Abstract
The placenta plays a critical role in the growth and survival of the fetus. Here we demonstrate that the Homologous to the E6-AP Carboxyl Terminus (HECT) domain E3 ubiquitin ligase, Hectd1, is essential for development of the mouse placenta. Hectd1 is widely expressed during placentation with enrichment in trophoblast giant cells (TGCs) and other trophoblast-derived cell subtypes in the junctional and labyrinth zones of the placenta. Disruption of Hectd1 results in mid-gestation lethality and intrauterine growth restriction (IUGR). Variable defects in the gross structure of the mutant placenta are found including alterations in diameter, thickness and lamination. The number and nuclear size of TGCs is reduced. Examination of subtype specific markers reveals altered TGC development with decreased expression of Placental lactogen-1 and -2 (Pl1 and Pl2) and increased expression of Proliferin (Plf). Reduced numbers of spongiotrophoblasts and glycogen trophoblasts were also found at the junctional zone of the Hectd1 mutant placenta. Finally, there was an increase in immature uterine natural killer (uNK) cells in the maternal decidua of the Hectd1 mutant placenta. Proliferation and apoptosis are differentially altered in the layers of the placenta with an increase in both apoptosis and proliferation in the maternal decidua, a decrease in proliferation and increase in apoptosis in the labyrinth layer and both unchanged in the junctional zone. Together these data demonstrate that Hectd1 is required for development of multiple cell types within the junctional zone of the placenta.
Keywords: HECT E3 ligase, Placenta, Trophoblast
Introduction
Development and survival of the mammalian embryo depends on the placenta, which is responsible for the exchange of nutrients and gases, removal of embryonic waste and secretion of hormones (Rossant and Cross, 2001). Abnormal placental development underlies a wide range of complications during pregnancy including intrauterine growth restriction (IUGR), preeclampsia and miscarriage, and has been implicated in predisposition to chronic diseases in adulthood (Cross, 2006; Fowden et al., 2008; Longtine and Nelson, 2011). Many similarities exist between the mouse and human placenta making the mouse an excellent model to elucidate the mechanisms of placental development in humans (Georgiades et al., 2002; Malassine et al., 2003). Mouse models have become indispensable for defining cell types, lineage relationships and molecular pathways required for normal placentation (Cross, 2005; Rossant and Cross, 2001).
The mature mouse placenta is a layered structure comprised of cells derived from both the conceptus (trophoectoderm and extra-embryonic mesoderm) and the maternal decidua (Rossant and Cross, 2001). Closest to the umbilical cord that connects the placenta to the embryo is the labyrinth zone, which consists of fetal and maternal blood spaces separated by vascular endothelium and specialized trophoblast cell types. The labyrinth zone provides the interface for exchange between maternal and fetal circulations. The junctional zone separates the labyrinth from the maternal decidua and consists of spongiotrophoblasts, glycogen trophoblasts and a discontinuous layer of trophoblast giant cells (TGCs) with large polyploid nuclei. TGCs are the main endocrine cells of the placenta secreting a number of hormones necessary for modulating maternal physiology and placental development (Malassine et al., 2003). While spongiotrophoblasts and glycogen trophoblasts are essential for normal function of the placenta, their specific function remains unknown. Uterine natural killer (uNK) cells are recruited to the implantation site and interact with trophoblasts influencing their invasion into the maternal decidua (Bilinski et al., 2008; Croy et al., 2003). Together uNK and trophoblast cells remodel spiral arteries to increase maternal blood flow to the placenta (Harris, 2010; Wallace et al., 2012).
Hectd1 is a HECT domain E3 ubiquitin ligase that plays an important role in development of the neural tube (Sarkar and Zohn, 2012; Zohn et al., 2007) and regulation of Wnt signaling (Tran et al., 2013). Ubiquitin conjugated substrates are widely distributed in the endometrium and decidua in rodents and primates (Bebington et al., 2000), and studies of mouse mutants demonstrate that ubiquitin dependent modification plays an indispensable role in placental development. For example, mutation of mouse Cul4B, an E3 ligase of the cullin ring family results in an abnormal placenta and early embryonic lethality (Jiang et al., 2012). Here we demonstrate that homozygous Hectd1 mutant embryos exhibit midgestation lethality and growth deficiencies associated with defects in placental development. Our data indicate Hectd1 is transcribed and translated in multiple trophoblast-derived cell types throughout placental development. Loss of Hectd1 function results in altered placenta architecture, apoptosis and proliferation. Furthermore, the number, size and/or distribution of TGCs, spongiotrophoblasts, glycogen trophoblasts and uNK cells are affected in homozygous Hectd1 mutant placentas.
Materials and methods
Mouse breeding and placental tissue collection
The Hectd1opm and Hectd1Gt(XC266)Byg (Hectd1XC) mouse lines used in this study have been previously described (Kasarskis et al., 1998; Zohn et al., 2007). All experiments were done on congenic 129S1/SvImJ. C57Bl/6-Hectd1opm or 129S1/SvImJ.129P2/OlaHsd-Hectd1XC backgrounds. For phenotypic analysis Hectd1opm/+ mice were intercrossed and placentas collected at the indicated developmental stages. Hectd1XC/+ mice were also intercrossed for determination of Mendelian ratios of survival and expression analysis. Heterozygous Hectd1XC mice were also bred to wildtype mice for analysis of Hectd1 expression. Only placentas serving live embryos were analyzed. Placentas were fixed by immersion in 4% paraformaldehyde fixative for 24 h, embedded in Optimal Cutting Temperature compound (OCT, Tissue-Tek) and serial 10 μm sections cut using a cryostat. Genotyping was performed using the yolk sac as described (Sarkar and Zohn, 2012).
Phenotypic and histological analyses
Embryos were weighed on a chemical balance and placental diameters measured using a dissecting scope with the aid of a graduated slide. The size of TGC nuclei was measured using ImageJ software. To assess placental thickness and histology, sections were stained with hematoxylin and eosin (H&E). To analyze Hectd1 expression, sectioned Hectd1XC/+ conceptuses or isolated placentas were subjected to LacZ staining and counterstained with eosin (Nagy, 2003). In situ hybridization on sectioned placentas was carried out as described (Zohn et al., 2006) and counterstained with eosin. The following probes were used: Hand1 (Scott et al., 2000), Prl3d1/Pl1 (Faria et al., 1991), Prl3b1/Pl2 (Jackson et al., 1986), Prl2c2/Plf (Linzer et al., 1985), Tpbpa (Carney et al., 1993), Cdx2 (Strumpf et al., 2005) and Mash2 (Guillemot et al., 1994). Immunofluorescence and immunohistochemical analyses were performed as described (Sarkar and Zohn, 2012) using the following antibodies: Hectd1 (Novus Biologicals, #H00025831-M03), Pecam1 (Becton-Dickinson, #550274), FK2 (mono- and polyubiquitin; Biomol, #PW0150-0100), Cdx2 (Cell Signaling, #3977), Cleaved caspase-3 (CC3; Cell Signaling, #9661), Phospho-Histone H3 (PH3; Cell Signaling, #9713) and Perforin (Cell Signaling, #3693). In immunofluorescence experiments, nuclei were stained with Hoechst (Promega, P5541). To stain glycogen-containing cells, a Periodic Acid-Schiff (PAS) staining kit (Polyscience Inc., #24200) was used. Glycogen content of placentas was determined as described (Tesser et al., 2010). Stereological analyses were performed essentially as described (Coan et al., 2004) using the Stereo Investigator software system (MBF Biosciences, Vermont, USA).
Western blotting and quantitative PCR (qPCR) analyses
Placentas and tissue fragments were collected from time-mated adult female mice and analyzed by western blotting with anti-Hectd1 (Novus Biologicals, #H00025831-M03) and anti-GAPDH (Cell Signaling, #2118) antibodies. For determination of ubiquitinated proteins in wildtype and mutant placentas, Hectd1opm mice were intercrossed and placentas dissected at E12.5. Ubiquitinated-proteins were enriched from whole placenta extracts using the ubiquitin protein enrichment kit (EMD-Millipore, #662200) then visualized by immunoblotting with the FK2 antibody (Biomol, #PW0150-0100). qRT-PCR on E10.5 placentas was performed as described (Maynard et al., 2013) using primers listed in Parast et al. (2009). All statistical analyses were performed using Microsoft Excel software. Student’s t-test with Bonferroni correction was used to determine statistical significance between groups.
Results
Mutation of Hectd1 disrupts placental development
Two Hectd1 mutant mouse lines were used for these studies: Hectd1opm and Hectd1Gt(XC266)Byg (Hectd1XC). Hectd1opm is an N-ethyl-N-nitrosourea (ENU) induced allele and contains a missense mutation that results in truncation of the 2611 amino acid Hectd1 protein after amino acid 144 (Zohn et al., 2007). This results in an effectively null allele as all protein–protein interaction and catalytic domains are deleted. The Hectd1XC allele contains a genetrap insertion disrupting the C-terminal HECT domain and the ubiquitin ligase activity of the protein. The mutant protein is expressed and other protein–protein interaction domains of the protein remain intact (Sarkar and Zohn, 2012; Zohn et al., 2007). Hectd1opm and Hectd1XC fail to complement each other and exhibit identical phenotypes with regard to neural tube closure (Zohn et al., 2007). Homozygous Hectd1opm and Hectd1XC mutant embryos begin to die after E12.5 and expected Mendelian ratios of mutants are not found from E13.5 to E18.5 (Table 1). This mid-gestation lethality is not due to the neural tube defect, as exencephaly is not fatal until after birth (Copp et al., 2003). Homozygous Hectd1opm mutant embryos that survive to E13.5 are pale, developmentally delayed and occasionally exhibit edema (data not shown). Furthermore, the weight of Hectd1opm mutant embryos at E11.5 and E12.5 is significantly reduced (Fig. 1A).
Table 1.
Distribution of genotypes from Hectd1 heterozygous crosses (NS = not significant).
| Hectd1opm/+ × Hectd1opm/+ | +/+ | +/opm | opm/opm | χ2 (df=1) | p |
|---|---|---|---|---|---|
| E18.5 | 7 | 29 | 1 | 9.835 | <0.0025 |
| E17.5 | 14 | 33 | 1 | 13.564 | <0.0005 |
| E16.5 | 17 | 31 | 2 | 8.82 | <0.005 |
| E15.5 | 47 | 63 | 3 | 22.568 | <0.0005 |
| E14.5 | 12 | 46 | 10 | 2.882 | <0.1(NS) |
| E13.5 | 35 | 33 | 10 | 4.628 | <0.05 |
| Total (394) | 132 | 235 | 27 | 51.901 | <0.0005 |
| Actual % | 33.5 | 59.64 | 6.85 | ||
| Expected % | 25 | 50 | 25 |
| Hectd1XC/+ × Hectd1XC/+ | +/+ | XC/+ | XC/XC | χ2 (df=1) | p |
|---|---|---|---|---|---|
| E18.5 | 9 | 17 | 1 | 7.25 | <0.01 |
| E17.5 | 7 | 18 | 0 | 6.25 | <0.02 |
| E16.5 | 22 | 25 | 0 | 11.75 | <0.001 |
| E15.5 | 26 | 55 | 2 | 16.94 | <0.0005 |
| E14.5 | 14 | 48 | 2 | 15.06 | <0.0005 |
| E13.5 | 23 | 43 | 2 | 15.01 | <0.0005 |
| Total (314) | 101 | 206 | 7 | 65.124 | <0.0005 |
| Actual % | 32.71 | 65.60 | 2.23 | ||
| Expected % | 25 | 50 | 25 |
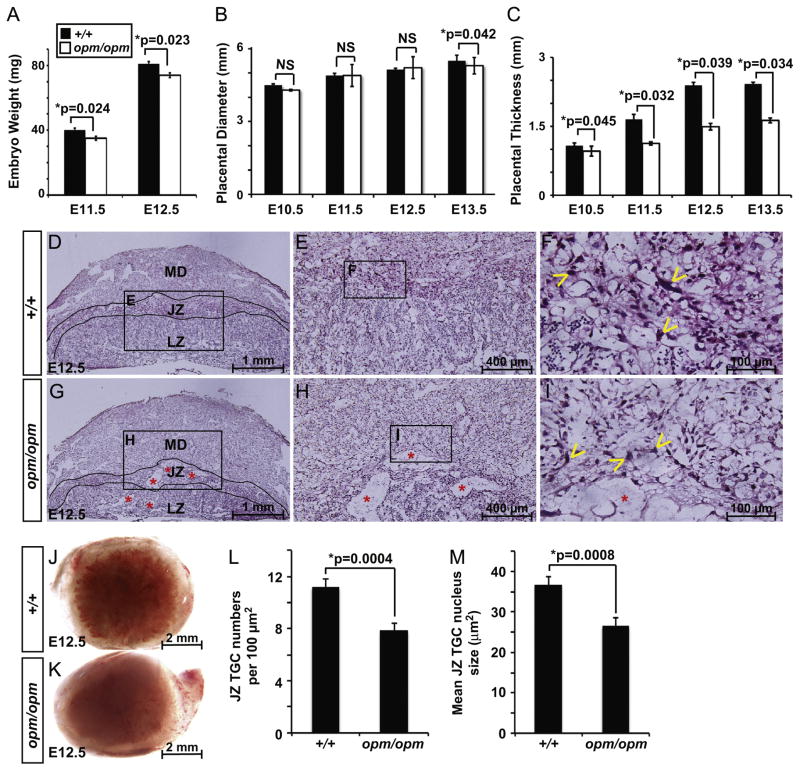
Fig. 1.
Mutation of Hectd1 disrupts placental development: (A) weight of Hectd1opm/opm mutant embryos (white bar) is significantly reduced at E11.5 and E12.5 compared to littermate controls (Hectd1+/+; black bar; n=15 at each stage). (B) Placental diameter is significantly reduced at E13.5 but not from E10.5 to E12.5 in Hectd1opm mutant placentas. Ten wildtype and mutant placentas were analyzed at each stage. NS=not significant. (C) Placental thickness is significantly reduced in Hectd1opm mutant placentas from E10.5 to E13.5 (n=10 at each stage). (D–I) H&E stained E12.5 wildtype (+/+; D–F) and moderately affected Hectd1opm mutant (opm/opm; G–I) placentas. The layers of the placental architecture are labeled: maternal decidua (MD), junctional zone (JZ) and labyrinth zone (LZ). Higher magnification images of boxed regions are shown as indicated. Note the larger tissues spaces (red *) and reduced lamination of the mutant placenta (n=34). A few of the many TGCs are labeled with yellow arrowheads. Also, note that the size of TGC nuclei appears smaller in the mutant. (J, K) Whole placentas imaged from the embryonic side reveal paleness of the mutant (opm/opm; K) placenta versus wildtype (+/+; J) that was observed in 60% of 123 mutant placentas examined. (L, M) Quantification of the number of TGCs per 100 μm2 field (L) and TGC nuclear area (M) in the junctional zone of wildtype and mutant placentas. The number of cells in 10 fields or the size of 10 nuclei, respectively, were counted in 3 placentas. Data represent mean values and error bars, standard error of the mean (SEM).
Mid-gestation lethality is associated with abnormal development of multiple organ systems including the placenta. Since our previous studies demonstrate Hectd1 expression in the E12.5 placenta (Zohn et al., 2007), we analyzed the Hectd1opm mutant placenta for developmental defects to determine if placental defects possibly contribute to mid-gestation lethality. Hectd1opm mutant placentas exhibit a spectrum of phenotypic severities ranging from no apparent phenotype to a placenta that is severely reduced in diameter and thickness and does not show clear lamination (Fig. 1D–I and Supplemental Fig. 1). To quantify the effect of Hectd1 mutation on placental development, placental diameter and thickness were measured in wildtype and Hectd1 mutant placentas between ages E10.5 and E13.5. At E13.5, but not earlier, the diameter of Hectd1opm homozygous mutant placentas is significantly smaller than sibling controls (Fig. 1B) and thickness is reduced from E10.5 through E13.5 (Fig. 1C). To quantify phenotypic variability, we scored for the incidence of six factors in 123 mutant placentas: growth restriction of the embryo (31.7% of mutant placentas showed this phenotype) and placental hemorrhage (47.9%), paleness (Fig. 1J and K; 30.9%), reduced diameter (43.1%), thickness (66.7%) and weight (69.1%). A severe phenotype (all 6 parameters present) was found in 8% and no apparent phenotype in 10% of the Hectd1opm mutant placentas examined (Supplemental Fig. 1). All subsequent analyses focused on the majority (82%) of cases that exhibited a moderate phenotype with between one and five of these defects. This variable penetrance and expressivity of placenta defects is very common in mouse mutants (see Barak et al., 2002; Caspary et al., 1999; Catela et al., 2009; Eggenschwiler et al., 1997 for examples).
Hematoxylin and eosin (H&E) staining was used to examine the tissue architecture of Hectd1opm mutant placentas (Fig. 1D–I). Analysis of mutant placentas reveals larger tissue spaces in all layers compared to the wildtype (n=34; labeled with * in Fig. 1G–I). In roughly half of the mutant placentas, trophoblast layers were reduced and the maternal decidua was correspondingly enlarged in thickness. To quantify differences in placental structures, a stereological analysis was undertaken on three moderately affected E13.5 placentas. A significant reduction in the labyrinth zone volume was observed in mutants to 41% of the wildtype volume (51±6 mm3; mutant=21±2 mm3; p=0.042). The maternal decidual volume was increased 44% in the mutant (wild type = 18±3 mm3; mutant=26±3 mm3; p=0.023), while the junctional zone volume was not significantly different between wildtype (24±2 mm3) and mutant (17±4 mm3; p=0.61). The number and nuclear size of TGCs in the junctional zone were also significantly reduced (Fig. 1L and M). Taken together, these results demonstrate that the Hectd1 mutant placenta exhibits a range of structural abnormalities.
Hectd1 is expressed in multiple cell types during development of the placenta
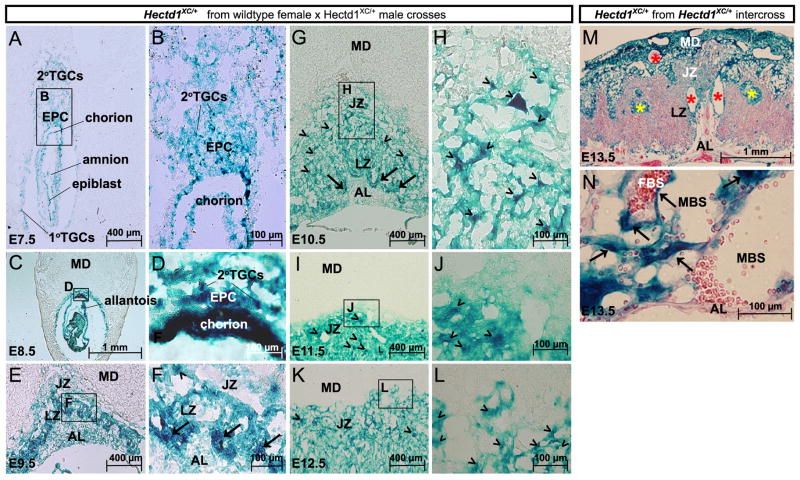
To better understand the role of Hectd1 in placentation, the spatiotemporal expression pattern of Hectd1 between E7.5 and 13.5 was examined in detail (Figs. 2, 3 and Table 2). To examine expression of the Hectd1 transcript, expression of beta-galactosidase was determined by LacZ staining in the heterozygous Hectd1XC/+ genetrap conceptus (Zohn et al., 2007). Samples from crosses between wildtype females and Hectd1XC/+ males were used to label Hectd1 expressing cells derived from the conceptus and not the maternal decidua. At E7.5, the implantation site is lined by primary and secondary TGCs that arise from the mural and polar trophectoderm, respectively. The polar trophectoderm contributes to the extra-embryonic ectoderm (ExE) and contains pluripotent trophoblast stem cells that differentiate into the different trophoblast cell types that will make up the chorioallantoic placenta. The ExE develops into the chorion and the ectoplacental cone (EPC), which gives rise to trophoblast cells that populate the labyrinth and junctional zones, respectively. The EPC also contributes secondary TGCs that invade the maternal uterine wall to promote decidualization. Hectd1 is expressed in the primary and secondary TGCs, chorion and EPC at E7.5 (Fig. 2A and B). By E8.5, Hectd1 expression intensifies in the chorion and scattered TGCs in the EPC (Fig. 2C and D). In the E9.5 conceptus, the allantois has fused with the chorion and chorionic villi have begun to undergo branching morphogenesis. Allantoic mesoderm-derived fetal vessels invade the branching villi to form the fetal vasculature. The EPC has expanded and flattened to form the spongiotrophoblast layer of the junctional zone. TGCs further invade the maternal decidua and around the maternal spiral arteries. Hectd1 is expressed in all of these cell types with higher levels in trophoblasts at the border of the allantoic mesoderm (arrows in Fig. 2F) and scattered trophoblasts within the junctional and labyrinth zones (arrowheads in Fig. 2F). Between E9.5 and 13.5 Hectd1 expression becomes progressively reduced in cells of the allantoic mesoderm, compared to the trophoblast layers (Fig. 2G–N). During these stages, Hectd1 continues to be expressed at higher levels in labyrinth trophoblasts at the junction of the chorion and allantoic mesoderm (arrows in Fig. 2G and data not shown). By E13.5, these cells will develop into labyrinth trophoblasts that separate fetal and maternal blood spaces that express high level of Hectd1 including syncytiotrophoblasts (long flat cells) and sinusoidal trophoblast giant cells (Fig. 2N; arrows). In addition, scattered clusters of cells in the junctional zone show high levels of Hectd1 expression (arrowheads in Fig. 2G–L). To compare expression of Hectd1 in the maternal decidua versus trophoblast and inner cell mass-derived lineages, heterozygous Hectd1XC mice were intercrossed and expression analyzed in heterozygous E13.5 Hectd1XC/+ placentas. Hectd1 is expressed at greater intensity in the maternal decidua and cells that separate maternal and fetal circulations in the labyrinth zone than in other cells derived from the conceptus (Fig. 2M and N).
Fig. 2.
Hectd1 is expressed in multiple cell types throughout placental development. (A–L) Hectd1 expression in concepti or placentas (n=8) at each of the indicated stages from wildtype females mated to Hectd1XC/+ males. This mating scheme resulted in LacZ staining in lineages derived from the conceptus but not the maternal decidua: (A, B) at E7.5, Hectd1 is expressed in the primary and secondary TGCs and the extraembryonic lineages (ectoplacental cone; EPC and chorion) that contribute to the mature placenta. Hectd1 is also expressed in the epiblast and amnion. (C, D) E8.5 Hectd1XC/+ conceptus showing widespread expression of Hectd1 with relatively higher levels in the chorion and TGCs in the EPC. (E, F) E9.5 Hectd1XC/+ conceptus showing widespread expression of Hectd1 in the developing placenta with higher levels in labyrinth trophoblasts at the border of the allantoic mesoderm (arrows in F) and scattered cells throughout the junctional zone (arrowheads in F). The layers of the placenta are labeled: maternal decidua (MD), junctional zone (JZ), labyrinth zone (LZ) and allantoic mesoderm (AL). (G–L) E10.5 (G, H), E11.5 (I, J) and E12.5 (K, L) Hectd1XC/+ placentas showing Hectd1 expression. Expression is markedly reduced in the allantoic mesoderm as compared to trophoblasts. High levels of expression are found in labyrinth trophoblasts bordering the allantoic mesoderm (arrows in G) and trophoblasts scattered throughout the junctional zone (arrowhead in G–L). B, D, F, H, J and L are magnifications of boxed regions in A, C, E, G, I and K, respectively. (M, N) E13.5 Hectd1XC/+ placentas (n=7) from Hectd1XC/+ intercrosses where expression of Hectd1 is highlighted in both the maternal- and fetal-derived placental lineages. Intensity of Hectd1 expression is relatively greater in the maternal decidua than in fetal trophoblast lineages where both maternal and fetal genotypes are Hectd1XC/+. High levels of Hectd1 expression are found in cells lining large maternal arteries (red *) and sinusoids (yellow *) in the junctional zone and maternal decidua. (N) In the labyrinth zone, Hectd1 is expressed at high levels in trophoblasts that separate maternal (MBS) and fetal (FBS) blood spaces including flat cells, which are likely syncytiotrophoblasts (arrows), but expression in the allantoic mesoderm is comparatively less.
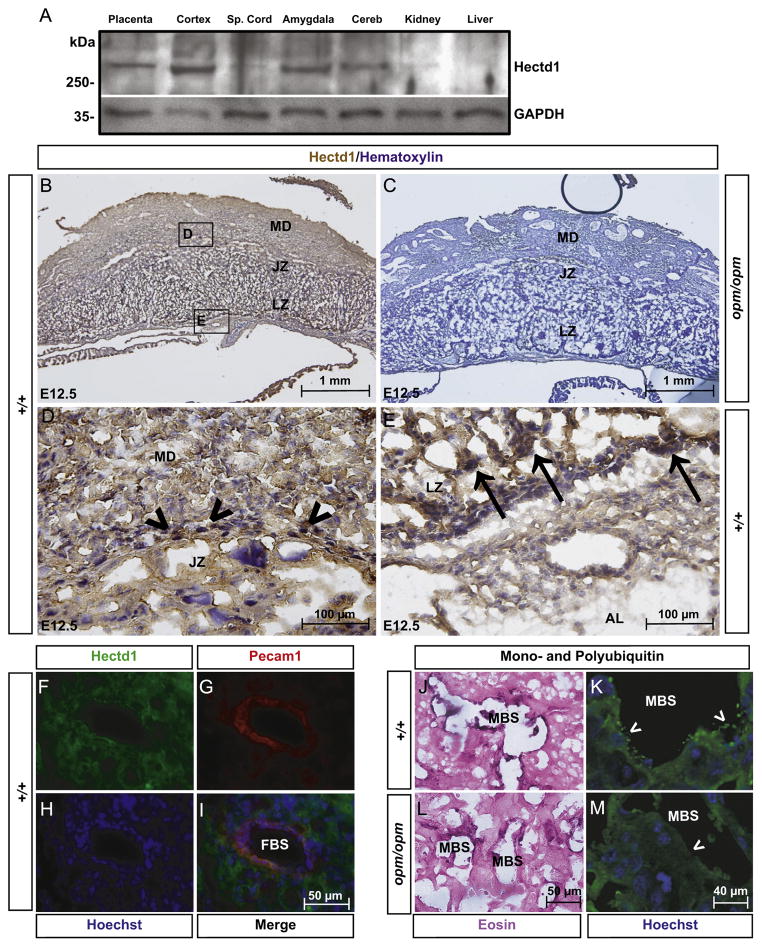
Fig. 3.
Hectd1 protein is ubiquitously expressed in the placenta: (A) Western blotting reveals Hectd1 protein is expressed in the E12.5 placenta and cerebral cortex (cortex), amygdala and cerebellum (cereb) of the adult female mouse (n=2). Minimal expression is found in the spinal cord (Sp. Cord), kidney and liver. (B–G) Immunohistochemistry using the anti-Hectd1 antibody on E12.5 wildtype (B, D, E) placentas reveals widespread distribution of Hectd1 protein (n=3). The layers of the placenta are labeled: maternal decidua (MD), junctional zone (JZ), labyrinth zone (LZ) and allantoic mesoderm (AL). (C) Absence of staining in the Hectd1opm/opm placenta confirms the specificity of the antibody. (D) and (E) are higher magnified images of boxed regions in (B). Unlike the transcript, Hectd1 is expressed at similar levels in the maternal decidua and junctional zone. Similar to the transcript, the protein is expressed at higher levels in scattered trophoblasts in the junctional zone (arrowheads) and trophoblasts that separate maternal and fetal blood spaces in the labyrinth zone (arrows). (F–I) immunofluorescence to detect Pecam1 (red) and Hectd1 (green) reveals Hectd1 is expressed in endothelial cells lining fetal blood spaces (FBS) in the labyrinth zone of E12.5 wildtype placentas. Nuclei were stained with Hoechst. (J–M) Immunohistochemistry (J, L) and immunofluorescence (K, M) staining in the labyrinth zone of E12.5 placenta with the FK2 antibody to label poly- and mono-ubiquitinated conjugates (n=3). Enhanced ubiquitin staining is concentrated in projections from cells lining the maternal blood spaces (MBS). Staining of ubiquitin conjugates here is reduced in Hectd1 mutants. Immunohistochemistry (J, L) was counterstained with eosin and immunofluorescence (K, M) with Hoechst.
Table 2.
Summary of Hectd1 expression.
| E7.5 | E8.5 | E9.5 | E10.5 | E11.5 | E12.5 | E13.5 | |
|---|---|---|---|---|---|---|---|
| EPC/TGCs | + + | + + | + + | + + | + + | + + | + + |
| Chorion/labyrinth trophoblasts | + + | + + + | + + + | + + + | + + + | + + + | + + + |
| Allantoic mesoderm | + + | + + | + + | + | + | + | −/+ |
| Maternal decidua | ND | ND | ND | ND | ND | + + + | + + + + |
Hectd1 protein is expressed in the E12.5 placenta (Fig. 3A). Additionally, in the adult female mouse, Hectd1 protein is present in specific regions of the brain with minimal expression in the spinal cord, kidneys and liver (Fig. 3A). Immunohistochemical analysis was used to determine the spatial expression of Hectd1 protein in the E12.5 placenta. Hectd1 protein is widely expressed with higher levels in certain cell types (Fig. 3B, D, and E); but unlike the transcript, relatively equivalent levels are found in the maternal decidua and junctional and labyrinth zones. Scattered trophoblasts in the junctional zone express high levels of Hectd1 (arrowheads in Fig. 3D). In the labyrinth zone, Hectd1 protein is highly expressed in trophoblasts bordering the allantoic mesoderm (arrow in Fig. 3E) and at low levels in the allantoic mesoderm itself (AL in Fig. 3E). Hectd1 is also expressed in endothelial cells lining fetal blood vessels, as it colocalizes with Pecam1 (Fig. 3F–I). As a negative control, the antibody was tested on homozygous mutant Hectd1opm placentas, since the antibody does not recognize the truncated Hectd1opm mutant protein (Sarkar and Zohn, 2012; Fig. 3C).
Our previous studies reported a decrease in total ubiquitin conjugated proteins in Hectd1 mutant embryos (Sarkar and Zohn, 2012), thus we expected that total ubiquitin labeling in the placenta might be similarly reduced. However, detection of total mono- and polyubiquitin conjugated protein using the FK2 antibody by immunohistochemistry showed no obvious difference between wildtype and mutant placentas (Supplementary Fig. 2A and B). Furthermore, the total amount of ubiquitin conjugated protein was unchanged between wildtype and Hectd1opm mutant placental lysates in biochemical pull down assays (Supplementary Fig. 2C). Interestingly, in the wildtype placenta, ubiquitin conjugates were enriched in cells that line maternal blood spaces in the labyrinth zone (Fig. 3J–M). In particular, ubiquitin was concentrated in projections from these cells. However, in mutant placentas, FK2 immunoreactivity was reduced in cells lining blood spaces and their projections. Quantitation reveals significant differences. In wildtype placentas, the average sinusoid had on average 26.7±4 ubiquitin-positive projections, whereas Hectd1 mutants had only 2.6±2 ubiquitin-positive projections per sinusoid (p=0.013). Thus, while loss of Hectd1 function does not significantly alter total ubiquitin levels in the placenta, differences in ubiquitin conjugated proteins are found in cells lining maternal blood spaces.
The Hectd1 mutant placenta shows altered TGC development
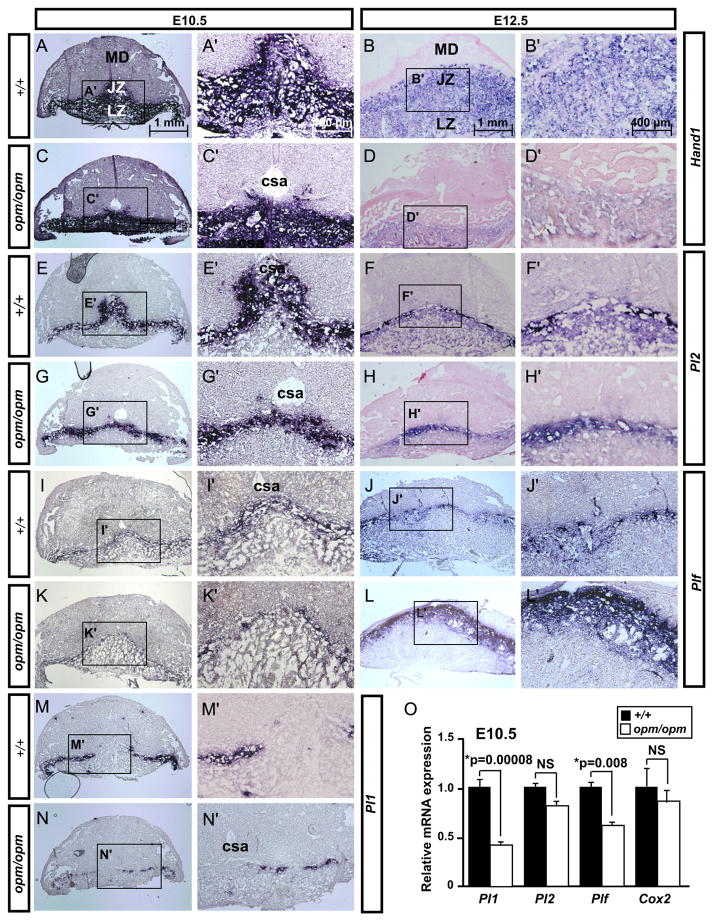
Defects in development of the Hectd1 mutant placenta may be caused by altered development and/or differentiation of the trophoblast lineage. To address this possibility, expression of a number of trophoblast cell markers was examined in Hectd1 mutant placentas (Figs. 4 and 5 and summarized in Table 3). Hand1 is expressed in all trophoblasts at E10.5 and is required for the differentiation of TGC subtypes (Riley et al., 1998). At E12.5, Hand1 is expressed in trophoblasts of the junctional and labyrinth zones (Scott et al., 2000). Analysis of Hand1 expression in E10.5 wildtype and Hectd1 mutant placentas showed no alterations in expression levels. However, no staining is detected around the central spiral arteries (csa in Fig. 4A and C) in mutants suggesting that these cells are either not properly localized and/or specified. By E12.5, Hand1 expression was markedly reduced throughout the mutant placenta.
Fig. 4.
Altered expression of TGC markers in Hectd1 mutants: (A–N′) Expression of Hand1 (A–D), Pl2 (E–H), Plf (I–L) and Pl1 (M–N) was examined in E10.5 and E12.5 wildtype and Hectd1opm mutant placentas by in situ hybridization and counterstained with eosin as indicated. Five placentas per stage were used for this analysis. Images in A′–N′ are magnifications of boxed regions in A–N. Cells surrounding the central spiral arteries (csa) do not express trophoblast markers in E10.5 Hectd1 mutant placentas. (O) Whole placental RNA extracts were prepared from E10.5 wildtype (black bars) or mutant (white bars) placentas and analyzed by qRT-PCR for expression of cell-type specific markers. Ten placentas were used for this analysis. Data is normalized to wildtype values. Expression of TGC markers, Pl1 and Plf are significantly decreased in E10.5 Hectd1opm mutant placentas, whereas changes in expression levels of the internal control Cox2 and Pl2 were not significant (NS; not significant).
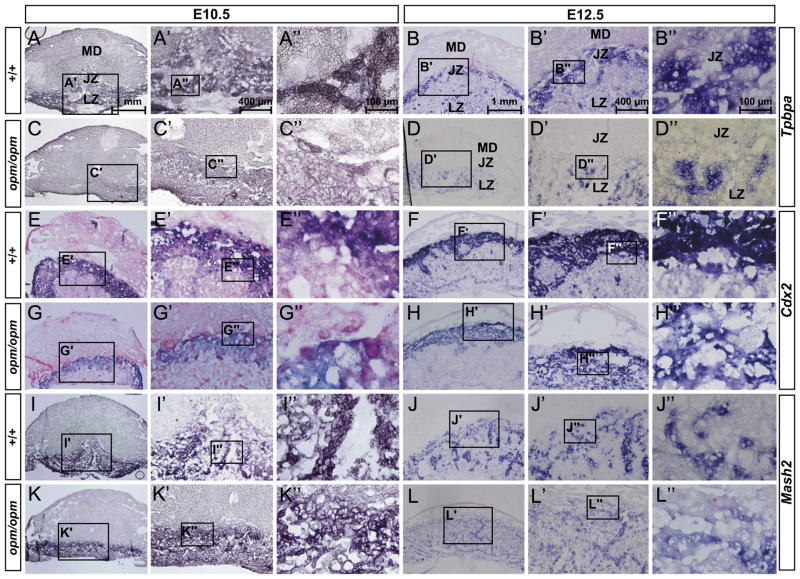
Fig. 5.
Reduced spongiotrophoblasts and glycogen trophoblasts at the junctional zone of the Hectd1 mutant placenta. (A–L) Expression of spongiotrophoblast markers Tpbpa (A–D), Cdx2 (E–H), and Mash2 (I–L) was examined in E10.5 and 12.5 wildtype and Hectd1 mutant placentas by in situ hybridization on sections and counterstained with eosin. Five placentas per stage were used for this analysis. Tpbpa is also expressed in glycogen trophoblasts. Images in A′–L′ and A″–L″ are magnifications of boxed region as indicated.
Table 3.
Summary of gene expression changes in Hectd1opm/opm placentas.
| Marker | Cell type | E10.5 | E12.5 |
|---|---|---|---|
| HAND1 | Trophoblast (E10.5); TGC, spongiotrophoblast, labyrinth trophoblast (E12.5) | NC | ↓↓↓ |
| PL1 | Parietal TGC | ↓↓ | N/A |
| PL2 | Parietal TGC, canal and sinusoidal associated TGC | NC | ↓ |
| Plf | Spiral artery and canal associated TGC | ↓ | ↑↑↑ |
| Tpbpa | Spongiotrophoblast and glycogen trophoblasts | ↓↓ | ↓↓↓ |
| Cdx2 | Spongiotrophoblast | ↓ | ↓↓ |
| Mash2 | Spongiotrophoblast | ↓ | ↓↓ |
Multiple subtypes of terminally differentiated TGCs have been reported based on gene expression, localization, morphology and ploidy (Table 3 and Simmons et al., 2007). Parietal TGCs are localized at the junctional zone forming a discontinuous border between the maternal decidua and spongiotrophoblast layers. These cells express Pl1, Pl2 and Plf. Spiral artery associated TGCs express only Plf. Maternal blood canal associated TGCs express Pl2 and Plf but not Pl1. TGCs associated with the labyrinth layer sinusoids express Pl2 but not Pl1 or Plf. Expression of these TGC subtype markers was determined in E10.5 wildtype and Hectd1opm homozygous mutant placentas by quantitative RT-PCR (qRT-PCR; Fig. 4O). This analysis revealed significant reductions in Pl1 and Plf but not Pl2 levels in the mutant placenta at E10.5.
To examine alterations in the spatial expression of these markers, in situ hybridization analyses was performed on placental sections from E10.5 and 12.5 wildtype and Hectd1opm mutants (Fig. 4E–N and summarized in Table 3). Pl1 is expressed at 10.5 but not 12.5 and is unique to parietal TGCs (Simmons et al., 2007). In the mutant placenta, Pl1 shows reduced and discontinuous expression at the junctional zone (Fig. 4M and N). Consistent with the qRT-PCR results, Pl2 expression levels were not altered in mutants at E10.5 but Pl2-positive cells failed to localize around the central spiral arteries (Fig. 4E and G, similar to the change seen in Hand1 expression). By E12.5, Pl2 expression is reduced throughout the junctional zone (Fig. 4F and H). Plf shows a reduced domain of expression at E10.5; but by E12.5 its expression is expanded at the junctional zone in Hectd1 mutants (Fig. 4I–L). Taken together these observations indicate that TGC subtypes are differentially and dynamically affected in the Hectd1 mutant placenta.
Reduced spongiotrophoblasts in the junctional zone of Hectd1 mutant placentas
To determine if Hectd1 is required for development of spongio-trophoblasts and glycogen trophoblasts at the junctional zone, expression of Tpbpa, Cdx2 and Mash2 was examined in E10.5 and E12.5 placentas of wildtype and Hectd1opm homozygous mutants (Fig. 5 and summarized in Table 3). At E10.5, Tpbpa is expressed in spongiotrophoblasts and by E12.5 also in glycogen cells of the junctional zone (Cross, 2005; Hu and Cross, 2011). Expression of Tpbpa was decreased in Hectd1 mutant placentas at both E10.5 and E12.5. Moreover, at E12.5, Tpbpa expression, while detectible in the labyrinth layer of mutants, was absent from the junctional zone (Fig. 5B and D), demonstrating a reduction in the spongiotrophoblasts and/or glycogen trophoblasts in this region. At E10.5 and 12.5, Cdx2 is expressed in spongiotrophoblasts of the junctional zone (van Nes et al., 2006). In Hectd1 mutant placentas, the expression domain as well as intensity of Cdx2 is reduced (Fig. 5E–H). In addition, immunohistochemistry at E10.5 shows reduced intensity of Cdx2 staining in the nuclei of spongiotrophoblasts at the junctional zone as well as less cells expressing the protein (Supplemental Fig. 3). Mash2 is expressed in spongiotrophoblasts in the junctional and labyrinth zones at E10.5 and 12.5 (Tanaka et al., 1997). In Hectd1 mutant placentas, the level of Mash2 expression is not affected at E10.5 (Fig. 5I and K), but at E12.5 expression of Mash2 is reduced (Fig. 5J and L). Taken together, these results indicate that Hectd1 is required for development of a subset of spongiotrophoblasts marked by Tpbpa, Mash2 and Cdx2, particularly at the junctional zone.
Reduced glycogen trophoblasts in the junctional zone of Hectd1 mutant placentas
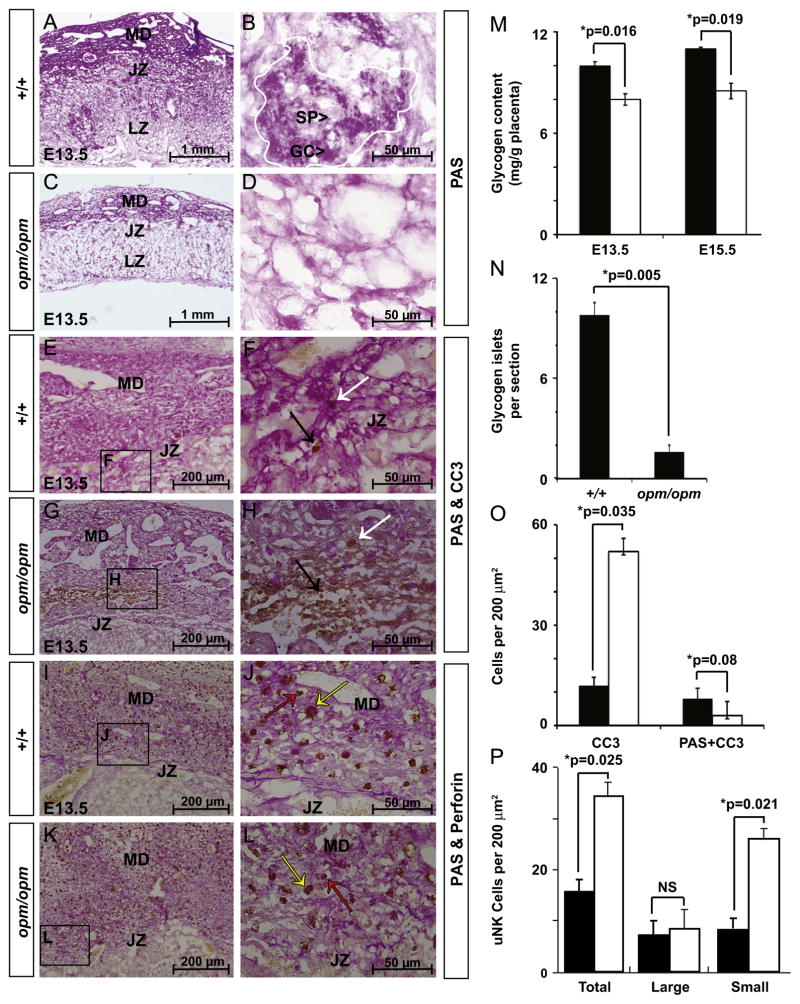
Glycogen trophoblasts differentiate from spongiotrophoblasts forming islets with in the junctional zone and migrate into the maternal decidua around E12.5 (Bouillot et al., 2006; Coan et al., 2006). Tpbpa also marks glycogen trophoblasts and its reduction at the junctional zone of E12.5 Hectd1 mutant placentas (Fig. 5A–D) suggests a possible loss of this cell type. To determine if glycogen trophoblasts are affected in the Hectd1 mutant placenta, the glycogen content of E13.5 wildtype and mutant placentas was determined by Periodic Acid-Schiff (PAS) staining on undigested and amylase digested control placental sections (Fig. 6 and data not shown). PAS staining is reduced in the maternal decidua, junctional zone and scattered glycogen cell islets in the spongiotrophoblast and labyrinth zones at E13.5 in Hectd1 mutants (Fig. 6A–D). There is a significant reduction in glycogen cell islets in the junctional zone of Hectd1 mutant placentas (Fig. 6N). Furthermore, biochemical quantification of total glycogen content in whole placentas shows a significant decrease in Hectd1 mutants at both E13.5 and E15.5 (Fig. 6M).
Fig. 6.
Reduced glycogen trophoblasts in Hectd1 mutant placentas: (A–D) reduced PAS staining of glycogen containing cells in E13.5 mutant (opm/opm; C, D) compared to wildtype (+/+; A, B) placentas (n=6). (B) A single glycogen cell islet (outlined in white) is composed of both spongiotrophoblasts (SP), and glycogen cells (GC). Few islets were found in the junctional zone of Hectd1opm mutants. (E–L) E13.5 sections were co-stained for PAS and cleaved caspase-3 (CC3; E–H) or PAS and Perforin (I–L). Perforin stained cells either had large cell bodies with intense cytoplasmic granules (yellow arrows) or small cell bodies with few cytoplasmic granules (red arrows). Three placentas per stage were used for these analyses. Panels F, H, J and L are higher magnification images of the boxed regions in E, G, I and K, respectively. (M) Glycogen content of the mutant placenta is significantly reduced (opm/opm, white bars) versus wildtype (+/+, black bars) at E13.5 and 15.5. Three placentas of each stage were used for analysis. (N) Average numbers of islets per section were counted in 10 sections each of wildtype (n=7) and Hectd1opm mutant (n=5) placentas. (O) Cleaved caspase-3 positive cells (black arrow points out an example in panel F and H) or cells co-stained with cleaved caspase-3 and PAS (white arrow points out an example in panels F and H) in the junctional zone and maternal decidua were counted in ten 200 μm2 fields in three placentas demonstrating an increase in apoptosis in the mutant but not of glycogen containing cells. (P) Quantification of uNK cells co-labeling with Perforin and PAS. Total uNK cells and small and large cells were counted separately. Ten 200 μm2 fields were counted in three placentas (NS: not significant). Data represent the mean and error bars standard error of mean (SEM).
To determine if alterations in apoptosis could explain the reduction of glycogen cell numbers in Hectd1opm mutant placentas, apoptosis of these cells was examined. At E13.5, the total number of cleaved caspase-3 positive cells were significantly increased in the mutant and clustered in the medial region of the decidua (Fig. 6E–H and Supplemental Fig. 4). However, no difference in apoptosis of glycogen containing cells was found between wild-type and mutant samples (Fig. 6E–H, O). Thus increased apoptosis is not likely responsible for the reduction in PAS positive cells observed in the mutant placenta.
Altered uNK cell development in the Hectd1 mutant placenta
uNK cells of the maternal decidua are marked by high glycogen content and undergo dramatic apoptosis after E12.5 (Bilinski et al., 2008; Croy et al., 2003). uNK cells play an essential role in spiral artery remodeling and promote TGC invasion around the spiral arteries (Harris, 2010; Wallace et al., 2012). Since TGCs around the central spiral arteries are decreased in Hectd1 mutants (Fig. 4), we examined uNK cells as a possible underlying mechanism. The maternal decidua of E13.5 mutant placentas showed a significant increase in uNK cells that co-label with PAS and the uNK marker Perforin (Fig. 6I–L and P). However, these cells were smaller and contained fewer cytoplasmic granules indicative of less mature uNK cells (Fig. 6P). No difference in uNK cell numbers was noted in the maternal decidua at E10.5 (Supplemental Fig. 5). Hence the reduction in the glycogen content of the mutant placenta could be attributed to both reduced number of glycogen islets in the junctional zone and mature uNK cells in the maternal decidua.
Altered proliferation and apoptosis in the Hectd1 mutant placenta
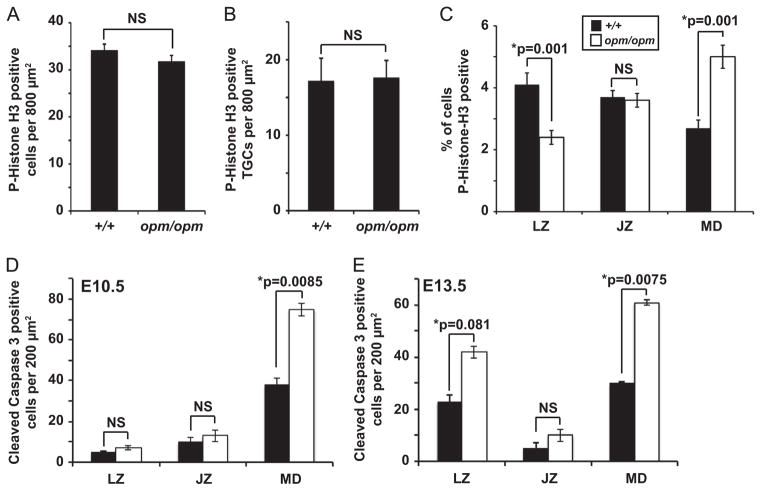
To determine if proliferation is affected in Hectd1 mutant placentas, the mitosis marker Phospho-Histone H3 was used. No significant differences in mitosis overall was observed in wildtype versus Hectd1 mutant placentas at E12.5 (Fig. 7A and Supplemental Fig. 6). Similarly, restricting the analysis to TGCs did not show a significant difference (Fig. 7B). Analysis of mitosis in different placental layers shows an increase in the number of mitotic cells in the maternal decidua, a decrease in the labyrinth zone and no significant difference in the junctional zone (Fig. 7C).
Fig. 7.
Cell proliferation and apoptosis are altered in the labyrinth zone and maternal decidua but not the junctional zone of Hectd1 mutant placentas. (A, B) Total Phospho-Histone H3/mitotic cell numbers (A) or mitotic TGCs (B) do not differ significantly between E12.5 wildtype (+/+) and Hectd1 mutant (opm/opm) placentas. The total number of Phospho-Histone H3 positive cells (A) or TGCs (B) identified by the presence of large polyploid nuclei were counted. (C) Mitotic cells were counted separately in the labyrinth zone (LZ), junctional zone (JZ) and maternal decidua (MD) and expressed as a percentage of Hoechst stained nuclei. For (A–C), ten random 800 μm2 fields were counted in three placentas each. (D, E) Quantitation of cleaved caspase-3 positive cells in 10 random 200 μm2 frames each in three placentas. An increase in apoptotic cells was found in the maternal decidua (MD) of Hectd1 mutant (white bars) compared to wildtype (black bars) placenta at E10.5 (D) and the maternal decidua and labyrinth zone (LZ) at E13.5 (E). No change in the number of apoptotic cells was found in the junctional zone (JZ) at either stage. Data represent mean values and error bars, standard error of the mean (SEM).
Apoptosis is increased in the Hectd1 mutant placenta (Fig. 6O). To determine if regional specific differences are found, apoptotic cells were counted in each layer of the placenta. This analysis reveals a significant difference in apoptotic cells in the maternal decidua at E10.5 but not in the junctional or labyrinth zones (Fig. 7D and Supplemental Fig. 4). At E13.5, apoptosis was significantly increased in both the labyrinth zone and maternal decidua but not the junctional zone (Fig. 7E and Supplemental Fig. 4). Since no change in junctional zone proliferation or apoptosis was observed, alterations in TGC and spongiotrophoblast populations in this region are not explained by disruption of cell division or death. On the other hand, reduced proliferation and increased apoptosis may account for the reduced thickness of the labyrinth layer.
Discussion
Here, we describe a novel requirement for the E3 ubiquitin ligase Hectd1 in placentation. Mutant placentas show reduced size and lamination of trophoblast-derived layers with a corresponding increase in the thickness of the maternal decidua. Analysis of cell type specific markers reveals defects in TGC development. Spongiotrophoblasts and glycogen trophoblasts at the junctional zone are also reduced. Finally, the numbers of immature uNK cells were increased. Placental zone specific alterations in proliferation and apoptosis were also identified. Namely, in the labyrinth zone proliferation was decreased and apoptosis increased. In the maternal decidua both proliferation and apoptosis was increased, while in the junctional zone both were unchanged. Together these data demonstrate complex changes in development of the Hectd1 mutant placenta. Placental defects likely contribute to midgestation lethality and IUGR observed in Hectd1 mutants. However, other causes of mid-gestation lethality and IUGR also likely play an important role. For example, complex cardiac defects have been reported in a different N-ethyl-N-nitrosourea (ENU)-induced allele of Hectd1 (MGI:5313700). Since alterations in cardiac function could exacerbate or even cause placental defects, additional experiments are required to determine if placental abnormalities are due to cell autonomous defects in trophoblast development or are secondary to embryonic defects in Hectd1 mutants.
Hectd1 is highly expressed in scattered TGCs in the EPC that contain progenitors for TGCs and TGC development is abnormal in Hectd1 mutants. The number and nuclear size of TGCs is reduced in Hectd1 mutants and altered expression and localization of TGC markers are found in the mutant. In addition, TGCs migrate to positions around the maternal central canal and medial spiral arteries during decidualization. In the Hectd1 mutant placenta, the lack of Hand1-, Pl1- and Pl2-expressing cells around the central canal and spiral arteries suggests a possible requirement for Hectd1 in TGC invasion as well as differentiation. Additionally, the number and morphology of uNK cells is altered and these play an essential role in spiral artery remodeling.
The EPC also contains progenitors for spongiotrophoblasts and glycogen trophoblasts, which are affected in Hectd1 mutant placentas. The specific functions of these cells are not known; but mouse mutants with defects in development of these cell types show abnormal placentation (reviewed in Cross, 2005). Multiple mouse mutants have documented defects in both cell types suggesting common developmental programs (for example Frank et al., 2002; Withington et al., 2006; Yang et al., 2003). Significant defects in development of both spongiotrophoblast and glycogen trophoblast populations were found in the Hectd1 mutant placenta. Expression of Tpbpa, Cdx2 and Mash2 is reduced at E12.5 in the junctional zone of Hectd1 mutant placenta. PAS staining is reduced in mutant placental sections, as well as the number of glycogen cell islets and the total glycogen content of the placenta. Reduction of these cell-types could occur with alterations in trophoblast differentiation, survival or proliferation. Since neither apoptosis nor proliferation were affected in the junctional zone of Hectd1 mutants, it is likely that Hectd1 is required for differentiation of spongiotrophoblasts and glycogen trophoblasts.
The maternal decidua is also abnormal in Hectd1 mutants. The decidua compartment of the Hectd1 mutant placenta is larger with respect to the other layers. Our data suggest that both apoptosis and mitosis in the maternal decidual compartment is increased. In addition, immature uNK cells are increased in numbers in the mutant. uNK cells play an essential role in uterine vascular remodeling, induce apoptosis in the decidualized uterus and are themselves removed by cell death (Bilinski et al., 2008; Croy et al., 2003; Harris, 2010; Wallace et al., 2012). Interestingly, the increase of uNK cells in the decidua occurs coincident with an increase in cleaved caspase-3 positive cells in the medial region of the placenta. Together alterations in the invasive cell types (uNK cells, glycogen trophoblasts, canal and spiral artery associated TGCs) in the Hectd1 mutant placenta indicate a possible mis-regulation of the coordination between the invasive trophoblasts and the decidualizing maternal endometrial cells.
Hectd1 is highly expressed in the chorion and these cells differentiate into labyrinth trophoblasts. This continued pattern of expression in this lineage suggests a role for Hectd1 in development of the labyrinth trophoblast. Hectd1 is an E3 ubiquitin ligase that we have previously shown regulates total cellular ubiquitination in the mouse embryo and embryonic fibroblasts (Sarkar and Zohn, 2012). Interestingly, while no obvious changes in overall ubiquitination are found in the mutant placenta, the presence of ubiquitin conjugates in labyrinth trophoblasts that line maternal blood spaces is decreased in the mutant. The requirement for Hectd1 in development of the labyrinth zone will be further explored in another study where we show that Hectd1 mutant placentas have defects in branching morphogenesis in the labyrinth zone. In addition, Hectd1 mutant placentas show compromised integrity of the endothelium that lines fetal vessels, aberrant expression of labyrinth trophoblast markers and altered allocation of trilaminar labyrinth trophoblast cell types (Sarkar and Zohn, in preparation).
Placental defects in Hectd1 mutants show variable penetrance and expressivity. This is common among placental defects (see Barak et al., 2002; Caspary et al., 1999; Catela et al., 2009; Eggenschwiler et al., 1997 for examples). Variable penetrance has been attributed to a number of factors including genetic background, modifier effects and environmental conditions (Barak et al., 2008). However, our previous studies were done on a congenic mouse line under relatively controlled environmental conditions. On the other hand, there is inherent variability in placental development possibly due to stochastic fluctuations in gene expression, epigenetic regulation, differentiation and morphogenesis that underlie development of the placenta (Coan et al., 2004; Yuen and Robinson, 2011). One interesting hypothesis proposed to explain how phenotypic variation is normally suppressed is that molecular chaperones act to repress diversity (Ruden et al., 2003; Yeyati and van Heyningen, 2008). Molecular chaperones, such as heat shock proteins, assist in protein folding and have the potential to buffer variations in gene function (Bergman and Siegal, 2003; Queitsch et al., 2002; Rohner et al., 2013; Rutherford and Lindquist, 1998; Sollars et al., 2003). Our studies indicate that Hectd1 binds to and affects the function of Hsp90 (Sarkar and Zohn, 2012). Thus it is possible that altered Hsp90 function in Hectd1 mutants contributes to variable penetrance and expressivity with respect to placental defects.
Known substrates of Hectd1 include heat shock protein 90 (Hsp90) and adenomatous polyposis coli (APC) involved in Wnt signaling; both of which regulate placental development (Sonderegger et al., 2010; Voss et al., 2000). Hsp90beta (Hsp90ab1) mutants show defects in formation of the labyrinth layer due to a requirement of Hsp90beta in the allantois for induction of trophoblast differentiation (Voss et al., 2000). Wnt signaling is required at multiple steps in placentation including early determination of trophoblast lineages and chorioallantoic fusion. However, with regard to spongiotrophoblast, glycogen trophoblast, uNK and TGC development, these mutant phenotypes differ significantly from Hectd1 mutants, suggesting Hsp90 and Wnt pathways are unlikely to mediate all of these defects. Our ongoing studies to elucidate the pathways regulated by Hectd1 indicate that the ubiquitination of numerous substrates is affected in Hectd1 mutants (Li et al., 2013; Sarkar and Zohn, 2012; Tran et al., 2013 and AAS and IEZ unpublished). Since, multiple molecular pathways are implicated in development of the placenta, the role of Hectd1 is likely very complex.
Supplementary Material
Acknowledgments
We are grateful to Drs. Anne Croy, Anna Penn and Joshua Corbin for suggestions and critical reading of the manuscript. Lee Niswander for support of early studies. Trevor Williams and James Cross for in situ hybridization probes. This study was supported by R01-HD058629 to I.E.Z.
Abbreviations
- HECT
homologous to E6-AP carboxyl terminus
- Ub
ubiquitin
- IUGR
intrauterine growth restriction
- TGC
trophoblast giant cell
- H&E
hematoxylin and eosin
- PAS
periodic acid-Schiff
- uNK
uterine natural killer cells
Appendix A. Supplementary Information
Supplementary data associated with this article can be found in the online version at: http://dx.doi.org/10.1016/j.ydbio.2014.05.007.
Footnotes
Author contributions
T.M. and A.S.L. performed the qRT-PCR experiment. All other experiments were conceived and conducted by A.A.S. with oversight by I.E.Z. and technical assistance by S.J.N. Initial studies were performed by F.G. and J.T.H. The manuscript was written by A.A.S. and I.E.Z.
References
- Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci USA. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Sadovsky Y, Shalom-Barak T. PPAR signaling in placental development and function. PPAR Res. 2008:142082. doi: 10.1155/2008/142082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebington C, Doherty FJ, Fleming SD. Ubiquitin and ubiquitin–protein conjugates are present in human cytotrophoblast throughout gestation. Early Pregnancy. 2000;4:240–252. [PubMed] [Google Scholar]
- Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- Bilinski MJ, Thorne JG, Oh MJ, Leonard S, Murrant C, Tayade C, Croy BA. Uterine NK cells in murine pregnancy. Reprod Biomed Online. 2008;16:218–226. doi: 10.1016/s1472-6483(10)60577-9. [DOI] [PubMed] [Google Scholar]
- Bouillot S, Rampon C, Tillet E, Huber P. Tracing the glycogen cells with protocadherin 12 during mouse placenta development. Placenta. 2006;27:882–888. doi: 10.1016/j.placenta.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Carney EW, Prideaux V, Lye SJ, Rossant J. Progressive expression of trophoblast-specific genes during formation of mouse trophoblast giant cells in vitro. Mol Reprod Dev. 1993;34:357–368. doi: 10.1002/mrd.1080340403. [DOI] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Perlman EJ, Zhang P, Elledge SJ, Tilghman SM. Oppositely imprinted genes p57(Kip2) and igf2 interact in a mouse model for Beckwith–Wiedemann syndrome. Genes Dev. 1999;13:3115–3124. doi: 10.1101/gad.13.23.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Models Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan PM, Conroy N, Burton GJ, Ferguson-Smith AC. Origin and characteristics of glycogen cells in the developing murine placenta. Dev Dyn: Off Publ Am Assoc Anat. 2006;235:3280–3294. doi: 10.1002/dvdy.20981. [DOI] [PubMed] [Google Scholar]
- Coan PM, Ferguson-Smith AC, Burton GJ. Developmental dynamics of the definitive mouse placenta assessed by stereology. Biol Reprod. 2004;70:1806–1813. doi: 10.1095/biolreprod.103.024166. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice—a review. Placenta. 2005;26(Suppl A):S3–9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Cross JC. Placental function in development and disease. Reprod Fertil Dev. 2006;18:71–76. doi: 10.1071/rd05121. [DOI] [PubMed] [Google Scholar]
- Croy BA, He H, Esadeg S, Wei Q, McCartney D, Zhang J, Borzychowski A, Ashkar AA, Black GP, Evans SS, et al. Uterine natural killer cells: insights into their cellular and molecular biology from mouse modelling. Reproduction. 2003;126:149–160. doi: 10.1530/rep.0.1260149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith–Wiedemann and Simpson–Golabi–Behmel syndromes. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria TN, Ogren L, Talamantes F, Linzer DI, Soares MJ. Localization of placental lactogen-I in trophoblast giant cells of the mouse placenta. Biol Reprod. 1991;44:327–331. doi: 10.1095/biolreprod44.2.327. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Forhead AJ, Coan PM, Burton GJ. The placenta and intrauterine programming. J Neuroendocrinol. 2008;20:439–450. doi: 10.1111/j.1365-2826.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, Li CM, Reik W, Ludwig T, Tycko B. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci USA. 2002;99:7490–7495. doi: 10.1073/pnas.122039999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- Harris LK. Review: trophoblast–vascular cell interactions in early pregnancy: how to remodel a vessel. Placenta. 2010;31(Suppl):S93–S98. doi: 10.1016/j.placenta.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Hu D, Cross JC. Ablation of Tpbpa-positive trophoblast precursors leads to defects in maternal spiral artery remodeling in the mouse placenta. Dev Biol. 2011;358:231–239. doi: 10.1016/j.ydbio.2011.07.036. [DOI] [PubMed] [Google Scholar]
- Jackson LL, Colosi P, Talamantes F, Linzer DI. Molecular cloning of mouse placental lactogen cDNA. Proc Natl Acad Sci USA. 1986;83:8496–8500. doi: 10.1073/pnas.83.22.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Zhao W, Yuan J, Qian Y, Sun W, Zou Y, Guo C, Chen B, Shao C, Gong Y. Lack of Cul4b, an E3 ubiquitin ligase component, leads to embryonic lethality and abnormal placental development. PloS One. 2012;7:e37070. doi: 10.1371/journal.pone.0037070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc Natl Acad Sci USA. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou Q, Sunkara M, Kutys ML, Wu Z, Rychahou P, Morris AJ, Zhu H, Evers BM, Huang C. Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I gamma by HECTD1 regulates focal adhesion dynamics and cell migration. J Cell Sci. 2013;126:2617–2628. doi: 10.1242/jcs.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer DI, Lee SJ, Ogren L, Talamantes F, Nathans D. Identification of proliferin mRNA and protein in mouse placenta. Proc Natl Acad Sci USA. 1985;82:4356–4359. doi: 10.1073/pnas.82.13.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Nelson DM. Placental dysfunction and fetal programming: the importance of placental size, shape, histopathology, and molecular composition. Semin Reprod Med. 2011;29:187–196. doi: 10.1055/s-0031-1275515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malassine A, Frendo JL, Evain-Brion D. A comparison of placental development and endocrine functions between the human and mouse model. Hum Reprod Update. 2003;9:531–539. doi: 10.1093/humupd/dmg043. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Gopalakrishna D, Meechan DW, Paronett EM, Newbern JM, LaMantia AS. 22q11 Gene dosage establishes an adaptive range for sonic hedgehog and retinoic acid signaling during early development. Hum Mol Genet. 2013;22:300–312. doi: 10.1093/hmg/dds429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Parast MM, Yu H, Ciric A, Salata MW, Davis V, Milstone DS. PPARgamma regulates trophoblast proliferation and promotes labyrinthine trilineage differentiation. PloS One. 2009;4:e8055. doi: 10.1371/journal.pone.0008055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Ruden DM, Garfinkel MD, Sollars VE, Lu X. Waddington’s widget: Hsp90 and the inheritance of acquired characters. Semin Cell Dev Biol. 2003;14:301–310. doi: 10.1016/j.semcdb.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Sarkar AA, Zohn IE. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. J Cell Biol. 2012;196:789–800. doi: 10.1083/jcb.201105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AA, Zohn IE. Hectd1 is required for development of the placental labyrinth. in preparation. [Google Scholar]
- Scott IC, Anson-Cartwright L, Riley P, Reda D, Cross JC. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol Cell Biol. 2000;20:530–541. doi: 10.1128/mcb.20.2.530-541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol. 2007;304:567–578. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- Sonderegger S, Pollheimer J, Knofler M. Wnt signalling in implantation, decidualisation and placental differentiation—review. Placenta. 2010;31:839–847. doi: 10.1016/j.placenta.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gertsenstein M, Rossant J, Nagy A. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev Biol. 1997;190:55–65. doi: 10.1006/dbio.1997.8685. [DOI] [PubMed] [Google Scholar]
- Tesser RB, Scherholz PL, do Nascimento L, Katz SG. Trophoblast glycogen cells differentiate early in the mouse ectoplacental cone: putative role during placentation. Histochem Cell Biol. 2010;134:83–92. doi: 10.1007/s00418-010-0714-x. [DOI] [PubMed] [Google Scholar]
- Tran H, Bustos D, Yeh R, Rubinfeld B, Lam C, Shriver S, Zilberleyb I, Lee MW, Phu L, Sarkar AA, et al. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC–axin interaction. J Biol Chem. 2013;288:3753–3767. doi: 10.1074/jbc.M112.415240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nes J, de Graaff W, Lebrin F, Gerhard M, Beck F, Deschamps J. The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development. 2006;133:419–428. doi: 10.1242/dev.02216. [DOI] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P. Mice lacking HSP90beta fail to develop a placental labyrinth. Development. 2000;127:1–11. doi: 10.1242/dev.127.1.1. [DOI] [PubMed] [Google Scholar]
- Wallace AE, Fraser R, Cartwright JE. Extravillous trophoblast and decidual natural killer cells: a remodelling partnership. Hum Reprod Update. 2012;18:458–471. doi: 10.1093/humupd/dms015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withington SL, Scott AN, Saunders DN, Lopes Floro K, Preis JI, Michalicek J, Maclean K, Sparrow DB, Barbera JP, Dunwoodie SL. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev Biol. 2006;294:67–82. doi: 10.1016/j.ydbio.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, Hemmings BA. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J Biol Chem. 2003;278:32124–32131. doi: 10.1074/jbc.M302847200. [DOI] [PubMed] [Google Scholar]
- Yeyati PL, van Heyningen V. Incapacitating the evolutionary capacitor: Hsp90 modulation of disease. Curr Opin Genet Dev. 2008;18:264–272. doi: 10.1016/j.gde.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Robinson WP. Review: a high capacity of the human placenta for genetic and epigenetic variation: implications for assessing pregnancy outcome. Placenta. 2011;32(Suppl 2):S136–141. doi: 10.1016/j.placenta.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Zohn IE, Anderson KV, Niswander L. The Hectd1 ubiquitin ligase is required for development of the head mesenchyme and neural tube closure. Dev Biol. 2007;306:208–221. doi: 10.1016/j.ydbio.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125:957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.