and amygdala-dependent fear memory (Phillips and LeDoux, 1992), mice were exposed to a foot shock associated with a particular environment (a foot shock chamber; complex and polymodal stimuli) and exposed to the same environment 24 hr or 7 days after the memory acquisition. Salm3−/− mice showed normal levels of context-dependent freezing at both time points (Figures 7K and 7L).
Finally, in the contextual fear extinction test, in which the hippocampus plays a critical role (Ji and Maren, 2007), Salm3−/− mice showed normal fear extinction over the course of 5 days after memory acquisition (Figure 7M). Therefore, Salm3−/− mice display unaltered hippocampus-dependent learning and memory in the tests administered in the present study.
DISCUSSION
Our study identifies that SALM3 interacts with all three LAR-RPTPs in a splicing-dependent manner and provides in vivo support for the roles of SALM3 in excitatory synapse development and locomotion behavior.
SALM3 interacts with and requires all three known LAR-RPTPs (LAR, PTPσ, and PTPδ) for presynaptic differentiation. This LAR-RPTP-binding spectrum of SALM3 is most similar to that of NGL-3 and IL-1RAcP (Kwon et al., 2010; Woo et al., 2009; Yoshida et al., 2012). However, whether NGL-3 requires all three LAR-RPTPs for presynaptic differentiation has not been tested. In addition, NGL-3 binds to the first two FNIII domains of LAR-RPTPs (Kwon et al., 2010; Woo et al., 2009), whereas SALM3 binds to the Ig1–3 domains. IL-1RAcP slightly differs from SALM3 in that it binds much more strongly to PTPδ than to PTPσ or LAR; consistent with this, IL-1RAcP-dependent presynaptic differentiation is substantially reduced in PTPδ−/− neurons (Yoshida et al., 2012). SALM3 differs greatly from other LAR-RPTP-interacting postsynaptic organizers such as TrkC, Slitrk3, and IL1RAPL1, which interact selectively with PTPσ, PTPδ, and PTPδ, respectively (Takahashi et al., 2011, 2012; Valnegri et al., 2011; Yoshida et al., 2011). Therefore, SALM3 appears to be unique in its LAR-RPTP-binding spectrum and mechanism.
SALM3 displays a splice B code for interactions with LAR-RPTPs. The presence of the meB splice insert in the Ig domain region of all three LAR-RPTPs is required for their SALM3 interactions; the meA insert, though not required, enhances these interactions. This property is somewhat similar to but distinct from the requirement of meB, but not meA, for the interaction of Slitrk1/2 with LAR-RPTPs (Takahashi et al., 2012; Um et al., 2014; Yamagata et al., 2015a), and the enhancement of the interaction between IL-1RAcP and PTPδ involving both meA and meB (Yamagata et al., 2015b; Yoshida et al., 2012). In addition, this property is dissimilar to the meA splice code of IL1RAPL1, in which meA is required and meB enhances the interaction with PTPδ (Yamagata et al., 2015b; Yoshida et al., 2011), and the splice code of TrkC, in which meA or meB tends to inhibit the interaction with PTPσ (Takahashi et al., 2011). Among the LAR-RPTPs, PTPδ ranks highest both in fraction containing a B insert and in effect of knockdown on presynaptic induction by SALM3. However, although only 10% of LAR mRNAs contains the B insert, knockdown also revealed a functional role of LAR as a SALM3 interactor, perhaps through high protein level, surface expression, or affinity, as suggested by the cell-based SALM3-Fc binding assay using equal amounts of LAR-RPTP expression vectors.
Although the abovementioned overall characteristics of SALM3 appear to be most similar to IL-1RAcP, SALM3 has several features that distinguish it from IL-1RAcP. SALM3 contains a C-terminal type I PDZ-binding motif that binds to PSD-95 (Ko et al., 2006; Morimura et al., 2006; Nam et al., 2011; Wang et al., 2006), whereas IL-1RAcP does not. Instead, IL-1RAcP contains a cytoplasmic TIR domain that is absent in SALM3. A splice variant of IL-1RAcP, termed IL-1RAcPb (Smith et al., 2009), that contains a type II PDZ-binding motif, however, has not been demonstrated to bind PSD-95. In terms of brain distribution, SALM3 is widespread, as revealed by X-gal staining, similar to the mRNA distribution of IL-1RAcP (Smith et al., 2009). However, in the hippocampus, SALM3 is more abundant in the CA3 and CA1 regions, whereas IL-1RAcP expression mainly maps to the dentate gyrus region (Allen Brain Atlas). Therefore, SALM3 and IL-1RAcP are likely to have distinct functions in the brain.
The Salm3−/− mice data indicate that SALM3 is important for excitatory synapse development in the hippocampal CA1 region, as supported by substantial reductions in mEPSC frequency (~52.3%) and PSD density (~14.5%). The lentiviral rescue experiment, in which only sparse CA1 neurons were transduced in a Salm3−/− background, including Salm3−/− SC inputs, indicates that SALM3 functions cell autonomously in the postsynaptic cell to control the density of excitatory inputs. Several mouse lines lacking LAR-RPTP-binding postsynaptic organizers have been reported. TrkC−/− mice are perinatally lethal (Tessarollo et al., 1997), making it difficult to assess its specific role in synapse development, although TrkC knockdown in sparse cortical neurons reduced dendritic spine density (Takahashi et al., 2011). Il1rapl1−/− mice display decreased excitatory synapse and spine density in the hippocampal CA1 region (synapse, ~25% by EM; spine, ~12% and 24% in apical and basal dendrites, respectively) (Pavlowsky et al., 2010). These mice also show suppressed GABAergic network in the cerebellum (Gambino et al., 2009) and reduced excitatory transmission in the lateral amygdala (Houbaert et al., 2013). Slitrk5−/− mice show reduced levels of AMPAR and NMDAR subunits in the striatum and decreased amplitude of corticostriatal population spikes (Shmelkov et al., 2010), whereas Slitrk3−/− mice display reduced frequency of mIPSCs and weakened signals of GAD65 (inhibitory synapse marker) in the hippocampal CA1 region (Takahashi et al., 2012). Therefore, LAR-RPTP-interacting postsynaptic organizers seem to regulate excitatory and inhibitory synapse development in diverse brain regions, and SALM3 and IL1RAPL1, in particular, appear important for hippocampal excitatory synapse development.
Although Salm3−/− hippocampal CA1 region displays significantly reduced excitatory synapse number, the remaining synapses seem to display apparently normal synaptic transmission and plasticity. For instance, WT and Salm3−/− synapses show comparable paired pulse ratio and AMPA-NMDA ratio. In addition, Salm3−/− synapses show normal levels of LTP, LTD, and late LTP. This contrasts with the reported regulation of LTP by neuroligin-1 and LRRTM1/2 (Blundell et al., 2010; Jedlicka et al., 2015; Jung et al., 2010; Kim et al., 2008; Shipman and Nicoll, 2012; Soler-Llavina et al., 2013), but is similar to the minimal LTP regulation by neuroligin-3 (Shipman and Nicoll, 2012).
Behaviorally, Salm3−/− mice perform normally in T-maze, Morris water maze, object recognition, contextual fear conditioning, and extinction tests, which is apparently in line with the lack of changes in synaptic plasticity in these animals. This contrasts with the impairments in cued fear memory and amygdalal theta-burst stimulation-induced LTP in Il1rapl1−/− mice (Houbaert et al., 2013), which show reduced excitatory synapse and spine densities in the hippocampus (Pavlowsky et al., 2010).
Salm3−/− mice display hypoactivity in both novel and familiar environments, and this seems to be caused by frequent immobility rather than increased immobility duration or decreased movement velocity. It has been suggested that ~1.56% and 0.75% of genes in the whole genome cause hyperactivity and hypoactivity, respectively, when dysregulated (Mignogna and Viggiano, 2010; Viggiano, 2008). In addition, hypoactivity can be caused by genetic or pharmacological interventions that suppress dopamine or histamine activity or that enhance acetylcholine activity in the brain (Mignogna and Viggiano, 2010; Viggiano, 2008). Glutamate and GABA have also been implicated in the modulation of locomotor activity, although their alterations mainly cause hyperactivity. A detailed understanding of how hypoactivity manifests in Salm3−/− mice will require further exploration.
In conclusion, our data suggest that postsynaptic SALM3 interacts with presynaptic LAR-RPTPs (LAR, PTPσ, and PTPδ) in a splice variant-dependent manner and provide in vivo support for the roles of SALM3 in the regulation of excitatory synapse development and locomotor behavior.
EXPERIMENTAL PROCEDURES
Constructs
The following constructs have been described: LAR-Ecto-A−B−-pDis, PTPσ-Ecto-A−B−-pDis, PTPδ-Ecto-A+B+ (Kwon et al., 2010) and SALM3-Ecto-pDis (Mah et al., 2010), mPTPσ A+B+-CFP, mPTPσ A+B−-CFP, mPTPσ A−B+-CFP, and mPTPσ A−B−-CFP (Takahashi et al., 2011). PTPσ-Ecto-A+B+-pDis was a kind gift from Dr. Homin Kim at KAIST. The entire coding sequences of mouse PTPδ, PTPδA−B+, PTPδA+B− (A9), PTPδA−B−, LAR A6B+, LAR A6B−, LARA−B+, LARA−B−in pcDNA3 vector were kind gifts from Dr. Yoshida at University of Tokyo (Yoshida et al., 2011).
Antibodies
SALM3 (1929) guinea pig polyclonal antibodies were generated using a synthetic peptide as immunogen (aa 594–608 of mouse SALM3; CYGYARRLGGAWARR). Peptides mimicking the last 30 aa of mouse SALM1, SALM2, and SALM4 were used to generate guinea pig polyclonal antibodies (2022, 2058, and 2026, respectively). The following antibodies have been described: PSD-95 (1690) (Han et al., 2010), NGL-3 (2020) (Lee et al., 2014), SALM3 (1816), and SALM5 (1907) (Mah et al., 2010). The following antibodies were purchased: α-tubulin, synapsin I (Sigma), synaptophysin, GluA1, GluA2 (Santa Cruz), slitrk1, slitrk3 (Abcam), GluN1 (Invitrogen), GluN2A (Zymed), GluN2B (NeuroMab), PTPσ (17G7.2, mouse, Medimabs), and IL-1RAcP (Millipore). IL1RAPL1 antibody was a kind gift from Dr. Carlo Sala.
Generation and Characterization of Salm3−/− Mice
Mouse sperms carrying the Salm3 gene, in which exons 2 and 3 were replaced with a cassette containing lacZ-neomycin gene and a polyadenylation signal by homologous recombination, were obtained from KOMP. Mice were obtained by in vitro fertilization. Heterozygotes were crossed to obtain SALM3 deficiency mice.
Virus Infection
Lentivirus carrying the SALM3 rescue construct was purchased (Applied Biological Materials). Briefly, mouse SALM3 cDNA was subcloned into the pLenti-GIII-CMV-GFP-2A–Puro vector. 293T cells were used for virus packaging of lenti-EGFP (control) and lenti-SALM3. Lentiviral titer for EGFP and SALM3 was ~3 × 108 IU/ml. Mice at P9–10 were anesthetized with xylazine/ketamine and transcranially injected of virus into the hippocampal CA1 region using glass pipettes and a syringe pump (KD Scientific) at a rate of 75 nl/min with a 30-s delay. Electrophysiological experiments were made 2 weeks after virus injection.
Supplemental Experimental Procedures
The Supplemental Experimental Procedures contain details on Cell Aggregation Assays, Production of Soluble Fc-fusion Proteins, Pull-down of PTPδ-Fc and Mass Spectrometry, Surface Binding Assay, Knockdown of LAR-PTPs Family in Co-culture, Electron Microscopy, Field Potential Recording, Patch Analysis, and Animal Behavioral Tests.
Supplementary Material
Highlights.
SALM3 interacts with presynaptic LAR-RPTPs in a mini-exon B-dependent manner
SALM3 requires all three LAR-RPTPs for presynaptic differentiation
Salm3−/− neurons display reduced excitatory synapse number
Salm3−/− mice exhibit hypoactivity in novel and familiar environments
ACKNOWLEDGMENTS
We thank Haram Park for the help with the graphical abstract. This study was supported by the Institute for Basic Science (IBS-R002-D1 to E.K.), the National Research Foundation (MSIP, 2008-0062282 to Y.C.B.), the Michael Smith Foundation for Health Research Postdoctoral Fellowship (to P.Z.), and the NIH (MH-070860 to A.M.C.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.08.002.
AUTHOR CONTRIBUTIONS
J.D.R., D.L., R.K., and H.P. acquired and analyzed mEPSC, mIPSC, and late LTP data. S.K.P. acquired and analyzed EM data. W.M. generated SALM3 antibodies. T.-Y.C, D.L, and E.-J.L acquired and analyzed field recording data. H.K. and Y.K. acquired and analyzed mouse behavioral data. P.Z. designed, acquired, and analyzed proteomics, cell-based binding, and neuron knockdown experiments/data and drafted and revised the manuscript. Y.L designed, acquired, and analyzed cell aggregation, mouse behavioral, staining, and immunoblot data and drafted and revised the manuscript. Y.C.B., S.-Y.C, A.M.C., and E.K. conceived, designed, supervised, analyzed the experiments/data, and drafted and revised the manuscript.
REFERENCES
- Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J. Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Stagi M. Signaling by synaptogenic molecules. Curr. Opin. Neurobiol. 2008;18:261–269. doi: 10.1016/j.conb.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Suüdhof TC, Powell CM. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat. Rev. Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn DR, van Dijk AH, Pfundt R, Hoischen A, Merkx GF, Gradek GA, Lybæk H, Stray-Pedersen A, Brunner HG, Houge G. Severe progressive autism associated with two de novo changes: A 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1;p11.2) translocation with long-range epigenetic silencing of LRFN5 expression. Mol. Syndromol. 2010;1:46–57. doi: 10.1159/000280290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Ghosh A. Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 2014;37:539–550. doi: 10.1016/j.tins.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu. Rev. Cell Dev. Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- Deacon RM, Rawlins JN. T-maze alternation in the rodent. Nat. Protoc. 2006;1:7–12. doi: 10.1038/nprot.2006.2. [DOI] [PubMed] [Google Scholar]
- Dix SL, Aggleton JP. Extending the spontaneous preference test of recognition: evidence of object-location and object-context recognition. Behav. Brain Res. 1999;99:191–200. doi: 10.1016/s0166-4328(98)00079-5. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Hueske E, Wyszynski M, Hoogenraad CC, Jaworski J, Pak DT, Simonetta A, Liu G, Sheng M. LAR receptor protein tyro-sine phosphatases in the development and maintenance of excitatory synapses. Nat. Neurosci. 2005;8:458–467. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’arcy M, deBerardinis R, Frackelton E, Kim C, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennaceur A, Delacour J. A new one-trial test for neurobiological studies of memory in rats. 1: Behavioral data. Behav. Brain Res. 1988;31:47–59. doi: 10.1016/0166-4328(88)90157-x. [DOI] [PubMed] [Google Scholar]
- Gambino F, Kneib M, Pavlowsky A, Skala H, Heitz S, Vitale N, Poulain B, Khelfaoui M, Chelly J, Billuart P, Humeau Y. IL1RAPL1 controls inhibitory networks during cerebellar development in mice. Eur. J. Neurosci. 2009;30:1476–1486. doi: 10.1111/j.1460-9568.2009.06975.x. [DOI] [PubMed] [Google Scholar]
- Han S, Nam J, Li Y, Kim S, Cho SH, Cho YS, Choi SY, Choi J, Han K, Kim Y, et al. Regulation of dendritic spines, spatial memory, and embryonic development by the TANC family of PSD-95-interacting proteins. J. Neurosci. 2010;30:15102–15112. doi: 10.1523/JNEUROSCI.3128-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Feliu-Mojer MI, Spangler SA, Milstein AD, Dunah AW, Hung AY, Sheng M. Liprinalpha1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev. Cell. 2007;12:587–602. doi: 10.1016/j.devcel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Houbaert X, Zhang CL, Gambino F, Lepleux M, Deshors M, Normand E, Levet F, Ramos M, Billuart P, Chelly J, et al. Target-specific vulnerability of excitatory synapses leads to deficits in associative memory in a model of intellectual disorder. J. Neurosci. 2013;33:13805–13819. doi: 10.1523/JNEUROSCI.1457-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Vnencak M, Krueger DD, Jungenitz T, Brose N, Schwarzacher SW. Neuroligin-1 regulates excitatory synaptic transmission, LTP and EPSP-spike coupling in the dentate gyrus in vivo. Brain Struct. Funct. 2015;220:47–58. doi: 10.1007/s00429-013-0636-1. [DOI] [PubMed] [Google Scholar]
- Ji J, Maren S. Hippocampal involvement in contextual modulation of fear extinction. Hippocampus. 2007;17:749–758. doi: 10.1002/hipo.20331. [DOI] [PubMed] [Google Scholar]
- Johnson KG, Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 2003;83:1–24. doi: 10.1152/physrev.00016.2002. [DOI] [PubMed] [Google Scholar]
- Jung SY, Kim J, Kwon OB, Jung JH, An K, Jeong AY, Lee CJ, Choi YB, Bailey CH, Kandel ER, Kim JH. Input-specific synaptic plasticity in the amygdala is regulated by neuroligin-1 via postsynaptic NMDA receptors. Proc. Natl. Acad. Sci. USA. 2010;107:4710–4715. doi: 10.1073/pnas.1001084107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung SY, Lee YK, Park S, Choi JS, Lee CJ, Kim HS, Choi YB, Scheiffele P, Bailey CH, et al. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc. Natl. Acad. Sci. USA. 2008;105:9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Kim S, Chung HS, Kim K, Han K, Kim H, Jun H, Kaang BK, Kim E. SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Tuffy LP, Papadopoulos T, Brose N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr. Opin. Neurobiol. 2012;22:412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Kwon SK, Woo J, Kim SY, Kim H, Kim E. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J. Biol. Chem. 2010;285:13966–13978. doi: 10.1074/jbc.M109.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee EJ, Song YS, Kim E. Long-term depression-inducing stimuli promote cleavage of the synaptic adhesion molecule NGL-3 through NMDA receptors, matrix metalloproteinases and presenilin/γ-secretase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130158. doi: 10.1098/rstb.2013.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Grace AA, Duzel E. A neoHebbian framework for episodic memory; role of dopamine-dependent late LTP. Trends Neurosci. 2011;34:536–547. doi: 10.1016/j.tins.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah W, Ko J, Nam J, Han K, Chung WS, Kim E. Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J. Neurosci. 2010;30:5559–5568. doi: 10.1523/JNEUROSCI.4839-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mander A, Hodgkinson CP, Sale GJ. Knock-down of LAR protein tyrosine phosphatase induces insulin resistance. FEBS Lett. 2005;579:3024–3028. doi: 10.1016/j.febslet.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Mignogna P, Viggiano D. Brain distribution of genes related to changes in locomotor activity. Physiol. Behav. 2010;99:618–626. doi: 10.1016/j.physbeh.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rut-ledge SL, Korf BR, Carroll AJ. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am. J. Med. Genet. A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- Missler M, Südhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb. Perspect. Biol. 2012;4:a005694. doi: 10.1101/cshperspect.a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura N, Inoue T, Katayama K, Aruga J. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Nam J, Mah W, Kim E. The SALM/Lrfn family of leucine-rich repeat-containing cell adhesion molecules. Semin. Cell Dev. Biol. 2011;22:492–498. doi: 10.1016/j.semcdb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Pavlowsky A, Gianfelice A, Pallotto M, Zanchi A, Vara H, Khelfaoui M, Valnegri P, Rezai X, Bassani S, Brambilla D, et al. A postsynaptic signaling pathway that may account for the cognitive defect due to IL1RAPL1 mutation. Curr. Biol. 2010;20:103–115. doi: 10.1016/j.cub.2009.12.030. [DOI] [PubMed] [Google Scholar]
- Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schormair B, Kemlink D, Roeske D, Eckstein G, Xiong L, Lichtner P, Ripke S, Trenkwalder C, Zimprich A, Stiasny-Kolster K, et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat. Genet. 2008;40:946–948. doi: 10.1038/ng.190. [DOI] [PubMed] [Google Scholar]
- Seabold GK, Wang PY, Chang K, Wang CY, Wang YX, Petralia RS, Wenthold RJ. The SALM family of adhesion-like molecules forms heteromeric and homomeric complexes. J. Biol. Chem. 2008;283:8395–8405. doi: 10.1074/jbc.M709456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu. Rev. Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman SL, Nicoll RA. A subtype-specific function for the extracellular domain of neuroligin 1 in hippocampal LTP. Neuron. 2012;76:309–316. doi: 10.1016/j.neuron.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, Shmelkov E, Kushner JS, Baljevic M, Dincheva I, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat. Med. 2010;16:598–602. 1p, 602. doi: 10.1038/nm.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Craig AM. Synaptic organizing complexes. Curr. Opin. Neurobiol. 2011;21:132–143. doi: 10.1016/j.conb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Lipsky BP, Russell C, Ketchem RR, Kirchner J, Hensley K, Huang Y, Friedman WJ, Boissonneault V, Plante MM, et al. A central nervous system-restricted isoform of the interleukin-1 receptor accessory protein modulates neuronal responses to interleukin-1. Immunity. 2009;30:817–831. doi: 10.1016/j.immuni.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Llavina GJ, Arstikaitis P, Morishita W, Ahmad M, Südhof TC, Malenka RC. Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation. Neuron. 2013;79:439–446. doi: 10.1016/j.neuron.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YS, Kim E. Presynaptic proteoglycans: sweet organizers of synapse development. Neuron. 2013;79:609–611. doi: 10.1016/j.neuron.2013.07.048. [DOI] [PubMed] [Google Scholar]
- Stryker E, Johnson KG. LAR, liprin alpha and the regulation of active zone morphogenesis. J. Cell Sci. 2007;120:3723–3728. doi: 10.1242/jcs.03491. [DOI] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Craig AM. Protein tyrosine phosphatases PTPδ, PTPσ, and LAR: presynaptic hubs for synapse organization. Trends Neurosci. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Arstikaitis P, Prasad T, Bartlett TE, Wang YT, Murphy TH, Craig AM. Postsynaptic TrkC and presynaptic PTPσ function as a bidirectional excitatory synaptic organizing complex. Neuron. 2011;69:287–303. doi: 10.1016/j.neuron.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Katayama K, Sohya K, Miyamoto H, Prasad T, Matsumoto Y, Ota M, Yasuda H, Tsumoto T, Aruga J, Craig AM. Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat. Neurosci. 2012;15:389–398. S1–S2. doi: 10.1038/nn.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarollo L, Tsoulfas P, Donovan MJ, Palko ME, Blair-Flynn J, Hemp-stead BL, Parada LF. Targeted deletion of all isoforms of the trkC gene suggests the use of alternate receptors by its ligand neurotrophin-3 in neuronal development and implicates trkC in normal cardiogenesis. Proc. Natl. Acad. Sci. USA. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Ko J. LAR-RPTPs: synaptic adhesion molecules that shape synapse development. Trends Cell Biol. 2013;23:465–475. doi: 10.1016/j.tcb.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Um JW, Kim KH, Park BS, Choi Y, Kim D, Kim CY, Kim SJ, Kim M, Ko JS, Lee SG, et al. Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion. Nat. Commun. 2014;5:5423. doi: 10.1038/ncomms6423. [DOI] [PubMed] [Google Scholar]
- Valnegri P, Montrasio C, Brambilla D, Ko J, Passafaro M, Sala C. The X-linked intellectual disability protein IL1RAPL1 regulates excitatory synapse formation by binding PTPδ and RhoGAP2. Hum. Mol. Genet. 2011;20:4797–4809. doi: 10.1093/hmg/ddr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnegri P, Sala C, Passafaro M. Synaptic dysfunction and intellectual disability. Adv. Exp. Med. Biol. 2012;970:433–449. doi: 10.1007/978-3-7091-0932-8_19. [DOI] [PubMed] [Google Scholar]
- Viggiano D. The hyperactive syndrome: metanalysis of genetic alterations, pharmacological treatments and brain lesions which increase locomotor activity. Behav. Brain Res. 2008;194:1–14. doi: 10.1016/j.bbr.2008.06.033. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Yoo HJ. Current progress and challenges in the search for autism biomarkers. Dis. Markers. 2013;35:55–65. doi: 10.1155/2013/476276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Chang K, Petralia RS, Wang YX, Seabold GK, Went-hold RJ. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J. Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Kwon SK, Choi S, Kim S, Lee JR, Dunah AW, Sheng M, Kim E. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, Gogos JA, Karayiorgou M. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc. Natl. Acad. Sci. USA. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR, Weiner JA. Synaptic adhesion molecules. Curr. Opin. Cell Biol. 2003;15:621–632. doi: 10.1016/s0955-0674(03)00107-8. [DOI] [PubMed] [Google Scholar]
- Yamagata A, Sato Y, Goto-Ito S, Uemura T, Maeda A, Shiroshima T, Yoshida T, Fukai S. Structure of Slitrk2-PTPδ complex reveals mechanisms for splicing-dependent trans-synaptic adhesion. Sci. Rep. 2015a;5:9686. doi: 10.1038/srep09686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata A, Yoshida T, Sato Y, Goto-Ito S, Uemura T, Maeda A, Shiroshima T, Iwasawa-Okamoto S, Mori H, Mishina M, Fukai S. Mechanisms of splicing-dependent trans-synaptic adhesion by PTPδ-IL1RAPL1/IL-1RAcP for synaptic differentiation. Nat. Commun. 2015b;6:6926. doi: 10.1038/ncomms7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li L, Yang R, Shen GQ, Chen Q, Foldvary-Schaefer N, Ondo WG, Wang QK. Family-based and population-based association studies validate PTPRD as a risk factor for restless legs syndrome. Mov. Disord. 2011;26:516–519. doi: 10.1002/mds.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim YS, Kwon Y, Nam J, Yoon HI, Lee K, Kim DG, Kim E, Kim CH, Ko J. Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA. 2013;110:4057–4062. doi: 10.1073/pnas.1209881110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Yasumura M, Uemura T, Lee SJ, Ra M, Taguchi R, Iwakura Y, Mishina M. IL-1 receptor accessory protein-like 1 associated with mental retardation and autism mediates synapse formation by trans-synaptic interaction with protein tyrosine phosphatase δ. J. Neurosci. 2011;31:13485–13499. doi: 10.1523/JNEUROSCI.2136-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Shiroshima T, Lee SJ, Yasumura M, Uemura T, Chen X, Iwakura Y, Mishina M. Interleukin-1 receptor accessory protein organizes neuronal synaptogenesis as a cell adhesion molecule. J. Neurosci. 2012;32:2588–2600. doi: 10.1523/JNEUROSCI.4637-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki M. Cbln1 and its family proteins in synapse formation and maintenance. Curr. Opin. Neurobiol. 2011;21:215–220. doi: 10.1016/j.conb.2011.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.