Abstract
Autism spectrum disorders (ASD) are a group of neurodevelopmental disorders, characterized by impairment in communication and social interactions, and by repetitive behaviors. ASDs are highly heritable, and estimates of the number of risk loci range from hundreds to > 1000. We considered 7 extended families (size 12 – 47 individuals), each with ≥ 3 individuals affected by ASD. All individuals were genotyped with dense SNP panels. A small subset of each family was typed with whole exome sequence (WES). We used a 3-step approach for variant identification. First, we used family-specific parametric linkage analysis of the SNP data to identify regions of interest. Second, we filtered variants in these regions based on frequency and function, obtaining exactly 200 candidates. Third, we compared two approaches to narrowing this list further. We used information from the SNP data to impute exome variant dosages into those without WES. We regressed affected status on variant allele dosage, using pedigree-based kinship matrices to account for relationships. The p-value for the test of the null hypothesis that variant allele dosage is unrelated to phenotype was used to indicate strength of evidence supporting the variant. A cutoff of p=0.05 gave 28 variants. As an alternative third filter, we required Mendelian inheritance in those with WES, resulting in 70 variants. The imputation and association based approach was effective. We identified four strong candidate genes for ASD (SEZ6L, HISPPD1, FEZF1, SAMD11), all of which have been previously implicated in other studies, or have a strong biological argument for their relevance.
Keywords: whole exome sequencing, imputation, family-based association, genome scan, pedigree
Background
Autism spectrum disorders (ASD) are a group of behaviorally defined neurodevelopmental disorders. They are characterized by varying degrees of impairment in communication, social interactions, and repetitive behaviors, sometimes accompanied by intellectual disability. As most broadly defined, ASDs affect 1 in 68 children at 8 years of age(CDC, 2014). The care of an individual with an ASD represents a significant economic burden to both the family and society, whether in terms of medical care, special education, supportive living accommodation or loss of parental productivity. The lifetime cost of care for an individual with an ASD without intellectual disability is estimated at US$1.4 million(Buescher et al. 2014).
ASDs are considered among the most heritable of neuropsychiatric disorders. Evidence includes concordance rates between monozygotic twins of 50% to 90% and between siblings of 3% to 26% (Berg and Geschwind 2012), and a risk of ASD to subsequent siblings of children with autism of 18.7% (Ozonoff et al. 2011). However, high heritability does not imply a simple genetic explanation. Recent estimates of the number of ASD risk loci range from the hundreds(O’Roak et al. 2012; Sanders et al. 2011) to as high as one thousand(Sanders et al. 2012). Such extreme heterogeneity means that replication for any particular gene is expected to be rare. Studies of de-novo variation in singleton probands generate large numbers of potential candidate genes(Neale et al. 2012; O’Roak et al. 2012; Sanders et al. 2012), and in such data sets it is hard to distinguish true risk loci from false positives(Gratten et al. 2013). While broad categories of genes can be identified amongst the candidates, relatively few genes have strong statistical evidence for causality based on their frequency in cases and controls(Neale et al. 2012; O’Roak et al. 2012; Sanders et al. 2012).
One approach to identifying ASD strong candidate genes with more solid support is to consider inherited variation in rare families with more than one member affected with ASD. Four recent studies have used whole exome sequencing (WES) in multiplex families to identify inherited variants that are associated with ASD risk(Chahrour et al. 2012; Cukier et al. 2014; Shi et al. 2013; Toma et al. 2014). In all cases, variants were filtered according to frequency in population databases, and all four required that variants be predicted to be damaging by various bioinformatics approaches. Furthermore, all groups made assumptions about the mode of inheritance of ASD in their families, in order to further reduce the number of variants under consideration. Two groups(Chahrour et al. 2012; Shi et al. 2013) required that variants in their families must be recessive, in one case because 2 of 8 children were affected(Shi et al. 2013) and in the other because the families were chosen for excess homozygosity(Chahrour et al. 2012). In these two studies, relatively small numbers of genes were implicated, when any variants survived filtering at all. In studies where dominant inheritance was considered, a group of 10 nuclear families with 2 or 3 affected individuals(Toma et al. 2014) and a group of 40 extended families(Cukier et al. 2014) with a minimum of two affected cousins were analyzed. Analysis requiring a strictly dominant mode of inheritance, coupled with filtering on population frequency and bioinformatics predictions resulted in identification of 220 and 745 candidate genes, respectively. Such a large number of candidate genes makes it difficult to evaluate the importance of each individual gene and perpetuates issues of multiple testing in further analysis.
We recruited families with a minimum of three affected individuals with ASD or the broader autism phenotype (BAP), including an affected sib pair and at least one affected cousin. We hypothesize that in these families, ASD and BAP are caused by inherited variation, perhaps specific to each family. We obtained SNP genotypes on all available individuals in the families, and WES on a small subset. We first filtered WES variants using family-specific linkage analysis to identify regions of interest. Similar to other groups, we then filtered variants by frequency and predicted function. Third, family-based genotype imputation to infer variant allele dosage in individuals not explicitly typed with WES allowed family-based association testing of variants, using more information than is available on only the directly-sequenced subjects. In comparison to the simpler approach of requiring segregation consistent with Mendelian inheritance in individuals with WES, this approach allowed for a marked reduction in the number of prioritized variants and candidate genes within families, thus avoiding the problem of long lists of variants with no clear way to prioritize them. This approach would be useful in other complex disorders where familial forms exist, but there is heterogeneity between families.
Methods
Families
Extended families were identified by adding new subjects to families previously collected as part of the National Institutes of Health Collaborative Programs of Excellence in Autism(Chapman et al. 2011). The original families all had two or more children with an ASD(Schellenberg et al. 2006). We collected the Broader Phenotype Autism Symptom Scale(Dawson et al. 2007) (BPASS), and focused recruitment efforts on the side of the family where the BPASS social score was inflated, indicating presence of the broader autism phenotype and greater autistic symptomatology in family members. Amongst the families that were extended, we found 7 families with new subjects with ASD in additional sibships. Four individuals did not meet diagnostic threshold criteria on the Autism Diagnostic Observation Schedule(Lord et al. 2012) (ADOS) and Autism Diagnostic Interview-Revised(Rutter 2003)(ADI-R), but did have elevations on the BPASS indicative of BAP. Phenotype data for the affected individuals is summarized in Online Resource 1. Total family sizes range from 12 – 47 individuals. Each family has a minimum of three individuals affected with ASD or BAP, including the original sibship with two or more affected individuals and one or more affected cousins. Table 1 shows the number and sex of affected individuals in each family, and the number of unaffected individuals without children in each family. AU119 was ascertained through three sibships, all of whom had two or more affected individuals. Connections between the sibships were found in the quality control stage of analysis, and examination of family members identified three more affected individuals in related singleton sibships. Pedigree drawings are not provided, in order to protect the anonymity of participants. All families have European ancestry. This study was approved by the University of Washington Institutional Review Board, and informed consent was obtained from all participants and/or their parents.
Table 1.
Family characteristics and number of individuals with genotype data. ASD=autism spectrum disorder; BAP=broader phenotype; OE=Illumina OmniExpress chip, CE=Illumina Human Core Exome chip, WES=whole exome sequencing.
| Family | Number affected | Number affected by dx Total (M,F)
|
Number unaffected ** Total (M,F) | Number genotyped
|
Total size | ||||
|---|---|---|---|---|---|---|---|---|---|
| Proband sibship | Cousin sibship(s) | ASD | BAP | OE | CE | WES | |||
| AU119 | 2,4,2* | 1,1,1* | 11 (8,3) | - | 14 (3,11) | 43 | 18 | 5 | 47 |
| AU625 | 2 | 1 | 3M | - | 3 (2,1) | 6 | 8 | 4 | 18 |
| AU366 | 2 | 1 | 3 (1,2) | - | 3 (2,1) | - | 12 | 4 | 13 |
| AU071 | 3 | 1 | 3 (2,1) | 1M | 3 (1,2) | - | 10 | 5 | 13 |
| AU754 | 4 | 1 | 2F | 3 (2,1) | 3 (0,3) | 6 | 5 | 6 | 16 |
| AU599 | 2 | 2 | 4M | - | 5 (1,4) | 6 | 6 | 5 | 15 |
| AU113 | 2 | 1 | 3M | - | 3 (1,2) | - | 10 | 4 | 12 |
In AU119, three multiplex sibships were independently ascertained and connected during relationship checking using genotype data. Singleton affected cousins were identified in three other sibships.
with available SNP data and no children in the pedigree
Phenotyping
Diagnostic status was determined through assessment by trained clinicians with expertise in ASD, as described previously(Schellenberg et al. 2006). Diagnoses of ASD(American Psychiatric Association 2013) were confirmed by administration of the ADOS, ADI-R, and expert clinical judgment using all available information.
Phenotypic characterization of both affected and unaffected participants included assessment of (1) ASD related symptoms, from diagnostic level symptomatology through the broader autism phenotype to normative range of functioning (Family History Interview(Rutter and Folstein 1995); SRS(Constantino 2012);Social Competence Questionnaire(Sarason et al. 1985); BPASS), (2) cognitive functioning (age appropriate Wechsler test(Wechsler 1981; Wechsler 1989; Wechsler 1992)), (3) social language ability (Communication Checklist, Children’s Communication Checklist(Bishop 1998)), (4) face memory skills (age appropriate Face Memory Subtests from Children’s Memory Scales(Cohen 1997) or Wechsler Memory Scales(Wechsler 1997)), (5) sensory sensitivities/aversions (Sensory Profile(Dunn 1999)), (6) phonological processing ability (Nonword Repetition from CTOPP(Wagner et al. 1999)), (7) physical measurements (height, weight, and orbital frontal head circumference), and (8) parent/self report of comorbid medical diagnoses. Additionally, assessment of affected individuals included administration of the Vineland Adaptive Behavior Scales-2nd Edition(Sparrow 2005) to assess adaptive functioning.
Genotyping and Sequencing
We obtained Illumina HumanOmniExpress(OE) genotypes on 61 people in 4 families and Illumina HumanCoreExome(CE) genotypes on 69 people in 7 families (see Table 1). Of the 7 families, 3 had genotypes for only the CE chip, and 4 had subjects genotyped on both platforms. WES was performed on at least one affected representative of each sibship and a common ancestor of as many affected individuals as possible. Due to cost restrictions and family size, WES in AU119 was restricted to representatives of 4 of 6 sibships with affected members, and a common ancestor of all but 3 of the 11 individuals with ASD.
Genotypes were obtained for Illumina HumanOmniExpress and Illumina HumanCoreExome beadchips per manufacturer’s instructions at the University of Washington. Capture of the exome and surrounding regions was done using Nimblegen SeqCap EZ Human Exome Library v2.0 kit (Roche, Basel, Switzerland) following manufacturer’s recommended instruction. The capture kit targets 28,858 genes with total size of the target regions 36.5 Mb, resulting in probes covering 44.1 Mb. The sequencing library clusters were generated on Illumina flowcells using cBlot (Illumina, Inc.) and paired-end 50bp (AU119 & AU599 – batch 1) or 101bp (remaining families – batch 2) sequencing was performed on the Illumina HIseq2000 sequencing platform at Genome Sciences, University of Washington. Mean read depths of 131.6 and 57.2 were obtained for batches 1 and 2 respectively, and 92% and 94% of exome targets were covered with read depth greater than 8. The raw base calling was performed with CASAVA (Illumina, Inc.). Sequenced reads were aligned to NCBI human reference genome GRCh37 (hg19) using Burrows-Wheeler Aligner(Li and Durbin 2010) and BAM files were generated using SAMtools(Li et al. 2009). PCR duplicates were marked using Picard(Picard Tools:A set of Java command line tools for manipulating high-throughput sequencing data and formats. http://broadinstitute.github.io/picard/ 2014). After base recalibration the sequence reads were realigned around indels and mapped. The Genome Analyzer Toolkit(McKenna et al. 2010) (GATK) was used for SNP calling.
Exome variants were removed from consideration if there were fewer than 6 reads, if they failed the following sequencing quality metrics: GATK quality score ≤ 50, quality by depth less than 5, or reference allele proportion in heterozygotes greater than 0.75. Additional filters included homopolymer run (greater than 3 in batch 1 or greater than 4 in batch 2) and strand bias ≥ 0.10 in batch 2.
Selected exome variants were validated using Sanger sequencing. Primer sets were designed using Primer3 (v 4.0.0). DNA amplification by PCR was carried out in 20μl reactions in the presence of 200μM dNTP, 2.5U Hot Start Taq DNA polymerase, 0.5μM MgCl, 0.01μg/μL DNA and 0.5μM of each primer. PCR amplification was performed under the following thermal conditions: 95°C for 15 minutes, 35 cycles of 95°C for 30 seconds, 57°C for 30 seconds and 72°C for 1 minute followed by a 72°C hold for 10 minutes using a T100 thermal cycler from BioRad. A 5μl aliquot of ExoSAP-IT was added to the PCR product and cycled at 37°C for 45 minutes followed by enzyme deactivation for 15 minutes at 80°C. The ExoSAP-IT treated PCR product was then cycle sequenced using BigDye Terminator v3.1 on a BioRad T100 thermal cycler. The conditions were 95°C for 5 minutes and 31 cycles of 95°C for 10 seconds, 55°C for 7 seconds and 60°C for 3 minutes. The 10μl reaction consisted of 1.5μl BigDye, 4pmol primer and 2μl of 5M betaine. The samples were analyzed by capillary sequencing on an ABI 3730 DNA analyzer with a 50cm array. The sequencing results were read manually for mutations using Sequencher program (Gene Codes Corp.).
Statistical Analysis
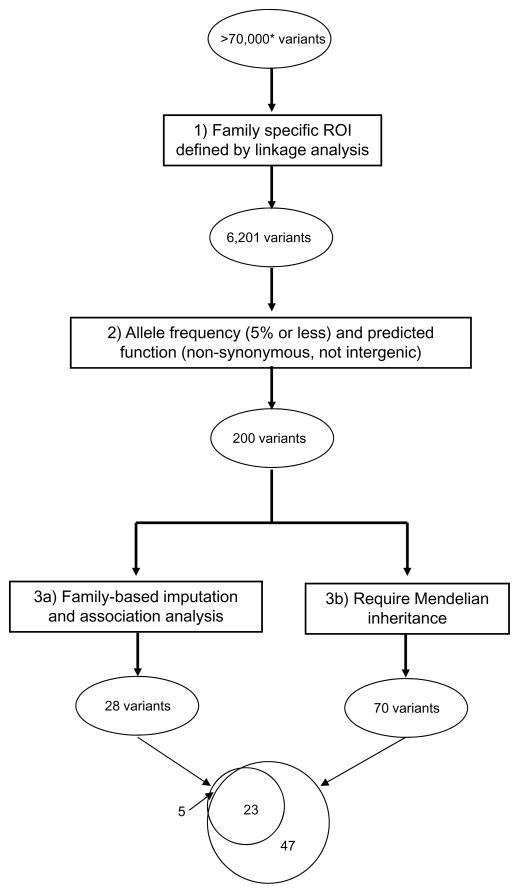
We used a three-step approach, applied to each family in turn, to reduce the set of potentially causal exome variants in each family. First, we performed family-specific parametric linkage analysis in order to identify genomic regions of interest, and removed variants that were not in these regions. Second, in these family-specific regions of interest, we filtered variants on function (removing synonymous and intergenic variants) and by frequency in the 1000 Genomes Project Europeans (1KGP-EUR) (removing variants with allele frequency exceeding 0.05). After this stage, we considered two alternatives to further narrow the lists of variants. First, we used family-based genotype imputation to infer the number of copies of each exome variant present in each of the unaffected individuals in the family. After imputation,we regressed phenotype (affected/unaffected) on the variant allele dosage, using the pedigree-based kinship matrix to account for the relationships between individuals. The nominal p-value for the test of the null hypothesis that variant allele dosage is unrelated to phenotype was then used as an indicator of strength of evidence supporting the variant as an ASD susceptibility allele, using a cutoff of p=0.05. As an alternative third filter, we required variants to appear in all individuals with exome sequencing data who were either affected or obligate carriers, assuming a Mendelian dominant mode of inheritance. An overview of this approach is presented in Figure 1. The numbers on the filtering steps correspond to the sections below. Note that filters 3a and 3b are applied in parallel to the variants that survive filter 2.
Figure 1.
Schematic representation of the variant filtering procedure. *The variant counts at each stage of filtering are summed over all seven families. The overlapping circles at the bottom are a Venn diagram of the relationship between variants passing the imputation and association filter, and those passing the Mendelian filter.
1) Filtering based on linkage analysis
The first step of variant filtering focused on variants existing in family-specific regions of interest identified by linkage analysis of SNP data. Multipoint linkage analysis requires that a sparser set of SNPs be selected from the available high-density SNP chips. PBAP (pedigree-based analysis pipeline) is a suite of programs that perform marker selection for an informative linkage panel, pedigree-based quality control, and file manipulation for family-based downstream analyses(Nato et al. 2013). Details of the parameters we specified for PBAP can be found in the Appendix. A subpanel was generated from the CE chip for use in the three families who were typed with this chip exclusively. Because the other four families have a mixture of individuals typed on both chips, analyses in these families used a subpanel chosen from the set of SNPs that are represented on both the CE and OE chips.
Family specific linkage analyses were performed using glauto from the MORGAN(MORGAN: A package for Markov chain Monte Carlo in genetic analysis (version 3.1.1) http://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan.shtml 2012) package followed by FASTLINK(O’Connell and Weeks 1995). Details of this approach can be found in the Appendix. A dominant model is appropriate in these families because there are pairs of affected individuals more distantly related than siblings in each, and therefore a recessive model is not adequate to explain all cases. Reduced penetrance is required because under the dominant model, a parent of each affected must be a carrier of the causal variant, and their phenotype is mild enough for them to reproduce. We assumed a risk allele frequency of 1%, penetrance of 50%, and a high phenocopy rate (1% in non-carriers). Individuals with ASD or BAP were coded as affected, and all others were coded as unaffected, unless they were deceased, in which case they were coded as having unknown phenotype. The regions of interest for further consideration of variants were defined by lod scores ≥ 1 or ≥ 0.75 in families where lod scores did not reach 1.These cutoffs were chosen to include the most prominent lod score in each family, without implicating an overly large number of regions. Because these families are so well genotyped, the boundaries of the regions of interest were generally clearly defined by a substantial drop in lod score. We restricted attention to exome variants in the regions of interest so defined, and required a variant to be present in a minimum of two affected individuals, thereby potentially segregating in the family.
2) Filtering on frequency and function
In the second step, we filtered on frequency and function. We used variant allele frequencies from the 1KGP-EUR samples, and restricted our attention to variants whose frequency was ≤ 0.05, in order to allow for the involvement of more common variants in ASD susceptibility. We removed variants predicted to be synonymous or intergenic by both dbSNP137 and GVS as reported by SeattleSeq137, a resource provided by NHLBI(Ng et al. 2009).
3a) Filtering on results of imputation and association testing
These families all have subjects for whom exome data were not available, but for whom SNP chip genotype data were available. We imputed into all subjects without WES data the expected dosage of each exome variant remaining after the second filtering step, using the program GIGI(Cheung et al. 2013). GIGI uses the inheritance information for the whole family as inferred from the SNP data, in addition to the exome variant genotypes in available individuals, to calculate genotype probabilities and thus the expected dosage of the variant allele in every member of the family. In contrast to population-based genotype imputation, GIGI does not require a population sample or assumptions about linkage disequilibrium in a matched population. Details of the parameters we used for GIGI can be found in the Appendix.
In each family, for each variant of interest, we compared affected family members to unaffected family members, using the observed dosage (0, 1, or 2) of the variant allele in those individuals with exome data, and the expected dosage (potentially non-integer) of the variant allele imputed by GIGI in those individuals without exome data. We considered only individuals without children, to avoid analyzing both parent and child. We used the function lmekin from the R library kinship to regress phenotype (affected/unaffected) on the variant allele dosage, using the pedigree-based kinship matrix to account for the relationships between individuals. This model assumes a linear effect of dosage on disease risk, and is thus an additive model rather than the dominant one used for the linkage analysis. Because we have already filtered out variants that are more common (greater than 0.05), homozygous variants are almost never seen in this data set, and therefore we would not expect a practical difference between the conclusions of testing based on this additive model and those based on a dominant model. The p-value for the test of the null hypothesis that variant allele dosage is unrelated to phenotype was then used as an indicator of strength of evidence supporting the variant as an ASD susceptibility allele. To be considered a candidate variant, we required both that the variant have nominal p-value ≤ 0.05, and that the mean dosage of the variant in the affected individuals be greater than the mean expected dosage in the unaffected individuals. Since we already selected for regions with evidence of linkage, and we required that the variant must be present in at least two affected individuals, we would not expect the p-values obtained to be exact. Similarly, we did not correct for multiple testing, since p-values for variants on the same haplotype will be highly correlated or identical. Nonetheless these nominal p-values can be used to prioritize long lists of variants.
3b) Filtering on segregation consistent with a Mendelian dominant model
For comparison purposes, we also took the simpler approach of requiring that variants of interest be present in a pattern consistent with a rare dominant Mendelian locus. More specifically, we required that a variant be present in all individuals who were certainly affected ASD, and also in any available common ancestor, unless exome data were not available on the common ancestor’s spouse (since in this case the variant could have been introduced in the spouse).
The thresholds used in the filtering steps described here (lod scores of 1 or 0.75, allele frequency of 0.05, nominal p-value of 0.05) are all somewhat arbitrary, but reasonable in the experience of the investigators. The goal is not to produce an exact statistical test for the significance of each variant, but rather to identify a subset of variants for further examination in terms of their biological relevance.
Annotation
Variants were annotated using SeattleSeq 137, a resource provided by NHLBI(Ng et al. 2009). SeattleSeq includes a measure of position specific evolutionary conservation, the Genomic Evolutionary Rate Profiling (GERP) score(Cooper et al. 2005), and a measure of the impact of amino acid substitution as predicted by PolyPhen-2 class(Adzhubei et al. 2010). Sorting Intolerant from Tolerant (SIFT) predictions of amino acid substitution effect(Kumar et al. 2009a) and measures of protein expression in human brain(Uhlen et al. 2010) were obtained directly from the SIFT website(SIFT http://sift.jcvi.org 2014) and Human Protein Atlas(Human Protein Atlas http://proteinatlas.org 2014) respectively.
Results and discussion
1) Filtering based on linkage analysis
We identified between 2 and 10 genomic regions of interest per family (Table 2 and Online Resource 2) using family specific linkage analysis of SNP chip data. The total length of regions implicated ranged from 26.1 Mb, in the largest family with a large number of affected and unaffected individuals (AU119), to 140.7 Mb in the smallest family (AU113). In order to pass filtering on linkage regions, variants must be present in 2 or more copies in the family in question, and be in one of the linkage regions implicated in that family. Table 3, column 1 shows the number of variants in these regions. These counts vary from 185 in the largest family, to 1,610 in the smallest family.
Table 2.
Linkage regions defined by lod score > 1
| # regions | Total length (Mb) | Average length (Mb) | |
|---|---|---|---|
|
|
|||
| AU119 | 2 | 26.1 | 13.1 |
| AU625 | 4 | 60.1 | 15.0 |
| AU366* | 3 | 40.5 | 13.5 |
| AU071 | 10 | 104.5 | 10.5 |
| AU754 | 7 | 117.7 | 16.8 |
| AU599 | 8 | 112.1 | 14.0 |
| AU113* | 7 | 140.7 | 20.1 |
lod score cutoff of 0.75 was used due to weaker signals in these pedigrees.
Table 3.
Variant counts after sequential stages of filtering
| Stage of Filtering
|
||||
|---|---|---|---|---|
| 1) Linkage | 2) Frequency* & Function** | 3a) Imputation & Association*** | 3b) Mendelian Pattern | |
|
|
||||
| AU119 | 185 | 4 | 2 | 1 |
| AU625 | 504 | 8 | 1 | 3 |
| AU366 | 420 | 20 | 1 | 2 |
| AU071 | 1,124 | 40 | 1 | 18 |
| AU754 | 1,129 | 43 | 9 | 12 |
| AU599 | 1,229 | 32 | 9 | 11 |
| AU113 | 1,610 | 53 | 5 | 23 |
|
| ||||
| Total | 6,201 | 200 | 28 | 70 |
frequency of variant alleles less than or equal to 0.05 in 1KGP-EUR
synonymous or intergenic regions excluded
p-value less than or equal to 0.05
2) Filtering based on frequency and function
Our frequency filtering required that the alternate allele frequency was ≤ 0.05 in 1KGP-EUR. In addition, we removed variants that were predicted to be synonymous or intergenic by both dbSNP137 and GVS as reported by SeattleSeq137. Variant counts per family after application of these filters (Table 3, column 2) range from 4 in the largest family, to 53 in the smallest family. Five of the seven families have 20 or more variants at this stage of filtering, which motivated the imputation and association analyses. After this stage of filtering, there were no variants or genes that were implicated in more than one family.
3a) Filtering on results of imputation and association testing
As described in the Methods, we computed the expected dosage of the variant allele in individuals for whom WES was not available. We then regressed phenotype (affected/unaffected) on the variant allele dosage, testing the hypothesis that phenotype and variant allele dosage are unrelated. Table 3, column 3a) reports the number of variants with a p-value ≤ 0.05. Between 1 and 9 variants per family passed this association-based filtering step. The total across families of 28 variants surviving this step of filtering is a substantial decrease from the 200 variants that were present after the frequency and function filter. The 28 variants remaining after this filter was applied are shown in Table 4.
Table 4.
Candidate variants, by family, with association test p-value ≤ 0.05 in seven families where imputation and association analyses were possible. p-values and means are shown for dosages determined by imputation and whole exome sequence. Pattern denotes whether the variant was present in a pattern consistent with Mendelian inheritance, based on WES only.
| Gene | chr | Pos | rsname | Pattern | pvalue | Mean dose
|
1KGP-EUR | GVS | dbSNP | Family | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| affecteds | unaffecteds | ||||||||||
| FEZF1 | 7 | 121,944,239 | . | yes | 0.01 | 0.87 | 0.44 | 0 | synonymous | missense | AU119 |
| LGALS1 | 22 | 38,071,707 | rs75882122 | no | 0.03 | 0.49 | 0.14 | 0.05 | utr-5 | utr-5-prime | AU119 |
|
| |||||||||||
| HISPPD1 | 5 | 102,490,411 | rs35671301 | yes | 0.00 | 1.00 | 0.02 | 0.01 | missense | missense | AU625 |
|
| |||||||||||
| SEZ6L | 22 | 26,688,831 | rs137203 | yes | 0.00 | 1.00 | 0.01 | 0.01 | missense | missense | AU366 |
|
| |||||||||||
| SAMD11 | 1 | 877,523 | rs200195897 | No*** | 0.01 | 0.82 | 0.04 | 0 | missense | missense | AU071 |
|
| |||||||||||
| CENPF** | 1 | 214,816,297 | rs3795514 | yes | 0.03 | 1.00 | 0.35 | 0.04 | missense | missense | AU754 |
| USH2A** | 1 | 216,062,273 | rs189748047 | yes | 0.03 | 1.00 | 0.34 | 0 | missense | missense | AU754 |
| TBL2 | 7 | 72,992,858 | rs76029572 | No*** | 0.04 | 0.97 | 0.35 | 0.04 | missense | missense | AU754 |
| BLNK | 10 | 98,031,160 | rs7916154 | yes | 0.03 | 1.00 | 0.33 | 0.01 | utr-5 | utr-5-prime | AU754 |
| TLL2 | 10 | 98,155,678 | rs41291628 | yes | 0.03 | 1.00 | 0.34 | 0.02 | missense | missense | AU754 |
| C10orf12 | 10 | 98,742,043 | rs112594620 | yes | 0.03 | 1.00 | 0.33 | 0.02 | missense | missense | AU754 |
| PKD2L1 | 10 | 102,089,635 | rs117403721 | yes | 0.03 | 1.00 | 0.33 | 0.01 | missense | missense | AU754 |
| ADAMTS1 | 21 | 28,210,457 | rs71317487 | yes | 0.03 | 1.00 | 0.34 | 0.004 | missense | missense | AU754 |
| ETS2 | 21 | 40,190,408 | rs61735785 | yes | 0.03 | 1.00 | 0.34 | 0.01 | missense | missense | AU754 |
|
| |||||||||||
| KIAA1009 | 6 | 84,904,604 | rs17790493 | No | 0.01 | 1.51 | 0.22 | 0.04 | missense | missense | AU599 |
| GABRR2 | 6 | 90,024,862 | . | Yes | 0.01 | 1.00 | 0.20 | 0 | missense | missense | AU599 |
| PIP | 7 | 142,836,646 | rs75076193 | Yes | 0.00 | 1.00 | 0.00 | 0.01 | missense | missense | AU599 |
| SSPO** | 7 | 149,522,165 | . | Yes | 0.01 | 1.00 | 0.20 | 0.01 | notMod3 | missense | AU599 |
| GIMAP2 | 7 | 150,389,751 | . | Yes | 0.01 | 1.00 | 0.20 | 0 | synonymous | missense | AU599 |
| MLL3** | 7 | 151,945,007 | rs2479172 | Yes | 0.01 | 1.00 | 0.20 | 0 | synonymous | missense | AU599 |
| MLL3** | 7 | 151,949,735 | rs77652527 | Yes | 0.01 | 1.01 | 0.21 | 0.05 | missense | missense | AU599 |
| MLL3** | 7 | 151,970,856 | rs10454320 | Yes | 0.01 | 1.00 | 0.20 | 0 | synonymous | missense | AU599 |
| MLL3** | 7 | 151,970,931 | rs56850341 | Yes | 0.01 | 1.00 | 0.20 | 0 | synonymous | missense | AU599 |
|
| |||||||||||
| PIGG | 4 | 520,973 | rs201269761 | yes | 0.00 | 1.00 | 0.02 | 0 | missense | missense | AU113 |
| FAM53A | 4 | 1,656,767 | rs146433325 | No*** | 0.00 | 1.00 | 0.01 | 0 | missense | missense | AU113 |
| OTOP1* | 4 | 4,198,948 | rs112623841 | yes | 0.00 | 1.00 | 0.04 | 0.04 | coding | cds-indel | AU113 |
| MAP3K4 | 6 | 161,508,880 | rs35533223 | yes | 0.00 | 1.00 | 0.03 | 0.03 | missense | missense | AU113 |
| DNAH9 | 17 | 11,543,588 | rs17600516 | yes | 0.01 | 1.00 | 0.50 | 0.03 | near-splice | synonymous | AU113 |
this variant was not confirmed by Sanger sequencing
indicates genes in known artifact lists (Fajardo et al 2011, Detecting false positive signals in exome sequencing, Human Mutation 33(4):609–613).
genotype(s) was/were missing due to low depth in WES (resolved by Sanger sequencing for selected variants)
3b) Filtering on segregation consistent with a Mendelian dominant model
An alternative approach, after application of linkage, frequency, and function filters, is to require a segregation pattern consistent with Mendelian transmission in affected individuals and their ancestors. The variant counts after applying this filter are shown in Table 3, column 3b), and range from 1 variant to 23 per family, with a total of 70 across all families. This is more than the double the number of variants that remained after filtering based on imputation and association analysis.
The Venn diagram at the bottom of Figure 1 shows the relationship between the variants identified by imputation and association analysis, and those identified by the simple Mendelian filter. The imputation and association analysis variants are a subset of the Mendelian variants, with five exceptions. Three of the exceptions were cases where problems with genotype quality or read depth led to missingness in the WES data. The LGALS1 variant in AU119 did not pass the Mendelian filter because it was present in only two of the five exomed individuals. In the case of the KIA1009 variant in AU599, it was seen in two copies in an affected individual, which is not consistent with a rare dominant. Thus the imputation and association analysis identifies a subset of the Mendelian variants which have stronger evidence of association with the autism phenotype in these families.
Validation of WES genotypes and imputation accuracy
Table 4 shows the 28 variants across all families that survived filtering step 3a. The columns show the mean imputed dose over the affected and unaffected individuals in the family and the p-value for the association test, in addition to variant allele frequencies in 1KGP-EUR and functional annotation from GVS and dbSNP. The column “pattern” indicates a variant that survived filtering with the alternate filter 3b. In order to validate WES genotypes and confirm the accuracy of imputation, fourteen variants were chosen for Sanger sequencing from these. We chose a subset comprised of one variant per family per chromosome region, and each selected variant was sequenced in all available members of the family in which it appeared. One variant (in OTOP1 in family AU113) was not confirmed as it was likely a misalignment artifact due to an indel in the region. All 13 remaining variants were confirmed, for a total of 219 genotypes. In order to explore the accuracy of our imputation method, we defined as “ambiguous” imputed variant dosages in the range of 0.2–0.8, and 1.2–1.8 (28 of 219 imputed dosages, 12.8%). Imputed variant dosages less than 0.2, between 0.8 and 1.2, and over 1.8 correspond to variant dosages of 0, 1 and 2, respectively, and are considered unambiguous. Table 5 shows each confirmed exome variant, rates of ambiguity in imputation, accuracy in imputation, and the p-value for the association test based on directly sequenced Sanger genotypes. Ambiguity rates were very low for most variants, with the exception of FEZF1 in AU119, DNAH9 in AU113, and SAMD11 in AU071. This is likely the result of poor information in the SNP panel in the vicinity of these variants, since two of these families had other variants with much lower ambiguity. Ambiguities in these variant dosages were resolved by Sanger genotype. In all 191 genotypes where the imputed dosage was unambiguous, the imputed dosage agreed with the Sanger genotype. The only variant for which there is a difference compared to the imputation-based p-values shown in Table 5 is for DNAH9 (Sanger p=0.12, imputation p=0.01). This difference is due to an unaffected individual in whom the imputed dosage was ambiguous, but the Sanger genotype confirmed the variant was present in one copy.
Table 5.
Results of Sanger sequencing to confirm accuracy of imputation. The p-value shown is for the family-based association test comparing the mean dosage (from Sanger genotyping) in affected and unaffected individuals.
| Gene | chr | position | Family | Number of individuals with
|
p-value | ||
|---|---|---|---|---|---|---|---|
| Sanger genotypes | Ambiguous imputation results | Unambiguous imputation results matching Sanger | |||||
| FEZF1 | 7 | 121,944,239 | AU119 | 45 | 13 | 32 | 0.02 |
| LGALS1 | 22 | 38,071,707 | AU119 | 44 | 3 | 41 | 0.03 |
| HISPPD1 | 5 | 102,490,411 | AU625 | 13 | 0 | 13 | 0 |
| SEZ6L | 22 | 26,688,831 | AU366 | 11 | 0 | 11 | 0 |
| SAMD11 | 1 | 877,523 | AU071 | 10 | 5 | 5 | 0 |
| USH2A | 1 | 216,062,273 | AU754 | 12 | 0 | 12 | 0.03 |
| TBL2 | 7 | 72,992,858 | AU754 | 12 | 0 | 12 | 0.03 |
| BLNK | 10 | 98,031,160 | AU754 | 12 | 0 | 12 | 0.03 |
| ADAMTS1 | 21 | 28,210,457 | AU754 | 12 | 0 | 12 | 0.03 |
| GABRR2 | 6 | 90,024,862 | AU599 | 13 | 0 | 13 | 0.01 |
| PIP | 7 | 142,836,646 | AU599 | 13 | 0 | 13 | 0 |
| MAP3K4 | 6 | 161,508,880 | AU113 | 11 | 0 | 11 | 0 |
| DNAH9 | 17 | 11,543,588 | AU113 | 11 | 7 | 4 | 0.12 |
| Total | 219 | 28 | 191 | ||||
In four families, only one or two variants remain after filtering based on imputation and association testing (see Table 4). After filtering in AU119, two variants achieve p≤ 0.05, but evidence is much stronger for the variant in FEZF1 (FEZ family zinger finger 1). The variant is rare (0.6% in NHLBI- ESP) and exists in multiple transcript types, including missense and regulatory region variants(Hubbard et al. 2007). It is predicted by PolyPhen2 to be benign, but occurs at an evolutionarily conserved location (GERP = 3.5). The variant in LGALS1 has a much higher frequency, is in the 5′ UTR region, and is present in many fewer of the affected individuals, so the FEZF1 variant is a more interesting candidate in this family. In AU625, evidence is strongest for a missense variant with 1% frequency in 1KGP-EUR in the gene HISPPD1, an inositol pyrophosphate molecule that is involved in cell signaling. This variant is predicted by PolyPhen2 to be benign, but occurs at an evolutionarily conserved location (GERP = 3.4). In AU366 evidence is strongest for a missense variant with 1% frequency in 1KGP-EUR, in the gene SEZ6L (seizure related 6 homolog (mouse) –like). The position of the specific variant in this family is conserved (GERP score = 3.7), and PolyPhen2 predicts that the change to the protein is probably-damaging. After filtering in AU071, one variant that is not observed in 1KGP-EUR achieves p≤ 0.05. The variant in the gene SAMD11 (sterile alpha motif domain containing 11) did not present the Mendelian segregation pattern in WES because of missing genotypes (due to low read depth) in two affected individuals. Sanger sequencing confirmed the absence of the variant in the unaffected individuals and its presence in all affected individuals. While the variant itself is not predicted to be deleterious, it is very rare (0.03% in NHLBI-ESP). In the other three families, variant lists were reduced compared to the simpler Mendelian filter, but we were still left with a number of variants and no clear way to choose the one most likely to be causative. Variants in 9, 6, and 5 genes were left in families AU754, AU599 and AU113, respectively (see Table 4).
Conclusions
Filtering of WES variants by linkage analysis, frequency and function, and family-based imputation and association testing was an effective tool for prioritizing exome variants. In seven families, exome sequencing followed by linkage analysis and bioinformatics filtering based on frequency and function identified 200 candidate variants (between 4 and 53 per family). When we applied filtering based on a Mendelian pattern of inheritance this candidate list was narrowed to 70 variants (between 1 and 23 per family). An alternate filter, imputing exome variants and performing association testing, resulted in further reduction of number of candidate variants in such families to 28 variants (between 1 and 9 per family). The evidence using this filtering approach is stronger than in studies where the simpler Mendelian filter is used, because it incorporates information from unaffected individuals. This process provided four strong candidate genes for ASD in extended families, all of which have either been previously implicated in other studies, or have a strong biological argument for their relevance.
SEZ6L is an exceptionally strong candidate gene. It is a member of the SEZ6 family of proteins with cell adhesion properties, and shares high protein similarity with SEZ6 and SEZ6L2. SEZ6L2 was identified as an ASD candidate gene based on both an association study of genes in the 16p11.2 region(Kumar et al. 2009b), where duplications and deletions are known to be associated with ASD, and in large scale genomic analyses(Glessner et al. 2009; Marshall et al. 2008). However, a later study did not detect enrichment of non-synonymous variation in SEZ6L2 in ASD compared with controls(Konyukh et al. 2011). The members of the SEZ6 family affect excitatory synaptic transmission and are important for the achievement of balance between elongation and branching during dendritic arborization in mice(Anderson et al. 2012; Gunnersen et al. 2009). In addition, SEZ6 null mice, although not specifically tested for ASD associated phenotypes, demonstrate abnormalities in motor coordination (Rota-Rod testing), behave differently under novel potentially threatening conditions (Elevated Plus Maze) and might have long-term memory deficits (Morris Water Maze)(Gunnersen et al. 2007). mRNA transcripts of SEZ6L are observed in the human cerebral cortex, and variants in this gene have been associated with bipolar disorder(Xu et al. 2013). Interestingly, SEZ6 was originally identified as an upregulated gene following chemically induced seizures(Shimizu-Nishikawa et al. 1995). Seizures are often comorbid with ASD(Spence and Schneider 2009). Mutations in SEZ6, in particular a recurrent frame-shift mutation, were reported to be associated with febrile seizures in a Chinese population(Yu et al. 2007), but this finding was not replicated in a Caucasian one(Mulley et al. 2011).
Our implication of HISPPD1 is particularly exciting, because a variant within this gene was observed to segregate with ASD in another multiplex family(Cukier et al. 2014). The extreme heterogeneity of ASD makes such a replication very unlikely to occur by chance. HISPPD1 codes for an inositol pyrophosphate molecule, a class of cell signaling molecules that have roles in cellular migration and differentiation, both important to brain development. Differential alternative splicing of this gene, which is expressed in many tissues including the brain, has been observed between subgroups of individuals with ASD based on total cranial volume(Stamova et al. 2013).
FEZF1 is interesting because it resides in one of the first regions linked to ASD, in addition to being biologically plausible. It is located in AUTS1, a region originally implicated in several linkage studies (IMGSAC 2001; Schellenberg et al. 2006), including one which used a smaller, earlier version of the AU119 family. Case-control analysis of SNP variation in the IMGSAC families further implicated FEZF1 and other genes in the region(Maestrini et al. 2010). Additionally, FEZF1 is involved in patterning of the diencephalon(Shimizu and Hibi 2009) in mice. The diencephalon is a component of the neural tube that gives rise to the thalamus, a region of the brain which is thought to be affected in ASD(Nair et al. 2013). ENCODE (Rosenbloom et al. 2013) annotation in the UCSC Genome Browser(Kent et al. 2002) indicates the variant observed in AU119 is located within a DNaseI hypersensitivity cluster and transcription factor binding site in multiple relevant cell lines. We have positional, functional, and statistical evidence supporting this gene as a candidate for ASD risk.
Finally, SAMD11 is a gene whose function is not well understood in humans, but it is evolutionarily conserved from zebrafish to humans(Jin et al. 2013). The Gene Ontology(Ashburner et al. 2000) term for SAMD11 includes the negative regulation of transcription from RNA polymerase II promoters. ENCODE annotation in the UCSC Genome Browser provides evidence that the variant identified in AU071 sits on a DNaseI hypersensitivity cluster and transcription factor binding site in multiple relevant cell lines. In mice, it is expressed in retinal photoreceptors and in the adult pineal gland. In humans, SAMD11 is expressed in neuronal cells in the cerebral cortex and Purkinje cells in the cerebellum, in addition to numerous other tissues. The cerebellum is thought to be important in language, executive function, and regulation of affect(Becker and Stoodley 2013), processes that are abnormal in ASD. Altered anatomy of the cerebellum is also seen in ASD(Fatemi et al. 2012).
In addition to the four candidate genes described above, this work yields three important lessons about the analysis of exome variants in family based samples for a complex trait. First, it is important to sample both affected and unaffected individuals whenever possible, because this allows for variant prioritization based on a comparison of variants dosages in affected and unaffected individuals from the same family. The importance of unaffected individuals should not be a surprise, given the well known value of unaffected individuals in parametric linkage analysis(Wijsman 2012). Extending our pedigrees through careful recruitment and the inclusion of unaffected subjects allowed us to dramatically reduce the number of candidate variants per family. Second, exome variant dosages need not be directly observed in all individuals. In our families, we obtained WES on one or more members of each sibship, and whenever possible, on a common ancestor of all affected individuals. Software is also available to help select optimal subjects for sequencing, with the goal of maximal imputation(Cheung et al. 2014). Using both these data and SNP chip data on most of the family, we reliably imputed variant dosages in individuals who were not directly sequenced. As WES is on the order of 5–7 times the cost of SNP chip typing, this is a cost effective approach to obtaining variant dosages. Additionally, imputation can be used when DNA is unavailable or is in short supply, and avoids the problem of batch effects that can occur when individuals are sequenced at different times(Derkach et al. 2014). Finally, when unaffected individuals are sampled and either imputed or directly-observed variant dosages are available, within-family association testing (using a model that accounts for family relationships) is useful to prioritize variants and genes for further study, and can increase the information gleaned from the pedigree over analysis of only the sequenced subjects(Saad and Wijsman 2014). The p-value from the association test for each variant can be used as a measure of strength of support for that variant as an ASD susceptibility allele in that family. One may take a qualitative approach and consider only variants that have a p-value below a preset cut-off, or one can take a “top N” approach, where the N variants with the lowest p-value are prioritized for continued investigation. Either method is an improvement on the simpler approach of following up all variants that segregate in affected individuals in a manner consistent with Mendelian inheritance. This is especially important for complex traits where heterogeneity can be expected and some affected individuals may not share the variant of interest.
The family-based approach described here results in relatively few candidate genes, but the statistical and biological evidence behind each is strong. In contrast, WES studies using samples of unrelated affected individuals generate long lists of potential candidate genes (e.g.(Neale et al. 2012; O’Roak et al. 2012; Sanders et al. 2012)), with little ability to discriminate between causal and incidental variation(Gratten et al. 2013). Samples of unrelated affected individuals are often easier to collect than large families, but the lack of inheritance information means that WES must be generated on all individuals, leading to high costs. The choice of the best design to use will depend on the exact characteristics of phenotype in question, but this family based approach should be useful in other complex disorders where familial forms exist (and large families can be ascertained), but there is extreme heterogeneity between families
Supplementary Material
Online Resource 1: ADOS and IQ data for affected individuals.
Online Resource 2: Genome scans using SNP panels and multipoint parametric linkage analysis, by family. SNP panels were chosen from the Illumina HumanCoreExome chip (AU366, AU071, AU113), or the set of SNPs that were on both chips (remaining families).
Online Resource 3: Variants with association test p-value ≤ 0.05, or Mendelian inheritance pattern, by family.
Acknowledgments
Research reported in this publication was supported by funding from the National Institute of Mental Health, and the National Institute on Aging, under award numbers R01MH092367, R01MH094293, R01MH094400, and R00AG040184 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank those with ASD and their families, because without their participation this research would not be possible.
List of abbreviations
- ASD
autism spectrum disorder
- WES
whole exome sequencing
- ADOS
Autism diagnostic observational schedule
- ADI-R
Autism diagnostic interview - revised
- BPASS
Broader Phenotype Autism Symptom Scale
- BAP
broader phenotype
- OE
Illumina HumanOmniExpress
- HCE
Illumina Human Core Exome
- 1KGP-EUR
1,000 genome project Europeans
- IV
inheritance vectors
- MCMC
Markov chain Monte Carlo
Appendix
Marker selection for linkage analysis
We used the marker subpanels program of the PBAP suite(Nato et al. 2013), with the following parameters: minimum intermarker distance 0.5 cM, marker completion threshold 80%, minor allele frequency > 20%, maximum linkage disequilibrum (r2) between markers 0.04. We used allele frequencies and linkage disequilibrium estimates from the 1KGP-EUR population, and maps based on the sex-averaged Rutgers map(Matise et al. 2007). Map locations were converted from those based on the Kosambi map function in the Rutgers map to those based on the Haldane map function. This was necessary because of the implicit assumptions in the multipoint analysis imposed by use of the Lander-Green algorithm(Lander and Green 1987).
Linkage analysis
Our approach to linkage analysis in these families was to use sampled inheritance vectors (IVs) as a basis for analysis. The set of IVs at a particular genomic position represents possible paths of descent of the chromosomes at that position through the pedigree. The sampled IVs are drawn from the posterior distribution of IVs, conditional on marker information, family structure and genetic map, at each marker location along each chromosome. For the smallest pedigree, we sampled IVs from the exact posterior distribution, and for the larger pedigrees we used Markov chain Monte Carlo (MCMC) sampling. Using the sampled IVs as a basis for analysis enabled us to perform chromosome wide multipoint pedigree based linkage analysis, even in our larger families. The same samples of IVs were also used for genotype imputation from the sequence data. Linkage analysis from the IVs followed three steps. First, we sampled IVs for each chromosome and family combination using the program gl_auto from the MORGAN(MORGAN: A package for Markov chain Monte Carlo in genetic analysis (version 3.1.1) http://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan.shtml 2012) package, saving 1,000 IV samples for analysis. For MCMC sampling, 50,000 scans were performed with each run using sequential imputation for setup, and the LMM sampler with 50% L-sampler. We used allele frequencies based on each dataset, except in the families typed with CE alone, where we used the 1KGP-EUR frequencies. These families are generally well genotyped, and therefore we do not expect the results to be sensitive to the source of allele frequencies(Huang et al. 2004). Second, we identified equivalence classes among the sampled IVs at each marker, using the program IBDgraph(IBDgraph 2.0: another C-library add-on for MORGAN 3 http://www.stat.washington.edu/thompson/Genepi/pangaea.shtml 2010). Identifying equivalence classes allows computations to be performed on one representative of each class, rather than on all 1,000 samples. Finally, we performed linkage analyses using FASTLINK(O’Connell and Weeks 1995) for one representative of each equivalence class, and calculated likelihoods by a weighted average over equivalence classes, where the weights are the sampled probabilities of the classes. Since these analyses were done, the MORGAN program gl_lods has been released(MORGAN: A package for Markov chain Monte Carlo in genetic analysis (version 3.2) http://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan.shtml 2013), which carries out the analyses directly from the output of gl_auto. In future it will not be necessary to use FASTLINK for this type of analysis.
Imputation of exome variant dosages
GIGI uses the IVs generated by gl_auto for the entire family based on the SNP genotypes, in addition to exome variant genotypes in available individuals, to calculate the expected dosage of the variant allele in each person in the family. The frequencies of the alternate alleles were taken from the 1KGP-EUR population, unless the allele was absent from that dataset, in which case it was set to 0.01. Sex-averaged map positions were converted to positions based on the Haldane map function, as described above. In cases where variants did not appear in the Rutgers map, map position was interpolated based on physical position.
Example of imputation and association results
Table 6 shows the results of imputation and subsequent association tests for family AU071, as an example. AU071 is interesting because we imputed variant dosages in multiple unaffected individuals, and there are examples of the imputed dosage being very clear, as well as examples where the dosage was more ambiguous. Variant allele dosages are italicized if they are based on imputation, and not if they are directly observed. The table lists the 18 variants that pass the filters based on linkage analysis, frequency and function, and also have a Mendelian segregation pattern. There is an additional variant where the observed segregation pattern was not obviously Mendelian (pattern=‘no’), due to missing genotypes because of low read depth in affected individuals. For this variant (chr 1 pos 877,523), no copies of the alternate allele are imputed in unaffected individuals, but the status of two affected individuals (A3 and A4) remains ambiguous (imputed dosage = 0.5). Sanger sequencing clarified that both A3 and A4 carry a single copy of the variant, making this variant a very good candidate gene for ASD in this family. Even without taking into account the Sanger results, this is the only variant where the p-value is ≤ 0.05, so the imputation and association based results represent a substantially reduced set of variants relative to those based on requiring a Mendelian segregation pattern in affected individuals with WES only. Online Resource 3 shows detailed results similar to Table 6 for each of the seven families studied.
Table 6.
Imputation and association testing in AU071. Shown are all variants with a segregation pattern consistent with Mendelian inheritance in directly exomed individuals and/or p ≤ 0.05 for association testing on imputed data. Imputed dosages are italicized, and dosages directly observed (by WES) are not. Pattern indicates whether the variant segregates in a manner consistent with Mendelian inheritance, based solely on WES.
| chr | pos | Variant dosage (expected and observed) | pattern | p-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Unaffected individuals (without offspring) | Affected individuals | |||||||||||
| U1 | U2 | U3 | mean | A1 | A2 | A3 | A4 | mean | ||||
| 1 | 877,523 | 0.03 | 0.03 | 0.05 | 0.04 | 1 | 1 | 0.48 | 0.46 | 0.82 | No* | 0.012 |
| 1 | 3,697,663 | 0.03 | 1.00 | 0.00 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 1 | 3,755,638 | 0.01 | 1.00 | 0.00 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 4 | 663,916 | 1.02 | 0.07 | 0.04 | 0.38 | 1 | 1 | 1 | 1 | 1 | Yes | 0.126 |
| 4 | 166,388,900 | 0.01 | 0.01 | 1.00 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 4 | 169,317,237 | 0.01 | 0.01 | 1.00 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 4 | 173,730,541 | 0.01 | 0.01 | 1.00 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 10 | 27,497,191 | 1.00 | 0.31 | 0.05 | 0.45 | 1 | 1 | 1 | 1 | 1 | Yes | 0.126 |
| 12 | 16,342,622 | 1.00 | 0.02 | 0.01 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 12 | 18,865,819 | 1.00 | 0.02 | 0.01 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 12 | 20,876,168 | 1.00 | 0.02 | 0.01 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 12 | 21,196,367 | 1.00 | 0.02 | 0.01 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 12 | 21,196,466 | 1.00 | 0.02 | 0.01 | 0.34 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
| 12 | 27,648,704 | 1.01 | 0.02 | 1.00 | 0.68 | 1 | 1 | 1 | 1 | 1 | Yes | 0.381 |
| 12 | 29,630,166 | 1.00 | 0.01 | 1.00 | 0.67 | 1 | 1 | 1 | 1 | 1 | Yes | 0.376 |
| 12 | 40,740,686 | 1.02 | 0.04 | 1.00 | 0.69 | 1 | 1 | 1 | 1 | 1 | Yes | 0.387 |
| 12 | 49,168,798 | 1.02 | 0.04 | 1.00 | 0.69 | 1 | 1 | 1 | 1 | 1 | Yes | 0.387 |
| 12 | 52,579,331 | 1.01 | 0.07 | 1.00 | 0.69 | 1 | 1 | 1 | 1 | 1 | Yes | 0.381 |
| 19 | 57,184,265 | 1.00 | 0.08 | 0.04 | 0.37 | 1 | 1 | 1 | 1 | 1 | Yes | 0.116 |
in this case, the variant did not show a Mendelian segregation pattern because of low read depth, which resulted in missing genotypes in affected individuals.
Footnotes
Ethical approval: “All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.”
Contributor Information
Nicola H. Chapman, Division of Medical Genetics, School of Medicine, University of Washington, Seattle, WA
Alejandro Q. Nato, Jr., Division of Medical Genetics, School of Medicine, University of Washington, Seattle, WA
Raphael Bernier, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA.
Harkirat Sohi, Division of Medical Genetics, School of Medicine, University of Washington, Seattle, WA.
Jeff Munson, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA. Center on Child Development and Disability, University of Washington, Seattle, WA.
Ashok Patowary, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA.
Marilyn Archer, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA.
Elizabeth M. Blue, Division of Medical Genetics, School of Medicine, University of Washington, Seattle, WA
Sara Jane Webb, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA. Center on Child Development and Disability, University of Washington, Seattle, WA.
Hilary Coon, Department of Internal Medicine, University of Utah, Salt Lake City, UT. Department of Psychiatry, School of Medicine, University of Utah, Salt Lake City, UT.
Wendy H. Raskind, Division of Medical Genetics, School of Medicine, University of Washington, Seattle, WA. Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA. Department of Genome Sciences, University of Washington, Seattle, WA
Zoran Brkanac, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA.
Ellen M. Wijsman, Division of Medical Genetics, School of Medicine, University of Washington, Seattle, WA. Department of Biostatistics, University of Washington, Seattle, WA. Department of Genome Sciences, University of Washington, Seattle, WA
References
- Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual. 5. American Psychiatric Association; Washington, DC: 2013. [Google Scholar]
- Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Sudhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci U S A. 2012;109:18120–18125. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EB, Stoodley CJ. Autism spectrum disorder and the cerebellum. Int Rev Neurobiol. 2013;113:1–34. doi: 10.1016/B978-0-12-418700-9.00001-0. [DOI] [PubMed] [Google Scholar]
- Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13:247. doi: 10.1186/gb4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DVM. Development of the Children’s Communication Checklist (CCC): A method for assessing qualitative aspects of communicative impairment in children. Journal of Child Psychology and Psychiatry and Allied Disciplines. 1998;39:879–891. [PubMed] [Google Scholar]
- Buescher AV, Cidav Z, Knapp M, Mandell DS. Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA Pediatr. 2014;168:721–728. doi: 10.1001/jamapediatrics.2014.210. [DOI] [PubMed] [Google Scholar]
- CDC. Morbidity and Mortality Weekly Report: Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2010. 2014;63(SS02) [PubMed] [Google Scholar]
- Chahrour MH, et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 2012;8:e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NH, et al. Genome-scan for IQ discrepancy in autism: evidence for loci on chromosomes 10 and 16. Human Genetics. 2011;129:59–70. doi: 10.1007/s00439-010-0899-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CYK, Blue EM, Wijsman EM. A statistical framework to guide sequencing choices in pedigress. American Journal of Human Genetics. 2014;94:257–267. doi: 10.1016/j.ajhg.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CYK, Thompson EA, Wijsman EM. GIGI: An approach to effective imputation of dense genotypes on large pedigrees. American Journal of Human Genetics. 2013;92:504–516. doi: 10.1016/j.ajhg.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. Children’s Memory Scale. Pearson; 1997. [Google Scholar]
- Constantino JN. Social Responsiveness Scale. 2. Western Psychological Services; Los Angeles: 2012. [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. gr.3577405 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier HN, et al. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism. 2014;5:1. doi: 10.1186/2040-2392-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Estes A, Munson J, Schellenberg G, Bernier R, Abbott R. Quantitative assessment of autism symptom-related traits in probands and parents: Broader Phenotype Autism Symptom Scale. J Autism Dev Disord. 2007;37:523–536. doi: 10.1007/s10803-006-0182-2. [DOI] [PubMed] [Google Scholar]
- Derkach A, et al. Association analysis using next-generation sequence data from publicly available control groups: the robust variance score statistic. Bioinformatics. 2014;30:2179–2188. doi: 10.1093/bioinformatics/btu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. Sensory Profile. Pearson; 1999. [Google Scholar]
- Fatemi SH, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Visscher PM, Mowry BJ, Wray NR. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nature Genetics. 2013;45:234–238. doi: 10.1038/ng.2555. [DOI] [PubMed] [Google Scholar]
- Gunnersen JM, et al. Sez-6 proteins affect dendritic arborization patterns and excitability of cortical pyramidal neurons. Neuron. 2007;56:621–639. doi: 10.1016/j.neuron.2007.09.018. S0896-6273(07)00716-7 [pii] [DOI] [PubMed] [Google Scholar]
- Gunnersen JM, Kuek A, Phipps JA, Hammond VE, Puthussery T, Fletcher EL, Tan SS. Seizure-related gene 6 (Sez-6) in amacrine cells of the rodent retina and the consequence of gene deletion. Plos One. 2009;4:e6546. doi: 10.1371/journal.pone.0006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QQ, Shete S, Amos CI. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. American Journal of Human Genetics. 2004;75:1106–1112. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Research. 2007;35:D610–617. doi: 10.1093/nar/gkl996. gkl996 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Protein Atlas. 2014 http://proteinatlas.org.
- IBDgraph 2.0: another C-library add-on for MORGAN 3. 2010 http://www.stat.washington.edu/thompson/Genepi/pangaea.shtml.
- IMGSAC. A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. American Journal of Human Genetics. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, et al. Identification and characterization of novel alternative splice variants of human SAMD11. Gene. 2013;530:215–221. doi: 10.1016/j.gene.2013.08.033. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konyukh M, et al. Variations of the candidate SEZ6L2 gene on Chromosome 16p11.2 in patients with autism spectrum disorders and in human populations. Plos One. 2011;6:e17289. doi: 10.1371/journal.pone.0017289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009a;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kumar RA, et al. Association and mutation analyses of 16p11.2 autism candidate genes. Plos One. 2009b;4:e4582. doi: 10.1371/journal.pone.0004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green PJ. Construction of multilocus genetic maps in humans. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore PC, Risi S, Gotham K, Bishop SL. ADOS-2: Autism diagnostic observation schedule. 2. Western Psychological Services; Torrance: 2012. [Google Scholar]
- Maestrini E, et al. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry. 2010;15:954–968. doi: 10.1038/mp.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. American Journal of Human Genetics. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, et al. A second-generation combined linkage-physical map of the human genome. Genome Research. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORGAN: A package for Markov chain Monte Carlo in genetic analysis (version 3.1.1) 2012 http://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan.shtml.
- MORGAN: A package for Markov chain Monte Carlo in genetic analysis (version 3.2) 2013 http://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan.shtml.
- Mulley JC, Iona X, Hodgson B, Heron SE, Berkovic SF, Scheffer IE, Dibbens LM. The Role of Seizure-Related SEZ6 as a Susceptibility Gene in Febrile Seizures. Neurol Res Int. 2011;2011:917565. doi: 10.1155/2011/917565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A, Treiber JM, Shukla DK, Shih P, Muller RA. Impaired thalamocortical connectivity in autism spectrum disorder: a study of functional and anatomical connectivity. Brain. 2013;136:1942–1955. doi: 10.1093/brain/awt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nato AQ, Chapman NH, Cheung CYK, Brkanac Z, Wijsman EM. PBAP: A pipeline for family-based quality control of pedigre structures and dense genetic marker data. Paper presented at the ASHG 63rd Annual Meeting; Boston, MA. 2013. [Google Scholar]
- Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272-U153. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nature Genetics. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard Tools: A set of Java command line tools for manipulating high-throughput sequencing data and formats. 2014 http://broadinstitute.github.io/picard/
- Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Folstein S. Modified autism family history interview for developmental disorders of cognition and social functioning. The Johns Hopkins University School of Medicine; Baltimore, MD: 1995. [Google Scholar]
- Rutter M, LeCouteur A, Lord C. Autism Diagnostic Interview Revised: WPS Edition Manual. Western Psychological Services; Los Angeles: 2003. [Google Scholar]
- Saad M, Wijsman EM. Power of family-based association designs to detect rare variants in large pedigrees using imputed genotypes. Genetic Epidemiology. 2014;38:1–9. doi: 10.1002/gepi.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarason B, Sarason I, Hacker A, Basham R. Concomitants of social support: Social skills, physical attractiveness, and gender. Journal of Personality and Social Psychology. 1985;49:469–480. [Google Scholar]
- Schellenberg GD, et al. Evidence for multiple loci from a genome scan of autism kindreds. Molecular Psychiatry. 2006;11:1049–1060. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- Shi L, et al. Whole-genome sequencing in an autism multiplex family. Mol Autism. 2013;4:8. doi: 10.1186/2040-2392-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Nishikawa K, Kajiwara K, Kimura M, Katsuki M, Sugaya E. Cloning and expression of SEZ-6, a brain-specific and seizure-related cDNA. Brain Res Mol Brain Res. 1995;28:201–210. doi: 10.1016/0169-328x(94)00203-q. 0169328X9400203Q [pii] [DOI] [PubMed] [Google Scholar]
- Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev Growth Differ. 2009;51:221–231. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- SIFT. 2014 http://sift.jcvi.org.
- Sparrow S, Cichetti D, Balla D. Vineland Adaptive Behavior Scales. 2. Pearson; Bloomington, IN: 2005. [Google Scholar]
- Spence SJ, Schneider MT. The role of epilepsy and epileptiform EEGs in autism spectrum disorders. Pediatr Res. 2009;65:599–606. doi: 10.1203/PDR.0b013e31819e7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamova BS, Tian Y, Nordahl CW, Shen MD, Rogers S, Amaral DG, Sharp FR. Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders. Mol Autism. 2013;4:30. doi: 10.1186/2040-2392-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C, et al. Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol Psychiatry. 2014;19:784–790. doi: 10.1038/mp.2013.106. [DOI] [PubMed] [Google Scholar]
- Uhlen M, et al. Towards a knowledge-based Human Protein. Atlas Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Wagner R, Torgesen J, Rashotte C. Comprehensive test of phonological processing (CTOPP) Western Psychological Services; Los Angeles, CA: 1999. [Google Scholar]
- Wechsler D. Wechsler adult intelligence scale - revised (WAIS-R) 1981. [Google Scholar]
- Wechsler D. WPPSI-R manual: Wechsler preschool and primary scale of intelligence, revised. 1989. [Google Scholar]
- Wechsler D. Wechsler intelligence scale for children - third edition (WISC-III) 1992. [Google Scholar]
- Wechsler D. Wechsler memory scale for adults. 3. The Psychological Corporation; San Antonio, TX: 1997. [Google Scholar]
- Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Human Genetics. 2012;131:1555–1563. doi: 10.1007/s00439-012-1190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, et al. Polymorphisms in seizure 6-like gene are associated with bipolar disorder I: evidence of gene x gender interaction. J Affect Disord. 2013;145:95–99. doi: 10.1016/j.jad.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Yu ZL, et al. Febrile seizures are associated with mutation of seizure-related (SEZ) 6, a brain-specific gene. J Neurosci Res. 2007;85:166–172. doi: 10.1002/jnr.21103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 1: ADOS and IQ data for affected individuals.
Online Resource 2: Genome scans using SNP panels and multipoint parametric linkage analysis, by family. SNP panels were chosen from the Illumina HumanCoreExome chip (AU366, AU071, AU113), or the set of SNPs that were on both chips (remaining families).
Online Resource 3: Variants with association test p-value ≤ 0.05, or Mendelian inheritance pattern, by family.