Abstract
Background
Camel contagious ecthyma (CCE) is an important viral disease of camelids caused by a poxvirus of the genus parapoxvirus (PPV) of the family Poxviridae. The disease has been reported in west and east of the Sudan causing economical losses. However, the PPVs that cause the disease in camels of the Sudan have not yet subjected to genetic characterization. At present, the PPV that cause CCE cannot be properly classified because only few isolates that have been genetically analyzed.
Methods and results
PCR was used to amplify the B2L gene of the PPV directly from clinical specimens collected from dromedary camels affected with contagious ecthyma in the Sudan between 1993 and 2013. PCR products were sequenced and subjected to genetic analysis. The results provided evidence for close relationships and genetic variation of the camel PPV (CPPV) represented by the circulation of both Pseudocowpox virus (PCPV) and Orf virus (ORFV) strains among dromedary camels in the Sudan. Based on the B2L gene sequence the available CPPV isolates can be divided into two genetic clades or lineages; the Asian lineage represented by isolates from Saudi Arabia, Bahrain and India and the African lineage comprising isolates from the Sudan.
Conclusion
The camel parapoxvirus is genetically diverse involving predominantly viruses close to PCPV in addition to ORFVs, and can be divided into two genetically distant lineages. Based on sequences of the B2L gene it is not possible to suggest that the viruses that cause CCE form a monophylogenetic group or species within the PPV phylogeny.
Keywords: Camel contagious ecthyma, B2L gene, Phylogenetic analysis
Background
Camel contagious ecthyma (CCE), also known as Auzdik, Orf in camels and Pustular dermatitis is a contagious skin disease of camelids caused by a pox virus of the genus parapoxvirus (PPV), subfamily Chordopoxvirinae of the family Poxviridae. The disease has a worldwide distribution and has been reported in Mongolia [1], Kenya [2], Kazakhstan and Turkmenistan [3], Somalia [4], Sudan [5–7], Libya [8], Saudi Arabia [9, 10], Bahrain [11] and India [12]. The disease is endemic in affected areas with variations in intensity of infection, morbidity and mortality rates and tends to occur annually in the rainy season, as the situation in Sudan and Saudi Arabia, affecting mostly young animals. The age group at risk are those less than one year of age [6] including cases reported in month-old camel calves in Saudi Arabia [10]. The marked seasonality associated with the rainy season may be due to the optimum condition for the survival and perpetuation of the virus and skin abrasions caused by browsing thorny trees [7]. In most cases, the disease caused no mortality, but when camel calves are severely affected the pox-lesions interferes with the calves’ ability to suckle or graze and extends to eyelids leading to blindness, particularly at the Savanah belt in the Sudan, leading to mortality rates that can reach 9 % [7]. Clinically, the pox-lesions first appeared on the lips of affected animals as small papules that progressively developed into scabs on the lips, muzzle, nares and eyelids culminating into fissured crusts on the lips. Swelling of the head and sometimes the neck have been observed in the field. The lesion is proliferative and highly vascularized and may extend into gum, palate and tongue. The major factors associated with increased likelihood of CCE occurrence are season of the year, camel age, camel movements and location and their association with thorny trees [7].
Camel contagious ecthyma is a sparsely studied disease and the causative virus have only recently genetically characterized. According to Abubakr et al [11] sequence homologies and phylogenetic analysis of the major envelope gene (B2L) of 2 CCE skin scabs (two from Bahrain and one from Saudi Arabia) showed that these viruses are closely related to the pseudocowpox virus (PCPV) species of the PPV genus of the family poxviridae. In addition, Nagarajan et al [12] showed that CCE in Indian dromedary camels is also caused by a PCPV.
Currently, there are four established species within the PPV genus of the family Poxviridae; Orf virus (ORFV), the type species of the genus, which causes disease mainly in sheep and goats, PCPV and bovine papular stomatitis virus (BPSV) both infect cattle, in addition to Parapoxvirus of red deer in New Zealand (PVNZ), which has only been isolated from red deer in New Zealand. BPSV and PCPV affect mainly cattle, but differ from ORFVs in the site of the pox lesion, as BPSV is restricted to the muzzle and PCPV the teat. Tentative species include the camel PPV (CPPV), reindeer parapox virus, musk ox and seal parapoxvirus [13–15]. At present, the CPPV that cause CCE cannot be properly classified because only few isolates that have been genetically analyzed.
Camel contagious ecthyma is believed to be endemic in the Sudan since long time, but was first diagnosed in the disease outbreaks west of the country in 1986 [5]. Later on, the disease was reported in eastern Sudan [6, 7]. However, the PPVs that cause the disease in dromedary camels of the Sudan have not yet subjected to genetic characterization.
In the present study, we investigated lesion-derived DNAs from eight skin scabs collected from dromedary camels in the Sudan presented with CCE lesions and showed for the first time, that the CPPV does not form a monophyletic clade within the PPV phylogeny and the disease is caused by viruses from more than one PPV species.
Results
In this investigation, we screened archive scab sample collected between 1993 and 2013 in addition to the new scab collection from Sudan in August 2013. The 2013 outbreak of the disease involved 17 camel herds around Showak area of eastern Sudan, causing morbidity and mortality rates of 20 and 1.5 %, respectively. Pustular dermatitis lesion characteristic for CCE were observed (Fig. 1). Out of 10 scab materials, eight gave positive specific amplification in real-time PCR and gel-based PCR (data not shown). Table 1 shows the origin of positive samples used in this study. To characterize the DNA fragments generated by PCR, the PCR products were sequenced and subjected to genetic analysis.
Fig. 1.

Contagious ecthyma lesions on the lips of a 14 month-old dromedary camel, Sudan
Table 1.
Details of scab specimens collected from dromedary camels in Sudan
| No | Sample | Date of collection | Area | Age of affected camel (month) |
|---|---|---|---|---|
| 1 | SD-V4 | Aug. 1993 | Showak, east Sudan | 12 |
| 2 | SD-V8 | Aug. 1993 | Showak, east Sudan | 11 |
| 3 | SD-V13 | Sept. 1994 | Obeid, west Sudan | 13 |
| 4 | SD-V34 | Jul. 2000 | Showak, east Sudan | 10 |
| 5 | SD-V20 | Jul. 2005 | Showak, east Sudan | 10 |
| 6 | SD-K1 | Aug. 2013 | Showak, east Sudan | 12 |
| 7 | SD-K2 | Aug. 2013 | Showak, east Sudan | 11 |
| 8 | SD-K3 | Aug. 2013 | Showak, east Sudan | 11 |
The B2L gene fragment from the eight DNAs has a high G + C content of 62.6 to 63.9 % (Table 2). The seven isolates (SD-V4, V8, V13, V20, K1, K2 and K3) shared 96–98 % homology with viruses in the species PCPV while one isolate (V34-SD) shared 96–99 % homology with viruses in the ORFV species of the genus PPV (Table 2). Seven CPPV Sudanese isolates shared 96–98 % nucleotide sequence identity with virus strains in PCPV species, while one isolate (V34-SD) shared only 94–95 % identity with PCPV, but 96–99 % identity with ORFV.
Table 2.
Nucleotide percent identity and G + C content based on B2L gene after BLAST search involving eight CPPVs analyzed in the present study
| Percent Identity with PCPV | Percent Identity with ORFV | G + C Content | ||
|---|---|---|---|---|
| 1 | SD-V4 | 96–79 | 92–94 | 62.6 |
| 2 | SD-V8 | 97–98 | 93–94 | 63.2 |
| 3 | SD-V13 | 96–98 | 92–94 | 63.0 |
| 4 | SD-V20 | 97–98 | 92–94 | 63.0 |
| 5 | SD-V34 | 94–95 | 96–99 | 63.9 |
| 6 | SD-K1 | 97–98 | 92–94 | 63.0 |
| 7 | SD-K2 | 96–98 | 92–94 | 62.7 |
| 8 | SD-K3 | 96–98 | 92–94 | 63.0 |
Results of multiple sequence alignment and per cent identity matrix based on 160 amino acids sequences (Table 3) revealed that the Sudanese CPPVs from 1993 to 2013 examined in the present study, shared variable amino acid sequence identity ranged from 95.04 to 100 % (intra-group similarity). Five isolates (V4-SD, V20-SD, K1-SD, K1-SD and K3-SD) possessed 100 % identity, isolate V13-SD has a less homology of 95.04 to 99.69 % with the rest Sudanese isolates. On the other hand, isolate V34-SD, which has the closest identity to ORFV species, shared a relatively lower per cent identity of 95.04–96.21 with the rest seven isolates (Table 3). As shown in Table 3, the previously published five CPPV isolates Judhpur-IN and Cam/09 from India, BH-1 and BH-3 from Bahrain and SA-98 from Saudi Arabia shared intra group similarity of 98.94 to 100 % homology and a 95.04 to 99.69 % homology with the eight Sudanese isolates analysed in the present study. On the other hand, isolate CE41, which was classified as BPSV shared only 81.21 to 82.55 % homology with all the CPPVs.
Table 3.
Percent Identity Matrix based on 160 amino acids created by Clustal 2.1 for all published CPPV B2L gene sequences
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Jodhpur-IN | 98.94 | 99.38 | 99.38 | 99.38 | 81.88 | 95.04 | 99.40 | 99.69 | 99.74 | 99.69 | 100.0 | 99.69 | 99.69 | |
| 2 | Cam/09 | 98.94 | 100.0 | 100.0 | 100.0 | 81.21 | 95.04 | 99.40 | 99.69 | 99.21 | 99.69 | 99.39 | 99.69 | 99.69 | |
| 3 | 1-BH | 99.38 | 100.0 | 100.0 | 100.0 | 81.21 | 95.45 | 98.75 | 99.38 | 99.38 | 99.38 | 99.38 | 99.38 | 99.38 | |
| 4 | 3-BH | 99.38 | 100.0 | 100.0 | 100.0 | 81.21 | 95.45 | 98.75 | 99.38 | 99.38 | 99.38 | 99.38 | 99.38 | 99.38 | |
| 5 | 98-SA | 99.38 | 100.0 | 100.0 | 100.0 | 81.21 | 95.45 | 98.75 | 99.38 | 99.38 | 99.38 | 99.38 | 99.38 | 99.38 | |
| 6 | CE41 | 81.88 | 81.21 | 81.21 | 81.21 | 81.21 | 84.85 | 82.55 | 81.88 | 81.88 | 81.88 | 81.88 | 81.88 | 81.88 | |
| 7 | V34-SD | 95.04 | 95.04 | 95.45 | 95.45 | 95.45 | 84.85 | 95.04 | 95.42 | 95.42 | 95.42 | 96.21 | 95.42 | 95.42 | |
| 8 | V13-SD | 99.40 | 99.40 | 98.75 | 98.75 | 98.75 | 82.55 | 95.04 | 99.69 | 99.70 | 99.69 | 99.39 | 99.69 | 99.69 | |
| 9 | V8-SD | 99.69 | 99.69 | 99.38 | 99.38 | 99.38 | 81.88 | 95.42 | 99.69 | 100.0 | 100.0 | 100.0 | 100.0 | 100. | |
| 10 | K2-SD | 99.74 | 99.21 | 99.38 | 99.38 | 99.38 | 81.88 | 95.42 | 99.70 | 100.0 | 100.0 | 100.0 | 100.0 | 100. | |
| 11 | K1-SD | 99.69 | 99.69 | 99.38 | 99.38 | 99.38 | 81.88 | 95.42 | 99.69 | 100.0 | 100.0 | 100.0 | 100.0 | 100. | |
| 12 | V4-SD | 100.00 | 99.39 | 99.38 | 99.38 | 99.38 | 81.88 | 96.21 | 99.39 | 100.0 | 100.0 | 100.0 | 100.0 | 100. | |
| 13 | V20-SD | 99.69 | 99.69 | 99.38 | 99.38 | 99.38 | 81.88 | 95.42 | 99.69 | 100.0 | 100.0 | 100.0 | 100.0 | 100. | |
| 14 | K3-SD | 99.69 | 99.69 | 99.38 | 99.38 | 99.38 | 81.88 | 95.42 | 99.69 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
A phylogenetic tree was created for the amino acids sequences of the B2L gene of 40 PPV strains, including the eight strains described in the present study using the Neighbor-Joining method. The result revealed that the Sudanese CPPVs clustered with other PPVs published earlier and that the analyzed PPVs displayed distinct clusters representing BPSV and PVNZ species (Fig. 2). Members of the other two PPV species, namely ORFV and PCPV are distributed between the two main branches shown in the phylogenetic tree. Seven of the eight Sudanese CPPV isolates sequenced in the present study clustered together closer to PCPV isolates from Finland and Germany. One Sudanese isolate (V34-SD) clustered in a different sub-branch that contains ORFV isolates from Finland and Germany and a PCPV isolate from Finland. On the other hand, the five previously published sequences representing CPPV isolates from India, Saudi Arabia and Bahrain clustered at the other branch of the phylogenetic tree together with PCPV isolates from Finland and ORFV isolates from Asian countries (India, Korea, China and Taiwan). Isolate CE41 isolated from a dromedary camel in the Sudan clustered with the BPSV species of PPV.
Fig. 2.
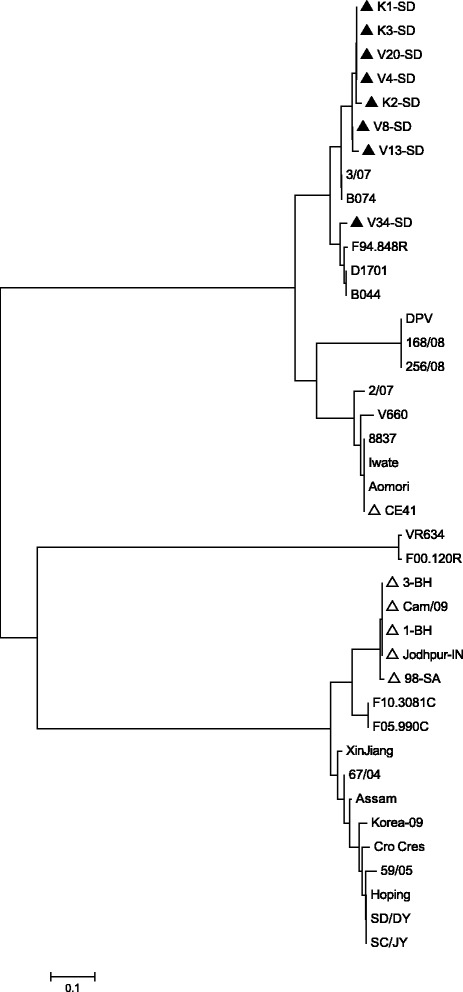
Molecular Phylogenetic analysis of 40 PPV amino acid sequences based on B2L gene. The evolutionary history was inferred using the Neighbor-Joining method [16]. The optimal tree with the sum of branch length = 3.16641726 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [17] and are in the units of the number of amino acid substitutions per site. The analysis involved 40 amino acid sequences. All positions containing gaps and missing data were eliminated. There were 85 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [18]. Black triangle dots represent the eight Sudanese CPPV isolates analyzed in the present study and empty triangles CPPV published sequences
The obtained nucleotide sequences were submitted to GenBank and assigned the accession numbers KR231664, KR231665, KR231666, KR231667, KR231668, KR231669, KR231670 and KR231671 for isolates V4-SD, V8-Sd, V13-Sd, V20-SD, V34-SD, K1-SD, K2-SD and K3-SD, respectively.
Discussion
A preferred target gene to generate PCR amplified DNA fragments for sequence analysis and comparison of PPV DNA is the open reading frame (ORF) 011 (B2L gene), the PPV homologue of the vaccinia virus Copenhagen (VACV) gene F13L, which encodes the major envelope antigen p37K [19].
In the present study, a genetic analysis of eight Sudanese isolates of the CPPV was performed. BLAST results unveiled that seven isolates are genetically nearly related to PCPV and one is closer to ORFV. The B2L gene fragment from the eight DNAs has a high G + C content of 62.6 to 63.9 % in agreement with that of the whole PPV genome [20].
The findings of the present study provide evidence for a close relationship of seven Sudanese strains despite the different geographic location and year of collection. Interestingly, five isolates collected at the same area of eastern Sudan, but during a time span between 1993 and 2013 have 100 % amino acid homology pointing to a circulation of genetically stable virus and a stable genetic makeup and a wide distribution in the country.
The dominant CPPV circulating in the Sudan is found to be genetically close to PCPV species of the PPV and a genetic variability in this country, of at least two PPV species: PCPV (n = 7) and ORFV (n = 1). Three isolates, which were collected over 20 years, possessed 100 % genetic homology.
Search of the literature showed that five isolates have been antecedently classified as PCPV [11, 12]. The results of this study ascent the total number of CPPV that belong to PCPV species to 12. However, the study also identified one isolate that genetically closer to ORFV than PCPV. Besides, we also corroborated previous classification of isolate CE 41 as BPSV [21]. Taken together, the present study shows heterogeneity among the CPPVs and suggest that clearly, these viruses do not form a monophyletic species within PPV phylogeny, at least based on the B2L gene analysis. A search of the literature revealed the presence of only one CPPV strain (Cam/09) that has been classified as ORFV (accession # GU460370.1) as it has 99 % nucleotide homology with an Indian ORFV from sheep [22]. Here we confirm previous research work and present grounds that CCE in camels can be caused by ORFV. This is what has been anticipated before any genetic analysis was made, as lesions produced by the causative virus is characterized by a progressive nodule-vesicle-pustule-scab formation found mainly on the lips that resembles lesions induced by ORFV in sheep and goats, but not PCPV or BPSV [9]. The findings also might points to a transmission from sheep or goats, as it is common for camels, sheep, and goats to share the same pasture in the Sudan. Contrary to such speculation, Abu Elzein et al [9], in Saudi Arabia failed to experimentally infect sheep and goats with an isolate of CPPV. Unfortunately, the PPV circulating in sheep and goats in the Sudan has not yet genetically analyzed. Therefore, molecular analysis of ORFV circulating in small ruminants in the same area and experimental infection of sheep and goats with a CPPV genetically closer to ORFV is expected to rule out such possibility.
Previous reports from Saudi Arabia [11] and India [12] implicated PCPV as the cause of the disease in camels. Of note, Abubakr et al [11] on the bases of their first connection of the CPPV to PCPV postulated a relatively recent introduction of the virus to camels, possibly from humans, that have been exposed to PCPV-infected ruminants. Yet, no PCPV infection in human or animals has been reported in the Sudan or Arabian Peninsula, but only one case of ORFV infection of a man in United Arab Emirates [23]. Of interest is to determine the reservoir host that maintain the virus in nature during the inter-epizootic period. A serological survey involving domestic and wild species of animals in the affected area of eastern Sudan may identify such reservoir host and contribute to its control.
On the other hand, a nucleotide sequence of the B2L gene from a cell culture- adapted CPPV (CE41) was found to be genetically closely related to BPSV [21]. This genetic placement might be caused by genomic rearrangements known to occur when PPVs are passaged in cell culture [24–26]. Nevertheless, taking into consideration the data presented in this report, it can be postulated that contagious ecthyma in dromedary camels is caused by viruses from different species of PPV. This situation is not uncommon as some PPVs obtained from wild Japanese serows are genetically closer to ORFV, and some are closer to BPSV [27]. However, a possibility still exists that a separate virus species of PPV genetically related to both PCPV and ORFV based on the sequence of the B2L gene, but differ in other genomic domains causes this disease. To this end whole genome sequencing and comparison with known PPV species is a requisite and anticipated to bring out a better approximation to reality.
At present 11 out of the 14 published CPPV sequences are grouped along with PCPV, two are ORFV related and one belonged to the BPSV species of PPV indicating genetic variability of the CPPV. Further investigations should examine other genes of the virus and perform complete genome sequencing in order to better describe and classify the CPPV.
According to previous phylogenetic analysis based on the B2L gene of PPV, PCPV and ORFV strains are usually characterized by a certain level of variability, while PVNZ and BPSV strains may possess 100 % identity [21]. In line with that, the phylogenetic tree generated in this study for 40 PPV sequences showed distinct clustering of BPSV and PVNZ strains, with ORFV and PCPV strains bundling in more than one subgroup mostly with a link to the geographic location of respective disease outbreaks. Similarly Oem et al [28] showed that some PPVs of a particular classification clustered with different species in the phylogenetic tree. Therefore, the present study supports the contention by Oem et al [28] that phylogenetic analysis using partial B2L gene sequences may not be suited owing to the recent increase in the publication of more sequences of PPV from different countries and hosts. Besides, it should also be kept in mind that infections with multiple PPV strains of a single species of PPV can occur in one animal as recently described [29].
CCE is a sparsely studied disease of camels and presently does not have to be reported to the World Organization of Animal Health (OIE) despite several publications reporting wide distribution of the disease. The disease is economically important and a recent publication from Iran [30] described severe cases of CCE that caused a mortality rate of 6 %, therefore, this important camel disease has to be looked at seriously.
Conclusion
We genetically analyzed based on sequences of the B2L gene eight strains of CPPV collected from two areas of the Sudan between 1993 and 2013. The prevailing virus is genetically close to PCPV species of the PPV and a genetic variability in this country, of at least two PPV species exists: PCPV (n = 7) and ORFV (n = 1). Based on sequences of the B2L gene it is not possible to suggest that the viruses that cause CCE form a monophylogenetic group or species within the PPV phylogeny.
Methods
Origin of specimens
This study was approved by the Institutional Review Board of the Veterinary Research Institute, Sudan. This study was approved by the Institutional Review Board of the Veterinary Research Institute, Sudan Skin scab specimens (n = 6) were collected from dromedary camels (camelus dromedarius) showing symptoms of contagious ecthyma in east and west Sudan during field epidemiological investigations between 1993 and 2005. Diagnosis of the disease was made on the bases of clinical evidence and electron microscopy and later by PCR [31]. Details on disease symptoms and epidemiology were published [6]. Additional four specimens were collected in 2013. Data on the clinical picture, morbidity and mortality rates resulted in the 2013 disease outbreaks in Showak area of eastern Sudan were collected. Table 1 shows some data on positive specimens analyzed in the present study.
Tissue homogenization and DNA extraction
A 20 % suspension (weight/volume) was made of the scab material in tris-EDTA (TE) buffer (pH 7.4), freeze-thawed at −30 °C, mechanically homogenized using a mechanical Homogenizer (TissueRuptor, Qiagen, Hilden, Germany) and centrifuged at 1500 × g for 10 min at 4 °C. Total viral DNA was extracted from supernatants using a QiaAmp DNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions.
PCR
To check its PPV identity each extracted DNA was first tested using the quantitative real-time PCR assay described by Nitsche et al [32]. The amplification was carried out on Mx3000P qPCR system (Agilent Technologies, USA) using Brilliant II QPCR Master Mix (Agilent Technologies, USA). Second, with DNA of known PPV identity, a PCR protocol was used to obtain DNA fragment for sequencing. This PCR assay targets the commonly used B2L gene (open reading frame 011) which is homologue of vaccinia virus Copenhagen (VACV) gene F13L encoding the major envelop antigen p37K [19]. PCR amplification was done using forward primer (5′-TTAATTTATTGGCTTGCAGAACTCCGAGCGC-3′) and reverse primer (5′-ATGTGGCCGTTCTCCTCCATC-3′) [33] that amplify 1200 bp DNA sequence. PCR amplification of the envelope gene was performed using the following thermal profiles initial denaturation at 94 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 30 s, extension at 72 °C for 1.5 min, and final extension at 72 °C for 10 and DNA was amplified with a thermal cycler TP3000 (Biometra, Germany). The PCR products were then checked in agarose gel, purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and then sent for sequencing. Sequencing was completed using the BigDye® Terminator v3.1 cycle sequencing kit chemistry and each primer pairs. Nucleotide positions were confirmed based on two independent sequencing reactions in both directions.
BLAST and phylogenetic analysis
The terminal unreliable nucleotides of the obtained B2L gene sequences were first trimmed-off automatically and both forward and reverse sequences aligned in the Geneious® 8.1.4 package. The biologically correct sequences of the eight CPPV isolates were subjected to basic local alignment search was compared with nucleotide sequence in the GenBank database using the online BLASTN program on the NCBI website [34].
Phylogenetic tree based on amino acid sequences was constructed for 40 PPV strains (Table 4), including sequences from the four PPV species; ORFV (n = 9), PCPV (n = 10), BPSV (n = 6) and PVNZ (n = 3) and eight strains from this study, using the Neighbor-Joining method in MEGA6 [18]. The amino acid substitution model used was the Johen-Tylor-Thornton model [45]. The significance of all deduced phylogenetic trees was verified by bootstrap analysis of 1000 replicates.
Table 4.
Information on Parapoxviruses used for the phylogenetic analysis of the B2L gene
| No | Species | Virus identification | Host | Origin | GenBank accession | Reference |
|---|---|---|---|---|---|---|
| 1 | ORFV | D1701 | Reindeer | Finland | AY453654.1 | Tikkanen et al (2004) [35] |
| 2 | ORFV | Korea 09 | Goat | Korea | GQ328006.1 | Oem et al (2013) [28] |
| 3 | ORFV | 59/05 | Sheep | India | DQ263304.1 | Hosamani et al (2006) [36] |
| 4 | ORFV | Hoping | Goat | Taiwan | EU935106.1 | Chan et al (2009) [37] |
| 5 | ORFV | B044 | Goat | Germany | KF478798.1 | Friederichs et al (2014) [38] |
| 6 | ORFV | Cam/09 | Dromedary | India | GU460370.1 | Venkatesan (unpublished) |
| 7 | ORFV | Assam | Goat | India | JQ040300.1 | Bora et al. (2012) [39] |
| 8 | ORFV | Xinjiang | Sheep | China | JN565694 | Li et al. (2013) [22] |
| 9 | ORFV | Jodhpur | Dromedary camel | India | GQ390365 | Nagarajan et al (2010) [12] |
| 10 | ORFV | SD/DY | Sheep | China | JQ904794.1 | Zhang et al (2014) [40] |
| 11 | ORFV | Cro_Cres | Sheep | Croatia | HQ215589.1 | Lojkic et al (2010) [41] |
| 12 | ORFV | SC/JY | Goat | China | JQ904792.1 | Zhang et al (2014) [40] |
| 13 | ORFV | 67/04 | Sheep | India | DQ263305.1 | Hosamani et al (2006) [36] |
| 14 | PCPV | F94.848R | Reindeer | Finland | AY453661.1 | Tikkanen et al (2004) [35] |
| 15 | PCPV | F05.990C | Reindeer | Finland | JF773694.1 | Hautaniemi et al (2011) [42] |
| 16 | PCPV | F10.3081C | Reindeer | Finland | JF773695.1 | Hautaniemi et al (2011) [42] |
| 17 | PCPV | 3/07 | Cattle | Germany | KF478804.1 | Friederichs et al (2014) [38] |
| 18 | PCPV | B074 | Man | Germany | KF478803.1 | Friederichs et al (2014) [38] |
| 19 | PCPV | SA 98 | Dromedary camel | Saudi Arabia | EF555793.1 | Abubakr et al (2007) [11] |
| 20 | PCPV | BH 1 | Dromedary camel | Bahrain | EF555792.1 | Abubakr et al (2007) [11] |
| 21 | PCPV | BH 3 | Dromedary camel | Bahrain | EF555791.1 | Abubakr et al (2007) [11] |
| 22 | PCPV | F00.120R | Reindeer | Finland | GQ329669 | Hautaniemi et al (2011) [42] |
| 23 | PCPV | VR634 | Man | New Zealand | GQ329670.1 | Gassmann et al. (1985) [43] |
| 24 | BPSV | V660 | Cattle | Germany | KF478805.1 | Friederichs et al (2014) [38] |
| 25 | BPSV | 2/07 | Cattle | Germany | KF478806.1 | Friederichs et al (2014) [38] |
| 26 | BPSV | CE 41 | Dromedary camel | Sudan | JN171861.1 | Dal Pozzo et al. (2011) [21] |
| 27 | BPSV | Aomori | Cattle | Japan | AB044797.1 | Inoshima et al. (2001) [27] |
| 28 | BPSV | 8837 | Cattle | Korea | JX968998.1 | Oem et al. (2013) [28] |
| 29 | BPSV | Iwate | Cattle | Japan | AB538385 | Oem et al (2013) [28] |
| 30 | PVNZ | DPV | Red Deer | New Zealand | AY453655.1 | Tikkanen et al (2004) [35] |
| 31 | PVNZ | 168/09 | Red Deer | Italy | HQ239068.1 | Scagliarini et al (2011) [44] |
| 32 | PVNZ | 256/08 | Red Deer | Italy | HQ239069.1 | Scagliarini et al (2011) [44] |
Acknowledgements
This research was partially supported by a research grant no 140105 from the Deanship of Scientific Research, King Faisal University, Kingdom of Saudi Arabia. We thank Prof. Marzook M Al-Eknah, Director Camel Research Center for the continuous support.
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
AIK and YHA initiated and participated in the planning of the study and collected the field specimens. IME, KAA and AIA participated in the planning of the study and in the lab work. AIK wrote the first draft of the manuscript. All authors contributed to data analysis and overseen manuscript revision.
Contributor Information
Abdelmalik I. Khalafalla, Email: abdokhlf@yahoo.co.uk
Ibrahim M. El-Sabagh, Email: ibsabagh@kfu.edu.sa
Khalid A. Al-Busada, Email: kbusadah@kfu.edu.sa
Abdullah I. Al-Mubarak, Email: aialsheikhmubarak@gmail.com
Yahia H. Ali, Email: y_beik@hotmail.com
References
- 1.DashtserenTs SF, Varejka F, Khokho A. Camel contagious ecthyma (Pustular dermatitis) Acta Virol. 1984;28:122–127. [PubMed] [Google Scholar]
- 2.Munz E, Schillinger D, Reimann M, Mahnel H. Electron microscopical diagnosis of ecthyma contagiosum in camels (camelus dromedarius). First report of the disease in Kenya. J Vet Medicine B. 1986;33:73–77. doi: 10.1111/j.1439-0450.1986.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 3.Buchnev KN, Tulebaev SZ, Sansyzbaev AB. Infectious diseases of camels in the USSR. Revue Sci Tech Off Int Epi. 1987;6(2):487–495. doi: 10.20506/rst.6.2.297. [DOI] [PubMed] [Google Scholar]
- 4.Moallin ASM, Zessin KH. Outbreak of camel contagious ecthyma in central Somalia. Trop Anim Health Prod. 1988;20:185–186. doi: 10.1007/BF02240091. [DOI] [PubMed] [Google Scholar]
- 5.Ali AO, Kheir AM, Abdulamir H, Barri MES. Camel contagious ecthyma in Sudan. A case report. Rev Elev Med Vet Pays Trop. 1991;44:143–145. [PubMed] [Google Scholar]
- 6.Khalafalla AI, Mohamed MEH. Epizootiology of camel contagious ecthyma in eastern Sudan. Revue Elev Vet Pays Trop. 1997;50:99–103. [Google Scholar]
- 7.Khalafalla AI. Camel contagious ecthyma: risks in young calves. Revue Elev Vet Pays Trop. 2000;53(2):173–176. [Google Scholar]
- 8.Azwai SM, Carter SD, Woldehiwet Z. Immune responses of the camel (Camelus dromedarius) to contagious ecthyma (Orf) virus infection. Vet Microbiol. 1995;47(1 –2):119–31. doi: 10.1016/0378-1135(95)00055-F. [DOI] [PubMed] [Google Scholar]
- 9.Abu Elzein EME, Housawi FMT, Al-Afaleq AI, Ramadan RO, Gameel AA, Al-Gundi O. Clinicopathological response of dromedary camels and sheep to cross-experimental infection with two virulent orf viruses originating from camels and sheep. J Camel Pract Res. 2004;11(No. 1):15–19. [Google Scholar]
- 10.Housawi FM, Abu Elzein EME, Gameel A, Mustafa M, Al-Afaleq A, Gilary J, et al. Severe Auzdyk infection in one-month old camel calves (camelus dromedarius) Vet Archives. 2004;74:467–474. [Google Scholar]
- 11.Abubakr MI, Abu-Elzein EME, Housawi FM, Abdelrahman AO, Fadlallah ME, Nayel MN, Adam AS, Moss S, Forrester NL, Coloyan E, Gameel A. Pseudo cowpox virus: The etiological agent of contagious ecthyma (Auzdyk) in camels (Camelus dromedarius) in the Arabian Peninsula. Vector Borne Zoonotic Dis. 2007;7(2):257–60. doi: 10.1089/vbz.2006.0627. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan G, Ghorui SK, Kumar S, Pathak KM. Complete nucleotide sequence of the envelope gene of pseudocowpox virus isolates from Indian dromedaries (Camelus dromedarius) Arch Virol. 2010;155(10):1725–8. doi: 10.1007/s00705-010-0784-z. [DOI] [PubMed] [Google Scholar]
- 13.Mercer A, Fleming S, Robinson A, Nettleton P, Reid H. Molecular genetic analyses of parapoxviruses pathogenic for humans. Arch Virol Suppl. 1997;13:25–34. doi: 10.1007/978-3-7091-6534-8_3. [DOI] [PubMed] [Google Scholar]
- 14.Becher P, Koenig M, Mueller G, Siebert U, Thiel H-J. Characterization of sealpox virus, a separate member of the parapoxviruses. Arch Virol. 2002;147:1133–1140. doi: 10.1007/s00705-002-0804-8. [DOI] [PubMed] [Google Scholar]
- 15.Vikøren T, Lillehaug A, Akerstedt J, Bretten T, Haugum M, Tryland M. A severe outbreak of contagious ecthyma (orf) in a free-ranging musk ox (Ovibosm oschatus) population in Norway. Vet Microbiol. 2008;127:10–20. doi: 10.1016/j.vetmic.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 17.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York: Academic; 1965. pp. 97–166. [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan JT, Mercer AA, Fleming SB, Robinson AJ. Identification and characterization of an orf virus homologue of the vaccinia virus gene encoding the major envelope protein p37K. Virology. 1994;202:471–478. doi: 10.1006/viro.1994.1420. [DOI] [PubMed] [Google Scholar]
- 20.Delhon G, Tulman ER, Afonso CL, Lu Z, De la Concha-Bermejillo A, Lehmkuhl HD, et al. Genomes of the parapoxviruses ORF virus and bovine papular stomatitis virus. J Virol. 2004;78:168–177. doi: 10.1128/JVI.78.1.168-177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dal Pozzo F, Martinelle L, Gallina L, Mast J, Sarradin P, Thir E, et al. Original findings associated with two cases of bovine papular stomatitis. J Clin Microbiol. 2011;49:4397–4400. doi: 10.1128/JCM.05281-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Zhu X, Zheng Y, Wang S, Liu Z, Dou Y, et al. Phylogenetic analysis of two Chinese orf virus isolates based on sequences of B2L and VIR genes. Arch Virol. 2013;158(7):1477–85. doi: 10.1007/s00705-013-1641-7. [DOI] [PubMed] [Google Scholar]
- 23.Al-Salam S, Nowotny N, Sohail MR, Kolodziejek J, Berger TG. Ecthyma contagiosum (orf)—report of a human case from the United Arab Emirates and review of the literature. J Cutan Pathol. 2008;35(6):603–7. doi: 10.1111/j.1600-0560.2007.00857.x. [DOI] [PubMed] [Google Scholar]
- 24.Cottone R, Büttner M, Bauer B, Henkel M, Hettich E, Rziha HJ. Analysis of genomic rearrangement and subsequent gene deletion of the attenuated Orf virus strain D1701. Virus Res. 1998;56:53–67. doi: 10.1016/S0168-1702(98)00056-2. [DOI] [PubMed] [Google Scholar]
- 25.Fleming SB, Lyttle DJ, Sullivan JT, Mercer AA, Robinson AJ. Genomic analysis of a transposition-deletion variant of orf virus reveals a 3.3 kbp region of non-essential DNA. J Gen Virol. 1995;76:2969–2978. doi: 10.1099/0022-1317-76-12-2969. [DOI] [PubMed] [Google Scholar]
- 26.McInnes CJ, Wood AR, Nettleton PE, Gilray JA. Genomic comparison of an avirulent strain of Orf virus with that of a virulent wild type isolate reveals that the Orf virus G2L gene is non-essential for replication. Virus Genes. 2001;22:141–150. doi: 10.1023/A:1008117127729. [DOI] [PubMed] [Google Scholar]
- 27.Inoshima Y, Murakami K, Yokoyama T, Sentsui H. Genetic heterogeneity among parapoxviruses isolated from sheep, cattle and Japanese serows (Capricornis crispus) J Gen Virol. 2001;82:1215–20. doi: 10.1099/0022-1317-82-5-1215. [DOI] [PubMed] [Google Scholar]
- 28.Oem JK, Lee EY, Lee KK, Kim SH, Lee MH, Hyun BH. Bovine papular stomatitis virus (BPSV) infections in Korean native cattle. J Vet Med Sci. 2013;75(5):675–8. [DOI] [PubMed]
- 29.Huang T, Tulman ER, Diel DG, Khatiwada S, Sims W, Edwards JF, Wen X, Kutish GF, Rock DL, Delhon G. Coinfection with multiple strains of bovine papular stomatitis virus. Arch Virol. 2015;160(6):1527–32. doi: 10.1007/s00705-015-2394-2. [DOI] [PubMed] [Google Scholar]
- 30.Mombeni EG, Mombeini MG, Varshovi HR, Khalaj M, Kenarkohi M, Goudarzi M, et al. Outbreak of contagious ecthyma in camels (Camelus dromedarius and Camelus bactrianus) in Southwest Iran. Rev Elev Med Vet Pays Trop. 2013;66(4):113–5. [Google Scholar]
- 31.Khalafalla AI, Büttner M, Rziha HJ. Polymerase chain reaction (PCR) for rapid diagnosis and differentiation of para-and orthopox virus infections in camels. In: H.P.S.Makkar and G.J.Viljoen (eds). Applications of Gene-Based Technologies for Improving Animal Production and Health in Developing Countries. FAO/IAEA Publications, SPRINGER; 2005: p.335–42.
- 32.Nitsche A, Büttner M, Wilhelm S, Pauli G, Meyer H. Real-time PCR detection of Parapoxvirus DNA. Clin Chem. 2005;52:316–319. doi: 10.1373/clinchem.2005.060335. [DOI] [PubMed] [Google Scholar]
- 33.Guo J, Rasmussen J, Wünschmann A, de La Concha-Bermejillo A. Genetic characterization of orf viruses isolated fromvarious ruminant species of a zoo. Vet Microbiol. 2004;99(2):81–92. doi: 10.1016/j.vetmic.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tikkanen MK. Recent isolates of parapoxvirus of Finnish reindeer (Rangifer tarandus tarandus) are closely related to bovine pseudocowpox virus. J Gen Virol. 2004;85:1413–1418. doi: 10.1099/vir.0.79781-0. [DOI] [PubMed] [Google Scholar]
- 36.Hosamani M, Bhanuprakash V, Scagliarini A, Singh RK. Comparative sequence analysis of major envelope protein gene (B2L) of Indian orf viruses isolated from sheep and goats. Vet Microbiol. 2006;116:317–324. doi: 10.1016/j.vetmic.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 37.Chan KW, Hsu WL, Wang CY, Yang CH, Lin FY, Chulakasian S, et al. Differential diagnosis of orf viruses by a single-step PCR. J Virol Methods. 2009;160:85–89. doi: 10.1016/j.jviromet.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 38.Friederichs S, Krebs S, Blum H, Wolf E, Lang H, von Buttlar H, Büttner M. Comparative and retrospective molecular analysis of Parapoxvirus (PPV) isolates. Virus Res. 2014;181:11–21. doi: 10.1016/j.virusres.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 39.Bora DP, Barman NN, Das SK, Bhanuprakash V, Yogisharadhya R, Venkatesan G, et al. Identification and phylogenetic analysis of orf viruses isolated from outbreaks in goats of Assam, a northeastern state of India. Virus Genes. 2012;45(1):98–104. doi: 10.1007/s11262-012-0740-y. [DOI] [PubMed] [Google Scholar]
- 40.Zhang K, Liu Y, Kong H, Shang Y, Liu X. Comparison and phylogenetic analysis based on the B2L gene of orf virus from goats and sheep in China during 2009-2011. Arch Virol. 2014;159(6):1475–1479. doi: 10.1007/s00705-013-1946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lojkic I, Cac Z, Beck A, Bedekovic T, Cvetnic Z, Sostaric B. Phylogenetic analysis of Croatian orf viruses isolated from sheep and goats. Virol J. 2010;7(1):314. doi: 10.1186/1743-422X-7-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hautaniemi M, Vaccari F, Scacliarini A, Laaksonen S, Huovilainen A, McInnes CJ. Analysis of deletion within the reindeer pseudocowpoxvirus genome. Virus Res. 2011;160:326–332. doi: 10.1016/j.virusres.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Gassmann U, Wyler R, Wittek R. Analysis of parapoxvirus genomes. Arch Virol. 1985;83:17–31. doi: 10.1007/BF01310961. [DOI] [PubMed] [Google Scholar]
- 44.Scagliarini A, Vaccari F, Turrin F, Bianchi A, Cordioli P, Lavazza A. Para-poxvirus infections of Red Deer, Italy. Emerg Infect Dis. 2011;17:684–687. doi: 10.3201/eid1704.101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]


