Abstract
Background
Infections of Echinococcus granulosus sensu stricto (s.s), E. multilocularis and E. shiquicus are commonly found co-endemic on the Qinghai-Tibet plateau, China, and an efficient tool is needed to facilitate the detection of infected hosts and for species identification.
Methodology/Principal Findings
A single-tube multiplex PCR assay was established to differentiate the Echinococcus species responsible for infections in intermediate and definitive hosts. Primers specific for E. granulosus, E. multilocularis and E. shiquicus were designed based on sequences of the mitochondrial NADH dehydrogenase subunit 1 (nad1), NADH dehydrogenase subunit 5 (nad5) and cytochrome c oxidase subunit 1 (cox1) genes, respectively. This multiplex PCR accurately detected Echinococcus DNA without generating nonspecific reaction products. PCR products were of the expected sizes of 219 (nad1), 584 (nad5) and 471 (cox1) bp. Furthermore, the multiplex PCR enabled diagnosis of multiple infections using DNA of protoscoleces and copro-DNA extracted from fecal samples of canine hosts. Specificity of the multiplex PCR was 100% when evaluated using DNA isolated from other cestodes. Sensitivity thresholds were determined for DNA from protoscoleces and from worm eggs, and were calculated as 20 pg of DNA for E. granulosus and E. shiquicus, 10 pg of DNA for E. multilocularis, 2 eggs for E. granulosus, and 1 egg for E. multilocularis. Positive results with copro-DNA could be obtained at day 17 and day 26 after experimental infection of dogs with larval E. multilocularis and E. granulosus, respectively.
Conclusions/Significance
The multiplex PCR developed in this study is an efficient tool for discriminating E. granulosus, E. multilocularis and E. shiquicus from each other and from other taeniid cestodes. It can be used for the detection of canids infected with E. granulosus s.s. and E. multilocularis using feces collected from these definitive hosts. It can also be used for the identification of the Echinococcus metacestode larva in intermediate hosts, a stage that often cannot be identified to species on visual inspection.
Author Summary
The canid adapted intestinal tapeworms, Echinococcus granulosus, E. multilocularis and E. shuiqucus are well known to be endemic in Northwestern China. The first two species can cause fatal disease in humans. Although E. shiquicus has not been reported to infect humans, all three species can be transmitted by dogs. The very close relationship between dogs and humans can readily lead to human infection. To aid the surveillance and management of echinococcosis, effective diagnostic approaches are urgently needed. We developed a single tube multiplex PCR assay for the accurate identification and discrimination of the three Echinococcus species for use in both clinical diagnosis and epidemiological studies.
Introduction
In the most recent taxonomic revision, nine species were recognized in the genus Echinococcus [1]. Of these, the most important and widespread are E. granulosus sensu stricto (genotypes G1-G3) and E. multilocularis, which cause cystic echinococcosis (CE) and alveolar echinococcosis (AE), respectively. The former is commonly associated with livestock and human infections worldwide whereas the latter is primarily found in voles and humans and is geographically limited to the northern hemisphere [2]. To date, E. granulosus s.s., E. canadensis (G6), E. multilocularis and E. shiquicus have been identified in China [3–5]. Both E. multilocularis and E. granulosus s.s. are particularly widespread in western China, including Qinghai, Ningxia, Gansu, Xinjiang and Sichuan provinces/autonomous regions, and are well known as major public health and medical threats. Unlike the other species, E. shiquicus has a very restricted distribution, being reported only from Qinghai Province, China. This species is not known to cause human echinococcosis. The intermediate hosts are plateau pikas (Ochotona curzoniae), in which unilocular cysts occur.
For Echinococcus species in general, dogs, wolves, other canids and cats are definitive hosts in which adult worms cause sub-clinical infections [6–9]. However, larval Echinococcus spp. can cause morbidity and mortality in their intermediate hosts which include cattle, sheep, small mammals (including rodents, plateau pikas, etc.) and humans [10, 11]. It can be difficult to discriminate morphologically adults of some Echinococcus species, such as E. multilocularis and E. shiquicus [12].
To replace traditional morphological methods, a number of molecular approaches targeting parasite DNA have been developed for identification/discrimination of different life stages of Echinococcus species in definitive and intermediate hosts [13–15]. Multiplex PCR approaches, simultaneously using multiple specific primers in a single tube and detecting more than one target species, are material- and time-saving, precise, efficient and cost-effective when DNA from a mixture of pathogens may be present in a sample. This approach is also suitable for mass-screening of samples that may be generated from epidemiological investigations in endemic areas. Several multiplex PCR methods have been developed for identifying certain Echinococcus species, but none for the identification of E. shiquicus [16–17].
Based on interspecific variation in mitochondrial genes of the genus Echinococcus, we designed a multiplex PCR assay with three pairs of specific primers in a single reaction tube for rapid identification of E. granulosus s.s., E. multilocularis and E. shiquicus originating from either intermediate or definitive hosts. Further assessment of the sensitivity and specificity of the multiplex PCR assay was performed using metacestode DNA and copro-DNA to determine the reliability and accuracy of the new diagnostic tool developed in this study.
Materials and Methods
Ethics statement
Dogs and mice used in this study were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People's Republic of China (Regulations for Administration of Affairs Concerning Experimental Animals, China, 1988). No endangered/protected species were involved in this study. The dogs and mice used were also treated in accordance with the institutional procedures and guidelines for animal husbandry issued by the Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Approval No. LVRIEC2010-005).
Sampling of Echinococcus material
Adult worms were collected from stray dogs during routine work of the endemic echinococcosis prevention and control program in Dari County, Qinghai Province, P.R. China. A total of 86 Echinococcus spp. metacestode samples from yaks, sheep, Qinghai voles (Microtus/Neodon fuscus) and plateau pikas were collected on the Qinghai-Tibet plateau, P.R. China. Ten yak lungs and 16 sheep livers harboring hydatid cysts were collected from abattoirs in Maqu County, Gansu Province and Xining City, Qinghai Province, respectively. Thirty Qinghai vole livers and 30 plateau pika lungs harboring hydatid cysts were provided by the epidemic prevention station of Dari County, Qinghai Province.
Parasite materials were dissected from the host tissue and stored either in 70% ethanol before molecular analyses, or temporarily stored at 4°C prior to experimental infections of dogs.
Experimental infection of dogs
Fifteen dogs (mixed breeds) aged 6–8 months were purchased in Lanzhou City, Gansu Province, China. These were de-wormed using praziquantel and confirmed to be free of intestinal parasites by examination of their feces two weeks later. Samples of these feces were retained as negative controls for the multiplex PCR assay. Live protoscoleces (100,000) of each Echinococcus spp. were fed independently to five dogs after their viability for dog challenge was confirmed by microscopy.
Sampling of adults/eggs of Echinococcus spp. from challenged dogs
Dogs were euthanized three months after challenge with protoscoleces. Fecal samples were collected from the dogs each day prior to sacrifice. After removal of the coarse gut contents, the small intestine was cut into 15–20 cm lengths and opened to expose the mucosa. Samples, taken by scraping the mucosa with glass strips, were placed in petri dishes in bio-safety containers [18]. After addition of a small volume of sterile phosphate-buffered saline (PBS, pH 7.2), the contents were checked for the presence of worms (intact or fragmented) and/or eggs. Adult worms were removed using a glass needle and washed in PBS three times. All procedures were performed following appropriate bio-safety conditions [19].
Fecal sampling from non-experimented definitive hosts
Ten stray dogs, provided by the epidemic prevention station in Dari County, Qinghai Province, were processed as above to obtain mucosal samples, worms and eggs. Additionally, five fecal samples from captive foxes were collected from a fur farm in Lanzhou City, Gansu Province. All the collected fecal samples were frozen at -80°C for at least seven days for bio-safety reasons. Worm samples were preserved either in 70% ethanol or frozen at (-80°C) in PBS for further analyses.
Other helminths
DNA samples, extracted from a variety of cestodes (identities confirmed by sequencing and morphology), were used to determine the specificity of the newly developed multiplex PCR assay (Table 1). They were kindly provided by the Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, CAAS.
Table 1. Parasite species and their GenBank accession numbers for the nad1, nad5 and cox1 genes used in this study.
| Parasite | Sequence accession no. | Reference |
|---|---|---|
| E. granulosus s.s. | NC_008075/ AF297617 | Yang et al, 2005 [22]; Le et al, 2000 [35]. |
| E. multilocularis | NC_000928 | Nakao et al, 2002 [23]. |
| E. shiquicus | NC_009460 | Nakao et al, 2007 [24]. |
| E. oligarthrus | AB208545/ NC_009461 | Nakao et al, 2007 [24]. |
| E. canadensis | NC_011121/ AB235847 | Nakao et al, 2007 [24]. |
| E. ortleppi | NC_011122 | Nakao et al, 2007 [24]. |
| E. vogeli | NC_009462 | Nakao et al, 2007 [24]. |
| E. canadensis (G6)* | AB208063 | Nakao et al, 2007 [24]. |
| T. hydatigena* | GQ228819/ FJ518620 | Jia et al, 2010 [36]; Liu et al, 2011 [37]. |
| T. multiceps* | GQ228818/ FJ495086 | Jia et al, 2010 [36]; Liu et al, 2011 [37]. |
| T. pisiformis* | GU569096 | Jia et al, 2010 [36]. |
| T. asiatica | AF445798 | Jeon et al, 2005 [38]. |
| T. saginata | NC009938/ AY684274 | Jeon et al, 2007 [39]. |
| T. taeniaeformis* | JQ663994/ FJ597547 | Jia et al. 2012 [40]; Liu et al, 2011 [37]. |
| T. solium* | AB086256 | Nakao et al, 2003 [41]. |
| D. caninum* | AB732959 | Nakao et al, 2013 [42]. |
Note: DNA from parasites labeled with * were used to test the specificity of the multiplex PCR system in this study.
Host tissue sampling
DNA extracted from host tissues was used to check for nonspecific reactions or assay interference that might be caused by contamination of parasite samples with host DNA. Host tissues included dog intestines, and liver and lung samples from cattle, sheep, Qinghai voles and plateau pikas.
DNA extraction from samples
Two hundred mg of each metacestode sample was frozen in liquid nitrogen and ground to powder after removal of ethanol or PBS by rinsing with ddH2O. Total genomic DNA was extracted using a QIAGEN DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions and stored at -20°C until use.
To minimize the impact of inhibitors on PCR using copro-samples as template, an additional step of stool flotation in saturated zinc chloride solution was used before copro-DNA extraction [20]. Briefly, about 20 g (20 ml) fecal material was placed in a 50 ml centrifuge tube, which was then filled with zinc chloride solution. The tube was vortexed until the fecal material was completely broken up. The tube was then centrifuged at 1000 ×g for 5 min. Five hundred μl of the supernatant (usually containing helminth eggs, proglottids or cells of parasites) was transferred to a 2 ml centrifuge tube, 1.5 ml ddH2O was added to dilute the solution, and the tube was centrifuged at 12,000 ×g for 10 min. The supernatant was carefully discarded and 200 μl ddH2O added to suspend the sediment for DNA extraction. Total genomic DNA was extracted using a QIAGEN QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions, and the DNA concentration was determined using a spectrophotometer (Thermo, NanoDrop 2000, USA) after elution in 50 μl ddH2O for use in the PCR assay.
Genomic DNA was extracted from host tissues using a QIAGEN DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions, and stored at -20°C until use.
Primer design
The complete mt genomes (mtDNA) of various cestodes (Table 1) available in GenBank (http://www.ncbi.nlm.nih.gov/) were retrieved to facilitate design of primers specific for E. granulosus s.s., E. multilocularis and E. shiquicus (Table 1). The sequences were aligned automatically using Clustal in MEGA5.0 [21]. Primer pairs, expected to be specific for E. granulosus s.s. (S1 Fig), E. multilocularis (S2 Fig) and E. shiquicus (S3 Fig), were thus obtained. After some preliminary experimentation, one pair of primers specific for each Echinococcus spp. was selected for inclusion in the multiplex PCR assay. Sequences of these primers, target genes and other related information are presented in Table 2.
Table 2. Sequences of primers used in the multiplex PCR.
| Species | Gene | Primer | Sequence (5'-3') | Amplicon size (bp) | Concentration (nM) |
|---|---|---|---|---|---|
| E. granulosus s.s. | nad1 | F-Eg | GGTTTTATCGGTATGTTGGTGTTAGTG | 219 | 100 |
| R-Eg | CATTTCtTGAAGTTAACAGCATCACG | ||||
| E. multilocularis | nad5 | F-Em | CATTAATTATGGATGTTTCC | 584 | 50 |
| R-Em | GGAAATACCCCACTATCC | ||||
| E. shiquicus | cox1 | F-Es | GCTTTAAGTGCGTGACTTTTAATCCC | 471 | 100 |
| R-Es | CATCAAAACCAGCACTAATACTCA |
Multiplex PCR assay
PCR amplification was carried out in a 25 μl mixture containing 2 μl dNTPs (2.5 mM of each), 2.5 μl 10× ExTaq Buffer (Mg2+ free), 2 μl MgSO4 (25 mM), 0.25 μl ExTaq DNA polymerase (5U/μl) (TaKaRa, Dalian, Liaoning), 100 pg DNA template of each Echinococcus sample, and all three primer pairs were added according to the final concentrations given in Table 2. Fragments were amplified using the following optimized thermocycling conditions: 95°C/5 min for denaturation; 30 cycles of 94°C/30 sec, 55°C/30 sec, 72°C/40 sec; and 72°C/10 min extension. For all the multiplex PCR assays, positive DNA (DNA templates of the three Echinococcus spp.) and negative (no-DNA) controls were included.
Identification of PCR products
Amplicons were visualized by electrophoresis in 2.0% (w/v) agarose gels in 1×TAE (40 mM Tris-acetate, 2 mM EDTA, pH 8.5), stained with ethidium bromide (EB), and viewed under UV light. The fragments were purified using an agarose Gel DNA Purification Kit (TaKaRa, Dalian, Liaoning), and then cloned into pMD18-T Simple vectors using a TA cloning strategy. The recombinant vectors were identified by enzyme digestion and at least two clones for each DNA region were sequenced by the Shanghai Invitrogen Biotechnology Co. Ltd.
Controls for the multiplex PCR assay
Positive control for fecal sample tests
DNA from protoscoleces of the three Echinococcus spp. was added to a fecal sample as positive control. Another type of positive control was provided by the copro-samples that were directly collected from dogs successfully infected with E. multilocularis or E. granulosus s.s.
Negative controls
To exclude the possibility of contamination in the PCR amplification, two negative fecal samples (no-DNA) were used. Other negative controls included all reagents except for the addition of parasite DNA.
Fecal inhibitor controls
To test for potential inhibitors, DNA extracted from protoscoleces and identified by gene sequencing was added to a negative fecal sample, and subjected to the multiplex PCR in parallel with the negative fecal sample.
Host tissue controls
DNAs extracted from tissues of dogs and foxes as well as those from intermediate hosts were tested using the multiplex PCR assay to determine the minimum contamination level that could cause interference in the assay. Host DNA (0.1, 0.5, 1, 5, 10, 50, 100, 500 or 1000 ng) was mixed into each relevant parasite DNA sample prior to the assay.
Specificity and sensitivity
Specificity
Three pairs of primers were added to each PCR tube with the optimized multiplex PCR reaction conditions (described above) to test various parasite DNA samples as listed in Table 1.
Lowest/highest detection limit of DNA using Echinococcus larval tissue
DNA samples from protoscoleces of the three Echinococcus spp. were quantified by spectrophotometry using a NanoDrop 2000 (Thermo Scientiific, Wilmington, DE, USA). Serial dilutions of the DNA template (0.01, 0.02, 0.05, 0.1, 0.5, 1, 5, 10, 50, 100, 500 and 1000 ng) were used to assay the analytical sensitivity and potential nonspecific amplification of DNA in the multiplex PCR system. Amplification results were visualized by electrophoresis in a 2.0% (w/v) agarose gel.
Minimum numbers of eggs detectable in fecal samples
One to ten Echinococcus eggs were added to the diluted negative fecal samples. DNA extracted from these samples was used in the multiplex reaction to determine the minimum number of eggs that could yield a positive PCR outcome.
Earliest time post-infection on which dog fecal samples yielded positive PCR results
All copro-DNAs, extracted from fecal samples that had been collected every day from experimentally infected dogs, were tested using the multiplex PCR assay to determine the first day when a positive signal occurred.
Results
Infection outcomes of the experimentally challenged dogs and the examination of stray dogs
Infections of E. granulosus s.s. and E. multilocularis were successfully achieved in all the experimentally infected dogs with 5539, 8562, 12535, 18932 and 20775 E. granulosus s.s. and 2893, 3153, 3762, 3864 and 5322 E. multilocularis adult worms being recovered from each group of 5 dogs that were fed with protoscoleces of each species. No adult worms were found in any of the 5 dogs fed larval E. shiquicus. None of the stray dogs was found harboring E. shiquicus or E. multilocularis; only E. granulosus s.s. adult worms were found in their intestinal contents (identity confirmed by both morphology and cox1 sequencing). Worm burdens were relatively low (circa 100–200 worms) in the ten stray dogs examined.
Identification of PCR products
Expected PCR products of 219, 584 and 471 bp were obtained for E. granulosus s.s. (nad1), E. multilocularis (nad5) and E. shiquicus (cox1), respectively (Fig 1), and products of mixed templates of the three Echinococcus species are shown in Fig 2. The multiplex PCR products contained 3 DNA bands (219, 471 and 584 bp) with mixed DNA templates of E. granulosus s.s., E. multilocularis and E. shiquicus; 2 DNA bands (219 and 584 bp) with E. granulosus s.s. and E. multilocularis DNA templates; 2 DNA bands (219 and 471 bp) with E. granulosus s.s. and E. shiquicus DNA templates; and 2 DNA bands (471 and 584 bp) with E. multilocularis and E. shiquicus DNA templates. DNA sequences of these products corresponded in each case with the relevant reference sequences in GenBank: E. granulosus (G1) (NC_008075) [22], E. multilocularis (NC_000928) [23] and E. shiquicus (NC_009460) [24].
Fig 1. Amplicons of the target genes using the multiplex PCR assay.

Lanes 1, 2 and 3, Amplicons of E. granulosus s.s., E. multilocularis and E. shiquicus respectively; Lane 4, Negative control; M, DNA Marker DL 2000.
Fig 2. Amplicons of the mixed templates using the multiplex PCR assay.
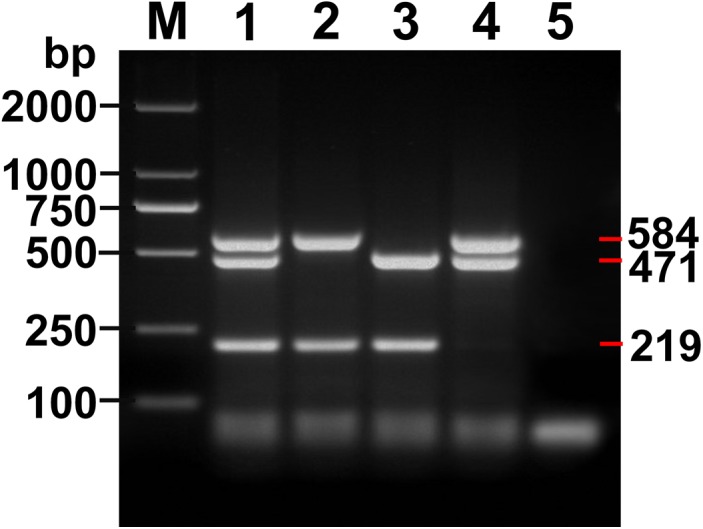
Lane 1, Amplicons of E. granulosus s.s., E. multilocularis and E. shiquicus; Lane 2, Amplicons of E. granulosus s.s. and E. multilocularis, Lane 3, Amplicons of E. granulosus s.s. and E. shiquicus; Lane 4, Amplicons of E. granulosus s.s., E. multilocularis and E. shiquicus; Lane 5, Negative control; M, DNA Marker DL 2000.
Specificity
Comparison of various sources of DNA
No PCR products were detected when DNAs from other cestodes (Table 1, labeled with an asterisk) were used in the multiplex assay (Fig 3). False positive results were never produced from confirmed negative samples. Further, no PCR products were obtained when DNA samples from various host tissues were used in the multiplex PCR. Therefore, the specificity of the multiplex PCR for E. granulosus s.s. (G1), E. multilocularis and E. shiquicus was shown to be 100%.
Fig 3. Specificity of the multiplex PCR assay.
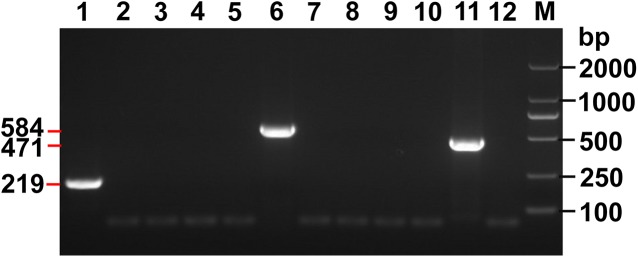
M, DNA Marker DL 2000; Lanes 1, 6 and 11, Amplicons of E. granulosus s.s., E. multilocularis and E. shiquicus respectively; Lane 2–5, 7–10 and 12, Other cestode samples: e.g. E. canadensis (G6), T. hydatigena, T. multiceps, T. pisiformis, T. taeniaeformis, T. solium, D. caninum, liver tissue of Qinghai vole, lung tissue of plateau pika.
Copro-DNA templates
Fecal samples, collected from dogs before experimental infection with Echinococcus spp. and from captive foxes (confirmed parasite-free by microscopy and DNA analysis), were negative in the multiplex PCR whereas fecal samples from dogs after experimental infection with either larval E. granulosus s.s. or E. multilocularis were positive. Furthermore, the PCR products obtained were of the expected sizes, matching those that were also obtained for all positive controls. No false positive signals were obtained with any negative control sample.
Fecal inhibitors and the copro-DNA test
To test for the presence of potential inhibitors, parallel multiplex PCR assays were performed with positive Echinococcus spp. DNA, negative fecal samples, and mixtures of Echinococcus spp. DNA and fecal sample DNA as templates. Positive signals were detectable in the multiplex PCRs with mixtures of the Echinococcus spp. DNAs and fecal sample DNA as templates. We infer from this that no adverse fecal inhibitors affected the integrity of the multiplex PCR assay.
Effect of host tissue DNA on the multiplex PCR test
To test for interference due to host DNA contamination in samples, we added various quantities of host DNA to known quantities (100 pg) of parasite DNA. Quantities below 500 ng of host tissue DNA (from intestinal, hepatocyte or pulmonary cells) did not affect PCR outcomes: clear bands of expected sizes were present in gels. However, smeared bands appeared in the gels if the amount of host DNA exceeded 500 ng.
Sensitivity
Minimum/maximum quantity of Echinococcus metacestode DNA
The lower limit for the detection of metacestode DNA was 20 pg for E. granulosus s.s., 10 pg for E. multilocularis, and 20 pg for E. shiquicus, respectively. Clear bands could be visualized up to a maximum quantity of 500 ng template DNA. Smearing of bands occurred if this amount was exceeded. Accordingly, the optimum amount of template DNA used was 100 pg for the multiplex PCR as this quantity produced clear amplicon bands and provided savings on template DNA and PCR primers.
Minimum number of eggs detectable in the fecal samples
Positive PCR products were obtained in reactions using DNA from as few as two eggs of E. granulosus s.s. and one egg of E. multilocularis (Fig 4).
Fig 4. Sensitivity of the multiplex PCR assay.
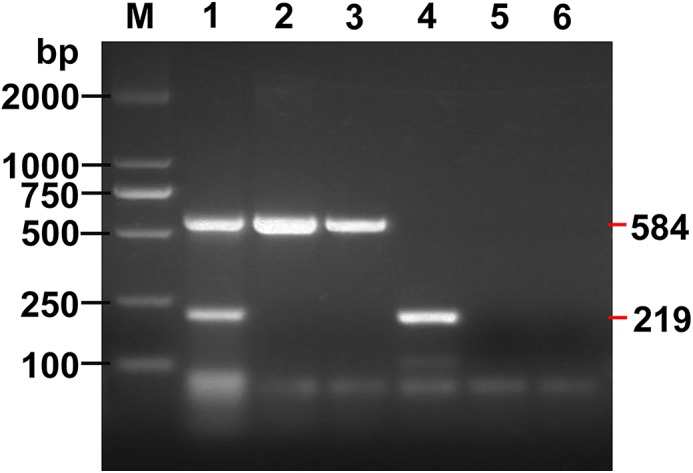
Lane 1, Amplicons of E. granulosus s.s. and E. multilocularis with 2 eggs and 1 egg respectively, mixed in fecal sample; Lanes 2 and 3, Amplicons of E. multilocularis from 2 eggs and 1 egg respectively, mixed in fecal sample; Lanes 4 and 5, Amplicon of E. granulosus s.s. amplified with 2 eggs and 1 egg mixed in fecal sample; Lane 6, Negative control fecal sample; M, DNA Marker DL 2000.
Earliest time for a positive multiplex PCR assay after experimental infection
Eggs of E. granulosus s.s. and E. multilocularis were visualized under microscopy at days 47–56 and days 36–44 post-challenge, respectively. The multiplex PCR assay yielded positive results from copro-DNA 17 days after experimental infection of dogs with larval E. multilocularis and 26 days after infection with larval E. granulosus s.s.
Assessment of feces from stray dogs
Copro-DNA from all the stray dogs infected with E. granulosus s.s. tested positive in the multiplex PCR.
Discussion
China is the most severe pandemic country for cystic echinococcosis (CE), in humans and livestock, due mainly to E. granulosus s.s., and for alveolar echinococcosis (AE) due to E. multilocularis in humans and small wild mammals. E. shiquicus is also endemic although it has not been reported to infect humans. Dual infections of animal hosts with different Echinococcus spp have been reported in the eastern Qinghai-Tibet plateau region of China [4, 25]. The very close relationship between dogs and humans can lead readily to human infection. The increasing number of human AE and CE cases in northwestern China, where considerable numbers of dogs are present, places a heavy burden on public health and veterinary services. To aid surveillance, management and diagnosis, effective methods are needed for rapid and accurate detection and identification of different life cycle stages of the three Echinococcus spp. simultaneously. The multiplex PCR assay developed in this study provides such a method.
Traditional epidemiological surveys for tapeworms often involve recovery of eggs from feces of potential definitive hosts. However, morphological identification of Echinoccocus eggs to species level is practically impossible, prompting the development of several molecular approaches [26, 27]. Inhibitors present in fecal material that co-purify with parasite DNA extracted from feces often present a problem for PCR-based methods [28]. In this study, the QIAGEN QIAamp DNA Stool Mini Kit, containing InhibitEX tablets for removing inhibitors in fecal samples, was used to purify copro-DNA. The sieving-flotation method was helpful in overcoming this problem due to its enrichment of worm eggs [29]. The positive control (protoscolex DNA in fecal samples) used in this study demonstrated the lack of inhibitor effects in our copro-multiplex PCR assay.
E. granulosus s.s. has been reported as having a pre-patent period of 6 weeks (42 days) [30, 31], while E. multilocularis eggs have been observed in feces at 42–46 days post infection [32]. However, in the current study we first identified eggs of E. granulosus s.s. at 47–56 days post-challenge and those of E. multilocularis at 36–44 days post-challenge by microscopy similar to reports by others [30, 33]. The discrepancies between these studies may be due to the use of different dog-breeds, ages, nutrient status or the conditions under which the dogs were maintained. We were unable to experimentally infect dogs with E. shiquicus although the viability of the challenge sample of protoscoleces was confirmed by microscopy.
PCR-positive signals in this study were obtained from dog fecal samples much earlier (17 days for E. multilocularis and 26 days for E. granulosus) than any other previous studies using microscopy as a method of detecting infected canid hosts. The much earlier detection of an Echinococcus infection by the multiplex PCR method compared with egg recovery from feces and microscopic examination is a marked improvement that can aid surveillance programs aimed at preventing echinococcosis transmission.
The method developed in this study has achieved high species specificity because it produced no amplicon from any other helminth (including several that might dual infect with Echinococcus species in dogs) or from the negative copro-samples (no-DNA). The primer set (three pairs of primers) multiplex reaction in a single tube worked well with all templates tested and yielded specific amplicons of the expected length for each of the three Echinococcus spp. examined.
E. granulosus s.s and E. multilocularis are of major public health concern in many endemic countries globally [34]. A cost effective diagnostic tool is required for echinococcosis surveillance of definitive and intermediate hosts, and for monitoring the effectiveness of control programs. The multiplex PCR assay developed in this study provides an effective method that can be applied in both clinical and epidemiological settings for the identification of Echinococcus spp in diverse hosts, and would be particularly useful for identifying infected hosts in areas co-endemic for AE and CE.
In this study, we focused on Echinococcus samples collected from the Qinghai-Tibet plateau region of China, where three species (E. granulosus, E. multilocularis and E. shiquicus) are known to be endemic. In total, nine species are now recognized in the genus Echinococcus, including E. granulosus sensu stricto (genotypes G1-G3), E. equinus, E. canadensis (genotypes G6, G7, G8 and G10), E. ortleppi, E. multilocularis, E. shiquicus, E. vogeli, E. oligarthrus and E. felidis [1]. None of the three specific pairs of primers developed in this study produced a PCR-amplified product using DNA isolated from E. canadensis (G6 genotype) showing in Fig 3 (the lane 2 with non-band as a negative result). This is supported by inspection and comparison of the primer target sequence for the G6 genotype with those of the three Echinococcus spp., which showed six base pair differences between them (S1 Fig, S2 Fig and S3 Fig in the Supporting Supplementary Information). Furthermore, six or more base pair differences are apparent between the target sequences for E. equinus, E. canadensis (genotypes G7, G8 and G10), E. ortleppi, E. vogeli, E. oligarthrus and E. felidis. Therefore, it is highly unlikely that any amplicon would be produced from these species during the multiplex PCR due to its high species specificity.
Supporting Information
(DOC)
(DOCX)
(PDF)
(PDF)
(PDF)
Acknowledgments
We would like to acknowledge Mr. Naizhi Jiancuo and Ms. Ma Zhuo for their valuable help in providing tissues of yaks, sheep, Qinghai voles and plateau pikas, and fecal samples from stray dogs.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the following: WZJ: Gansu Provincial Key Science and Technology Projects (Grant No. 1203NKDA039, http://www.gsstc.gov.cn/), ZZL: the Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201303037, http://www.moa.gov.cn), WZJ: the Special Fund for Agro-scientific Research in the Public Interest (Grant No. 200903036-07, http://www.moa.gov.cn), WZJ: the Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006, http://www.gsstc.gov.cn/), YRY: Natural Science Foundation of China (Grant No. 30960339, http://www.nsfc.gov.cn/), YRY: National Health and Medical Research Council (NHMRC) Project (Grant No. APP-1009539, http://www.nhmrc.gov.au/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nakao M, Lavikainen A, Yanagida T, Ito A (2013) Phylogenetic systematics of the genus Echinococcus (Cestoda: Taeniidae). Int J Parasitol 43: 1017–1029. 10.1016/j.ijpara.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 2. Eckert J and Deplazes P (2004) Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev 17: 107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang ZH, Wang XM, and Liu XQ (2008) Echinococcosis in China, a Review of the Epidemiology of Echinococcus spp. EcoHealth 5: 115–126. 10.1007/s10393-008-0174-0 [DOI] [PubMed] [Google Scholar]
- 4. Boufana B, Qiu JM, Chen XW, Budke CM, Campos-Ponce M, et al. (2013) First report of Echinococcus shiquicus in dogs from eastern Qinghai-Tibet plateau region, China. Acta Trop 127: 21–24. 10.1016/j.actatropica.2013.02.019 [DOI] [PubMed] [Google Scholar]
- 5. Zhang LH, Chai JJ, Jiao W, Osman Y, McManus DP (1998) Mitochondrial genomic markers confirm the presence of the camel strain (G6 genotype) of Echinococcus granulosus in north-western China. Parasitology, 116 (Pt 1): 29–33. [DOI] [PubMed] [Google Scholar]
- 6. Eckert J, Muller B, Partridge AJ (1974) The domestic cat and dog as natural definitive hosts of Echinococcus (Alveococcus) multilocularis in southern federal republic of Germany. Tropenmed Parasitol 25: 334–337. [PubMed] [Google Scholar]
- 7. Deplazes P, Alther P, Tanner I, Thompson RC, Eckert J (1999) Echinococcus multilocularis coproantigen detection by enzyme-linked immunosorbent assay in fox, dog, and cat populations. J Parasitol 85: 115–121. [PubMed] [Google Scholar]
- 8. Armua-Fernandez MT, Castro OF, Crampet A, Bartzabal A, Hofmann-Lehmann R, et al. (2014) First case of peritoneal cystic echinococcosis in a domestic cat caused by Echinococcus granulosus sensu stricto (genotype 1) associated to feline immunodeficiency virus infection. Parasitol Int 63: 300–302. 10.1016/j.parint.2013.11.005 [DOI] [PubMed] [Google Scholar]
- 9. Sobrino R, Gonzalez LM, Vicente J, Fernandez DLD, Garate T, et al. (2006) Echinococcus granulosus (Cestoda, Taeniidae) in the Iberian wolf. Parasitol Res 99: 753–756. [DOI] [PubMed] [Google Scholar]
- 10. Hajialilo E, Harandi MF, Sharbatkhori M, Mirhendi H, Rostami S (2012) Genetic characterization of Echinococcus granulosus in camels, cattle and sheep from the south-east of Iran indicates the presence of the G3 genotype. J Helminthol 86: 263–270. 10.1017/S0022149X11000320 [DOI] [PubMed] [Google Scholar]
- 11. Guislain MH, Raoul F, Poulle ML, Giraudoux P (2007) Fox feces and vole distribution on a local range: ecological data in a parasitological perspective for Echinococcus multilocularis . Parasite 14: 299–308. [DOI] [PubMed] [Google Scholar]
- 12. Xiao N, Qiu JM, Nakao M, Li TY, Yang W, et al. (2005) Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China. Int J Parasitol 35: 693–701. 10.1016/j.ijpara.2005.01.003 [DOI] [PubMed] [Google Scholar]
- 13. Abbasi I, Branzburg A, Campos-Ponce M, Abdel HS, Raoul F, et al. (2003) Copro-diagnosis of Echinococcus granulosus infection in dogs by amplification of a newly identified repeated DNA sequence. Am J Trop Med Hyg 69: 324–330. [PubMed] [Google Scholar]
- 14. Jiang WB, Liu N, Zhang GT, Renqing P, Xie F, et al. (2012) Specific detection of Echinococcus spp. from the Tibetan fox (Vulpes ferrilata) and the red fox (V. vulpes) using copro-DNA PCR analysis. Parasitol Res 111: 1531–1539. 10.1007/s00436-012-2993-8 [DOI] [PubMed] [Google Scholar]
- 15. Boufana B, Umhang G, Qiu JM, Chen XW, Lahmar S, et al. (2013) Development of three PCR assays for the differentiation between Echinococcus shiquicus, E. granulosus (G1 genotype), and E. multilocularis DNA in the co-endemic region of Qinghai-Tibet plateau, China. Am J Trop Med Hyg 88: 795–802. 10.4269/ajtmh.12-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boubaker G, Macchiaroli N, Prada L, Cucher MA, Rosenzvit MC, et al. (2013) A multiplex PCR for the simultaneous detection and genotyping of the Echinococcus granulosus complex. PLoS Negl Trop Dis 7: e2017 10.1371/journal.pntd.0002017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trachsel D, Deplazes P, Mathis A (2007) Identification of taeniid eggs in the feces from carnivores based on multiplex PCR using targets in mitochondrial DNA. Parasitology 134: 911–920. 10.1017/S0031182007002235 [DOI] [PubMed] [Google Scholar]
- 18. Zare-Bidaki M, Mobedi I, Ahari SS, Habibizadeh S, Naddaf SR, et al. (2010) Prevalence of Zoonotic Intestinal Helminths of Canids in Moghan Plain, Northwestern Iran. Iranian J Parasitol 5: 42–51. [PMC free article] [PubMed] [Google Scholar]
- 19. Huang Y, Yang W, Qiu JM, Chen XW, Yang Y, et al. (2007) A modified coproantigen test used for surveillance of Echinococcus spp. in Tibetan dogs. Vet Parasitol 149: 229–238. 10.1016/j.vetpar.2007.08.026 [DOI] [PubMed] [Google Scholar]
- 20. Mathis A, Deplazes P, Eckert J (1996) An improved test system for PCR-based specific detection of Echinococcus multilocularis eggs. J Helminthol 70: 219–222. [DOI] [PubMed] [Google Scholar]
- 21. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang YR, Rosenzvit MC, Zhang LH, Zhang JZ, McManus DP (2005) Molecular study of Echinococcus in west-central China. Parasitology 131 (Pt 4): 547–555. 10.1017/S0031182005007973 [DOI] [PubMed] [Google Scholar]
- 23. Nakao M, Yokoyama N, Sako Y, Fukunaga M, Ito A (2002) The complete mitochondrial DNA sequence of the cestode Echinococcus multilocularis (Cyclophyllidea: Taeniidae). Mitochondrion 1: 497–509. 10.1016/S1567-7249(02)00040-5 [DOI] [PubMed] [Google Scholar]
- 24. Nakao M, McManus DP, Schantz PM, Craig PS, Ito A (2007) A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology 134 (Pt 5): 713–722. 10.1017/S0031182006001934 [DOI] [PubMed] [Google Scholar]
- 25. Xiao N, Nakao M, Qiu J, Budke CM, Giraudoux P, et al. (2006) Dual infection of animal hosts with different Echinococcus species in the eastern Qinghai-Tibet plateau region of China. Am J Trop Med Hyg 75: 292–294. [PubMed] [Google Scholar]
- 26. Deplazes P, Dinkel A, Mathis A (2003) Molecular tools for studies on the transmission biology of Echinococcus multilocularis . Parasitology 127 Suppl: S53–S61. [DOI] [PubMed] [Google Scholar]
- 27. Mathis A, Deplazes P (2006) Copro-DNA tests for diagnosis of animal taeniid cestodes. Parasitol Int 55 Suppl: S87–S90. 10.1016/j.parint.2005.11.012 [DOI] [PubMed] [Google Scholar]
- 28. Dinkel A, von Nickisch-Rosenegk M, Bilger B, Merli M, Lucius R, et al. (1998) Detection of Echinococcus multilocularis in the definitive host: coprodiagnosis by PCR as an alternative to necropsy. J Clin Microbiol 36: 1871–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monnier P, Cliquet F, Aubert M, Bretagne S (1996) Improvement of a polymerase chain reaction assay for the detection of Echinococcus multilocularis DNA in fecal samples of foxes. Vet Parasitol 67: 185–195. 10.1016/S0304-4017(96)01039-4 [DOI] [PubMed] [Google Scholar]
- 30. Gemmell MA, Lawson JR, Roberts MG (1986) Population dynamics in echinococcosis and cysticercosis: biological parameters of Echinococcus granulosus in dogs and sheep. Parasitology 92 (Pt 3): 599–620. [DOI] [PubMed] [Google Scholar]
- 31. Aminzhanov M (1975) [Duration of the life of Echinococcus granulosus in the organism of dogs]. Veterinariia 12: 70–72. [PubMed] [Google Scholar]
- 32. Ni XW, McManus DP, Yan HB, Yang JF, Lou ZZ, et al. (2014) Loop-Mediated Isothermal Amplification (LAMP) assay for the identification of Echinococcus multilocularis infections in canine definitive hosts. Parasit Vectors 7: 254 10.1186/1756-3305-7-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kapel CM, Torgerson PR, Thompson RC, Deplazes P (2006) Reproductive potential of Echinococcus multilocularis in experimentally infected foxes, dogs, raccoon dogs and cats. Int J Parasitol 36: 79–86. 10.1016/j.ijpara.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 34. McManus DP, Gray DJ, Zhang WB, Yang YR (2012) Diagnosis, treatment, and management of echinococcosis. Brit Med J 344: e3866 10.1136/bmj.e3866 . [DOI] [PubMed] [Google Scholar]
- 35. Le TH, Pearson MS, Blair D, Dai N, Zhang LH, et al. (2002) Complete mitochondrial genomes confirm the distinctiveness of the horse-dog and sheep-dog strains of Echinococcus granulosus . Parasitology 124 (Pt 1): 97–112. [DOI] [PubMed] [Google Scholar]
- 36. Jia WZ, Yan HB, Guo AJ, Zhu XQ, Wang YC, et al. (2010) Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics 11: 447 10.1186/1471-2164-11-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu GH, Lin RQ, Li MW, Liu W, Liu Y, et al. (2011) The complete mitochondrial genomes of three cestode species of Taenia infecting animals and humans. Mol Biol Rep 38: 2249–2256. 10.1007/s11033-010-0355-0 [DOI] [PubMed] [Google Scholar]
- 38. Jeon HK, Lee KH, Kim KH, Hwang UW, Eom KS (2005) Complete sequence and structure of the mitochondrial genome of the human tapeworm, Taenia asiatica (Platyhelminthes; Cestoda). Parasitology 130 (Pt 6): 717–726. [DOI] [PubMed] [Google Scholar]
- 39. Jeon HK, Kim KH, Eom KS (2007) Complete sequence of the mitochondrial genome of Taenia saginata: comparison with T. solium and T. asiatica . Parasitol Int 56: 243–246. 10.1016/j.parint.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 40. Jia WZ, Yan HB, Lou ZZ, Ni XW, Dyachenko V, et al. (2012) Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis . Acta Trop 123: 154–163. 10.1016/j.actatropica.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 41. Nakao M, Sako Y, Ito A (2003) The mitochondrial genome of the tapeworm Taenia solium: a finding of the abbreviated stop codon U. J Parasitol 89: 633–635. [DOI] [PubMed] [Google Scholar]
- 42. Nakao M, Lavikainen A, Iwaki T, Haukisalmi V, Konyaev S, et al. (2013) Molecular phylogeny of the genus Taenia (Cestoda: Taeniidae): Proposals for the resurrection of Hydatigera Lamarck, 1816 and the creation of a new genus Versteria. Int J Parasitol 43: 427–437. 10.1016/j.ijpara.2012.11.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


