Abstract
Objectives
Non-invasive biomarkers would be valuable for diagnosis and monitoring of eosinophilic esophagitis (EoE). The aim of this study was to determine the utility of a panel of serum biomarkers for the diagnosis and management of EoE.
Methods
We conducted a prospective cohort study of consecutive adults undergoing outpatient EGD. Incident cases of EoE were diagnosed per consensus guidelines; controls had GERD or dysphagia and did not meet EoE criteria. EoE cases were treated with topical steroids and had repeat endoscopy. Pre- and post-treatment serum samples were analyzed in a blinded fashion for: IL-4, IL-5, IL-6, IL-9, IL-13, TGF-α, TGF-β, TNF-α, eotaxin-1, -2, and -3, TSLP, major basic protein (MBP), and eosinophil-derived neurotoxin (EDN). Cases and controls were compared at baseline, and pre- and post-treatment assays were compared in cases.
Results
A total of 61 incident EoE cases and 87 controls were enrolled; 51 EoE cases had post-treatment serum analyzed. There were no significant differences in any of the biomarkers between EoE cases and controls at baseline. IL-13 and eotaxin-3 for cases and controls were 85 ±160 vs 43 ±161 pg/mL (p=0.12), and 41 ±159 vs 21 ±73 (p=0.30). There were no significant differences in assay values among cases before and after treatment. There were also no differences after stratification by atopic status or treatment response.
Conclusions
A panel of inflammatory factors known to be associated with EoE pathogenesis were not increased in the serum, nor were they responsive to therapy. None of these biomarkers are likely candidates for a serum test for EoE. Histologic analysis for diagnosis and management of EoE continues to be necessary, and novel, less invasive, biomarkers are needed.
Keywords: Eosinophilic esophagitis, biomarkers, serum, inflammation, cytokines
Introduction
The current diagnostic algorithm for eosinophilic esophagitis (EoE) requires upper endoscopy and biopsy, an invasive procedure, to assess for esophageal eosinophilia in patients with symptoms of esophageal dysfunction (1-3). In practice, several procedures are needed: the index endoscopy where the diagnosis is suspected, the follow-up endoscopy to confirm the diagnosis after a proton pump inhibitor (PPI) trial, and a third endoscopy to assess tissue response to therapy (1, 4). This approach is suboptimal due to high costs associated with the multiple procedures (5) as well as the possibility of procedural complications. Non-invasive biomarkers hold the potential to decrease costs and increase safety, but none have been clinically validated for routine use in EoE (6-8).
Candidate biomarkers could be selected from the pathogenesis of EoE, which is currently thought to involve a Th2-mediated response to allergens (9-12). A number of cytokines, including IL-4, IL-5, and IL-13 (13-18), chemokines such as eotaxin-3, which is the most highly upregulated gene in EoE (15, 19-21), and markers of eosinophil activation such as granule proteins (18, 22-24), have all been shown to be elevated in EoE as compared to controls. However, these findings have been generally reported in esophageal tissue, and data are primarily related to pathogenic studies in EoE (15, 19-21, 25-29). The true clinical utility of Th2-related cytokines, chemokines, and eosinophil granules as non-invasive serum biomarkers has yet to be demonstrated for either diagnosis or monitoring of treatment for EoE. Given the high cost of diagnosis and management of EoE using endoscopy findings, the translation of the research findings above into a viable serum test for the presence and/or severity of EoE would be of enormous value.
The aim of this study was to determine whether a panel of serum biomarkers based on the known pathogenesis of EoE could distinguish EoE from controls at baseline for diagnosis of EoE. We additionally sought to determine whether these biomarkers might have utility for monitoring EoE after treatment. We hypothesized that subjects with EoE would have significantly higher serum levels of one or more of these biomarkers, compared to clinically relevant non-EoE controls, and that these levels might decrease among the EoE cases after effective steroid therapy.
Methods
Study design, patients, clinical data, and follow-up
We conducted a prospective cohort study at University of North Carolina from July, 2011 through December, 2013. Consecutive adult patients (age 18-80 years) undergoing routine outpatient esophagogastroduodenoscopy (EGD) were approached if they had upper GI symptoms suggestive of esophageal dysfunction (e.g. dysphagia, food impaction, heartburn, reflux, chest pain). Subjects provided informed consent, including consent for future use of stored specimens, and were enrolled prior to the endoscopy. Subjects were excluded if they had a known (prevalent) diagnosis of EoE or a different eosinophilic gastrointestinal disorder (EGID), GI bleeding, active anticoagulation, known esophageal cancer, prior esophageal surgery, known esophageal varices, medical instability or multiple comorbidities precluding enrollment in the clinical opinion of the endoscopist, or inability to read or understand the consent form. This study was approved by the UNC Institutional Review Board and registered on clinicaltrials.gov (NCT 01988285).
Cases were diagnosed with EoE if they met consensus guidelines (1-3). Specifically, they were required to have at least one typical symptom of esophageal dysfunction; at least 15 eosinophils per high-power field (eos/hpf) on esophageal biopsy persisting after an 8 week PPI trial (20-40 mg twice daily of any of the available agents, prescribed at the discretion of the clinician); and other causes of esophageal eosinophilia excluded. Of note, baseline data for the EoE cases were obtained after the PPI trial, at the time of the confirmatory EGD, but prior to receiving the histologic results confirming the diagnosis or provision of EoE-specific treatment, so as to minimize potential recall bias. Controls were subjects who, after endoscopy and biopsy, did not meet clinical or histologic criteria for EoE. Subjects with PPI-responsive esophageal eosinophilia (PPI-REE) were not included in this study.
Clinical data were collected using a standardized case report form. Items recorded included demographics, symptoms, concomitant atopic diseases, indications for endoscopy, and endoscopic findings. Food allergy data was collected by patient self-report on a prospectively administered questionnaire, and could therefore include both food allergies and sensitizations. Systematic allergy testing was not a component of this study. During endoscopy, research-protocol esophageal biopsies were obtained (two from the proximal, one from the mid, and two from the distal esophagus) to maximize EoE diagnostic sensitivity (30, 31). Gastric and duodenal biopsies were also collected for research purposes to exclude concomitant eosinophilic gastroenteritis. Additional clinical biopsies were taken as indicated at the discretion of the endoscopist. Esophageal eosinophil counts were quantified by the study pathologists using our previously validated methods (32). In brief, slides were masked to case/control status, digitized, and reviewed with Aperio ImageScope (Aperio Technologies, Vista, CA). Five microscopy fields from each of the five biopsies were examined to determine the maximum eosinophil density (eosinophils/mm2 [eos/mm2]). So results could be compared to prior studies, eosinophil density was converted to an eosinophil count (eos/hpf) using a hpf size of 0.24 mm2, the most commonly reported field size in the literature (33).
EoE cases were treated for 8 weeks as clinically indicated with topical corticosteroids (either oval viscous budesonide 1 mg twice daily or fluticasone from a multi-dose inhaler, 880 mcg twice daily) (34-36). At the end of the treatment period, repeat upper endoscopy with biopsy was performed, with collection of a second set of blood and tissue samples as noted above. A second blood sample was also collected for a subset of control subjects at least 2 months after baseline samples were collected to assess for stability in biomarkers over time.
Serum data and biomarkers
Prior to each procedure, a blood sample was obtained from all subjects and centrifuged. Serum was separated and aliquots were frozen and stored at −80°C. All samples were labelled with a unique study ID that was blinded as to case/control status, as well as to pre- or post-treatment status. After patient enrollment and follow-up were complete, samples were removed from the freezer, arranged in random order, thawed only once, and analyzed in a batch.
The serum analytes measured were: IL-4, IL-5, IL-6, IL-9, IL-13, TGF-α, TGF-β, TNF-α, eotaxin-1, -2, and -3, TSLP, major basic protein (MBP), and eosinophil derived neurotoxin (EDN). These were chosen based on prior demonstration of elevated levels in the esophageal tissue and/or in a peripheral source (6, 7, 13-19, 21-29, 37-40). IL-4, IL-5, IL-6, IL-9, IL-13, TGF-α, TNF-α, and eotaxin-1 were measured using an 8-plex panel (cat # HCYTOMAG-60K, Millipore, St. Charles, MO). Eotaxin-2, eotaxin-3, and TSLP were measured using a 3-plex panel (cat# HCYP2MAG-62K, Millipore). TGF-β (cat# 559119, BD Biosciences, San Diego, CA) was measured individually via ELISA, as were MBP (cat# ABIN1115874) and EDN (cat# ABIN858221, ABO). Samples were run in duplicate on 96-well plates with standards and positive/negative controls per manufacturer instructions. A Bio-Plex 200 system (Bio-Rad Laboratories, Hercules, CA) was used to determine the mean fluorescence intensity of the multiplex assays. For a subset of EoE cases and non-EoE controls, immunohistochemical staining was used to measure tissue levels of MBP, eotaxin-3, and mast cell tryptase with methodology we have previously described (23, 24, 41).
Statistical analysis
Baseline clinical, endoscopic, and histologic characteristics of the cases and controls were described with bivariate analysis using Chi-square for categorical variables, and t-tests or Wilcoxon Rank-sum for continuous variables as appropriate. The mean baseline values of the individual serum biomarkers were compared between cases and controls using a 2 sample t-test, while baseline and follow-up values for the EoE cases were compared using a paired t-test or Wilcoxon Signed-rank as appropriate. Additional analyses were performed for the serum values after stratification for: 1) the presence of atopic diseases (ie asthma, atopic dermatitis, allergic rhinitis/sinusitis, and food allergy); and 2) histologic response to treatment for the EoE cases, defined as either <15 eos/hpf or <1 eos/hpf (4, 42). Because results were the same with both parametric and non-parametric testing, means and standard deviations are presented in the figures. Receiver operating characteristic (ROC) curves were constructed and areas under the curve (AUC) were calculated to determine the utility of the serum biomarkers, both individually and collectively, for distinguishing EoE cases from controls at baseline and for monitoring response following treatment. The sample size was determined based on the ROC analysis. Enrolling at least 60 cases EoE and 60 controls was expected to provide >80% power to conclude that the AUC for an individual marker was significantly greater than 0.50 (43).
Results
Patient characteristics
A total of 61 EoE cases and 87 non-EoE controls were included in this study. Compared to controls, EoE cases at diagnosis were more likely to be younger (39 vs 52 years; p < 0.001), male (58% vs 42%; p = 0.05), white (94% vs 82%), and have an atopic disorder (74% vs 54%; p = 0.01) (Table 1). Dysphagia was common in both groups, but more common in the cases (97% vs 80%; p = 0.002). Heartburn was less common among cases (16% vs 67%; p < 0.001), as was the presence of a hiatal hernia (13% vs 57%; p < 0.001). As expected, the typical endoscopic findings of EoE were also more common in cases (Table 1). The mean of the maximum eosinophil counts in the cases was 146 eos/hpf, compared to 3 for the controls (p < 0.001). After treatment of the EoE cases, the mean eosinophil count decreased to 55 eos/hpf (p < 0.001 compared to baseline), with 55% achieving a histologic response of <15 eos/hpf (mean post-treatment eosinophil count of 3 eos/hpf), and 28% achieving normalization of the biopsies (<1 eos/hpf).
Table 1.
Clinical characteristics of EoE cases and non-EoE controls
| Non-EoE controls (n = 87) |
EoE cases (n = 61) |
p | |
|---|---|---|---|
| Age at diagnosis (mean ± SD) | 51.9 ± 13.2 | 38.8 ± 13.2 | < 0.001 |
| Male (n, %) | 37 (42) | 36 (58) | 0.05 |
| White (n, %) | 73 (82) | 58 (94) | 0.04 |
| Symptoms/EGD indication (n, %) | |||
| Dysphagia | 71 (80) | 60 (97) | 0.002 |
| Heartburn | 60 (67) | 10 (16) | < 0.001 |
| Abdominal pain | 5 (6) | 7 (11) | 0.21 |
| Nausea/vomiting | 8 (9) | 1 (2) | 0.06 |
| Atopic disorders (n, %) | |||
| Asthma | 19 (21) | 19 (31) | 0.20 |
| Atopic dermatitis | 7 (8) | 4 (6) | 0.74 |
| Allergic rhinitis/sinusitis | 40 (45) | 40 (65) | 0.02 |
| Food allergies | 10 (11) | 28 (45) | < 0.001 |
| Any atopic disease | 48 (54) | 46 (74) | 0.01 |
| EGD findings at baseline (n, %) | |||
| Normal | 13 (15) | 3 (5) | 0.06 |
| Rings | 8 (9) | 47 (76) | < 0.001 |
| Stricture | 19 (21) | 15 (24) | < 0.001 |
| Narrowing | 2 (2) | 16 (26) | < 0.001 |
| Furrows | 3 (3) | 55 (89) | < 0.001 |
| Crêpe-paper | 3 (3) | 4 (6) | 0.38 |
| White plaques/exudates | 3 (3) | 27 (44) | < 0.001 |
| Decreased vascularity | 3 (3) | 37 (60) | < 0.001 |
| Erosive esophagitis | 17 (19) | 2 (3) | 0.004 |
| Schatzki’s ring | 13 (15) | 7 (11) | 0.55 |
| Hiatal hernia | 51 (57) | 8 (13) | < 0.001 |
| Baseline max eosinophil count (mean ± SD) | 2.8 ± 6.8 | 146.3 ± 128.4 | < 0.001 |
| Median eosinophil count (IQR) | 0 (0-2) | 111 (60-165) | < 0.001 |
| Follow-up max eosinophil count (mean ± SD)* | - | 54.5 ± 87.9 | - |
| Median eosinophil count (IQR) | - | 9 (0-84) | - |
| Histologic response rates | |||
| < 1 eos/hpf | - | 17 (28) | - |
| < 15 eos/hpf | - | 33 (55) | - |
n = 60 EoE cases with follow-up biopsy data
Baseline serum biomarkers
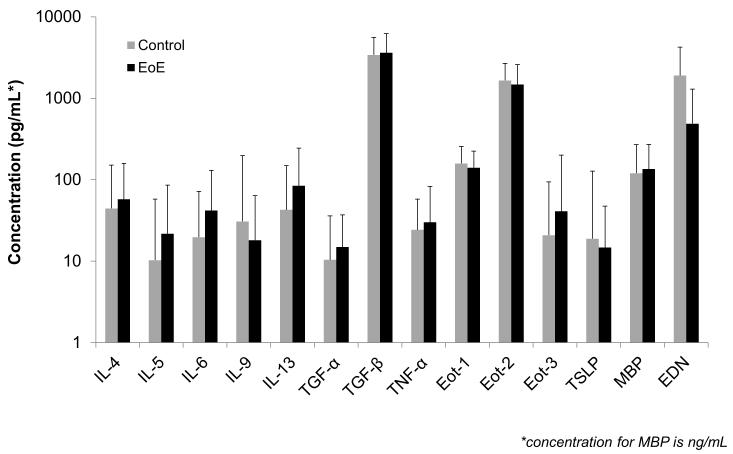
At baseline, there were no significant differences in any of the biomarkers between cases and controls (Figure 1). For example, mean values of IL-5, IL-13, eotaxin-3, and TSLP for cases and controls were 22 ±64 vs 10 ±47 pg/mL (p=0.21), 85 ±160 vs 43 ±161 (p=0.12), 41 ±159 vs 21 ±73 (p=0.30), and 15 ±33 vs 19 ±109 (p=0.77). Median values for the same comparisons did not change the results. ROC analysis confirmed that there was little diagnostic utility for the biomarkers either individually (AUCs ranging from 0.40 to 0.68) or in sum (AUC 0.69). There were also no differences between cases and controls after stratification by atopic status (data not shown). Of note, tissue levels of MBP, eotaxin-3, and mast cell tryptase were markedly elevated in esophageal biopsies from a subset of cases as compared to controls (Supplemental Table).
Figure 1.
Comparison of serum biomarkers for cases of EoE (black bars) and non-EoE controls (gray bars). The top of the bars represent the mean biomarker values, and the error bars represent the standard deviation. The y axis is on a log scale, and all concentrations are in pg/mL with the exception of major basic protein (MBP) which is in ng/mL.
Post-treatment serum biomarkers
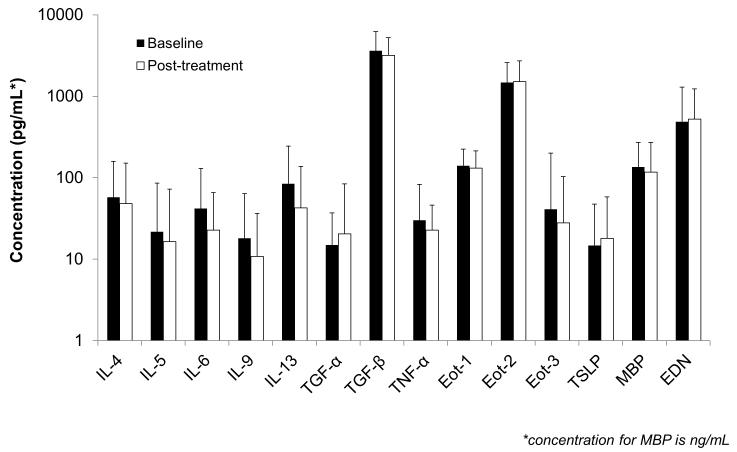
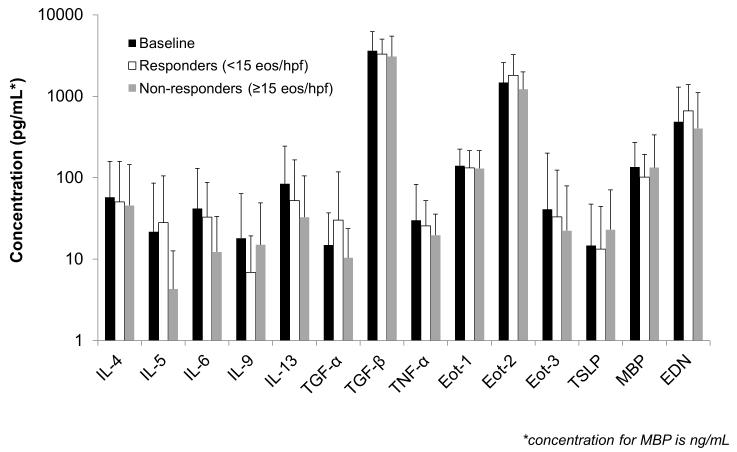
A total of 51 EoE cases had paired pre- and post-treatment serum available for analysis. There were no significant differences detected overall before and after treatment (Figure 2a). For example, values of IL-5, IL-13, eotaxin-3, and TSLP pre- and post-treatment were 19 ±57 vs 17 ±56 pg/mL (p=0.81), 71 ±152 vs 43 ±95 (p=0.18), 42 ±174 vs 28 ±76 (p=0.41), and 14 ±33 vs 18 ±40 (p=0.29). Median values for the same comparisons did not change the results. There were also no differences detected after stratification by treatment responder status, either at the <15 eos/hpf level (Figure 2b) or at the <1 eos/hpf level (data not shown). There were 17 controls with follow-up specimens, and there were no differences in any of the serum biomarker levels over time in this group (Supplemental Figure).
Figure 2.
(A) Comparison of serum biomarkers for cases of EoE before (black bars) and after (white bars) treatment with a topical corticosteroid. (B) Comparison for cases of EoE stratified by treatment response with pre-treatment (black bars), post-treatment responders at the <15 eos/hpf level (white bars), and non-responders at the ≥15 eos/hpf level (gray bars). For both panels, the top of the bars represent the mean biomarker values, the error bars represent the standard deviation, the y axis is on a log scale, and all concentrations are in pg/mL with the exception of major basic protein (MBP) which is in ng/mL.
Discussion
Given the serial upper endoscopies required for diagnosis and monitoring of EoE, current clinical practice is cumbersome for patients, invasive, and expensive. Non-invasive methods for diagnosis and monitoring would be extremely valuable in this condition, and a serum test for a biomarker panel would be ideal. In this prospective study that collected clinical, endoscopic, histologic, and biologic data on incident cases of EoE and non-EoE controls, we investigated the utility of a large number of the most promising serum biomarkers for both diagnosis and monitoring of EoE. Despite choosing markers shown in multiple studies to be increased in esophageal tissue in EoE, we were unable to demonstrate any difference in the serum measures between cases and controls, nor did we find any that were responsive to treatment. These results were unaffected by atopy status of the cases and controls, or by the level of treatment response measured histologically.
Previous studies have identified multiple biomarkers that are characteristic of EoE at the esophageal tissue level. These include Th2-related cytokines, most notably IL-5 and IL-13, chemokines such as eotaxin-3, eosinophil granule proteins, microRNAs, and mast cells (13-23, 25-29, 37, 38, 41, 44-49). However, data on serum biomarkers are sparse and conflicting, and primarily come from either small studies examining mechanisms of EoE or sub-analyses of clinical trials focusing on a few cytokines, chemokines, or granule proteins. The preponderance of the data are also in children.
Konikoff and colleagues studied 47 children, 16 of whom had active EoE, and assessed a number of plasma biomarkers, including IL-5, eotaxin-1, -2, and -3, and EDN (19). They found that plasma EDN and eotaxin-3 correlated with tissue eosinophil levels and were also significantly increased in active EoE compared with controls, though the differences were mild (50.3 vs 31.1 ng/mL for EDN and 37.7 vs 11.5 pg/mL for eotaxin-3). Notably, they did not report differences in IL-5, eotaxin-1, or -2. Changes in eotaxin-1 and -3 were also not observed following treatment with an anti-IL-13 antibody in a recent trial (50). In contrast, a mechanistic study of the role of fibroblast growth factor by Huang and colleagues showed elevated levels of plasma IL-5 and IL-13 in 35 pediatric EoE cases compared with 8 healthy controls (51). IL-5 was also noted to be elevated in EoE compared to controls in a study of eosinophil function in 12 adults with EoE (40). Subbarao and colleagues studied serum IL-5 and EDN in 60 children with EoE and 20 controls and found that while EDN levels were significantly higher in cases compared to controls (23.5 vs 2.7 ng/mL), IL-5 levels were not (18). They also found that serum EDN levels significantly decreased after treatment. Of studies that have examined serum eosinophil cationic protein (ECP) after treatment for EoE, two studies showed that ECP decreased after treatment (52, 53) while one found no change (54). We are unaware of any prior published studies examining serum MBP in EoE. Our study does not confirm the previous findings related to EDN or IL-5, and suggests that none of the panel of cytokines, chemokines, and eosinophil granules that we examined has utility as a biomarker panel.
When interpreting the data from this study, there are potential limitations to consider. Because the results were not positive, there is the possibility of a type II error. However, this is the largest study to date assessing biomarkers for diagnosis and monitoring of EoE, and it was powered to detect clinical meaningful differences based on ROC curve analysis. It is possible that different power calculation methods could have increased the sample size required, but given the lack of suggestive trends in our data, if any statistically significant differences between the assays could be detected with larger numbers of subjects, the clinical utility of such differences would be doubtful. Differences in specimen handling or degradation over time are unlikely to have contributed to our results, and analysis of a subset of controls with samples over time showed stability in the biomarker measures. All samples were handled identically, remained frozen until analysis, and were there to be degradation it would likely be non-differential for both the case and control groups. Finally, we did not include a PPI-REE group in the study, as the main goals were to develop biomarkers to detect EoE and monitor the response of EoE to treatment over time, not to distinguish EoE from PPI-REE. Therefore, we cannot comment on the utility of these serum biomarkers in patients with PPI-REE. While it has been recently reported that PPIs have anti-inflammatory and anti-eosinophilic effects independent of their anti-acid effect (55, 56), because the EoE cases in the present study all had a high tissue level of eosinophilic inflammation despite PPI use, it is not likely that PPIs impacted serum biomarker levels, but we are not able to test this directly with our study design.
Given the lack of signal in the serum for our biomarker panel, it may be that the brisk esophageal inflammation is not reflected systemically. While we did not measure the levels of the serum biomarkers in the esophageal biopsy specimens of the subjects included in this study either at baseline or after steroid treatment, we feel that it would be unlikely for baseline tissue levels to be low in these patients. For example, the EoE cases were highly inflamed, with an average peak esophageal eosinophil count of 146/hpf, and a subset of the EoE subjects who did have staining for MBP, eotaxin-3, and mast cell tryptase had markedly elevated tissue levels compared to controls. Minimally invasive diagnostic techniques are under development to sample the esophagus (57, 58), and these might be more amenable to a biomarker panel.
Our study also has a number of strengths. It was a prospective study specifically designed to evaluate biomarkers in a population of well-characterized incident EoE cases and non-EoE controls. The omission of prevalent cases makes it impossible that previous medical or dietary therapy for EoE would account for any observed differences between cases and controls. It is the largest study to date focusing on biomarkers for diagnosis and monitoring of EoE, and the only one of its kind to be done in an adult population. Uniform methods were used for case/control identification, sample handling, and analysis, and all baseline samples were obtained prior to the EoE diagnosis being known. Follow-up samples were obtained with identical methods as at baseline, and outcomes (eosinophil counts; biomarker levels) were quantified in blinded fashion, using rigorous, previously validated methods. These methodologic strengths make the data reliable and valid.
In conclusion, in this large prospective study, a panel of inflammatory markers associated with EoE pathogenesis and known to be elevated in esophageal tissue of patients with EoE were not increased in the serum of EoE patients at baseline compared to non-EoE controls. These markers also were unresponsive to treatment, even in the face of marked decrements in the esophageal eosinophil count. Therefore, none of these biomarkers are likely candidates for a serum test. Instead, focus should move to novel blood-based biomarkers for diagnosing and monitoring EoE, as well as the development of more economical, non-endoscopic methods of sampling the esophageal mucosa.
Supplementary Material
Study highlights.
What is current knowledge?
Diagnosis and management of eosinophilic esophagitis (EoE) currently requires repeated upper endoscopy with biopsies, an invasive and expensive procedure.
Previous studies have suggested that inflammatory factors can be detected at elevated levels in the blood of patients with EoE, but their clinical utility for diagnosis and monitoring EoE is not established.
What is new here?
This large prospective cohort study of adults with EoE analyzed a panel of serum biomarkers based on the known pathogenesis of EoE.
There was no difference in serum biomarker levels of IL-4, IL-5, IL-6, IL-9, IL-13, TGF-α, TGF-β, TNF-α, eotaxin-1, -2, and -3, TSLP, major basic protein (MBP), or eosinophil-derived neurotoxin (EDN) in EoE cases compared to non-EoE controls at baseline.
There was also no difference in the biomarker levels before or after treatment of EoE cases with topical corticosteroids.
Novel serum biomarkers are needed for less invasive detection and monitoring of EoE.
Acknowledgement
We gratefully acknowledge Carlton W. Anderson of the Advanced Analytics Core of the UNC Center for Gastrointestinal Biology and Disease for assay support and technical services, and Mervi M. Eeva and Bentley R. Midkiff of the UNC Translational Pathology lab for their technical assistance.
Financial support:
Grant support: This research was conducted with support, in part, by NIH awards K23DK090073 (ESD) and K24DK100548 (NJS). It also utilized the Histology and Advanced Analytics Cores of the UNC Center for Gastrointestinal Biology and Disease which is funded by NIH P30DK034987, and the UNC Translational Pathology lab which is funded by NIH P30CA016086.
The study sponsors had no role in the study design, collection, analysis, or interpretation of the data.
Dr. Dellon has received research funding from AstraZeneca and Meritage, an educational grant from Diagnovus, and is a consultant for Aptalsis, Novartis, Receptos, and Regeneron.
Footnotes
Guarantor of the article: Evan Dellon
Specific author contributions (all authors approved the final draft):
Dellon: project conception/design; data acquisition/analysis/interpretation; drafting of the article; critical revision
Rusin: data acquisition (slide review for eosinophil counts); critical revision
Gebhart: patient recruitment; data acquisition/management; critical revision
Covey: data acquisition (slide review for eosinophil counts); critical revision
Higgins: patient recruitment; data acquisition/management; critical revision
Beitia: patient recruitment; data acquisition/management; critical revision
Speck: data acquisition (slide review for eosinophil counts); critical revision
Woodward: data acquisition (slide review for eosinophil counts); critical revision
Woosley: pathology supervision; eosinophil count review; and critical revision
Shaheen: project conception and design; supervision; data interpretation; critical revision
Potential competing interests:
None of the other authors have any relevant conflicts of interest to disclose.
References
- 1.Dellon ES, Gonsalves N, Hirano I, et al. ACG Clinical Guideline: Evidence based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis. Am J Gastroenterol. 2013;108:679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 2.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20.e6. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 3.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–63. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Dellon ES, Liacouras CA. Advances in Clinical Management of Eosinophilic Esophagitis. Gastroenterology. 2014;147:1238–1254. doi: 10.1053/j.gastro.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen ET, Kappelman MD, Martin CF, et al. Health-Care Utilization, Costs, and the Burden of Disease Related to Eosinophilic Esophagitis in the United States. Am J Gastroenterol. 2014 doi: 10.1038/ajg.2014.316. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta SK. Noninvasive markers of eosinophilic esophagitis. Gastrointest Endosc Clin N Am. 2008;18:157–67. xi. doi: 10.1016/j.giec.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Menard-Katcher C, Furuta GT. Non- and semi-invasive methods of monitoring eosinophilic esophagitis. Dig Dis. 2014;32:102–6. doi: 10.1159/000357295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhardwaj N, Ghaffari G. Biomarkers for eosinophilic esophagitis: a review. Ann Allergy Asthma Immunol. 2012;109:155–9. doi: 10.1016/j.anai.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology. 2009;137:1238–49. doi: 10.1053/j.gastro.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wechsler JB, Bryce PJ. Allergic mechanisms in eosinophilic esophagitis. Gastroenterol Clin North Am. 2014;43:281–96. doi: 10.1016/j.gtc.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherrill JD, Rothenberg ME. Genetic and epigenetic underpinnings of eosinophilic esophagitis. Gastroenterol Clin North Am. 2014;43:269–80. doi: 10.1016/j.gtc.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng E, Souza RF, Spechler SJ. Tissue remodeling in eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1175–87. doi: 10.1152/ajpgi.00313.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straumann A, Bauer M, Fischer B, et al. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–61. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 14.Straumann A, Kristl J, Conus S, et al. Cytokine expression in healthy and inflamed mucosa: probing the role of eosinophils in the digestive tract. Inflamm Bowel Dis. 2005;11:720–6. doi: 10.1097/01.mib.0000172557.39767.53. [DOI] [PubMed] [Google Scholar]
- 15.Gupta SK, Fitzgerald JF, Kondratyuk T, et al. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2006;42:22–6. doi: 10.1097/01.mpg.0000188740.38757.d2. [DOI] [PubMed] [Google Scholar]
- 16.Noel RJ, Putnam PE, Collins MH, et al. Clinical and immunopathologic effects of swallowed fluticasone for eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2004;2:568–75. doi: 10.1016/s1542-3565(04)00240-x. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita Y, Furuta K, Ishimura N, et al. Elevated plasma cytokines in Japanese patients with eosinophilic esophagitis and gastroenteritis. Digestion. 2012;86:238–43. doi: 10.1159/000341421. [DOI] [PubMed] [Google Scholar]
- 18.Subbarao G, Rosenman MB, Ohnuki L, et al. Exploring potential noninvasive biomarkers in eosinophilic esophagitis in children. J Pediatr Gastroenterol Nutr. 2011;53:651–8. doi: 10.1097/MPG.0b013e318228cee6. [DOI] [PubMed] [Google Scholar]
- 19.Konikoff MR, Blanchard C, Kirby C, et al. Potential of blood eosinophils, eosinophil-derived neurotoxin, and eotaxin-3 as biomarkers of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4:1328–36. doi: 10.1016/j.cgh.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya B, Carlsten J, Sabo E, et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol. 2007;38:1744–1753. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kephart GM, Alexander JA, Arora AS, et al. Marked deposition of eosinophil-derived neurotoxin in adult patients with eosinophilic esophagitis. Am J Gastroenterol. 2010;105:298–307. doi: 10.1038/ajg.2009.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dellon ES, Chen X, Miller CR, et al. Diagnostic utility of major basic protein, eotaxin-3, and leukotriene enzyme staining in eosinophilic esophagitis. Am J Gastroenterol. 2012;107:1503–11. doi: 10.1038/ajg.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellon ES, Speck O, Woodward K, et al. Markers of Eosinophilic Inflammation for Diagnosis of Eosinophilic Esophagitis and Proton Pump Inhibitor-Responsive Esophageal Eosinophilia: A Prospective Study. Clin Gastroenterol Hepatol. 2014;12:2015–22. doi: 10.1016/j.cgh.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teitelbaum JE, Fox VL, Twarog FJ, et al. Eosinophilic esophagitis in children: immunopathological analysis and response to fluticasone propionate. Gastroenterology. 2002;122:1216–25. doi: 10.1053/gast.2002.32998. [DOI] [PubMed] [Google Scholar]
- 26.Zuo L, Fulkerson PC, Finkelman FD, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha2-inhibited pathway. J Immunol. 2010;185:660–9. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanchard C, Stucke EM, Burwinkel K, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–41. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucendo AJ, De Rezende L, Comas C, et al. Treatment with topical steroids downregulates IL-5, eotaxin-1/CCL11, and eotaxin-3/CCL26 gene expression in eosinophilic esophagitis. Am J Gastroenterol. 2008;103:2184–93. doi: 10.1111/j.1572-0241.2008.01937.x. [DOI] [PubMed] [Google Scholar]
- 29.Blanchard C, Mingler MK, Vicario M, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Gonsalves N, Policarpio-Nicolas M, Zhang Q, et al. Histopathologic variability and endoscopic correlates in adults with eosinophilic esophagitis. Gastrointest Endosc. 2006;64:313–9. doi: 10.1016/j.gie.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 31.Dellon ES, Speck O, Woodward K, et al. Distribution and variability of esophageal eosinophilia in patients undergoing upper endoscopy. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dellon ES, Fritchie KJ, Rubinas TC, et al. Inter- and intraobserver reliability and validation of a new method for determination of eosinophil counts in patients with esophageal eosinophilia. Dig Dis Sci. 2010;55:1940–9. doi: 10.1007/s10620-009-1005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellon ES, Aderoju A, Woosley JT, et al. Variability in diagnostic criteria for eosinophilic esophagitis: A systematic review. Am J Gastroenterol. 2007;102:2300–13. doi: 10.1111/j.1572-0241.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 34.Dellon ES, Sheikh A, Speck O, et al. Viscous Topical is More Effective than Nebulized Steroid Therapy for Patients with Eosinophilic Esophagitis. Gastroenterology. 2012;143:321–324. doi: 10.1053/j.gastro.2012.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander JA, Jung KW, Arora AS, et al. Swallowed Fluticasone Improves Histologic but Not Symptomatic Responses of Adults with Eosinophilic Esophagitis. Clin Gastroenterol Hepatol. 2012;10:742–9.e1. doi: 10.1016/j.cgh.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Butz BK, Wen T, Gleich GJ, et al. Efficacy, Dose Reduction, and Resistance to High-dose Fluticasone in Patients with Eosinophilic Esophagitis. Gastroenterology. 2014;147:324–33.e5. doi: 10.1053/j.gastro.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchard C, Stucke EM, Rodriguez-Jimenez B, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. 217, e1–7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aceves SS, Chen D, Newbury RO, et al. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–204. e4. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 39.Aceves SS, Newbury RO, Chen D, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65:109–16. doi: 10.1111/j.1398-9995.2009.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnsson M, Bove M, Bergquist H, et al. Distinctive blood eosinophilic phenotypes and cytokine patterns in eosinophilic esophagitis, inflammatory bowel disease and airway allergy. J Innate Immun. 2011;3:594–604. doi: 10.1159/000331326. [DOI] [PubMed] [Google Scholar]
- 41.Dellon ES, Chen X, Miller CR, et al. Tryptase staining of mast cells may differentiate eosinophilic esophagitis from gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:264–71. doi: 10.1038/ajg.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf WA, Green DJ, Hughes JT, et al. What cut-point should be used to define a histologic response to topical steroid use in eosinophilic esophagitis? A data-driven approach using symptoms and endoscopic findings. Gastroenterology. 2014;146(Suppl 1):S665–666. (Mo1832) [Google Scholar]
- 43.Hintze J. PASS. NCSS, LLC; Kaysville, Utah: 2008. www.ncss.com. 2008. [Google Scholar]
- 44.Lu TX, Lim EJ, Wen T, et al. MiR-375 is downregulated in epithelial cells after IL-13 stimulation and regulates an IL-13-induced epithelial transcriptome. Mucosal Immunol. 2012;5:388–96. doi: 10.1038/mi.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu TX, Sherrill JD, Wen T, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129:1064–75. e9. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Protheroe C, Woodruff SA, de Petris G, et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:749–755. e11. doi: 10.1016/j.cgh.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirsch R, Bokhary R, Marcon MA, et al. Activated mucosal mast cells differentiate eosinophilic (allergic) esophagitis from gastroesophageal reflux disease. J Pediatr Gastroenterol Nutr. 2007;44:20–6. doi: 10.1097/MPG.0b013e31802c0d06. [DOI] [PubMed] [Google Scholar]
- 48.Lucendo AJ, Arias A, De Rezende LC, et al. Subepithelial collagen deposition, profibrogenic cytokine gene expression, and changes after prolonged fluticasone propionate treatment in adult eosinophilic esophagitis: a prospective study. J Allergy Clin Immunol. 2011;128:1037–46. doi: 10.1016/j.jaci.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Colombo JM, Neilan NA, Schurman JV, et al. Validation of methods to assess potential biomarkers in pediatric patients with esophageal eosinophilia. World J Gastrointest Pharmacol Ther. 2013;4:113–9. doi: 10.4292/wjgpt.v4.i4.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 51.Huang JJ, Joh JW, Fuentebella J, et al. Eotaxin and FGF enhance signaling through an Extracellular signal-related kinase (ERK)-dependent pathway in the pathogenesis of Eosinophilic Esophagitis. Allergy Asthma Clin Immunol. 2010;6:25. doi: 10.1186/1710-1492-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlag C, Pfefferkorn S, Brockow K, et al. Serum Eosinophil Cationic Protein is Superior to Mast Cell Tryptase as Marker for Response to Topical Corticosteroid Therapy in Eosinophilic Esophagitis. J Clin Gastroenterol. 48:600–6. doi: 10.1097/01.mcg.0000436439.67768.8d. 201. [DOI] [PubMed] [Google Scholar]
- 53.Straumann A, Conus S, Grzonka P, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut. 2010;59:21–30. doi: 10.1136/gut.2009.178558. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Sanchez J, Gomez-Torrijos E, De-la-Santa-Belda E, et al. Effectiveness of serological markers of eosinophil activity in monitoring eosinophilic esophagitis. Rev Esp Enferm Dig. 2013;105:462–468. doi: 10.4321/s1130-01082013000800004. [DOI] [PubMed] [Google Scholar]
- 55.Cheng E, Zhang X, Huo X, et al. Omeprazole blocks eotaxin-3 expression by oesophageal squamous cells from patients with eosinophilic oesophagitis and GORD. Gut. 2013;62:824–32. doi: 10.1136/gutjnl-2012-302250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Cheng E, Huo X, et al. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS One. 2012;7:e50037. doi: 10.1371/journal.pone.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furuta GT, Kagalwalla AF, Lee JJ, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut. 2013;62:1395–405. doi: 10.1136/gutjnl-2012-303171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katzka DA, Geno DM, Ravi A, et al. Accuracy, saftey, and tolerability of tissue collection by cytosponge vs endoscocpy for evaluation of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:77–83.e2. doi: 10.1016/j.cgh.2014.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.