1. INTRODUCTION
As defined by the European Commission, nanomaterial is a natural, incidental or manufactured material containing particles in an unbound state or as an aggregate or agglomerate in which ≥ 50% of the particles in the number size distribution have one or more external dimensions in the size range 1 to 100 nm. In specific cases and where warranted by concerns for the environment, health, safety or competition, the number size distribution threshold of 50% may be replaced with a threshold between 1 and 50%.1 Engineered nanomaterials (ENMs) refer to man-made nanomaterials. Materials in the nanometer range often possess unique physical, optical, electronic, and biological properties compared with larger particles, such as the strength of graphene,2 the electronic properties of carbon nanotubes (CNTs),3 the antibacterial activity of silver nanoparticles4 and the optical properties of quantum dots (QDs).5 The unique and advanced properties of ENMs have led to a rapid increase in their application. These applications include aerospace and airplanes, energy, architecture, chemicals and coatings, catalysts, environmental protection, computer memory, biomedicine and consumer products. Driven by these demands, the worldwide ENM production volume in 2016 is conservatively estimated in a market report by Future Markets to be 44,267 tons or ≥ $5 billion.6
As the production and applications of ENMs rapidly expand, their environmental impacts and effects on human health are becoming increasingly significant.7 Due to their small sizes, ENMs are easily made airborne.8 However, no accurate method to quantitatively measure their concentration in air currently exists. A recently reported incident of severe pulmonary fibrosis caused by inhaled polymer nanoparticles in seven female workers obtained much attention.9 In addition to the release of ENM waste from industrial sites, a major release of ENMs to environmental water occurs due to home and personal use of appliances, cosmetics and personal products, such as shampoo and sunscreen.10 Airborne and aqueous ENMs pose immediate danger to the human respiratory and gastrointestinal systems. ENMs may enter other human organs after they are absorbed into the bloodstream through the gastrointestinal or respiratory systems.11,12 Furthermore, ENMs in cosmetics and personal care products, such as lotion, sunscreen and shampoo may enter human circulation through skin penetration.13 ENMs are very persistent in the environment and are slowly degraded. The dissolved metal ions from ENMs can also revert back to nanoparticles under natural conditions.14 ENMs are stored in plants, microbes and animal organs and can be transferred and accumulated through the food chain.15,16 In addition to the accidental entry of ENMs into human and biological systems, ENMs are also purposefully injected into or enter humans for medicinal and diagnostic purposes.17 Therefore, interactions of ENMs with biological systems are inevitable.
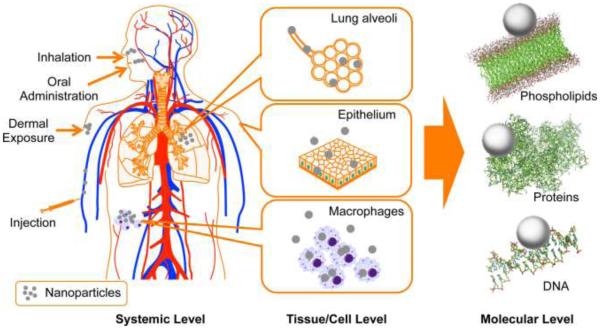
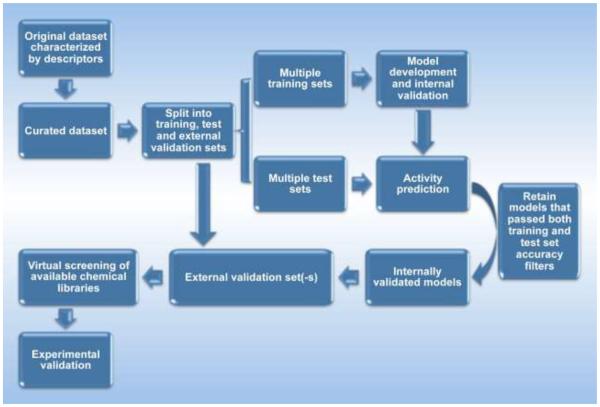
In addition to engineered nanomaterials, there are also naturally existing nanomaterials such as proteins and DNA molecules, which are key components of biological systems. These materials, combined with lipids and organic and inorganic small molecules, form the basic units of living systems –cells.18 To elucidate how nanomaterials affect organs and physiological functions, a thorough understanding of how nanomaterials perturb cells and biological molecules is required (Figure 1). Rapidly accumulating evidence indicates that ENMs interact with the basic components of biological systems, such as proteins, DNA molecules and cells.19-21 The driving forces for such interactions are quite complex and include the size, shape and surface properties (e.g., hydrophobicity, hydrogen-bonding capability, pi-bonds and stereochemical interactions) of ENMs.22-25
Figure 1.
Interactions of nanoparticles with biological systems at different levels. Nanoparticles enter the human body through various pathways, reaching different organs and contacting tissues and cells. All of these interactions are based on nanoparticle-biomacromolecule associations.
Evidence also indicates that chemical modifications on a nanoparticle’s surface alter its interactions with biological systems.26-28 These observations not only support the hypothesis that basic nano-bio interactions are mainly physicochemical in nature but also provide a powerful approach to controlling the nature and strength of a nanoparticle’s interactions with biological systems. Practically, a thorough understanding of the fundamental chemical interactions between nanoparticles and biological systems has two direct impacts. First, this knowledge will encourage and assist experimental approaches to chemically modify nanoparticle surfaces for various industrial or medicinal applications. Second, a range of chemical information can be combined with computational methods to investigate nano-biological properties and predict desired nanoparticle properties to direct experiments.29-31
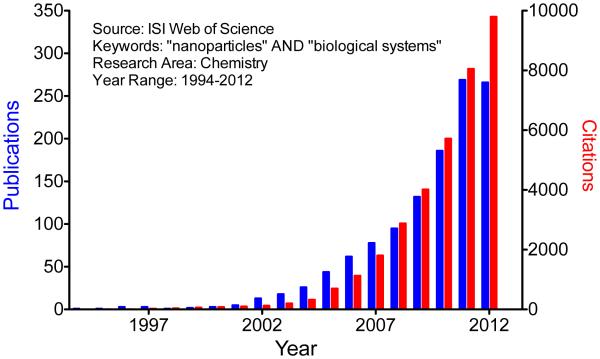
The literature regarding nanoparticle-biological system interactions has increased exponentially in the past decade (Figure 2). However, a mechanistic understanding of the chemical basis for such complex interactions is still lacking. This review intends to explore such an understanding in the context of recent publications.
Figure 2.
An analysis of literature statistics indicates growing concern for the topics that are the focus of this review. The number of publications and citations were obtained using the keywords “nanoparticles” and “biological systems” in the subject area of “Chemistry” when searching the Thomson Reuters (ISI) Web of Knowledge for the period 1994-2012.
A breakthrough technology cannot prosper without wide acceptance from the public and society; that is, it must pose minimal harm to human health and the environment. Nanotechnology is now facing such a critical challenge. We must elucidate the effects of ENMs on biological systems (such as biological molecules, human cells, organs and physiological systems). Accumulating experimental evidence suggests that nanoparticles interact with biological systems at nearly every level, often causing unwanted physiological consequences. Elucidating these interactions is the goal of this review. This endeavor will help regulate the proper application of ENMs in various products and their release into the environment. A more significant mission of this review is to direct the development of “safe-by-design” ENMs, as their demands for and applications continue to increase.
2. FACTORS AFFECTING NANOPARTICLE-BIOLOGICAL SYSTEM INTERACTIONS
2.1. Nanoparticles
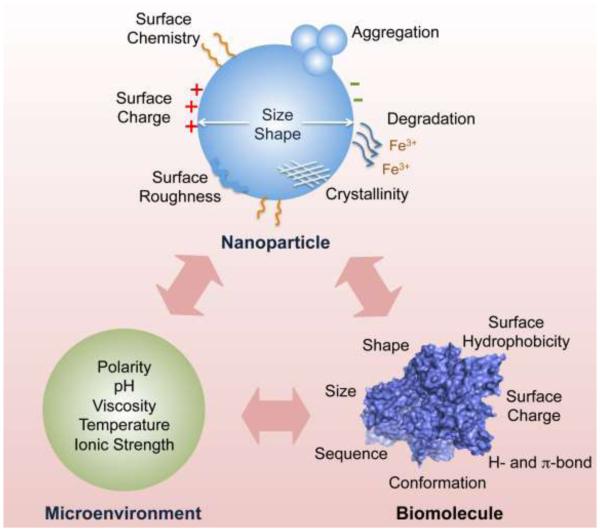
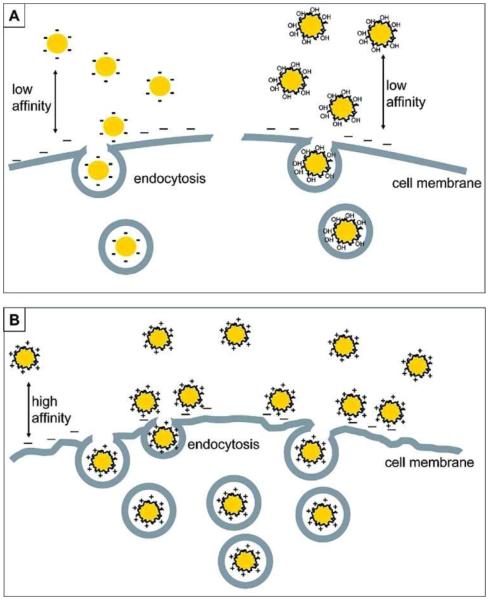
The extremely large surface area of nanoparticles results in the strong adsorption of various molecules to reduce nanoparticle surface energy. Completely “naked” nanoparticles are nearly non-existent. The interactions between nanoparticles and biological systems (nano-bio interactions) are actually driven by modified nanoparticles in biological media, such as nanoparticle complexes or their aggregates.32 The real identity of nanoparticles in biological systems is determined by their intrinsic properties, such as their size, shape, surface charge and hydrophobicity (Scheme 1). Therefore, a rigorous characterization of nanoparticles is essential prior to investigating nano-bio interactions.
Scheme 1.
Factors influencing nanoparticle-biomolecule interactions.
A very large collection of methods has been used to characterize nanoparticle physical and chemical properties. Transmission electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM) provide information on nanoparticle morphology.33 Dynamic light scattering (DLS) provides information on the hydrodynamic radii of nanoparticles in solution.34 The surface charge properties of nanoparticles can be determined with ζ-potential measurements.35 Auger electron spectroscopy, X-ray photoelectron spectroscopy (XPS), time-of-flight mass spectrometry (TOF MS) and elemental analyses reveal chemical composition details of nanoparticles.36,37 Surface ligands or adsorbed molecules can be identified with magic angle spinning nuclear magnetic resonance (MAS NMR), liquid chromatography mass spectroscopy (LC-MS) and Fourier-transform infrared spectroscopy (FTIR).38-40 Surface-enhanced Raman spectroscopy (SERS) is also among the most frequently applied analytical methods for characterizing nanoparticles.41
2.2. Biological Factors
In biological systems, nanoparticles are surrounded by excessive amounts of biological molecules, which may result in the saturation of the nanoparticle surface. This phenomenon occurs whether the nanoparticles are present in the body or in a cellular environment. Such biologically modified nanoparticles are the real participants in nano-bio interactions.32 These modifications also affect cell binding events and the internalization of nanoparticles. Inside the cell, nanoparticles tend to localize in certain cellular organelles and affect specific cell regions.42-44
In biological fluids, nanoparticles interact with phospholipids, proteins, DNA and small metabolites. Phospholipids are the main components in the pulmonary system and cell membranes, consisting of a negatively charged phosphate group (head) and a hydrophobic carbon chain (tail).18 Nanoparticles may interact with the charged head and/or the hydrophobic tail depending on their surface hydrophobicity. Nanoparticles spontaneously adsorb proteins in the bloodstream or biological fluid. Proteins are folded peptides with different sizes, shapes and net charges at physiological pH. Protein surfaces also exhibit varying hydrophobicity depending on the nature of the exposed amino acid residues.18 All of these properties may affect the stoichiometry and orientation of their bindings to nanoparticles. Nanoparticles may also bind DNA molecules in biological systems. DNA molecules carry negative charges from the phosphate groups and base π-systems.18 Single- or double-strand DNA molecules can bind to nanoparticles through electrostatic, π-π stacking and hydrophobic interactions (Section 3.3). Therefore, molecular interactions of nanoparticles in biological environments involve interactions with proteins, DNA, phospholipids, small molecules and inorganic ions. Such interactions are further complicated by the kinetics and thermodynamics of the nanoparticle associations with each component.
The organ from which a cell originates influences nanoparticle-cell interactions. First, different cell types have different shapes. Human erythrocytes exhibit a smooth, spherical shape, while ependymal cells demonstrate a long, slender shape.45,46 These differences produce in different cellular responses when internalizing foreign matter, such as nanoparticles. Second, plasma membranes from different cell types have different lipid compositions, which influence the physical properties of membranes and make them function differently. Third, different types of cells express different membrane receptors. For example, macrophages express a large number of scavenger receptors that help internalize foreign matter non-specifically.47 Fourth, different cellular organelles have various impacts on nanoparticles. Lysosomes are acidic organelles, while the cytoplasm is nearly neutral. Nanoparticles in lysosomes may undergo degradation, which may result in the release of ions and a toxic state.48 However, drug cargos may be preferably released in acidic environments for therapeutic purposes.49
3. NANOPARTICLE-BIOMOLECULE INTERACTIONS
Nanoparticles enter the human body via multiple routes, encountering various biomolecules regardless of the pathway by which they enter the human body. Airborne nanoparticles may enter the human body through inhalation. In the lungs, nanoparticles exhibit strong interactions with the lung surfactant system, which consists of proteins and phospholipids.50,51 Engineered medical nanoparticles are generally administered into the circulation system through intravenous injection. Injected nanoparticles first interact with blood proteins before being distributed in various organs.52 Some nanoparticles may also enter the cell nucleus and interact with nucleic acids, such as DNA.53 Furthermore, nanoparticles in the blood or cells also adsorb small molecules, such as amino acids, biotin and folic acid. The depletion of small metabolites may also contribute to the toxicity of nanoparticles.54 In this section, we discuss interactions of nanoparticles with proteins, lipids, DNA and small molecules. We focus on the characterization and mechanistic elucidation of these interactions.
3.1. Nanoparticle-Phospholipid Interactions
3.1.1. Interaction of Nanoparticles with Phospholipids in Pulmonary Surfactant Solutions
Phospholipids and proteins constitute pulmonary surfactant system. Inhaled nanoparticles inevitably interact with these components. The interactions between nanoparticles and pulmonary surfactant system determine nanoparticles’ pulmonary toxicity or drug delivery efficacy. Therefore, understanding these interactions and effectively regulate these interactions are imperative for safe nanotechnology and better nanomedicine. Pulmonary surfactant solutions have been used to mimic human pulmonary surfactant system for mechanistic studies. The nature of nanoparticle-phospholipid interactions can be investigated after isolating and identifying nanoparticle-bound surfactant molecules and characterizing the properties of nanoparticle-surfactant complexes. Nanoparticle-bound lipid molecules can be isolated by size exclusion chromatography55 or thin layer chromatography50 and structurally identified by NMR55 or MS.51 Nanoparticle-phospholipid interactions can also be measured by identifying alterations in nanoparticle physicochemical properties. Surfactant molecule coatings alter nanoparticle sizes and dispersion states, which can be measured using DLS,56 TEM56,57 or SEM.58 Surfactant molecules can also alter the surface charge status of nanoparticles, as determined with ζ-potential analysis.56 Binding of surfactant molecules onto the surfaces of nanoparticles may alter their surface plasma resonance (SPR) absorption and can also be determined using UV-Vis absorption spectroscopy.56 The surface properties of pulmonary surfactant solutions, such as equilibrium and dynamic surface tension behavior under the pulsation of samples, are also used to evaluate the effects of nanoparticles.57,59
Dipalmitoyl phosphatidylcholine (DPPC, the most abundant surfactant molecule in a pulmonary surfactant mixture; also termed Survanta) is strongly adsorbed onto the surface of nanoparticles (e.g., carbon-based nanoparticles), forming a rigid monolayer. The binding of long aliphatic hydrocarbon chains to carbon nanoparticle surfaces results in increased nanoparticle surface charge, size and stability. Adsorption onto a hydrophobic surface is also energetically favorable for DPPC.56 Although proteins bind to multi-walled CNTs (MWNTs) with different binding patterns depending on the surface functionalization of the CNTs, the surfactant lipid may nonspecifically adsorb onto different MWNTs.50 The adsorption of lipids onto the surface of carbon-based nanoparticles may affect their in vitro and in vivo behavior. When coated with DPPC, nanoscale carbon black and crocidolite asbestos increase reactive oxygen species (ROS) generation in primary bronchial epithelial cells and in A549 alveolar epithelial carcinoma cells, while fetal bovine serum protein-coated nanoparticles protect cells from oxidative insult.60 However, in another study, DPPC-coated, single-walled CNTs (SWNTs) did not generate cytotoxic or fibrogenic effects in vitro. Lung fibrosis in mice can be induced by SWNTs with or without DPPC coating, indicating that DPPC does not significantly mask the bioactivity of SWNTs in vivo.58 Surfactants and the nanoparticle system can also affect each other. Phospholipids have been shown to stabilize colloidal GNPs by forming lipid bilayers on the particle surface, while the presence of GNPs reduces the surface activity of surfactants, such as their ability to form a surface film or the ability of this film to reach low surface tension during compression.57 Size of particles also affects their interactions with lipids. TiO2 nanoparticles but not microsized particles increase the adsorption surface tension of the surfactant in a dose-dependent manner. These phenomena are also observed during surface area cycling in the presence of nanoparticles. TEM observations have demonstrated that lamellar body-like structures are deformed and decreased in size, with the formation of unilamellar vesicles.61
The binding events of proteins to nanoparticles most likely affect the surfactant interactions of nanoparticles. Interactions of polymeric nanoparticles with lipids in human plasma have been investigated using size exclusion chromatography, NMR and enzymatic assays. Nanoparticles bind cholesterol, triglycerides and phospholipids in human plasma. The binding events depend on the nanoparticle surface properties. A higher cholesterol content is present in the pellets of hydrophobic nanoparticles when compared to those of more hydrophilic nanoparticles.55
3.1.2. Interactions Between Nanoparticles and Model Membranes
Model membranes are organized lipids that mimic the arrangement of lipids in natural cell membranes. Interactions between nanoparticles and model membranes have been investigated using biophysical approaches to evaluate the cellular translocation of nanoparticles and the potential effects of nanoparticles on the integrity of the cell membrane. Lipid monolayers, bilayers and liposomes are three widely used model membranes.62
The surface pressure-area (π-A) isotherm is an indicator of the monolayer properties of an amphiphilic material and is obtained by measuring the surface pressure as a function of the water surface area available to each molecule.63 The isotherm is recorded by compressing the film at a constant rate while continuously monitoring the surface pressure. Although π-A isotherm measurements have been used to study the properties of model lipid monolayers in the presence of nanoparticles, this approach is limited to monolayer membranes that lack anchoring membrane proteins.64 AFM can be used to measure the effects of nanoparticles on morphological changes of Langmuir-Blodgett (LB) films. AFM measures subtle morphological details of soft matter at subnanometer levels, enabling the observation of the formation of lipid bilayers and their disruption by nanoparticles.65-68 A liquid crystal-based system has been developed to study the molecular interactions between protein-coated nanoparticles and a liquid crystal-supported model membrane. The spatial and temporal organization of the lipid monolayer is coupled to the orientation of the liquid crystal, permitting the direct observation of the dynamic characteristics of the lipid monolayer.69,70 Differential scanning calorimetry (DSC)68,71,72 and isothermal titration calorimetry (ITC)73 can also be used to measure nanoparticle-induced thermodynamic changes in supported membranes or liposomes. A quartz crystal microbalance (QCM) with dissipation monitoring can be used for real-time monitoring of nanoparticle-lipid membrane interactions.74 Fluorescence microscopy,65,75 fluorescence spectroscopy,68 fluorescence correlation spectroscopy (FCS),73 Föster resonance energy transfer,73,76 NMR77 and liposomal leakage assay78 have all been used to study the biophysical changes and disruptions of model lipid membranes or liposomes by nanoparticles.
Hydrophobic69,70 and electrostatic interactions74,78 may serve major roles in nanoparticle-lipid membrane interactions. The interactions of nanoparticles with the lipid membranes lead to changes in both components, such as disruption of the lipid membrane and the modification of the nanoparticle surface properties. Guanidinylated dendrimers that carry a positive surface charge first adhere to the liposomal membrane through binding to negatively charged phosphate groups. The dendrimer surfaces then become hydrophobic due to charge neutralization. The dendrimers are consequently transported through the hydrophobic liposomal membrane.68 Other polycationic polymers (i.e., polyamidoamine dendrimers, pentanol-core polyamidoamine dendrons, polyethyleneimine and diethylaminoethyl dextran) also induce the disruption of supported bilayers, including the formation of holes, membrane thinning and membrane erosion.79 Polycationic polymers disrupt the liquid phase but not the gel phase of dimyristoylphosphatidylcholine bilayers.77 Electrostatic interactions can also occur between protein-coated CNTs and liposomes, leading to liposome leakage.78 Surface charged polystyrene nanoparticles bind to liposomes and induce surface reconstruction of the phospholipid membranes, indicating a potential structural perturbation of the cellular membrane by nanoparticles (Figure 3).73 Nanoparticle-induced lipid bilayer rearrangements are limited to bubble-like liposomes, not supported bilayers.76 QDs can insert into lipid membranes and induce a current burst under an electric field, indicating their potential to perturb neuronal and muscle cells.80,81 Cationic phosphorus dendrimers can alter the thermotropic behavior of lipid bilayers by reducing the cooperativity of the phospholipids; this effect is membrane surface charge-dependent.71 A model has been proposed to describe the behavior of nanoparticles at the air-liquid interface of alveoli (Figure 4). Nanoparticles inhaled into the alveolar space during breathing may interact with continuous surfactant films. The nanoparticles then become wetted and lined with phospholipid bilayers.57
Figure 3.
A nanoparticle-induced surface reconstruction of phospholipid membranes. A phospholipid bilayer vesicle with bound nanoparticles is shown. Binding of anionic nanoparticles to the lipid bilayer in the fluid phase causes a transition to gel phase at the site at which the nanoparticle binds. Binding-induced reorientation of the phosphocholine (PC) head group causes lipids in the fluid phase to exhibit a lower density (A) than in the gel phase (B). In the PC head group, P− and N+ are denoted by blue and red, respectively. Reprinted with permission from ref 73. Copyright 2011 National Academy of Sciences, USA.
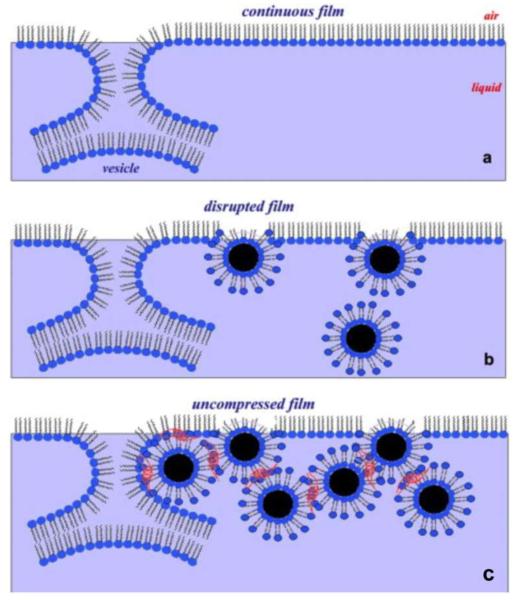
Figure 4.
The potential inhibitory effects of GNPs on pulmonary surfactants in the alveolar space. (a) A continuous surfactant film (monolayer and underlying multilayer) forms after the rupture of pulmonary surfactant vesicles at the air-liquid interface as the hypothetical rate-limiting intermediate structure between bilayer vesicles and the interfacial monolayer. (b) The disrupted interfacial surfactant film due to the entrapment of GNPs (from the air phase as pollutants) by pulmonary surfactant is depicted. (c) The self-aggregation of lipid-capped GNPs in the presence of surfactant protein B is shown in red. Reprinted with permission from ref 57. Copyright 2008 Cell Press.
In addition to the lipid membranes properties, the factors that influence nanoparticle-lipid interactions include nanoparticle dose, composition, size and surface chemistry. Hydrophobic polyorganosiloxane nanoparticles do not substantially affect the structural organization of a model pulmonary surfactant film at low concentrations (e.g., 10 μg/mL) but significantly affect such properties at high concentrations (e.g., 3000 μg/mL).82 Polystyrene nanoparticles with a diameter of 20 nm demonstrate greater interactions with the endothelial model membrane than those with a diameter of > 60 nm.64 Perturbations from nanoparticles may cause pore formation in lipid bilayers.65,66,79 Studies of polycationic polyamidoamine dendrimers and supported dimyristoylphosphatidylcholine bilayers have demonstrated that G7 dendrimers (8.0-8.2 nm) but not G3 dendrimers (3.5-4.1 nm) induce the formation of holes in lipid bilayers. G5 dendrimers (4.3-6.6 nm) have a reduced effect when compared to G7 dendrimers but had no effect when their amine end groups were replaced with acetamide groups, indicating the role of surface chemistry.67 The density of the functional groups also affects the interactions. Lightly guanidinylated dendrimeric derivatives act as “molecular glue” by adhering to liposomes, while highly guanidinylated dendrimeric derivatives induce liposome fusion.68 The crucial role of nanoparticle surface hydrophobicity has also been demonstrated. Hydrophobic, cholesterol-coated GNPs interact with DPPC vesicles and decrease their melting point, while hydrophilic nanoparticles increase the melting point of DPPC vesicles.72 The properties of model membranes, such as membrane surface charge71 and cholesterol content, also affect these interactions. A high cholesterol content in the phospholipid/cholesterol monolayer makes the lipids more susceptible to disruption by protein-coated GNPs.69
The development of efficient nanocarrier systems based on model membrane studies requires a good correlation between model systems and cellular studies. The effects of the size and surface chemistry of polymeric nanoparticles in model membranes correlate well with cell membrane disruption and cellular uptake.83-86
The interactions of nanoparticles with model membranes have also been investigated by molecular simulation approaches. Although computational methods cannot fully mimic complex biological systems, they facilitate an enhanced mechanistic understanding of their interactions and improved property predictions. The interactions of carbon nanoparticles, polymers and nanocrystals with lipid bilayers have been simulated by all-atom and coarse-grained approaches.87-94 The influences of nanoparticle size, surface chemistry, hydrophobicity, shape and bilayer properties on the interaction mechanisms have been studied, and the results have yielded explanations for some experimental outcomes.87,88,90,93,95-98 ENREF 70
In summary, the investigation of nanoparticle-lipid membrane interactions through molecular simulations, biophysical characterization and cellular studies may collectively unravel the mechanisms of these processes and elucidate regulation strategies. Such developments will benefit nanotechnology-based drug delivery and the rational reduction of nanotoxicity.
3.2. Nanoparticle-Protein Interactions
The interactions between nanoparticles and proteins are complex. Nanoparticles are generally surrounded by plasma proteins upon entering the circulatory system. As nanoparticles enter cells and are distributed into different organs, they are also in close contact with cellular proteins. Protein binding-induced changes in the surface properties of nanoparticles affect their biodistribution and clearance.99 Moreover, conformational changes in bound proteins may expose unexpected epitopes to activate cell signaling.100 The interactions of nanoparticles with proteins are dynamic processes and governed by properties of both components. The physicochemical changes in nanoparticles or proteins also allow the characterization of the thermodynamic and kinetic processes of their interactions.
3.2.1. Determination of Nanoparticle-Protein Interactions
Similar to drug-protein or protein-protein interactions, nanoparticle-protein interactions are also characterized by binding affinity, stoichiometry and kinetic properties. Determining these properties helps us understand the chemical and dynamic nature of these interactions. Modulating these properties by modified nanoparticles may prevent unwanted interactions or enhance specific binding events.
3.2.1.1. Determination of Binding Affinity and Stoichiometry and Identification of Binding Sites
Steady-state and time-resolved fluorescence spectroscopy are used to study nanoparticle-protein binding affinities, complex formation and binding-induced protein conformational changes.19 The dependence of protein fluorescence intensity or lifetime on quencher (nanoparticle) concentration follows the Stern-Volmer equation (1):
| (1) |
where F0, F, τ0 and τ represent the initial or modified fluorescence intensity or lifetime, respectively. Ksv is the Stern-Volmer constant, and [Q] is the quencher (nanoparticle) concentration.101
Fluorescence polarization permits the detection of interacting macromolecules in homogenous solutions and can quantify nanoparticle-protein interactions based on the change in rotational freedom of the fluorescent molecule upon complex formation.23,101 An advantage of fluorescence polarization is that it avoids complex separation, which may induce the denaturation of proteins or interrupt reaction equilibria. The binding ratio can be determined on the basis of fluorescence polarization measurements.
Stepwise photobleaching has been used to characterize nanoparticle-protein interactions. Both the protein and nanoparticles are labeled with fluorescence dyes. The protein-nanoparticle binding ratio is estimated by counting the steps of the nanoparticle photobleaching. The distribution of photobleaching steps is quantified to obtain the average number of proteins per nanoparticle. The photobleaching counts can be obtained by ensemble or single-molecule measurements.102
FCS measures the fluctuations of fluorescence intensity due to the Brownian motion of fluorescent particles in a small space. The hydrodynamic particle radii (RH) can be calculated through the average diffusion time (τD), the diffusion coefficient (D) and the Stokes–Einstein relationship. Using this approach, the KD of human serum albumin-nanoparticle binding (~5.1 μM) and the protein-nanoparticle thickness (~3.3 nm) were estimated.103
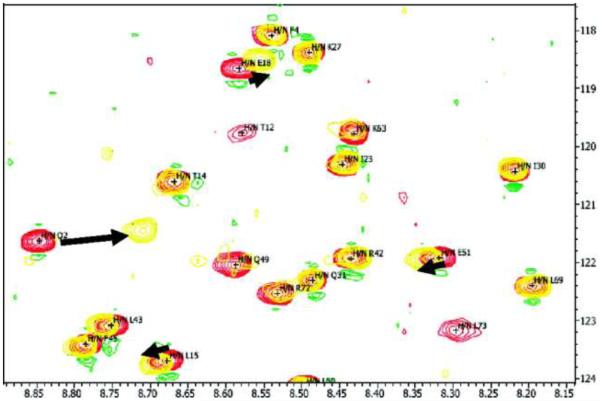
The binding sites of nanoparticles on a protein can be identified using NMR. 2D [15N-1H]-Heteronuclear single quantum coherence (HSQC) NMR was used to identify a specific binding domain of GNPs on the surface of human ubiquitin. The addition of GNPs changed the positions of certain peaks, while others were unaffected (Figure 5), indicating specific interactions. In the pre-assigned peak map, the largest chemical shift changes were from the NH backbone of Q2, L15 and E18. Based on a combination of the chemical shift changes within the protein crystal structure, a domain containing Q2-I3 and L15-E18 was proposed to be the specific binding site for GNPs in GNP-ubiquitin interactions.104
Figure 5.
Two-dimensional (2D) [15N-1H]-HSQC NMR spectra of free human ubiquitin (red) and human ubiquitin plus GNPs (yellow) at pH 7.7, showing chemical shift perturbation for some NH groups upon interaction with GNPs. The black arrows indicate the NH backbone groups with the largest chemical shift perturbations (Q2, L15, and E18) in this spectral window. Reprinted with permission from ref 104. Copyright 2010 American Chemical Society.
The binding sites of nanoparticles on a protein can also by identified using cross-linking chemistry and MS. As shown in Figure 6, the peptides located very close to polyacrylic acid-coated Fe3O4 nanoparticles in the binding site of human serum albumin (HSA) were cross-linked onto the nanoparticle surface with 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride. The attached peptides were identified by protein cleavage followed by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI TOF-MS). The nanoparticle binding site on HSA was therefore identified. The identification of binding sites helps determine the binding specificity and predict possible biological consequences of nanoparticle-protein interactions.105
Figure 6.
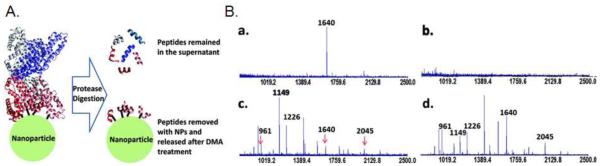
Identification of peptides associated with nanoparticle-protein interactions by cross-linking and MS. (A) A schematic illustration of the detection principle. (B) MALDI-MS spectra of released peptides when human serum albumin was (a) cross-linked and (b) adsorbed to the polyacrylic acid-Fe3O4 nanoparticles. The analysis of the supernatant from the cross-linked and noncross-linked samples is shown in (c) and (d), respectively. A peak of m/z 1640 in Ba indicates a peptide cross-linked onto the nanoparticle surface; in the supernatant, the peak intensity is significantly reduced (Bc). The arrows in Bc indicate three of thirteen identified peptides with consistently reduced peak intensity in supernatant samples. Reprinted with permission from ref 105. Copyright 2011 American Chemical Society.
DLS can be used to estimate nanoparticle-protein binding stoichiometry. The adsorption of proteins causes a small increase in the dynamic radius of nanoparticles. In one study, the adsorption of hexahistidine-tagged cytochrome P450BSβ molecules to the surface of QDs was investigated by DLS. The measurements revealed that six P450BSβ molecules bind to the surface of each QD.106
3.2.1.2. Measuring Nanoparticle-Protein Binding Kinetics
SPR technology is based on the change in the oscillation of surface plasmon waves caused by the adsorption of molecules onto a metal surface.107 Using this approach, polymeric nanoparticle-plasma protein binding kinetics have been investigated. Nanoparticles are anchored on the gold surface of the sensor chip, and proteins are injected to flow over the nanoparticle-modified surface.108 A QCM can be used to measure the resonant frequency shift, which can be correlated with a mass change at the oscillating quartz surface. Either proteins or nanoparticles can be immobilized onto the gold surface on the quartz crystal. The binding partner (nanoparticles or protein) is injected and flowed through a flow chamber over the quartz surface, and the frequency is monitored.109,110 Using either SPR or QCM, the real-time and quantitative binding profile can be obtained, and the association and dissociation constants can be determined by fitting to the Langmuir adsorption isotherm or Scatchard equation.
3.2.2. Chemical Origin of Nanoparticle-Protein Interactions
The interactions of nanoparticles with proteins are dominated by many chemical processes. The properties of the nanomaterials significantly influence these processes. Next, we discuss the mechanisms of interactions between proteins and different nanomaterials.
3.2.2.1. Carbon Nanoparticles
Carbon nanoparticles, such as CNTs, fullerene and graphene, contain only sp2 carbons, while nanodiamonds contain exclusively sp3 carbons.111-114 The uniqueness of sp2 carbon nanomaterials is due to their overwhelming π-π systems.114 Strong interactions between these materials and proteins are derived from π-π stacking, hydrophobic and electrostatic interactions.19,115,116 ENREF 36 Interactions between aromatic groups on carbon nanomaterials and aromatic or amine residues in proteins play a vital role in binding.117 Therefore, nanoparticle-protein binding events depend on the carbon nanoparticles, the nature of proteins and microenvironmental factors, such as pH and ionic strength.19,115,118-122 ENREF 38 Charge transfer between CNTs and streptavidin has been observed, indicating interactions between the amine groups of the protein and the aromatic surface of the CNTs.117 TEM and AFM observations have indicated that the bound proteins were found to form well-organized structures on the nanotube surface depending on the nanotube’s diameter and the bound proteins (Figure 7).52,123 CNT-bound proteins generally form multiple layers on the nanotube surface.124 This property can be used for nanotube debundling, stabilization, separation and protein sensing.115,120,125-127 ENREF 40 The bound proteins often undergo changes in structure and activity. Molecular simulation studies have demonstrated that proteins undergo stepwise conformational changes after binding to CNTs.128-130 For example, both soybean peroxidase and α-chymotrypsin bind to SWNTs; soybean peroxidase retains 30% activity, while α-chymotrypsin retains only 1% activity. This difference in activity occurs because soybean peroxidase partially maintains its secondary structure after binding, while α-chymotrypsin completely loses its secondary structure.118 CNTs enhance the rate of protein fibrillation by decreasing the lag time preceding nucleation, indicating the potential of CNTs to induce amyloid formation, as well as the assembly of novel nanoparticles.131
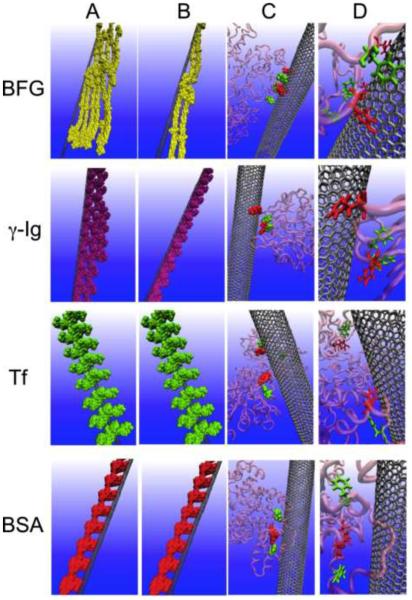
Figure 7.
Interactions between bovine fibrinogen, immunoglobulin, transferrin, bovine serum albumin and SWNTs. Molecular modeling illustrations of proteins (in bead representation) binding to SWNTs after incubation for 10 min (A) and 5 h (B). (C) The locations of the most preferred binding sites on proteins for SWNTs. The residues highlighted in the van der Waals representation correspond to tyrosine (red) and phenylalanine (green). The other regions of the protein are presented in pink. (D) The detailed orientations of the aromatic rings of the tyrosine and phenylalanine residues interacting with the six-membered rings of SWNTs are represented in silver. The tyrosine residues are rendered as licorice representations (red), while the phenylalanine residues are shown in green. Adapted from ref 52 with permission. Copyright 2011 National Academy of Sciences, USA.
The interactions of C60 with proteins have been studied using biochemical, structural and molecular simulation approaches.132-137 The possible chemical interactions responsible for C60-protein binding events include hydrophobicity, surface curvature, π-π stacking, uneven charge distribution, site fit and solvent displacement.135 Molecular dynamic studies have demonstrated that interactions such as hydrophobicity and π-π stacking represent the dominant factors.138 The binding site of C60 on its antibody has been identified using X-ray crystallography. C60 binds to the interface of the antibody light and heavy chains.139 The binding sites of C60 in human and bovine serum albumin (BSA) and HIV-protease are quite similar, but are different from that in the C60 antibody.132 Amino acid-modified C60s can penetrate the lipid membrane, bind to the hydrophobic domains of proteins and alter the function of membrane-bound enzymes.140
Graphene is another hexagonal carbon nanostructure. In contrast to CNTs and fullerenes, graphene consists of planar atomic thin layers.2,141 Because of the sp2 carbon structure, the driving forces for interactions between proteins and graphenes are mainly hydrophobicity and π-π stacking. Molecular dynamics simulation predicted that human insulin can be adsorbed onto graphene surfaces and that some proteins would be destroyed or partially destroyed.142 Water-soluble graphene oxide nanosheets (GO) are oxidation products of graphene nanosheets. GOs have a stronger serum protein adsorption capability than SWNT or MWNT. The thickness of the GO layer increased from ~1.0 nm to 4.0-18.0 nm after protein binding.143 The function of proteins adsorbed on the GO surface can be inhibited (e.g., the enzymatic activity of chymotrypsin144). The GO-protein binding equilibrium can be reached in 30 minutes, and protein adsorption mitigates the cytotoxicity of GO.143
In contrast to sp2 nanostructures, diamond nanoparticles consist exclusively of sp3 bonds. These nanoparticles bind protein molecules through hydrophobic interactions and physical adsorption and are used in tissue engineering (e.g., immobilization of bone morphogenetic protein 2 (BMP-2) to promote bone formation145) or as mass spectrometric matrices.146
3.2.2.2. GNPs
GNPs most likely interact with proteins through electrostatic, hydrophobic, and sulfur-gold interactions.23,147-151 GNPs with citrate as a stabilizing agent can bind to proteins according to the Langmuir isotherm.151,152 The size of nanoparticles influences the mechanism of protein binding. One study demonstrated that electrostatic interaction is the major binding force for nanoparticles with a diameter of 16 nm, while hydrophobic interaction is the major force for nanoparticles with a diameter of 2-4 nm.148 Because the gold surface forms strong bonds with thiol groups in cysteine residues, the GNPs bind BSA and heparin-binding growth factors.151,153 ENREF 4 The latter activity may be used to inhibit tumor angiogenesis.154 The perturbation of the conformation and activity of bound proteins is influenced by the size and surface properties of the nanoparticles, the characteristics of the protein and the ionic strength and pH of the solution.148,155-158 Cytochrome C is adsorbed onto anionic nanoparticles followed by conformational changes and proteolysis.147 Chymotrypsin binds to GNPs and undergoes a two-stage change: fast, reversible inhibition, followed by slow, irreversible inactivation.150 The nanoparticle surface chemistry can regulate GNP-protein interactions.155,159 Amino acid-modified GNPs can also tune the enzymatic activity of bound α-chymotrypsin.159 The nanoparticle size influences the denaturation kinetics of BSA on GNPs.157 Negatively charged poly(acrylic acid)-conjugated GNPs bind to and induce the unfolding of fibrinogen. The unfolded fibrinogen promotes MAC-1 receptor activation and inflammation.160
3.2.2.3. Iron Oxide Nanoparticles
The adsorption of BSA molecules onto magnetic iron oxide nanoparticles fits well to the Langmuir isotherm.161 The adsorption kinetics fit to a linear driving force mass-transfer model. Similar to other nanoparticles, iron oxide nanoparticles also induce protein conformational changes and alter protein activity. Human transferrin molecules are adsorbed onto the surface of superparamagnetic iron oxide nanoparticles and undergo conformational changes from a compact to an open structure. This transformation induces the release of iron.162
3.2.2.4. Quantum Dots (QDs)
Electrostatic interactions, sulfur-metal bond formation and spatial effects are believed to be major driving forces for QD-protein binding.163-165 CdS nanoparticles are trapped in the cylindrical cavity of chaperonin proteins and become thermally stable and tolerant to electrolytes.163 The binding between QDs and HSA occurs near a protein pocket centered at Lys199.164 Calculations reveal negative enthalpy (ΔH) and positive entropy (ΔS) values for this interaction, indicating that electrostatic interactions play a major role in the binding reaction. The stoichiometry of bound protein molecules on each QD is approximately 6. The bound proteins undergo substantial secondary and tertiary structural changes.164 In another study, the electrostatic adsorption of hemoglobin (Hb) to CdS nanoparticles was shown to be energetically favorable. Raman spectroscopic results indicated that the sulfur atoms of the cysteine residues form direct chemical bonds with the CdS QD surface. Although higher structures are disturbed, the spin state of the heme iron is not affected.166 In addition to native proteins, denatured BSA molecules also bind to the surface of CdTe QDs and form a shell-like complex.167 The protein shell structure results in the removal of Te atoms from the CdTe surface and the formation of a thermodynamically stable nanostructure. Similar to CNTs, QDs also enhance protein fibrillation.131
3.2.2.5. Silica Nanoparticles
Electrostatic interactions may also be a driving force for protein adsorption by silica nanoparticles, as determined by DSC.168 A decrease in ionic strength is accompanied by a reduction in protein binding enthalpy, indicating that there is a strong electrostatic attraction in silica nanoparticle/lysozyme and silica nanoparticle/RNase binding.168 Hydrophobic interactions and hydrogen bonding are also driving forces in silica nanoparticle-peptide binding.22 The bound proteins on silica nanoparticles assume a specific orientation. The adsorption of human carbonic anhydrase II onto negatively charged silica nanoparticles appears to be specific to limited regions at the N-terminal domain of the protein.169 In this study, the orientation of the bound proteins is also pH-dependent. At pH 6.3, a histidine-rich area around residue 10 is the dominant binding region. At higher pH values (e.g., pH 9.3), the protein is adsorbed near a region close to residue 37, which contains several lysine and arginine residues. This absorption behavior indicates that specific binding may be a result of electrostatic interactions between the positively charged areas on the protein surface and the negatively charged silica surface. The binding of protein onto nanoparticles occurs in two steps: the rapid formation of a protein-particle complex, followed by an irreversible protein conformational change.170 Nanoparticle-induced protein conformational changes vary depending on the nature of proteins. Unlike the “hard” (very stable) protein carbonic anhydrase II, the “soft” (less stable) protein carbonic anhydrase I undergoes a larger structural rearrangement when adsorbed onto silica nanoparticles, as shown by NMR.171 The size of silica nanoparticles determines the surface curvature, which influences protein adsorption and the stability of the adsorbed proteins. For instance, larger nanoparticles cause a greater loss of α helicity and enzymatic activity in bound lysozyme 172 and induce a greater decrease in the thermodynamic stability of RNase A.173 In another study, silica nanoparticles were shown to promote α-helical structures in a designed peptide of low helical content in solution.174 Consequently, the protein conformational changes altered their cell binding capability and reduced proliferation of human cells.175
3.2.2.6. Polymeric Nanoparticles
The adsorption of proteins onto polymeric nanoparticles also depends on the hydrophobicity and surface curvature of nanoparticles. The interactions of proteins with N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles with different monomer ratios and particle sizes demonstrate that proteins tend to bind to more hydrophobic nanoparticles by forming a single layer on the surface.176 Surface chemistry also influences protein binding. For instance, in dextran phenoxy/poly(ethylene oxide) copolymer-modified polystyrene latex nanoparticles, BSA can deform and penetrate the poly(ethylene oxide) layer to bind to the nanoparticle surface. In contrast, a densely packed dextran phenoxy layer prevents nonspecific adsorption.177 Similar to CNTs and QDs, polymer nanoparticles also promote protein fibrillation by enhancing the nucleation of protein fibrils from human β2-microglobulin.131 The promotion of fibrillation may be a common property for a large number of nanoparticles. Another study concluded that the first layer of bound proteins is much more stable than the secondary layer on the nanoparticle surface. The first layer was hence called the “hard corona”, indicating nearly irreversible binding, and the secondary layer was termed the “soft corona”, indicating greater dynamic exchange with the surrounding milieu.178
The above studies demonstrate that protein binding is a nature of all nanoparticles, regardless of material. The material origin, size, shape and surface chemistry of nanoparticles determine the chemical or physical forces involved in the interactions of nanoparticles with proteins. These interactions can be modified through adjusting the physicochemical properties of the nanoparticles.
3.2.3. Interactions Between Nanoparticles and Proteomes
Although the elucidation of the interactions between nanoparticles and single proteins facilitates an understanding of the chemical nature of such interactions, in biological systems, nanoparticles encounter proteomes, not a single protein. Blood and cellular proteomes consist of thousands of proteins with diverse functions, sequences, molecular weights, isoelectric points, surface hydrophobicities and concentrations. The protein corona composition is determined by the thermodynamic and kinetic properties of the interactions between the nanoparticles and proteins.108 The selectivity in protein binding is influenced by factors such as the size and surface chemistry of the nanoparticles and the properties of the proteins.179-182
The binding between polymer nanoparticles and plasma proteins is highly dynamic. Abundant yet low-affinity HSA molecules quickly bind to the surface of nanoparticles and are soon replaced by the high-affinity and slow-exchanging apolipoproteins AI, AII, AIV and E. These proteins remain associated with the particles.108,182,183 Although the formation of a protein monolayer is an acceptable model, nanoparticles can also attract additional layers of proteins. By analyzing the adsorption of fetal bovine serum proteins onto iron oxide nanoparticles by gel electrophoresis and LC-MS/MS, proteins that strongly bind to the surface of nanoparticles were identified. These proteins include complement factor H, antithrombin, complement factor I, α-1-antiproteinase and apolipoprotein E. These surface-bound proteins serve as linkers to further bind other serum proteins, such as BSA molecules, which adsorb with lower affinity onto iron oxide nanoparticles.184
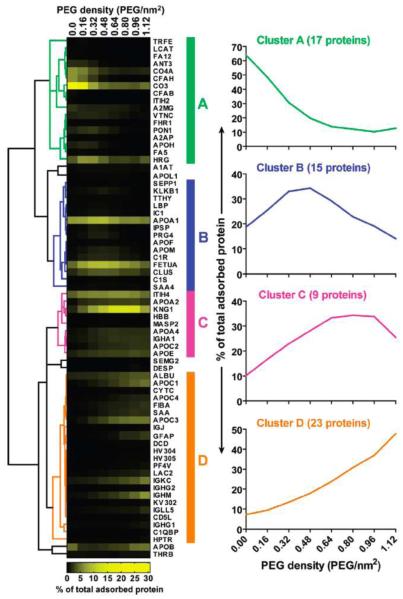
Like single-protein bindings, proteome binding by nanoparticles is strongly affected by the surface chemistry of nanoparticles. Low-density lipoprotein, very-low-density lipoprotein and high-density lipoprotein bind strongly to QDs with cholesterol and atheronal-B modifications when compared to amine-modified QDs.179 Polyethylene glycol (PEG)-modified nanoparticles reduce protein binding. PEG chain length and density on polymeric nanoparticle surfaces regulate the amount of human plasma protein adsorption but not the binding pattern. The main bound proteins identified by 2D polyacrylamide gel electrophoresis analyses are albumin, fibrinogen, IgG, Ig light chains and the apolipoproteins AI and E. The reduced protein binding on nanoparticles coincides with decreased nanoparticle internalization by cells.180 Stepwise increases in PEG density on nanoparticle surfaces shift the serum protein binding pattern in addition to the binding amount. (Figure 8) An increase in PEG density reduces the internalization of GNPs into cells, similar to polymeric nanoparticles.181 ENREF 54 Nanocapsules with longer PEG chains and higher PEG density also reduce complement system activation and uptake into J774A1 macrophage-like cells.185 The polysaccharide modifications on the surface of poly-(isobutylcyanoacrylate) nanoparticles reduce plasma protein binding and complement system activation.186 Magnetic iron oxide nanoparticles with different sizes and surface chemistries bind different serum proteins without changing the protein secondary structure.187 The PEG coating on these nanoparticles reduces both cell uptake and cytotoxicity. Smaller PEG-coated nanoparticles bind relatively more serum proteins and display reduced cellular uptake. The composition of cell culture media also affects protein adsorption and cellular effects. Using GNPs as a model, Dulbecco's modified Eagle’s medium elicited the formation of a protein corona, while the use of Roswell Park Memorial Institute medium resulted in different dynamics with reduced protein coating. Protein-nanoparticle complexes formed in the latter medium are more abundantly internalized by cells and cause greater cytotoxicity than those formed in the former medium.188 The heat inactivation of serum affects protein corona formation and cellular uptake of protein-nanoparticles complexes because heating induces changes in serum composition and protein conformation.189
Figure 8.
PEG density-based determination of the composition of the adsorbed serum protein corona on the surface of GNPs. Adsorbed serum proteins were isolated from 15-nm GNPs grafted with PEG at varying densities, purified, digested with trypsin and characterized by LC-MS/MS. Proteins were clustered into one of four groups: A, B, C or D (represented by the colored bars), based on the correlation of their relative abundance across PEG densities. PEG/nm2: number of PEG molecules per square nanometer. Reprinted from ref 181 with permission. Copyright 2012 American Chemical Society.
3.3. Nanoparticle-DNA Interactions
Nanoparticles bind nucleic acids (e.g., DNA) in biological systems. The features of intermolecular forces involved in DNA binding are similar to those involved in nanoparticle-protein binding. Such interactions can be exploited for biomedical applications, including DNA loading on nanoparticles for gene transfer and siRNA delivery. Nanoparticle-DNA interactions depend on the properties of both the DNA molecules and nanoparticles.
3.3.1. Determination of Nanoparticle-DNA Interactions
Nanoparticle-DNA interactions can be detected through determining complex formation, binding-induced DNA conformational changes and DNA degradation.
The binding of DNA molecules to nanoparticles alters the properties of both components. For example, binding alters nanoparticle dispersability and surface charge. Such changes can be detected with DLS and ζ-potential measurements.53,190,191 The nanoparticle-DNA complexes have altered electron absorption spectroscopic patterns, which can also be measured using UV-Vis spectroscopy.192-194 DNA binding may increase the near-IR fluorescence of some nanoparticles. This increase can be used to monitor the binding process.190 DNA molecules, when bound to nanoparticles, can be detected using XPS.20,195 By enhancing adenine and guanine ring breathing vibrations, surface-enhanced Raman spectroscopy (SERS) has been used to monitor DNA binding onto GNPs.196 AFM detects subnanometer scale phenomena and is used for the ultrastructural observation of nanoparticle-DNA complexes.190,193,197-199 Linear dichroism monitors the differential absorption of polarized light parallel and perpendicular to the orientation direction and enables the determination of the approximate orientations of the DNA molecules on CNTs.200,201
Fluorescence spectroscopy can be employed to determine DNA/nanoparticle binding affinity. Because of the weak intrinsic fluorescence emission of DNA, ethidium bromide (EB) or the low toxicity dye SYBR® Green is used to label DNA molecules.53,202 The resonance light scattering method has been used to detect metal ions and biomolecules.203,204 Resonance light scattering from nanoparticles and DNA molecules is weak, but the scattering intensity is enhanced dramatically when nanoparticles and DNA bind to each other, permitting the quantification of nanoparticle-DNA interactions.191,205 Real-time polymerase chain reaction (RT-PCR) can be used to amplify short DNA fragments attached to nanoparticles to quantify their surface coverage.206 The adsorption and desorption processes of nanoparticle-DNA complexes can also be measured using electrochemical approaches, such as cyclic voltammetry.207 In some cases, the nanoparticle-DNA complexes can be isolated by anion exchange chromatography.193
Circular dichroism (CD) can be used to detect DNA conformational changes induced by nanoparticles.53,191,197,202,208-213 B-A and B-Z transitions can be identified by the appearance of characteristic CD bands.197,211 NMR provides more detailed structural information than CD.208 Conformational information can also be obtained by surface-enhanced infrared spectroscopy, which measures the vibrational modes of DNA molecules.214 The structural stability of DNA can also be measured by its melting curve.53,197,206,208,210,212 DNA degradation can be evaluated by gel electrophoresis.198,201,208,209,215-217 In addition to experimental approaches, molecular simulations can predict nanoparticle-DNA binding patterns and nanoparticle-induced DNA conformational changes.218,219
3.3.2. Current Understanding of Nanoparticle-DNA Interactions
3.3.2.1. Carbon Nanoparticles
Due to the large surface area of CNTs, DNA molecules bind to the surfaces of nanoparticles almost spontaneously.218 This binding enhances the aqueous solubility of CNTs.193,220 Due to the unique shape and electronic properties of CNTs, the interaction modes between DNA and CNTs are different than those between DNA and other nanoparticles. DNA molecules can adsorb and wrap onto the nanotube through π-π stacking and hydrophobic interactions or even insertion into the central cavity.190,199,218,219 Such interactions also cause conformational changes in DNA molecules.190,197,208,211,214
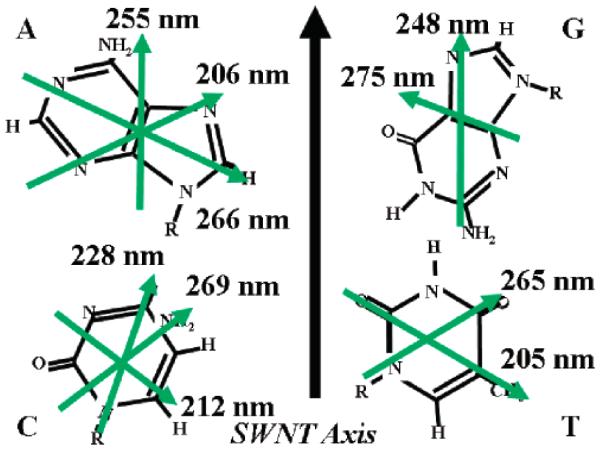
Both experimental evidence and molecular simulation indicate that DNA molecules form a helical structure before wrapping up CNTs.193,199 Optical absorbance measurements have revealed that anisotropic hypochromicity results from single-strand DNA/CNT interactions through π-π stacking interactions.194 Due to π-π stacking interactions, DNA bases tend to orient themselves (Figure 9) facing the tube, as determined by investigating the interactions between CNTs and DNA homopolymers.194 Although one study demonstrated that binding of CNTs with DNA is independent of DNA sequence,198 the sorting and separation of CNTs by DNA has been shown to be affected by DNA sequence.199 The separation of CNTs by d(GT)n (n = 10~45) is more efficient than separation by other sequences present in a single-strand DNA library. DNA molecules with GT-rich sequences wrap individual nanotubes more effectively and form a helical structure.199 Linear dichroism revealed that the DNA molecules lay flat on the nanotube surface with the backbone wrapping around the nanotube at an oblique angle of 45°.200
Figure 9.
Orientation of the bound DNA bases relative to the SWNT axis. The green arrows and wavelengths indicate the directions and wavelengths of optical dipole transitions, respectively. Reprinted with permission from ref 194. Copyright 2007 American Chemical Society.
DNA/CNT binding induces DNA conformational changes. Binding of SWNTs to DNA induces B-A and B-Z (Figure 10) transitions.197,211,214,218 The B-Z transition modulates the dielectric environment of the SWNTs and decreases their near-IR emission. These changes have been detected in whole blood, tissue and living cells.211 SWNTs inhibit DNA duplex association and induce telomeric i-motif formation by binding to the 5’-end major groove.208 In addition to CNT-induced DNA conformational changes, the binding of SWNTs affects DNA hybridization. DNA molecules adsorbed to SWNTs display much slower hybridization kinetics than free DNA.190 Pre-adsorption on the SWNT surface causes a decrease in the free energy of the DNA, which increases the energy required for the conversion of the DNA strands into the transition state. The process can be fitted well with a two-step Langmuir model.190 The binding of DNA to CNTs can also alter the optical properties of SWNTs. Racemic SWNTs exhibit circular dichroism only when wrapped by DNA molecules. This induced circular dichroism occurs when transition dipole moments of optically active electronic transitions in the DNA molecule are coupled to transition dipole moments of the SWNTs.213 The altered CD spectra can be used to characterize DNA-SWNT wrapping.
Figure 10.
Illustration of DNA undergoing a conformational transition from B form (top) to Z form (bottom) on a carbon nanotube. Reprinted with permission from ref 211. Copyright 2006 American Association for the Advancement of Science.
Other carbon nanoparticles also exhibit strong interactions with DNA molecules. For example, graphene oxide nanosheets and fullerene can cleave DNA strands when copper ions or visible light is present.209,217 The magnitudes of the binding energy of bases with graphene are similar to those found in SWNTs. Interactions are also driven by hydrophobic interactions and π-π stacking.221,222 The binding of a DNA duplex to graphene oxide causes a partial deformation of the double helix.223 Therefore, a high binding affinity to DNA molecules may be a common feature of carbon-based nanomaterials.
3.3.2.2. GNPs
Interactions between GNPs and DNA molecules are mainly driven by electrostatic interactions.192,216,224 The formation of a complex between DNA and tiopronin-modified GNPs occurs in three steps:53 the first step is the diffusion-controlled formation of an external adduct; the second step is the formation of DNA-GNP complex I based on their binding affinity; and the third step is the formation of a more compact DNA-GNP complex II after DNA conformational changes. DNA-GNP interactions are influenced by the properties of the DNA and nanoparticles and the microenvironment.202,225 Small Au55 clusters (size ~1.8 nm) attach to the major grooves of B-DNA and cause a B-A transition. Au55 nanoclusters are then degraded to 13-atom nanoclusters due to shrinkage of the major grooves. Au13 nanoclusters and A-DNA ultimately form a wire-like structure.24 In another study, GNPs with a diameter of 4.4 nm and coated with trimethyl(mercaptoundecyl)ammonium monolayer assembled into a one-dimensional (1D) chain along a DNA strand through electrostatic interactions between the cationic nanoparticle surface and anionic DNA bases.192 The surface coverage of thiol-capped oligonucleotides bound to GNPs decreases from 30 pmol/cm3 to 13 pmol/cm3 when the nanoparticle diameter is increased from 13 nm to 30 nm.206 In addition to nanoparticle size, nanoparticle surface chemistry, salt concentration and sonication affect DNA/GNP interactions.225
In addition to GNP-induced DNA conformational changes, nanoparticles also induce other changes in DNA molecules. Nanoparticles promote the relaxation of supercoiled DNA under X-ray radiation,226 inhibit hybridization and transcription,20,227 induce double strand separation228 and enhance Raman signals from double-stranded DNA molecules.196 These altered properties can be employed for DNA detection,229 protection of DNA from DNAase I digestion,230 inhibition of the amplification of mismatched primer-template pairs, and enhancement of the specificity of allele-specific PCR.231 Interactions between DNA and GNP can also be used to build novel nanostructures. For example, the hybridization of complementary DNA is used to construct discrete GNP-QD nanostructures.232 Organized DNA motifs are used to make ordered GNP arrays.233
3.3.2.3. Other Nanoparticles
Other nanoparticles also interact with DNA molecules through electrostatic and hydrophobic interactions. Linear QD chains are formed along double-stranded DNA through electrostatic interactions.234 Plasmid DNA bound to the QD surface can be released by intracellular glutathione (GSH) during transfection.195 Calf thymus DNA molecules are adsorbed onto zirconia nanoparticles.207 Titanium oxide nanoparticles are widely used in sunscreen products, and their potential toxicity has received considerable attention.235 These nanoparticles bind to DNA molecules and cause DNA degradation due to the generation of hydroxyl radicals on the nanoparticle surfaces.236,237 This DNA damages induce oxidative stress in living cells and animals.238 Similar to titanium oxide nanoparticles, copper nanoparticles cause DNA degradation through the generation of singlet oxygen.239 Ag and Pt nanoparticles inhibit DNA hybridization by disrupting hydrogen bonding between DNA double strands. This inhibition is weakened as the nanoparticle size increases.20 Binding of calf thymus DNA to Ag nanoparticles induces conformational changes in DNA molecules.210
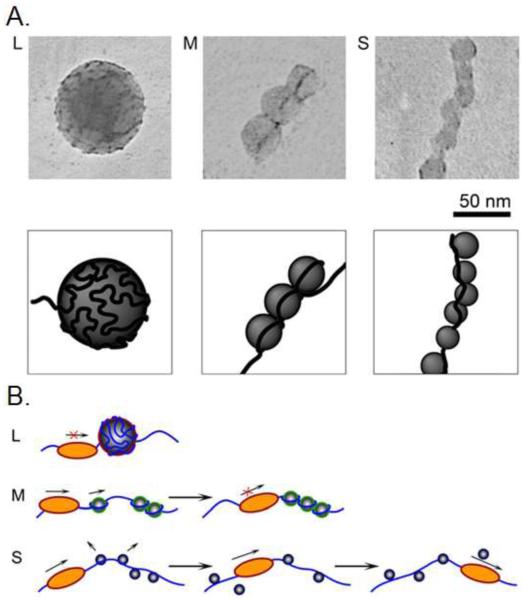
Both cationic and anionic polymer nanoparticles bind DNA molecules and cause DNA conformational changes.191,212 Cationic poly-L-lysine-modified silica nanoparticles inhibit DNA transcription, and this inhibition is size-dependent (Figure 11). DNA chains are adsorbed irreversibly to the surface of nanoparticles (40 nm), inhibiting polymerase function. However, smaller nanoparticles (10 nm) form chains along DNA strands and might eventually dissociate from the DNA strand. This allows the DNA molecules to become accessible to the polymerase, and transcription proceeds.240
Figure 11.
TEM of T4 DNA complexes with L (large, 40 nm), M (medium, 15 nm) and S (small, 10 nm) nanoparticles and schematic representations of the complexes. Reprinted with permission from ref 240. Copyright 2007 Cell Press.
The interactions between nanoparticles and DNA molecules share some common features with nanoparticle-protein interactions. In both cases, biomolecules are adsorbed to the nanoparticle surface, and the binding induces conformational changes in these molecules. Nanoparticle-DNA interactions can also be used to develop advanced nanoparticle-DNA hybrids to control cellular processes for biomedical and biosensing purposes.
3.4. Adsorption of Crucial Small Molecules by Nanoparticles
Due to their high surface energy, nanoparticles also adsorb various small molecules in biological systems. These molecules include but are not limited to carbohydrates, vitamins, hormones, amino acids and nucleic acid bases.54,241-245 These small molecules normally play essential roles in cell signaling and cell physiology. A sudden depletion of these molecules may influence proper cellular functions and cause toxicity.54,244,246 Small molecules are adsorbed onto nanoparticles through molecular interactions, such as hydrophobic, π-π stacking and electrostatic interactions.54,247 Although the chemical bases of nanoparticle/protein interactions and nanoparticle/amino acids interactions are similar, the latter is simpler, faster and does not involve conformational alterations.
SWNTs adsorb various amino acids (e.g., Arg, His, Met, Phe and Tyr), vitamins (folate, riboflavin and thiamine) and phenol red in cell culture media.54 An analysis of the structures of the adsorbed molecules indicates that π-π interactions and electrostatic interactions are likely the driving forces for adsorption. The SWNT-induced depletion of amino acids and vitamins in cell culture medium reduces cell viability. Spectroscopic studies have revealed that both SWNTs and MWNTs affect the cell culture medium in this manner.244,248 A study of the adsorption of folic acid and vitamin B1 by carbon fibers with modified nanopores revealed that the pore structure and surface modifications influenced the adsorption capacity and adsorption rate.245
Computational chemistry reveals that the curvature of nanoparticles and the polarizability of amino acids determine binding strength.247 Doping the carbon surface with calcium atoms results in a dramatic enhancement of binding of collagen amino acids (Gly, Pro and Hyp) to graphene. Electronic charge transfers from the calcium atoms (donor) to graphene (acceptor) and the carboxyl group of the amino acid (acceptor) may contribute to this enhancement.249
4. NANOPARTICLE-CELL INTERACTIONS
The cell is the elemental unit of all living organisms. Nanoparticles perturb living systems by interacting and altering live cells. Such perturbations occur at various levels. Nanoparticles are first adsorbed to the cell surface and are then internalized. As the nanoparticles are endocytosed and transported within cells, the cells undergo structural and functional changes. Some crucial processes, such as oxidative balance, are perturbed. Eventually, global alterations occur that involve cell signaling, genomic, proteomic and metabonomic processes.
4.1. Cellular Uptake and Intracellular Transport of Nanoparticles
Because of their sizes and surface properties, nanoparticles can enter various organisms and cells, including mammalian cells, plant cells, bacteria, fungi and viruses.250-254 These processes can be energy-dependent intake or energy-independent insertion.255,256 The former is also referred to as endocytosis, a process by which cells take up molecules or particles by engulfing them.257 Nanoparticles enter cells by taking advantage of various known endocytotic pathways, such as clathrin-mediated endocytosis, caveolae-mediated endocytosis, phagocytosis and macropinocytosis.256,258-261 Direct penetrations occur for 1D nanostructures or strongly positively charged nanoparticles.255,262 After entering cells, nanoparticles may localize in endosomes, lysosomes, the cytoplasm, mitochondria, the endoplasmic reticulum or the nucleus, depending on the nature of the nanoparticle.256,258-261,263-266 Various imaging techniques have been developed to monitor the cell uptake process, in addition to conventional TEM and confocal laser scanning microscopy (CLSM) analyses.267,268 For example, X-ray fluorescence microscopy is used to determine chemical element distribution of nanoparticles in cell.269 Magneto-photo-acoustic imaging permits the differentiation of membrane-adhered or endocytosed nanoparticles.270 Dynamic colocalization microscopy permits the spatio-temporal characterization of internalized nanoparticles.271 Raman spectral imaging maps vibrational bands of nanoparticles in live cells in the absence of an external label.272 Atomic force microscopy (AFM) measures the force between nanoparticles and the cell surface, indicating receptor binding and binding strength.273-275
4.1.1. Kinetics and Thermodynamics of the Cellular Uptake of Nanoparticles
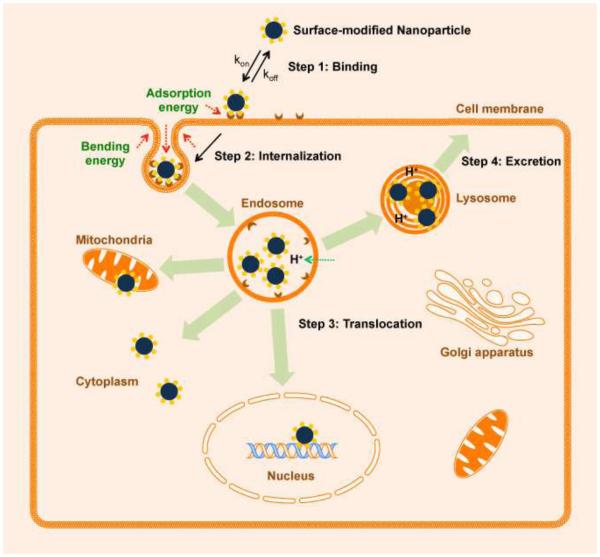
The endocytosis of nanoparticles is a dynamic process, and the endocytosis and exocytosis processes eventually reach thermodynamic equilibrium. Endocytosis likely involves at least two steps: binding of the nanoparticles to the cell surface, followed by the internalization of the nanoparticles into cells (Figure 12).276 Binding parameters derived from the binding of anionic magnetic nanoparticles to RAW macrophages and HeLa cells reveal that macrophages and tumor cells behave equally with respect to cell surface binding, although they differ with respect to internalization capability. The adsorption onto the cell surface is a slow step, and internalization is fast.277-279 Nanoparticles are eventually exocytosed from living cells. The exocytosis rate of SWNTs is reported to closely match their endocytosis rate with negligible temporal offset.280 However, the uptake of dye-labeled polystyrene nanoparticles is reported to increase with time and be essentially irreversible. The fluorescence signal decreases when cells divide.268 This controversy suggests that cell uptake kinetics may depend on both nanoparticle properties and cell type. The endocytosis of nanoparticles is also under thermodynamic control. Thermodynamic studies have revealed that adhesion strength governs receptor-mediated cell uptake. Endocytosis is determined by the enthalpic and entropic limits of the adhesion strength.281 Based on the energy requirements for a receptor-mediated uptake process, the optimal size of nanoparticles is 25~30 nm.282 When the nanoparticles are too small (less than 20 nm), endocytosis is limited because the adhesion energy is too low to compensate for the bending energy of the cell membrane. When the nanoparticles are too large (greater than 60 nm), the free receptors on the cell membrane are depleted, and nanoparticles may be only partially wrapped without endocytosis. Here, size refers to the apparent size after aggregation. A further understanding of the kinetics and thermodynamics of the cellular uptake of nanoparticles will assist future nanoparticle design and applications.
Figure 12.
Receptor-mediated cellular uptake and translocation of nanoparticles.
4.1.2. Determinants of the Cellular Uptake of Nanoparticles
Many physicochemical and biological factors govern the cellular uptake of nanoparticles. These factors include the size, shape, surface chemistry, surface charge and mechanical properties of nanoparticles, as well as cell type. The combination of these factors affects the cell surface binding strength,273,275 cellular entrance route,283,284 uptake amount278,285-290 and intracellular distribution.283,291,292 SWNTs with lengths of 100-200 nm can be internalized into cells through clathrin-coated pits. SWNTs with lengths of 50-100 nm enter cells through caveolae-mediated endocytosis, and some can enter the nucleus. The shortest (less than 50 nm) SWNTs can enter cells through direct insertion, which is energy independent.283 Studies of GNPs (4-17 nm) in HeLa cells have revealed that the uptake force increases with the size of the nanoparticles.275 The shape of the nanoparticle may also play a role. Gold nanospheres enter fibroblasts and tumor cells more efficiently than nanorods,293-295 while nanorods are internalized more efficiently than nanospheres in blood phagocytes.296 Particle replication in non-wetting template (PRINT) nanoparticles with a higher aspect ratio enter HeLa cells more readily than those with a lower aspect ratio.297 The surface charge on nanoparticles can play a significant role in cellular uptake. Due to the presence of phosphate groups on lipids, cell membranes are negatively charged. Nanoparticles with positive charges display strong interactions with cells compared with those with negatively charged or neutral surfaces.292,298
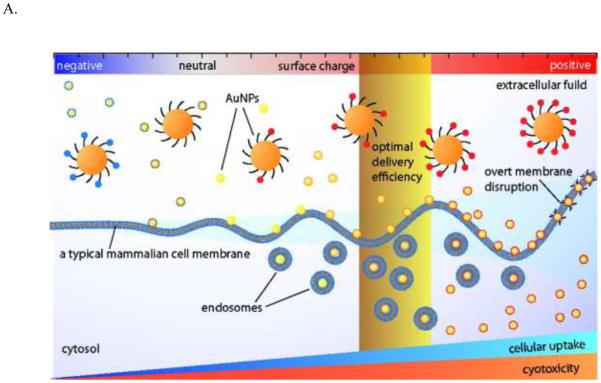
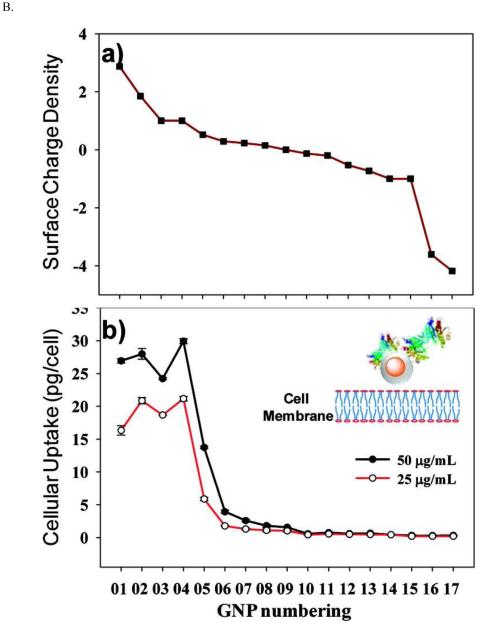
Furthermore, the structure, density and distribution of surface chemical groups all play significant roles in determining the cellular uptake of nanoparticles. For peptide-functionalized GNPs, an aromatic structure at the end of the peptide enhances their cellular uptake.290 A higher density of oligonucleotide loading on the surface of GNPs generates higher cellular uptake, while a higher density of PEG reduces GNP uptake.181,287 GNPs that have striations on the surface with alternating anionic and hydrophobic groups can penetrate the plasma membrane without disrupting it.299 The nanoparticle surface ligand induces protein corona misfolding and therefore indirectly enhances cellular uptake.300 A molecular simulation study has proposed that the effective surface charge density determines cell uptake.301 An experimental study of a surface-modified GNP array indicated that the effective surface charge density determines the electrostatic interactions between the positively charged nanoparticles and the negatively charged cell membrane (Figure 13).302
Figure 13.
(A) A schematic illustration of the effect of GNP surface charge on their cellular uptake by a typical mammalian cell. (B) The electrostatic attraction between nanoparticles and cells. (a) The surface charge density of a GNP library. (b) Cellular uptake of these GNPs in HeLa cells, as determined by ICP-MS. Reprinted with permission from ref 301 and 302. Copyright 2010 & 2012 American Chemical Society.
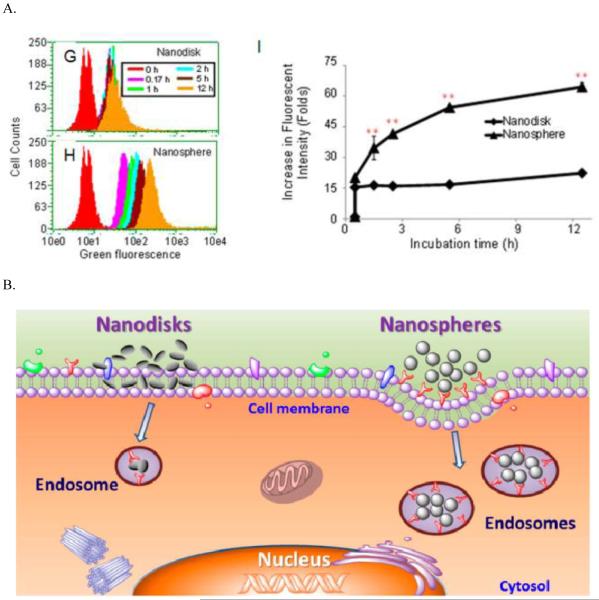
Shape is another significant factor in the cellular uptake process. For 1D nanotubes, the uptake process is spontaneous when the orientation favors insertion, while 2D nanosheets prefer to adhere to the cell surface.255,303 A nanosphere and nanodisk cell uptake comparison study demonstrated that nanodisks mainly adhere to the cell surface, while nanosphere are easily internalized into cells (Figure 14).25 A similar conclusion was reached in a subsequent study. Nonspherical polymeric nanoparticles exhibit reduced cellular uptake compared to their spherical counterparts. The larger average curvature radius of adsorbed non-spherical particles may be responsible for this difference.304 However, 1D superparamagnetic iron oxide nanoworms showed enhanced tumor-targeting efficiency compared to spherical nanoworms.305 This controversy indicates effects of nanoparticle shape on cell uptake are material-dependent.
Figure 14.
Changing polystyrene nanoparticles from a three-dimensional (3D) spherical shape to a 2D disk shape promotes their cell surface binding, with significant reduction of cell uptake. (A) Experimental evidence of a nanodisk reducing cellular uptake. (B) The model for reduced cellular uptake but enhanced cell surface binding by nanodisks. Reprinted from ref 25 with permission. Copyright 2012 American Chemical Society.
The mechanical properties of nanoparticles also influence the rate, route and amount of cellular uptake.306,307 Soft nanoparticles enter cells through macropinocytosis, while stiff nanoparticles enter through clathrin-mediated endocytosis. Nanoparticles with an intermediate stiffness have multiple internalization routes.306 The aggregation and sedimentation of nanoparticles in physiological environments also impacts cellular uptake. More nanoparticles are taken up in an upright cell culture configuration than in an inverted one, and nanoparticles with faster sedimentation rates display greater differences in uptake between the two configurations, suggesting that sedimentation needs to be considered for cellular uptake studies of heavy or large nanoparticles.278,308,309 The different protein binding properties of nanoparticles often indicate different nanoparticle-cell interactions.287,293,309 The pericellular matrix enhances the retention and cellular uptake of nanoparticles, due to the restriction and deceleration of diffusion by the entrapment and accumulation of nanoparticles.310 In summary, multiple factors collectively determine the cellular uptake of nanoparticles and their subsequent bioactivity in cells.
4.2. Cellular Substructural and Functional Alterations by Nanoparticles
Nanoparticles are translocated to different compartments in live cells and may interact with relevant subcellular structures and affect their function. Cell membranes, ion channels, the cytoskeleton, mitochondria and the nucleus have all been reported to be affected by nanoparticles.
4.2.1. Nanoparticle-Induced Cell Membrane Disruption
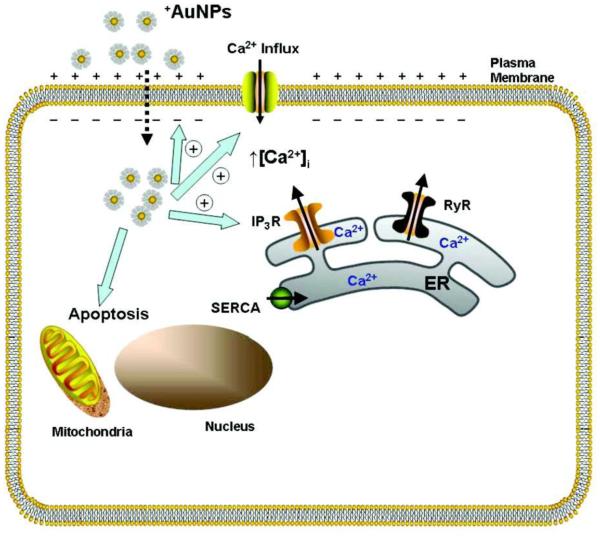
The cellular uptake of nanoparticles results in the disruption of the plasma membrane. Membrane disruption likely involves several physicochemical interactions. Electrostatic interactions of positively charged nanoparticles with negatively charged groups on the membrane surface alter cell membranes. Membrane penetration of positively charged nanoparticles into the cytoplasm causes the formation of holes within 1~100 ms, followed by a slow resealing in tens of seconds.311 Interactions between positively charged nanoparticles with negatively charged membrane also causes membrane depolarization and perturbation of membrane potential. Furthermore, an increase in [Ca2+] influx inhibits cellular proliferation.312 Membrane disruption of red blood cells and perturbation of their hemolytic activity by amorphous and mesoporous silica nanoparticles have been attributed to silanol groups that are accessible to the membrane.313 Nanoparticle-induced generation of ROS may also cause membrane damage. A water-soluble fullerene suspension disrupts cellular functions through lipid peroxidation by ROS. Upon addition of an antioxidant, the oxidative damage can be completely prevented.314 Nanoparticle shape may also serve a role in membrane perturbation. Needle-shaped nanoparticles, rather than sphere or flat nanoparticles, cause a transient disruption of the endothelial cell membrane.315 Stimulations such as near infrared (NIR) radiation cause cavitation of the membrane, followed by an increased influx of [Ca2+] and a degradation of the actin network.316 By expanding our understanding of the various mechanisms of interactions between nanoparticles and the cell membrane, reduction of membrane disruption will be achieved through the regulation of size, shape, surface geometry, surface charge and surface chemistry.46,313,317,318
4.2.2. Ion Channel Inhibition by Nanoparticles
Ion channels consist of pore-forming proteins that help establish and control a small voltage gradient across the cell membrane.319 When nanoparticles reach the cell surface, they may affect ion channel function by interacting with the extracellular domains of ion channel proteins. SWNTs reversibly block the K+ channel in a diameter-dependent manner, most likely by interacting with extracellular domains of the channel.320 In contrast to pristine SWNTs, SWNTs modified with 2-aminoethylmethane thiosulfonate inhibit the K+ channel irreversibly, suggesting possible bonding between functional groups on SWNTs and the cysteine groups in ion channels.321 Carboxylated MWNTs suppress the K+ channel in PC12 cells without induction of ROS.322 Molecular simulations indicate that there are multiple C60 binding sites on K+ channels. The energetically favorable bindings between C60 and K+ channels cause ion channel inhibition and cytotoxicity.323 Although direct interactions between nanoparticles and ion channels are observed, impurities may also perturb channel proteins. Contamination with yttrium, which is used as a catalyst in the production of CNTs, has been shown to inhibit ion channels.324 Thus, nanoparticle-ion channel interactions must be studied with care.
4.2.3. Cytoskeleton Alterations by Nanoparticles
The cytoskeleton maintains the cell shape and serves significant roles in intracellular transport, force generation, cell motility and division.18 The internalization of nanoparticles results in direct or indirect cytoskeletal alterations. The loss of proper cytoskeletal function further leads to reduced cell motility, division and proliferation.325-329 SWNTs induce actin bundling in both isolated actin filaments and HeLa cells. This induces actin-related cell division defects and retards cell proliferation. A weak binding enthalpy between SWNTs and actin filaments has been speculated to arise because both structures are anisotropic and similar in size. Such interactions induce dramatic anisotropic actin bundle formation both in cells and ex vivo.325 Nanoparticles may also affect the cytoskeleton through indirect interactions. Surface-modified iron oxide nanoparticles and gelatin nanoparticles cause a reorganization of the F-actin and β-tubulin cytoskeletons in human fibroblasts after endocytosis. Because the actin cytoskeleton plays important roles in receptor-mediated endocytosis, the different nanoparticle-induced alterations in cytoskeleton organization may indicate different mechanisms of internalization.327,328 Gold nanorods coated with hexadecyltrimethylammonium bromide cause actin aggregation and β-tubulin redistribution in the cytosol. Such effects reduce micromotion (fluctuations in cell shape) in MDCK-II kidney cells. Coating of nanorods with PEG diminishes such perturbations.326 High concentrations of surface-modified iron oxide nanoparticles affect cytoskeleton and focal adhesion formation and maturation. The reduced expression and decreased activity of focal adhesion kinase in turn alters the actin cytoskeleton. Large endosomes that trap nanoparticles may also affect the normal localization of actin fibers and disrupt the actin cytoskeleton through steric hindrance.329
4.2.4. Mitochondrial Alterations by Nanoparticles
Mitochondria serve as “cellular power plants” by supplying chemical energy (ATP) to cells.18 They are also involved in a range of other processes, such as signaling and cell differentiation. Nanoparticles enter the mitochondria and may affect their functions.330-333 SWNTs have been identified inside mitochondria.333 GNPs (3 nm) enter mitochondrial intermembrane spaces through voltage-dependent anion channels.334 The direct interaction of nanoparticles with mitochondria has been detected using SERS. SERS revealed that GNPs (13 nm) may bind to proteins, lipids and carbohydrates present on the mitochondrial membrane in live cells and in isolated mitochondria. The bands at 886, 875 and 1070 cm−1 are due to C-C and PO2− vibrations. Changes in these bands indicate interactions between nanoparticles and membrane lipids. However, these interactions do not cause damage to the mitochondria.331 GNPs with a much smaller size (1.4 nm) cause indirect damage by increasing the permeability of the inner mitochondrial membrane through the generation of oxidative stress.330
4.2.5. Nuclear Alterations by Nanoparticles
The nucleus maintains the integrity of genes and controls cellular activity cells by regulating gene expression. The nuclear structures include the nuclear envelope, which consists of a double lipid bilayer, the nuclear lumina, which consists of intermediate filaments and membrane-associated proteins, the chromosome, which consists of DNA and histones, and the nucleolus, which consists of rRNA, rDNA and proteins.49 Because the nuclear envelope keeps many macromolecules from entering the nucleus, nanoparticles entry into the nucleus is limited unless the nanoparticles are extremely small (< 5 nm) or engineered with nuclear targeting molecules.335-338 Unmodified QDs (3.2 nm) are able to accumulate in the nucleus and exhibit strong affinity to core histones and histone-rich cell organelles. Histone binding is driven by electrostatic interactions between negatively charged QDs and positively charged core histone proteins. The interaction causes a redshift of the steady-state photoluminescent emission of the QDs and a decrease in the fluorescence lifetime.335 GNPs (2.5 nm) may disturb the decondensation of nuclear chromatin in mouse sperm, indicating their potential spermatotoxicity. Such effects are likely driven by interactions between GNPs with DNA in the nucleus.337 The elucidation of nanoparticle-nucleus interactions provides crucial insight into the biological activity of nanoparticles; however, further investigation is still warranted.
4.3. Oxidative Stress and Nanoparticles
Cells maintain a homeostatic oxidative state and counterbalance external stimuli-induced oxidative stress through several regulation mechanisms.339 When these regulating mechanisms are overwhelmed, the excessive ROS or reactive nitrogen species (RNS) can damage cellular components, affect biomolecule function and ultimately induce cell death through apoptosis or necrosis. Nanoparticles may affect the cellular oxidative state in various ways. The generation of ROS in cells is considered a major factor in nanoparticle-induced cytotoxicity.340 However, nanoparticles with reductive properties may act as free radical scavengers to remedy oxidative damage in cells and tissues.341
4.3.1. Oxidative Stress Induced by Nanoparticles
The production of nanoparticle-induced ROS is triggered by electronically active nanoparticle surfaces, photoactivation, impurities, the dissolution of metal ions and the interactions of nanoparticles with biomolecules.342,343 Nonfluorescent dichlorofluorescin can be oxidized to fluorescent dichlorofluorescein inside the cell by various oxidants, and this reaction can be used to detect the total oxidative level in live cells.344 ROS generation can also be measured by free radical content, GSH level, mitochondrial membrane potential, stress-related gene/protein expression, microarray and stress-specific reporter assays.330,340,345-349
Most nanoparticles cause oxidative stress. Although the outcome is similar, the mechanisms may be material-dependent. First, GNPs with a diameter of 1.4 nm induce oxidative stress and reduce intracellular antioxidant levels. The oxidative stress causes mitochondrial damage, an upregulation of stress-related genes and cell necrosis.330 The generation of ROS by GNPs upon short-term (4 h) exposure can be counterbalanced by cells; however, longer exposure (48 h) can overwhelm antioxidant defenses.350 GNPs also catalyze nitrogen monoxide (NO) production from endogenous S-nitrosothiols in serum through the formation of Au-thiolate on the surface of nanoparticles.342 Second, TiO2 nanoparticles, which are widely used in cosmetics, cause oxidative stress with or without light activation. TiO2 nanoparticles absorb UV light and catalyze DNA damage in human cells.238 In the dark, TiO2 nanoparticles induce ROS generation, a decrease in GSH, upregulation of stress-related genes, activation of JNK and P53 and apoptosis.351,352 TiO2 nanoparticles generate O2· in sunlight and HO· free radicals in the dark.353 Third, carbon black (CB) nanoparticles induce oxidative stress by producing highly diffusible 4-hydroxynonenal. This species mediates cytotoxicity through the formation of 4-hydroxynonenal-protein adducts and GSH modification.354 CB nanoparticles enhance the oxidative stress induced by Fe2O3 nanoparticles in cell culture through co-exposure. However, surface-oxidized CB nanoparticles have no such effects, indicating that the oxidative stress is mediated by the surface reactivity of CB.348 Fourth, CNTs induce ROS-related inflammasome activation.355 Metallic impurities from the CNT synthesis process, if present, may be responsible for the generation of oxidative stress.356 Surface chemistry modifications by PEG reduce ROS and cytotoxicity.357 5). Fullerene (C60) causes membrane damage and cytotoxicity through ROS generation and lipid peroxidation. An antioxidant, L-ascorbic acid, completely prevents such oxidative damages.314 6). Other nanoparticles, including ZnO, silica, polystyrene, nickel ferrite, silver and nano-Co, have also been reported to induce oxidative stress, mitochondrial damage and apoptosis.346,347,358-361
A nanoparticle’s potential to generate oxidative stress depends on the nature of the materials. At the same dose, different nanomaterials cause different levels of oxidative stress. For example, ZnO, CuO and CdS nanoparticles generate a higher level of ROS than that of TiO2, MgO, SiO2, CNTs and Fe3O4 nanoparticles.362-366 CeO2 can even protect cells from oxidative damage.362 Due to the significance of nanomaterials’ intrinsic properties, a predictive paradigm for oxidative stress has been developed based on metal oxide nanoparticle band gaps.367 Critical physicochemical effectors are discussed below.
The crystal structure of nanoparticles appears to play a role in ROS generation. Rutile TiO2 nanoparticles cause the generation of ROS and cell apoptosis, while anatase TiO2 nanoparticles induce necrosis with reduced ROS generation.368 The size and surface area of nanoparticles affect ROS generation. Smaller nanoparticles with larger surface areas generate much more ROS than larger nanoparticles.369-371 Surface chemistry can regulate oxidative stress generation. Cellular ROS generation by surface-modified silicon nanoparticles occurs in the following order: positively charged > neutral > negatively charged.372 Surface-modified ZnO nanoparticles generate ROS in the following order: uncoated > oleic acid coated > poly(methacrylic acid) coated > protein coated.373 Nanoparticles with hydrophobic molecules on the surface cause oxidative stress in the cell. GNPs coated with longer hydrophobic alkyl chains cause much more ROS generation and cytotoxicity than those with shorter hydrophobic chains.374 PEG modification on the surface reduces oxidative stress caused by SWNTs.357 Some environmental factors may impact cellular ROS production. For instance, exposure to light enhances the generation of ROS in cells incubated with TiO2 nanoparticles.364 Fe2O3 or CB nanoparticles alone do not cause cellular oxidative stress. However, the co-application of these nanoparticles induced a two-fold increase in the oxidation of cellular proteins. Fe2O3 nanoparticles in endosomes or lysosomes are acidified to release Fe3+ ions, which do not generate hydroxyl radicals in the presence of H2O2. However, CB nanoparticles reduce Fe3+ ions to Fe2+ ions. Fe2+ ions can permeate the vesicle membrane, take part in the Fenton reaction and induce protein oxidation.375 In addition to these physicochemical factors, cells from different organs have different capabilities of internalizing nanoparticles and resisting oxidative stress.376
4.3.2. Reduction of Oxidative Stress by Nanoparticles
Several nanoparticles have free radical scavenging capabilities, which originate from their chemical structures. These effects may be potentially applied in antioxidative therapy. Cerium oxide (ceria) nanoparticles, fullerene and its derivatives, CNTs and heavy metal (Au and Pt) nanoparticles have been reported to reduce ROS through various mechanisms.341,377-386
Ceria nanoparticles inhibit inflammatory mediator production and reduce cell apoptosis. These nanoparticles protect rats against oxidative damage by scavenging free radicals.378,381,384 When ceria nanoparticles are doped with samarium, which decreases Ce3+ without affecting the oxygen vacancy (defect in oxides) contents, their antioxidant and anti-apoptotic abilities are reduced. This decrease indicates that the Ce3+/Ce4+ redox reaction and not oxygen vacancy is responsible for the antioxidant activity of ceria nanoparticles.381 Fullerene and its derivatives have the ability to quench superoxide radicals.377,379 CNTs are able to scavenge free radicals and retard the oxidation reaction.380,385 Doping of boron on CNTs enhances the efficiency of radical scavenging, indicating that electron affinity may be responsible for the effect. Au and Pt nanostructures have also been reported to display antioxidant activities.382,383,386 Pt nanoparticles exhibit anti-oxidant activity by scavenging O2·− and HO• with a second-order reaction rate.382 Au- and Pt-supported nanodiamonds exhibit a high antioxidant activity, as indicated by their peroxidase activity and the ability to trap carbon radicals.383 Apoferritin-coated Pt nanoparticles enter cells through receptor-mediated endocytosis, decrease hydrogen peroxide-induced oxidative stress and reduce cytotoxicity.386
4.4. Nanoparticle-Induced Systematic Cellular Biochemical Perturbations
Cells have complex biochemical networks that maintain homeostasis during proliferation, differentiation and apoptosis. Any extrinsic factors, such as nanoparticles, may interfere with these processes. Therefore, understanding nanoparticle-induced cellular biochemical perturbations to elucidate the chemical mechanisms of nanoparticle-biological system interactions is imperative.
4.4.1. Perturbation of Cell Signaling Pathways by Nanoparticles
Nanoparticles perturb many cellular signaling pathways. These pathways are involved in important biological processes that determine cell fate. The delineation of these mechanisms will facilitate an understanding of the cytotoxicity of nanoparticles and the development of safe nanomedicines and nanomaterials.
4.4.1.1. Mitogen-Activated Protein Kinase (MAPK) Signaling
MAPK signaling is a significant regulatory mechanism in eukaryotic cells. The MAPKs are serine/threonine-specific protein kinases that respond to external stimuli, such as mitogen, heat shock and oxidative stress. The MAPK signaling pathway regulates cell proliferation, apoptosis, morphogenesis, cell cycle and immunity.387
There are three groups of kinases in MAPK signaling. The first group, extracellular signal-related kinase (ERK), is activated by growth factors, mitogen and G protein coupled receptors (GPCRs). Carbon nanoparticles activate Erk1/2 in multiple cell lines.388 ERK is also activated by silica and iron oxide nanoparticles.389,390 Activation of Erk1/2 by iron oxide nanoparticles promotes neurite outgrowth in PC12 cells.390 The second group, p38 MAPK, and the third group, JNK, are activated by stress, GPCRs and inflammatory cytokines. GNPs can promote osteogenic differentiation through activation of p38 MAPK.21 TiO2 nanoparticles activate JNK and p53 in neuronal cells and cause G2/M cell cycle arrest and apoptosis.351 Silica nanoparticles activate p38 and JNK and reduce cell viability.391 Not all nanoparticles activate MAPK signaling. Carboxyl-modified fullerene nanoparticles enter oxidative stress-damaged cerebral microvessel endothelial cells, maintain cell integrity and inhibit apoptosis. Such effects are the result inhibiting the JNK pathway by nanoparticles.392
4.4.1.2. Nuclear Factor Kappa B (NF-κB) Pathway
The NF-κB pathway is an important cellular signaling pathway in response to external stimuli. Under normal circumstances, NF-κB forms a stable complex with IκB in the cytoplasm. When cells are exposed to stimuli, such as stress, free radicals or irradiation, the kinase IKK is phosphorylated (activated) by receptors. The activated IKK subsequently phosphorylates IκB. Phosphorylated IκB undergoes ubiquitous degradation, and NF-κB is released and translocated to the nucleus to initiate transcription of inflammatory factors.393 Silver nanoparticles, CNTs, silica nanoparticles, TiO2 and double hydroxide (Mg(OH)2 : Al(OH)3 1:1) nanoparticles have been reported to activate NF-κB signaling and induce the generation of pro-inflammatory cytokines including COX-2, TNF-α, IL-6, IL-12 and inducible nitric oxide synthase.47,389,394-398 The chemical basis for the activation of NF-κB signaling by different nanoparticles is that all of these nanoparticles generate ROS in cells. The activation of NF-κB finally leads to a reduction of cell proliferation, inflammation and apoptosis. Surface modifications on MWNTs direct the receptor recognition from mainly mannose receptors to scavenger receptors. The activation of NF-κB through the mannose receptor is alleviated, and the immunotoxicity of MWNTs is reduced.47 This study demonstrates that activation of cell signaling by nanoparticles can be regulated through modulating receptor recognition events by surface chemistry modifications.
4.4.1.3. DNA Damage and Repair
The cellular ROS generated by nanoparticles may cause DNA damage and activate DNA repair signals.399 Such damage may induce G2/M cell cycle arrest, apoptosis, mutagenesis and tumorigenesis.400-404 Therefore, an understanding of nanoparticle-induced DNA damage and its repair is critical. MWNTs increase the expression of the base excision repair protein OGG1, the double strand break repair protein Rad 51, the phosphorylation of p53 by the checkpoint protein kinase ChK2, the phosphorylation of histone H2AX at serine 139, and SUMO modification of XRCC4 (in response to DNA double-strand breakage) in mouse embryonic stem cells.402 Similarly, nanodiamonds and silver nanoparticles also induce DNA damage, as demonstrated by increased expression of OGG1 and Rad51 and phosphorylation of p53 and H2AX.400,401,403
4.4.1.4. Bone Morphogenetic Protein (BMP) and Transforming Growth Factor beta (TGF-β) Signaling
BMP receptors belong to the TGF-β receptor superfamily.405 CNTs suppress BMP signaling pathways and affect cell function (Figure 15).406-408 Suppression of BMP signaling reveals a non-apoptotic mechanism for the perturbation of cell proliferation by CNTs. A global examination of gene expression and Western blotting revealed that the Id (inhibitor of DNA binding) genes and proteins are suppressed by SWNTs and MWNTs. Id genes are direct targets of Smad-dependent BMP receptor signaling through activation of Smad 1/5/8 proteins. The activated Smad proteins translocate into the nucleus to initiate transcription. The phosphorylation and nuclear translocation of Smad 1/5/8 are inhibited by CNTs. Id genes and proteins are subsequently inhibited. The inhibition of the Id protein negatively regulates the cell cycle transition from G1 to S phase and cell proliferation.408 A further study revealed that the suppression of BMP signaling is due to binding of CNTs to BMP receptor type II (BMPR2) but not BMP receptor type I or the BMP ligand.406
Figure 15.
A schematic diagram showing the molecular interactions involved in MWNT 1-enhanced cell differentiation and apoptosis inhibition. Reprinted with permission from ref 405. Copyright 2012 Nature Publishing Group.
BMP regulates a class of bHLH transcription factor proteins. Id proteins are also bHLH proteins but lack the DNA binding domain. Id proteins function as negative regulators of transcription. This regulation is significant in cell differentiation, tumorigenesis and apoptosis. Suppression of BMP signaling in mesenchymal stem cells leads to inhibition of proliferation, myogenic differentiation and reduced apoptosis. Surface chemical modifications of CNTs can tune cell differentiation to different levels through the regulation of CNT/BMPR2 binding and BMP signaling.406
Graphene nanosheets activate MAPK and TGF-β signaling pathways in murine RAW 264.7 macrophages. Graphenes cause an increase in cellular ROS and the activation of phosphorylation of ERK, JNK and P38, indicating the activation of MAPK signaling. Furthermore, the expression of TGF-β receptor mRNA and phosphorylation of Smad2 are also enhanced, indicating the activation of TGF-β signaling. The ROS and activated TGF-β signaling pathways further induce cellular apoptosis.409
4.4.1.5. Other Signaling Pathways
GNPs bind to heparin-binding growth factors but not to non-heparin-binding growth factors and selectively inhibit heparin-binding growth factor-induced cell proliferation. This interaction is due to direct binding of gold surfaces with sulfur or amine groups present in the heparin-binding domain. The inhibition of heparin-binding growth factors causes a decrease in vascular endothelial growth factor (VEGF)-165-induced cell proliferation in vitro and angiogenesis in vivo.153,154 GNPs with a poly(acrylic acid) coating selectively bind fibrinogen and induce protein unfolding to expose the C-terminus of the γ chain. The newly exposed epitope interacts with the Mac-1 receptor and activates its signaling. Mac-1 activation induces the release of inflammatory cytokines, which is an alternative mechanism for the inflammatory response of cells to nanoparticles.160 Silver nanoparticles suppress glial cell line-derived neurotrophic factor signaling in spermatogonial stem cells and reduce cell proliferation. This inhibition may be due to interactions between nanoparticles and Fyn protein, a kinase in glial cell line-derived neurotrophic factor signaling.410 Graphene oxide nanosheets activate the toll-like receptor signaling cascades and trigger ensuing cytokine responses, which subsequently induce autophagy.411
The above reports indicate that nanoparticles may affect oxidative stress-dependent and -independent pathways. BMP, growth factors and Mac-1 cascades are affected by direct interactions between signaling proteins and nanoparticles (BMPR-CNT, VEGF-GNP Mac-1 receptor-GNP, respectively). These interactions do not necessarily induce oxidative stress and apoptosis but may affect proliferation or differentiation. Others signaling pathways such as MAPK, NF-κB and DNA damage are responsible for nanoparticle-induced oxidative stress. These events eventually cause cell death. All of these events are determined by both the nature of the nanoparticles and their surface chemistry. The control of nanoparticle-induced signaling events and related cellular outcomes are crucial subjects for future investigations.
4.4.2. Global Biochemical Alterations by Nanoparticles
In contrast to small molecule drugs, for which the intracellular targets and mechanisms of action have been at least partially elucidated, knowledge of the molecular events involved in nanoparticle-biological system interactions is limited. Nanoparticle-induced global biochemical alterations may provide useful insights into the molecular mechanisms and chemical basis of nanoparticle-biological systems interactions. The replication, transcription and translation of genes and synthesis of bioactive small molecules are biochemical processes that are controlled by tens of thousands of signals in response of intracellular demands or extracellular stresses. Mapping these global profiles will facilitate the identification of nanoparticle-generated biological outcomes and the eventual regulation of these outcomes through modulation of the physicochemical properties of nanoparticles.
4.4.2.1. Genomic Transcriptional Profile
Unlike small molecules, which may significantly regulate > 5000 genes in living cells, most nanoparticles usually affect much fewer (< 500) genes, indicating that they may be less active and more selective toward cells.330,408,412-415 The biological activity of nanoparticles can be investigated via genomic transcriptional profiling. Carboxylated SWNTs inhibit Id genes in HEK293 cells in a whole genome background. BMP signaling, which targets Id genes, was identified through multiple assays.408 Pristine SWNTs or MWNTs affect a large number of genes related to the cell cycle, signaling transduction, apoptosis, oxidative stress, metabolism, transport and immune responses in human cells.398,414,416 By contrast, multi-walled carbon nano-onions affect genes that fall into the category of external stimuli other than the immune response.416 PEG-modified SWNTs affect fewer genes, indicating a reduced biological activity.357 PEG-modified, silica-coated QDs have been reported to affect 38 genes at low doses (8 nM) and 12 genes at high doses (80 nM), with four shared genes. These genes are related to carbohydrate binding, intracellular vesicle localization and vesicular proteins involved in stress responses.412 Fe3O4 magnetic nanoparticles alter 711, 545 and 434 genes in mouse macrophage cells 4, 24 and 48 hours after treatment. MAPK, TLR and JAK-STAT signaling pathways are also affected by Fe3O4 nanoparticles. Bioinformatic analyses have revealed that these nanoparticles activate inflammatory and immune responses and inhibit biosynthesis and metabolism.415 In the absence of illumination, anatase TiO2 nanoparticles with diameters of 7, 20 and 200 nm upregulate the inflammatory response and cell adhesion genes but not oxidative stress genes in human keratinocytes.417 These effects indicate that TiO2 nanoparticles do not cause oxidative damage in the dark. Genomic expression profiles reveal that GNPs of smaller sizes (1.4 nm) upregulate heat shock and stress-related genes and lead to cell necrosis. However, larger GNPs (15 or 18 nm) affect fewer genes, indicating their superior biocompatibility.330,418 The toxicity of some metallic nanoparticles may be due to the release of the metal ions. Genomic transcriptional profiling reveals that silver nanoparticles affect 128 genes in human epithelial cells, similar to the response of cells exposed to silver ions.413 Treatment of epithelial cells with copper oxide nanoparticles upregulates genes in MAPK and downregulates cell cycle genes, also similar to cells treated with copper ions.419 These discoveries suggest that the release of metal ions from nanoparticles must be considered when evaluating the bioactivity of metallic nanoparticles.
4.4.2.2. Proteomic Expression
Protein synthesis is downstream of gene transcription and controlled by a number of regulatory factors. The evaluation of proteomic expression provides more relevant information than genomic transcriptome analyses. The global protein expression profile is generally examined using 2D gel electrophoresis combined with MS.420-422 Generally, total proteins extracted from cells are separated by 2D gel electrophoresis, and the protein spots on the gel are excised and digested, followed by MS analysis. The MS data are analyzed by searching databases to identify proteins. Another approach is the isobaric tags for relative and absolute quantitation (iTRAQ) technique.423 iTRAQ uses liquid chromatography to separate isotopically labeled peptides and MS/MS analysis to identify and quantitate peptides. Protein microarrays can be used for the high-throughput evaluation of protein expression/phosphorylation.420 Antibodies against protein targets can be spotted on nitrocellulose membranes, enabling the detection of multiple proteins in a single experiment. Using the above technologies, proteomic expression/phosphorylation profiles can be obtained. A total of 51 proteins in human hepatic carcinoma HepG2 cells are significantly affected by SWNTs, as detected by iTRAQ-coupled LC-MS/MS analyses. These proteins are involved in metabolic pathways, redox regulation, signaling pathways, cytoskeleton formation and cell growth.423 Using 2D gel electrophoresis with MALDI-TOF MS, 45 altered proteins were identified in human monoblastic leukemia U937 cells in response to MWNTs. These proteins participate in metabolism, biosynthesis, stress response and differentiation processes.422 Additionally, 2D gel electrophoresis/MS analyses and protein phosphorylation antibody array analyses revealed that GNPs activate endoplasmic reticulum (ER) stress response in human chronic myelogenous leukemia K562 cells.420 A similar analysis indicated that TiO2 nanoparticles significantly alter 46 proteins in human bronchial epithelial BEAS-2B cells. These proteins are involved in the stress response, metabolism, adhesion, cytoskeletal dynamics, cell growth, cell death and signaling pathways. Nanoparticles perturb approximately 20 proteomic pathways, consistent with pathways from genomic data analyses.421
4.4.2.3. Metabonomic Alterations
Nanoparticles cause physiological changes in living systems and generate altered metabolic profiles. Therefore, the investigation of global metabonomic profiles provides insight into the physiological processes that are perturbed by nanoparticles. Nanoparticle-induced metabonomic alterations are studied in live animals or in cells.424-427 Analytical methods that include 1H NMR, MS, HPLC-MS and gas chromatography-mass spectrometry (GC-MS) have been used in metabonomic analyses.424 A global metabolism study in live cells using high-resolution MAS NMR spectroscopy demonstrated that silver nanoparticles decrease the levels of GSH, lactate, taurine and glycine and increase the levels of most amino acids, choline analogues and pyruvate. These results indicate that the depletion of GSH by nanoparticles may induce the conversion of lactate and taurine to pyruvate, providing a biochemical mechanism for oxidative stress.425 HR-MAS 1H NMR was also used to study the metabonomic changes in RAW264.7 mouse macrophages following treatment with magnetic iron oxide nanoparticles. The nanoparticles caused a decrease in the levels of triglycerides, essential amino acids and choline metabolites while increasing glycerophospholipids, tyrosine, phenylalanine, lysine, glycine and glutamate. These changes reflect phagocytosis and cell membrane perturbation.427
In general, interactions between nanoparticles and biological systems include direct and indirect effects. Direct effects, such as the binding of nanoparticles to membrane phospholipids, receptors, cytosol proteins and DNA molecules, are modulated by the physicochemical properties of the biomacromolecules and nanoparticles and their microenvironment. Direct interactions are much more specific and material-dependent compared to indirect interactions and may have therapeutic potential. However, the investigation of direct interactions is challenging because of the complex cellular background. Indirect effects, such as signaling pathway activation by nanoparticle-induced ROS, cause cytotoxicity. Future investigations will enhance and regulate direct interactions while minimizing indirect interactions. In this endeavor, chemical technology may be very powerful for modulating the biological activities of nanomaterials, as discussed below.
5. CHEMICAL APPROACHES TO REGULATE AND UNDERSTAND NANOPARTICLE-BIOLOGICAL SYSTEM INTERACTIONS
Interactions between nanoparticles and biological systems are complex. The biological activities of nanoparticles depend on their size, shape, structure and surface properties. As nanoparticle sizes fall into the nanometer range, the surface-exposed atoms increase exponentially. Therefore, changing the properties of the surface molecules may significantly alter the biological activities of nanoparticles. In this section, we explore various surface chemistry modification methods, the biological impacts of such modifications and computational models to predict the relationships between nanoparticle surface chemistries and their biological activities.
5.1. One-at-a-Time Nanoparticle Surface Chemistry Modification
5.1.1. Modulation of the Surface Chemistry of Nanoparticles
Various molecules have been used to modify nanomaterial surfaces via coating chemistry and consequently, their biological activity. Surface modification strategies include covalently or noncovalently linking small organic ligands or biomolecules to nanoparticles or encapsulating nanoparticles in surface coatings, such as polymers, micelles, liposomes,428 graphene oxide,141 and silica shells.412,429 The ligand exchange method is effective for transferring nanoparticles synthesized in organic solvents to aqueous media by displacing hydrophobic ligands on nanoparticles with water-soluble ligands. For example, dithiocarbamate moieties are designed to replace trioctylphosphine oxide capped on CdSe/ZnS QDs,430 while (3-carboxypropyl)trimethylammonium chloride is used to replace lauric acid on Fe3O4 nanocrystals.431 This ligand exchange step is essential to apply these nanoparticles in biological systems. The covalent linkage strategy requires the aid of coupling reagents, such as 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccinimides, or Cu2+-mediated click chemistry432 to react with functional groups on nanoparticles. Some nanoparticles with an aliphatic shell can be encapsulated in polymer coatings or micelles through an intercalating effect with amphiphilic polymers.433 In addition, the polymer coating can be achieved through a layer-by-layer technique by sequentially trapping nanoparticles in layers of polymers of alternating charge.434
5.1.2 Modulation of the Surface Charge of Nanoparticles
An acidic group is often used to generate nanoparticles with negative charges on their surfaces. Similarly, an amine group is used to generate nanoparticles with a positively charged surface. Surface-charged nanoparticles differ greatly in their electrostatic and electrodynamic properties, which can be experimentally determined by ζ-potential measurements. Polystyrene nanoparticles with different surface functional groups, such as carboxylic acid, PEG, amine and amidine groups, have ζ-potential values ranging from −50 to 40 mV. The binding of serum proteins alters their ζ-potential values and narrows them to a range of −40 to −20 mV.435 BSA, myoglobin and cytochrome C are adsorbed to mercaptoundecanoic acid-protected GNPs, as shown by a quartz crystal microbalance study. All three proteins form adsorption layers consisting of an irreversibly adsorbed layer and a reversibly adsorbed layer.109 These data suggest that the charged surface of a nanoparticle is quite complicated in solution. Multiple layers of molecules are adsorbed and dissociated constantly. The charged surface of nanoparticles also affects the conformation of the adsorbed proteins. Cytochrome C maintains its conformation when adsorbed to neutral GNPs, and it is denatured on the surface of charged GNPs.436 GNPs surface modified with charged and neutral amino acids affect the conformation and activity of α-chymotrypsin differently. Charged GNPs exhibit more effects than neutral particles.437 To understand the influence of surface charge density on protein binding, a series of polystyrene latex nanoparticles with the same size were prepared using different amounts of Na-styrenesulfonate comonomer. As the charge density increased, there was no change in the bound protein species, while the amount of adsorbed protein increased.438
The different electrostatic and electrodynamic properties of nanoparticles also modulate their interactions with cells. Such alterations include effects on cell internalization, cell function and cell death. A better understanding of these effects will not only help us design nanomaterials for biomedicinal applications, but will also elucidate mechanisms of nanotoxicity. Due to the presence of phosphate groups from lipids, cell membranes are negatively charged. Positively charged nanoparticles generally exhibit higher cell binding and cellular uptake than negatively charged nanoparticles. To understand the electrostatic and electrodynamic interactions between nanoparticles and cells with minimal secondary interactions, a nanoparticle library with a continuous change in the surface charge density was synthesized and investigated. The nanoparticle/cell interaction, however, did not exhibit a continuous change. Instead, a sharp increase in cell binding was observed as the positive surface charge density increased. The effective surface charge density on the outermost layer of the nanoparticle determined the electrostatic attraction between the nanoparticles and the cells.439 Nanoparticles with different charges induce different perturbations of the cell membrane potential. Positively charged nanoparticles depolarize the membrane, and negatively charged nanoparticles have a negligible effect. Membrane potential perturbations result in an increase in [Ca2+] influx, which in turn inhibits the proliferation of cells (Figure 16).312 A method has been developed to differentiate GNPs bound to the cell surface from internalized GNPs. Neutral and negatively charged GNPs displayed lower cell membrane binding and cell internalization compared to positively charged nanoparticles.440
Figure 16.
Schematic depicting the surface charge effects of GNPs on cellular function. Reprinted from ref 311 with permission. Copyright 2010 American Chemical Society.
The surface charge effect seems applicable to a variety of nanomaterials. Positively charged GNPs, magnetic nanoparticles, silica nanoparticles and QDs all exhibit higher cellular uptake efficiency. (Figure 17).440 The internalization of negatively charged nanoparticles also occurs and is likely due to other interactions and subsequent endocytosis.441,442 QDs with a carboxylic acid surface coating are recognized by lipid rafts but not by clathrin or caveolae in human epidermal keratinocytes. QDs are internalized into early endosomes and then transferred to late endosomes or lysosomes. In addition, endocytic interfering agents have been used to investigate the mechanism by which QDs enter cells. The cellular uptake of QDs with negative surface charges is primarily regulated by pathways associated with G-protein coupled receptors, low-density lipoprotein receptors and scavenger receptors.443
Figure 17.
Schematics illustrating the interactions between cells and nanoparticles with different types of surface charges: (A) negatively and neutrally charged nanoparticles. (B) Positively charged nanoparticles. Reprinted from ref 439 with permission. Copyright 2009 American Chemical Society.
In a kinetics study, positively charged nanoparticles displayed a higher rate of endocytosis than negative charged materials.444 Positively charged core-shell silica nanoparticles coated with polyethyleneimine have been shown to escape from endosomes and enter the cytoplasm and nucleus.445 Positively charged nanoparticles destabilize cell membranes and form holes or expand holes at pre-existing defects.446 The holes led to the internalization of nanoparticles into cells, as well as the diffusion of cytosolic proteins out of cells. Further studies have demonstrated that the formation of holes in lipid bilayers by positively charged nanoparticles is a common property regardless of the shape, size and chemical composition of the nanoparticles.447
5.1.3. Modulation of the Surface Hydrophobicity of Nanoparticles
The hydrophobicity of nanoparticles influences their ability to interact with biomolecules and cells. To investigate the correlation between plasma protein adsorption and the surface hydrophobicity of nanoparticles, latex nanoparticles with varied surface hydrophobicity were synthesized and examined. The number of bound proteins increased with increasing particle hydrophobicity, as determined by 2D polyacrylamide gel electrophoresis analyses and ITC. A clear difference in protein affinity for polymeric particles of different hydrophobicity has been observed.108,448
A set of polymethacrylate nanoparticles (140 nm) with fluorescence labels was investigated to reveal the relationship between particle hydrophobicity and cellular uptake. The monomer chain length controls the hydrophobicity of the modified nanoparticles, as evaluated by measuring the interfacial tensions between the monomers and water. The length of the monomer chains influences the cellular uptake of nanoparticles. Increasing the side chain length increases cellular uptake significantly. This phenomenon can be explained by the increased hydrophobicity and the softness of the particles.449
The surface hydrophobicity of nanoparticles is also modified by peptide–GNP hybrids. By altering the end amino acids of the peptide ligands, the uptake of GNPs can be dramatically increased or suppressed. Nanoparticles with tryptophan end groups enhance cellular uptake due to the membrane anchoring and permeation effects of the aromatic group.450
5.1.4. Changes in the Interactions of Nanoparticles with Biological Systems
5.1.4.1. Making Toxic Nanoparticles More Biocompatible
Nanoparticles may induce toxicity in biological systems because of their specific properties.451,452 One, the large surface area to volume ratio leads to very strong nonspecific interactions with biomolecules. These interactions will alter the conformation of proteins or DNA, resulting in their malfunction.453 Second, some nanoparticles are composed of heavy metals, such as CdTe and CdHgTe QDs. Under some conditions, the free metal ions may be released into biological systems and induce toxicity.454 Third, nanoparticles may induce ROS, such as free radicals (hydroxyl radical, ·OH; superoxide, ·O2−) and singlet oxygen (1O2) (e.g., QDs455 and Ag nanoparticles456). ROS are known to cause irreversible damage to nucleic acids, enzymes and cellular components that included the mitochondria, plasma and nuclear membrane. Fourth, some capping ligands derived from the nanoparticle synthesis system may induce cytotoxicity.457 Surface chemistry modifications may diminish the above side effects and greatly improve the biocompatibility of potentially toxic nanoparticles. For example, the biocompatibility of QDs has been improved after surface modification with dendrimers,458,459 PEGylated dihydrolipoic acid,460,461 triblock copolymers,462 cyclodextran463 and alkyl-modified poly(acrylic acid).464 The coating of SWNTs with polyvinylpyrrolidone polymers,465 phospholipid derivatives466 on silver nanoparticles or human serum protein467 also prevents their toxicity. Hexadecyltrimethylammonium bromide (CTAB) released from CTAB-coated Au NRs is highly cytotoxic at a nanoparticle concentration as low as 0.05 mM (20% cell viability). However, when toxic CTAB was replaced by PEG-SH, the cell viability when subjected to nanoparticles was 95%, even at a concentration of 0.5 mM.16 Similar effects have also been reported after the replacement or overcoating of CTAB with poly(sodium 4-styrenesulfonate),468 copolymer poly(diallyldimethylammonium chloride)/polystyrene sulfonic acid469 or polyallylamine hydrochloride.470
5.1.4.2. Regulating Interactions with Cells
The interaction of nanoparticles with cells is affected by the surrounding biomolecules in physiological media. When bare nanomaterials (particularly inorganic nanoparticles) encounter biological macromolecules, they may form a nanoparticle–macromolecule (mainly protein) corona hybrid nanostructure.453 Moreover, a secondary protein corona on the hybrid nanoparticles may be further formed via protein–protein interactions. Some of these interactions do not occur in the native system in the absence of the nanoparticles, and an immunological response may be triggered. As a result, the nanohybrids are detected as foreign materials by the immune system. The nanohybrids are rapidly cleared from the blood stream and translocated to the liver and spleen,471 potentially limiting the biological applications of nanoparticles. Fortunately, the fate of nanoparticles in biosystems can be regulated. Two main requirements, cell targeting and in vivo stealth properties, can be modulated by proper nanoparticle surface chemical modifications.
When a nanoparticle is presented to a cell, the cell first recognizes the coating molecules at the outermost layer of the nanoparticle. For targeting purposes, linking specific ligands to nanoparticles is necessary. Certain molecules, such as antibodies, aptamers or small-molecule ligands, can bind to receptors overexpressed in tumor cells in a specific “lock-key” manner.428,472-475 After modification, nanohybrids are able to target specific cancer cells. This targeting strategy lays the foundation for designing nanomedicines for cancer diagnosis and therapy. The detection of circulating breast cancer cells in the blood was achieved by using a combination of an epithelial cell-specific, antibody-conjugated magnetic nanoprobes and anti-HER2 antibody-conjugated SERS tags.476 Because the HER2 receptors are highly expressed on the breast cancer cell membrane, the SERS tags recognized these tumor cells. Thus, the cancer cells were detected rapidly with good sensitivity. An anti-prostate antigen was conjugated to a multifunctional nanomedicine composed of loaded drugs, NIR QDs and a polystyrene matrix with internally embedded superparamagnetic Fe3O4 nanoparticles. Using this multifunctional nanoparticle, the specific detection of LNCaP prostate cancer cells was achieved.477
After being administered into the blood, most unmodified nanomaterials are taken up by the mononuclear phagocyte system within minutes or hours.478 Surface modification of nanomaterials is generally regarded as a practical strategy to prevent protein adsorption and, consequently, avoid triggering immune responses.474 Various biocompatible polymer surface coatings, such as PEG,479 hyaluronic acid,480,481 cyclodextran482,483 and polymers based on phosphorylcholine484 provide effective barriers against protein bindings and therefore reduce both protein adsorption and cellular uptake. These modifications are widely used to impart stealth properties to nanoparticles in biomedical applications. These surface modifications also change the in vivo pharmacokinetics of nanoparticles.485 QDs modified with mPEG 5000 display a significantly prolonged in vivo circulation time in addition to reduced nonspecific accumulation in various organs.486 After surface PEGylation, the blood half-life of gold nanorods is increased to 24 h.457,487 PEG-terminated phospholipid was used to encapsulate QDs488 and iron oxide nanocrystals489 for a longer blood circulation time in vivo. A mPEG-alkyl-thiol polymer with an alkyl linker between the PEG and the thiol moiety yielded a much stronger Au nanoparticle surface protection compared to commonly used methoxy-PEG-thiol in the presence of cysteine and cystine molecules at the physiological concentrations.490 Surface coating of lipid molecules has also improved the pharmacokinetics of the silica nanoparticles.491
5.2. Combinatorial chemistry for Nanoparticle Modifications
The discovery and optimization of novel nanoparticles with unique properties by conventional methods has always been challenging and time consuming. High-throughput approaches, including parallel and combinatorial synthesis, are more efficient than conventional linear synthesis, and in recent years, these techniques have had an increasing impact in nanotechnology.
5.2.1 Parallel Chemistry for Nanoparticle Modifications
A parallel chemistry approach provides the opportunity to rapidly synthesize a large amount of nanoparticles with similar surface chemistries. However, this approach has limitations, mostly based on the lack of a general method to modify nanoparticles rapidly and the availability of small molecules that can be used.
5.2.1.1 Parallel Nanoparticle Libraries for Regulating Gene Delivery
Numerous biomaterials have been studied as potential nonviral gene delivery vectors to improve DNA stability and the efficiency of cellular uptake. The tested materials include inorganic surfactants, cationic lipids, polysaccharides, cationic polymers and dendrimers. These materials display small cargo capacity, inefficient uptake, poor biostability and low safety. Using high-throughput synthesis and screening techniques, libraries of poly-β-amino esters (PBAEs) of > 2000 members were synthesized to evaluate their gene delivery efficiency.492-494 A total of 46 PBAEs were superior to polyethylenimine. A second-generation library of 486 polymers was synthesized to optimize the polymers and investigate their structure-function relationships.495 The best performing polymer nanoparticles were < 150 nm and had positive surface charges. PBAEs have been used to deliver DNA to primary human endothelial cells.496 To investigate a larger PBAE chemical space, libraries of end-modified PBAEs were also synthesized using various end-capping reagents.497,498 Two modified polymers with greatly improved transfection efficiency both in vitro and in vivo were discovered.
5.2.1.2 Parallel Nanoparticle Libraries for Regulating Cell Targeting
Magnetic nanoparticles (MNPs) that target tissues of interest have been developed by conjugating antibodies, peptides499 or small molecules to the surface of MNPs.26,28 Small molecules with anhydride, amine, hydroxyl carboxyl, thiol and epoxy handles were coupled to the MNPs via different synthetic routes. The nanoparticles were printed on glass slides and showed different binding properties towards different targeting proteins using fluorescence detection. A collection of MNPs comprising 146 nanoparticles modified by different synthetic small molecules was synthesized and screened against different cell lines (Figure 18). The In this screen, nanoparticles with high specificity for endothelial cells, pancreatic cancer cells or activated human macrophages were identified.28
Figure 18.
(A) Model of the cross-linked dextran coating modified with small molecules. Different classes of small molecules with amino, sulfhydryl, carboxyl or anhydride functionalities were anchored onto nanoparticles and stored in multi-well plates for testing. (B) Part of a heat map representing the cellular uptake of different nanoparticle preparations. Columns from right to left: 1, pancreatic cancer cells (PaCa-2); 2, macrophage cell line (U937); 3, resting primary human macrophages; 4, activated primary human macrophages; 5, human umbilical vein endothelial cells (HUVEC). Each column represents mean values from six different experiments. Red indicates the lowest accumulation, and green indicates the highest accumulation. Reprinted with permission from ref 28. Copyright 2005 Nature Publishing Group.
5.2.1.3 Parallel Nanoparticle Libraries for Imaging
Weissleder et al. have developed fluorescent nanoparticles derivatized with small molecules using a parallel chemistry approach. Cellular uptake was rapidly profiled in vitro via binding of those nanoparticle imaging probes in primary cell isolates. This approach selects nanoparticles that display the desired targeting effects across all tested members of a class of cells and decreases the likelihood that an idiosyncratic cell line will unduly skew the screening results. The identified nanoparticle imaging probes were validated in vivo via intravital microscopy using pancreatic cancer mouse model.500 Although the molecular target of the probes remains to be discovered, the specific localization of these probes validates their respective use as in vivo neural tracers and cell discriminating agents.
5.2.1.4 Parallel Nanoparticle Libraries for Biosensors
Six functionalized poly(p-phenyleneethynylene)s were used to construct a protein biosensor array containing six nanoprobes. These highly fluorescent polymer nanoparticles possessed various charge characteristics and molecular weights. In a proof-of-principle experiment, 17 protein analytes were successfully discriminated by their different fluorescence properties after nanoparticle binding. However, those probes were not tested in real samples, such as blood serum, containing complex mixtures of proteins with potentially interfering species.501 Parallel modifications of magnetic nanoparticles aided in designing biomimetic nanoparticle biosensors with selectivity for specific cell types. The attachment of fluorescent molecules to magnetic nanoparticles permits the sorting or targeting of cells using both fluorescence-based technologies and MR imaging.26,499 The conjugation of a lipoic acid-containing NIR active tricarbocyanine library to GNPs yielded probes with a strong SERS enhancement with 12-fold higher sensitivity than the current standard, 3,3'-diethylthiatricarbocyanine iodide. By further conjugating the probe to a scFv anti-HER2 antibody, this ultrasensitive SERS bioprobe was used for in vivo cancer imaging with the potential for use as a noninvasive diagnostic tool for HER-2-positive cancers.502
5.2.2 Nano-combinatorial Chemistry to Modify Nanoparticles
Compared with a parallel chemistry approach, combinatorial chemistry can provide a large number of different but structurally related molecules or materials. A series of nanoparticles with different surface coatings but the same core structure and different diversity points can be synthesized rapidly using nano-combinatorial chemistry. The number of compounds synthesized increases exponentially with the number of diversity points. Furthermore, because the nanoparticles obtained by nano-combinatorial chemistry have surface coatings with similar structures, structure-activity relationships can be obtained, which is critical for computer-aided design for predicting nano-bio interactions.
5.2.2.1. Combinatorial Surface Modifications to Regulate Biocompatibility
Drug-loaded nanoparticles with targeting ligands can grant them therapeutic, targeting or imaging functions. However, achieving a low blood clearance rate, low acute toxicity and a low immune response remains challenging. To discover biocompatible MWNTs without a priori knowledge of the related targets, our laboratory developed a computer-assisted, nano-combinatorial synthetic library strategy. Combinatorial modifications of nanotube surfaces allowed us to map the unknown chemical space more effectively and rapidly. Computer-aided design was used to select surface molecules to present the most diverse structural and physicochemical properties. As a result of multiple biological screenings, we identified nanomaterials with the most optimal biocompatibility and preliminary structure-activity relationship (Figure 19).27 Furthermore, to test the improved molecular recognition after surface modifications on MWNTs, the library was screened against α-chymotrypsin to compare the MWNT interactions with the enzyme catalytic site and other binding sites. Four of 80 MWNTs were found to significantly affect enzymatic catalysis with much less nonspecific bindings.503 The combinational chemistry modifications on MWNTs have also regulated immune perturbations in mice and in macrophages. In another example, a modified nanotube deterred the mannose surface receptor from binding to the scavenge receptor and reduced immunotoxicity.504
Figure 19.
(A) Surface molecular compositions of combinatorial MWNT library members. (B) The multi-assay score of the f-MWNT library from four protein binding assays, cytotoxicity and immune response assays (at 50 μg/mL) were ranked for all library members. The sum of their ranks was designated the multi-assay score and is shown as vertical bars in the graph. The structures of the optimal hits are shown. Reprinted from ref 27 with permission. Copyright 2008 American Chemical Society.
5.2.2.2. Combinatorial modifications to regulate cell targeting
Recently, we reported a dual-ligand gold nanoparticle library to discern the slight difference between cells that have different surface receptor profiles surrounding a common primary receptor.505 This library used folic acid as the primary targeting ligand. The secondary ligand was one of the ligands from a combinatorial library of 42 molecules. Cell recognition was investigated in four cell lines with various expressions of folate receptor (FR) and totally unknown secondary receptor profile. Cell binding and uptake in different cells were quite different, suggesting different receptor profiles surrounding the FRs. Furthermore, multivalent bindings of dual ligands can differentiate cells with high and low expression of FRs and enhance the contrast between these cells in radiation-induced cell death. This enhanced cell recognition and discrimination of cells with seemingly a common receptor provide an opportunity to treat patients at a personalized medicine level with a much better safety profile.505
5.2.2.3. Combinatorial Modifications to Regulate Cell Differentiation
Biomaterials can drive stem cells to an appropriate differentiation level and decrease the apoptosis of transplanted cells. We have observed that carboxylated MWNTs promote myogenic differentiation of mouse myoblast cells C2C12 and inhibit cell apoptosis under differentiation conditions.506 We explored the tuning of cell differentiation using combinatorial surface modifications on MWNTs. Different surface modifications conferred different modulations of binding to BMP receptor type II and caused different levels of suppression of the BMP signaling pathway. Modified MWNTs fine-tuned the downstream signaling through a series of bHLH proteins and myogenic differentiation. These results demonstrated control of biological activities through surface-modified nanomaterials and opened an avenue for the potential application of nanomaterials in regenerative medicine.507
5.3. Computational Approaches to Elucidate Nano-Bio Interactions
The recent emergence and proliferation of experimental studies on the biological and toxicological activities of nanoparticles has created new challenges for computational scientists interested in rationalizing and exploiting these data by generating predictive models. Similar to data-rich areas of chemistry and biology, where the development of high-throughput technologies and the accumulation of large amounts of data have spurred the establishment of cheminformatics and bioinformatics, we are beginning to witness the appearance of nanoinformatics, which naturally complements nanotechnology. For instance, a group of academic, governmental and industrial scientists interested in informatics opportunities in nanotechnology met at the first Nanoinformatics conference in 2010 (http://nanotechinformatics.org/2010/overview). Presentations and idea exchanges at the conference led to the creation of the Nanoinformatics 2020 Roadmap (http://eprints.internano.org/607/) as the “first broad-based community effort to articulate the comprehensive needs and goals in nanoinformatics”.
One of the most important components of nanoinformatics, which resonates with similar components of bio- and cheminformatics, concerns understanding the relationships between the structure and the physical and/or biological properties of nanoparticles. To this end, given that sufficient and organized data collections are available, well-established methods of quantitative structure activity relationship (QSAR) modeling, widely used in cheminformatics,508 could be expanded towards nanoparticles, as we have begun to advocate for recently.29,31 Herein, we provide a brief overview of the quantitative nanostructure-activity relationships (QNAR) modeling approach, which should help experimental scientists studying nanoparticle-biological systems interactions rationalize their data as well as employ QNAR models to enable the rational design of novel nanoparticles with the desired properties. We describe major principles and best practices in QSAR modeling, which serves as a basis for QNAR, and present a few recent examples of QNAR studies to illustrate the concept and its applicability to the computational modeling of nanoparticles.
5.3.1. Quantitative Structure-Activity Relationships (QSAR)
QSAR modeling has been a widely and successfully used approach in medicinal chemistry and drug discovery for nearly 50 years. The QSAR approach can be generally described as an application of data analysis methods and statistics to developing models for the accurate prediction of biological activities or properties of compounds based on their structures. Any QSAR method can be generally defined as an application of mathematical and statistical methods to the problem of finding empirical relationships (QSAR models) of the form , where Pi are the biological activities (or other properties of interest) of the molecules, D1, D2,…,Dn are calculated (occasionally experimentally measured) structural properties (molecular descriptors) of the compounds, and is some empirically established mathematical transformation that should be applied to descriptors to calculate the property values for all molecules. The goal of QSAR modeling is to develop a computational means of predicting compound target properties (e.g., specific bioactivities or toxicities) from their chemical structure, as characterized with computed chemical descriptors or, in some cases, by experimental physical chemical characteristics. All QSAR approaches directly or indirectly imply simple similarity principles, which for some time have provided a foundation for experimental medicinal chemistry: compounds with similar structures are expected to have similar biological activities. The detailed description of major tenets of QSAR modeling is beyond the scope of this paper; the overview of many popular QSAR modeling techniques, including statistical and data mining techniques and descriptor calculation approaches, can be found in many reviews and monographs).509,510
For brevity, we shall comment on the most critical general aspects of model development and, most importantly, validation, which are especially significant in the context of using QSAR models for identifying novel hit compounds by virtual screening. Additional essential information concerning both common errors and established practices in the QSAR modeling field can be found in other critical essays on the subject.511,512 We emphasize such aspects because they are generic for any QSAR-based approach application (i.e., QNAR) to enable rigorous model development.
The strategy for developing extensively validated and externally predictive QSAR models is summarized in Figure 20 (described in more detail elsewhere513). We begin by carefully curating chemical structures and, if possible, associated biological activities to prepare the dataset for subsequent calculations. Then, a fraction of compounds (generally, 10-20%) is selected randomly as an external evaluation set. A more rigorous n-fold external validation protocol can be employed when the dataset is randomly divided into n nearly equal parts. The n-1 parts are then systematically used for model development, and the remaining fraction of compounds is used for model evaluation. The remaining subset of compounds (the modeling set) is divided into multiple training and test sets. The training sets are used to establish correlations between values of chemical descriptors and those of biological activities or other target properties; the models are validated internally for their ability to accurately predict the target properties of compounds in the test sets. All of the models that display high prediction accuracy for both the training and the test set form an ensemble that is applied to predict target properties for compounds in the external evaluation set. The critical step of the external validation is the use of the applicability domains,514 which is defined uniquely for each model used in the consensus (ensemble) prediction of the external set. If external validation demonstrates the significant predictive power of the models, we employ the models for virtual screening of available chemical databases (e.g., ZINC515) to identify putative active compounds and work with collaborators who could experimentally validate such hits.
Figure 20.
General workflow for the development of validated and externally predictive QSAR models.
We shall emphasize two of the most important aspects of the QSAR modeling workflow that directly pertain to QNAR modeling. First, the most critical difference between any chemical datasets that the QSAR modeling strategy is applied to is the type of descriptors used to characterize the compounds. Thus, the selection of the most appropriate descriptors of nanoparticles is the main challenge for developing QNAR strategies. Second, models resulting from the predictive QSAR modeling workflow (Fig. 1) can be used in virtual screening to prioritize the selection of chemicals for the experimental validation. In fact, it is increasingly critical to include experimental validation as the ultimate assertion of the model-based prediction. This paradigm fully applies to the use of QNAR modeling in nanotechnology as described in the next section.
5.3.2. QNAR modeling: Applications to the Rational Design of Nanoparticles with Desired Bioactivity Profiles
The evaluation of both the desired and undesired biological effects of nanoparticles is of critical importance for the future of nanotechnology. Predictive QNAR modeling can be used, by analogy with QSAR, to establish statistically significant relationships between the measured biological effects of nanoparticles and their physical, chemical and geometrical properties. As follows from the previous section, modeling of the biological effects of nanoparticles is possible if they can be characterized by descriptors. Figure 21 illustrates two different categories of nanoparticle datasets: those comprising nanoparticles with diverse metal cores and organic decorations, for which only experimentally measured properties (i.e., size, shape, zeta potential, morphology, surface area, chemical reactivity, chemical composition and aspect ratio) can be used as descriptors and those involving nanoparticles possessing the same core (e.g., CNTs) but different surface-modifying organic molecules, for which descriptors can be calculated for a single representative of the surface-modifying molecule. Several commercially available software packages, such as Dragon,516 MOE517 and ISIDA,518 could compute hundreds to thousands of theoretical descriptors for any single compound directly from its molecular structure. To illustrate the possible applications of the QNAR method, we describe several case studies for which we constructed and validated models (Figure 22).
Figure 21.
The applications of QNAR methods to nanomaterials with either diverse cores (characterized by experimental properties) or similar cores but diverse surface modifiers (characterized by computed chemical descriptors of modifying organic molecules). Adapted from ref 31 with permission. Copyright 2010 American Chemical Society.
Figure 22.
Unsupervised hierarchical clustering of CNTs uncovered common subgroups for both protein binders (red) and non-binders (blue). (A) The heat map represents the protein binding activities of the CNTs, which are sorted according to the dendrogram. The red color indicates high activity, while the blue represents low activity. (B) The red cluster represents four compounds consisting of a common substituent (ethyl benzoate, highlighted in the red square) causing similar (high) protein binding responses as measured in four different assays. (C) The blue cluster represents five compounds consisting of a common substructure (benzene chloride) highlighted in the blue square. These compounds behave similarly (low) activity in four protein binding assays. BSA, bovine serum albumin; CA, carbonic anhydrase; CT, α-chymotrypsin; HB, hemoglobin.
5.3.2.1. Modeling of Cellular Effects Induced by Diverse Nanoparticles using Experimentally Measured Properties as Descriptors
In a study, 51 diverse nanoparticles were tested in various cell-based assays.30 All nanoparticles were tested in vitro against four cell lines in four different assays at four different concentrations, resulting in a 51 × 64 biological data matrix of experimental results. Each cell of this matrix reported the biological activity profile induced by a given nanoparticle at a certain concentration in a particular assay for a given cell line. Assay response values were expressed in units of standard deviations of the distribution obtained when control cells were treated with phosphate buffered saline (PBS) alone: ZNP = (μNP - μPBS) / σPBS, where μPBS is the mean of the control tests with PBS, and σPBS is its standard deviation. The authors also reported four experimentally measured descriptors for 44 of 51 tested nanoparticles: size, two types of relaxivities and zeta potential.
We have demonstrated the feasibility of deriving robust QNAR models using the following four experimental descriptors: size, two types of relaxivities and zeta potential. Four such structural descriptors were available for 44 of the 51 nanoparticles. To capture nanoparticles’ activity across a broader swath of biology, the Shaw group measured the effects of nanoparticles in four diverse cell line, at four doses and using four assays that interrogate different aspects of cellular physiology. Thus, the biological effects of each nanomaterial can be described as a 64-feature vector (4 cell lines × 4 assays × 4 doses).
In theory, 64 independent QNAR models could be developed, with each model attempting to reproduce the biological response induced by 44 nanoparticles for a given assay in a particular cell line at a given dose; however, we were unable to develop statistically significant models. Therefore, we combined the entire 64-dimensional vector for each nanoparticle into a single averaged response. We then defined two binary classes using an arbitrary threshold at Zmean = −0.40 to enable a binary classification of nanoparticles. Two groups, each containing the same number of nanoparticles, were thus defined: 22 nanoparticles belonged to class 1 (Zmean ≥ −0.40), and the remaining 22 were placed in class 0 (Zmean < −0.40).31
The WinSVM program (a Windows implementation of a support vector machine) developed in-house was used to derive QNAR classification models. A rigorous 5-fold external cross-validation was followed to validate the models; under this procedure, the program splits the entire dataset five times into a modeling set including 80% of nanoparticles and the external validation set, comprising the remaining 20% of nanoparticles. Only the modeling set (which is divided additionally into multiple training and test sets) was used to build and validate models, and models with reasonable training and test set prediction power were selected for predicting the class membership of the external set. Importantly, each nanoparticles was included in a validation set only once, allowing us to calculate the overall external prediction accuracy for the whole set.
The results demonstrated that the SVM models yielded an external prediction as high as 73% for the entire set of 44 nanoparticles. Within five independent external folds, prediction performances led from 56 to 88% accuracy. Apparently, the combination of biological profiles into one single binary classification scheme allowed us to detect the overall biological signal from noise and thus obtain QNAR models with both good internal fitness and external predictive power.
5.3.2.2. Modeling of Nanoparticle Uptake in PaCa2 Cancer Cells using Computational Descriptors
Whether the multivalent attachment of small organic molecules to the same nanoparticles can increase their specific binding affinity to certain cells without a priori knowledge of the cell target was investigated.519 A combinatorial library of 109 nanoparticles was synthesized in which a superparamagnetic nanoparticle (cross-linked iron oxide with amine groups, CLIO-NH2) was decorated with different synthetic small molecules. Nanoparticles were screened against different cell lines, including PaCa2 human pancreatic cancer cells, U937 macrophage cell lines, resting and activated primary human macrophages and human umbilical vein endothelial cells (HUVECs). In contrast to the other cell lines, the uptake of the nanoparticles in PaCa2 pancreatic cancer cells were diverse and highly dependent on surface modification, enabling the application of the QSAR modeling approach to these data. We represented each individual nanoparticle by the structure of a single, organic molecule decorating its surface. Thus, each nanoparticle was represented by a unique set of descriptor values determined for the conjugated small molecule.
Using this representation, we have developed statistically significant models by relying on chemical descriptors and nanoparticle cellular uptake. The models' external prediction power was shown to be as high as R2 = 0.72 and a mean absolute error of 0.18 under a 5-fold external validation procedure.31 This study suggests that the cellular behavior of a series of nanoparticles sharing a common core can be reasonably well predicted using QNAR models built with the descriptors of surface modifying ligands only.
5.3.2.3. QNAR Models as Tools for Designing CNTs with Desired Biological Properties
CNTs are among the most well-known nanoparticles, particularly in the area of drug delivery.520-523 We utilized a combinatorial synthesis technique to decorate the CNTs’ large surface area with a series of congeneric organic molecules and investigated the overall toxicity of this series of nanoparticles.ENREF 522 A total of 84 CNTs were tested in several in vitro toxicological assays, including protein binding assays (BSA, carbonic anhydrase, chymotrypsin and hemoglobin), acute toxicity assays (survival percentage) and immune toxicity assays (secretion of nitric oxide), all at two different doses (7.5 and 15 μg/mL). All of the CNTs shared the same core structure, and thus the variability of their biological/toxicological behavior could be interpreted only based on the surface modifications.27
Each CNT was represented with a unique organic molecule from its coating surface, similar to the study described above, and chemical descriptors were calculated for each molecule. We assigned a binary toxicity class (1 as toxic and 0 as non-toxic) to each CNT based on the acute toxicity assay and the toxicity measured for pristine CNTs (CNTs with no surface modifiers). Only strong and weak CNT toxicants were considered for the modeling: 38 strong toxicants (class 1) vs. 35 weak toxicants (class 0) were subjected to QNAR modeling using the Random Forest approach. Similarly to other case studies, we realized a 5-fold external cross-validation. The results were surprisingly good with an overall external prediction accuracy as high as 74% (n = 73, sensitivity = 79%, specificity = 69%). We also developed QNAR models using protein-binding profiles that yielded 77% external prediction accuracy for classifying strong vs. weak CNT protein binders.
These models have been employed for the virtual screening of a large library of 240,000 potential CNT surface modifiers in order to identify those with desired properties (low toxicity, low binding affinity for carbonic anhydrase). Ten compounds that were predicted to be toxic and ten compounds that were predicted to be non-toxic were selected for the experimental validation. The preliminary results suggest that all predictions for non-toxic compounds were accurate, and for six out ten compounds predicted to be toxic, the experimental results were in agreement with the computational annotation.
We conclude that analogous to conventional applications of QSAR modeling for the analysis of bioactive organic molecules, QNAR models can be useful for predicting activity profiles of novel nanoparticles, either from their experimental or computationally derived descriptors and for designing and manufacturing safer nanoparticles with desired properties. However, a shortage of systematic experimental data that characterize the constitutional, physical and chemical properties of nanoparticles remains. In addition, no standard set of biological assays has been clearly defined to objectively and relevantly evaluate the in vitro (and in vivo) effects of tested CNTs. The development of systematized data repositories through collective efforts of experimental and computational scientists will enable the development of new and highly valuable QNAR models capable of rationally guiding further experimental studies toward safer and more effective nanoparticles.
6. CONCLUDING REMARKS
A double-edged sword, nanotechnology provides us with enormous opportunities while posing a potential threat to human health. Therefore, understanding how engineered nanoparticles interact with biological systems is urgent and critical. Nanoparticles are known to cause organ damage in experimental animals and even in human. Such damages are the direct or indirect results of perturbations of cells and biological molecules such as proteins, lipids, DNA and small molecules by nanoparticles. The nature and magnitude of nanoparticle interactions with biological molecules are determined by both the biological molecules and engineered nanoparticles themselves.
The fundamental physicochemical properties of nanoparticles, such as their surface charge, hydrophobicity and chemical bonding interactions, play significant roles in nano-bio interactions. Nanoparticles with different physicochemical properties exhibit different interactions with biological systems. For example, they may be surrounded by protein coronas of different nature, bind to different cell membrane proteins, enter or perturb cells differently or exhibit diverse compatibility with physiological systems. A systematic understanding of such chemical interactions will help elucidate the QNAR of nano-bio interactions. Such an understanding also facilitates the generation of nanomaterials through safe-by-design processes.
Based on these considerations, the intentional surface modification of nanomaterials by one-at-a-time, parallel or combinatorial synthesis has been shown to alter nano-bio interactions. Such effects have encouraged the development of biocompatible or bio-targeting nanoparticles. Another significant development is that nano-bio interactions can now be predicted by computational chemistry. These experimental and theoretical approaches have strengthened our understanding of the chemical basis of nano-bio interactions and provide unprecedented methodologies to proactively design and manufacture safe nanomaterials for a sustainable nanotechnology era.
However, the investigation of nano-bio interactions is by no means complete. Many challenges remain. First, we must understand the mechanisms of nano-bio interactions with greater clarity via a better characterization of experimental systems, such as nanoparticles. Due to their chemical nature and their interactions with microenvironmental molecules, the chemical identity, biological identity or cellular identity of nanoparticles may differ. Comprehensive characterization will improve our understanding of such interaction mechanisms, and QNAR studies can be conducted with better accuracy. Moreover, diverse findings from different laboratories can be compared to form a systematic knowledge base. Second, at the cellular level, an interaction with a receptor protein leads to a network of cellular signaling and the release of numerous molecules for signal propagation, feedback and adaptation. Therefore, an in-depth understanding of nano-bio interactions at the cellular level relies on the future development of systems biology and related knowledge bases. Third and most significant, at the physiological level, the impact of nanoparticles on human health is an end result of complicated interplays of complex interactions. For example, nanoparticles always adsorb environmental pollutants and perturb cellular nutrient molecules and redox balance, yielding multipronged effects. Furthermore, such toxicant exposures are due to various sources and last a lifetime, beginning with a fertilized egg. Therefore, research aimed at understanding nano-bio interactions is an integral part of exploration in chemical biology and toxicology, having unique features. Therefore, it relies on the future progress of the chemical and biological sciences.
ACKNOWLEDGEMENTS
This work was supported by the National Basic Research Program of China (2010CB933504) and the National Natural Science Foundation of China (21077068, and 21137002).
LIST OF ABBREVIATIONS
- AFM
atomic force microscopy
- ATP
adenosine-5'-triphosphate
- BMP
bone morphogenetic protein
- BMPR2
bone morphogenetic protein receptor type II
- BSA
bovine serum albumin
- CB
carbon black
- CD
circular dichroism
- CLSM
confocal laser scanning microscopy
- CNTs
carbon nanotubes
- CTAB
hexadecyltrimethylammonium bromide
- DLS
dynamic light scattering
- DNA
deoxyribonucleic acid
- DPPC
dipalmitoyl phosphatidylcholine
- DSC
differential scanning calorimetry
- EB
ethidium bromide
- ENMs
engineered nanomaterials
- ER
endoplasmic reticulum
- FCS
fluorescence correlation spectroscopy
- FTIR
Fourier-transform infrared spectroscopy
- GC-MS
gas chromatography-mass spectrometry
- GNPs
gold nanoparticles
- GO
graphene oxide
- GPCRs
G protein coupled receptors
- GSH
glutathione
- Hb
hemoglobin
- HSA
human serum albumin
- HSQC NMR
heteronuclear single quantum coherence nuclear magnetic resonance
- HUVECs
human umbilical vein endothelial cells
- ITC
isothermal titration calorimetry
- iTRAQ
isobaric tags for relative and absolute quantitation
- LB
Langmuir-Blodgett
- LC-MS
liquid chromatography mass spectroscopy
- MALDI TOF-MS
matrix-assisted laser desorption-ionization time-of-flight mass spectrometry
- MAPK
mitogen-activated protein kinase
- MAS NMR
magic angle spinning nuclear magnetic resonance
- MNPs
magnetic nanoparticles
- MS
mass spectrometry
- MWNTs
multi-walled carbon nanotubes
- NF-κB
nuclear factor kappa B
- NIR
near infrared
- NMR
nuclear magnetic resonance
- PBAEs
poly-β-amino esters
- PBS
phosphate buffered saline
- PEG
polyethylene glycol
particle replication in non-wetting template
- QCM
quartz crystal microbalance
- QDs
quantum dots
- QNAR
quantitative nanostructure activity relationships
- QSAR
quantitative structure-activity relationships
- RNA
ribonucleic acid
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RT-PCR
reverse transcription polymerase chain reaction
- SEM
scanning electron microscopy
- SERS
surface-enhanced Raman spectroscopy
- SPR
surface plasma resonance
- SVM
support vector machine
- SWNTs
single-walled carbon nanotubes
- TEM
transmission electron microscopy
- TGF-β
transforming growth factor beta
- TLR
Toll-like receptor
- TOF MS
time-of-flight mass spectrometry
- UV-Vis
ultraviolet-visible
- VEGF
vascular endothelial growth factor
- XPS
X-ray photoelectron spectroscopy
REFERENCES
- (1).European Union Law 32011H0696. http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=CELEX:32011H0696:EN:NOT (accessed Sep 12, 2013)
- (2).Stankovich S, Dikin DA, Dommett GHB, Kohlhaas KM, Zimney EJ, Stach EA, Piner RD, Nguyen ST, Ruoff RS. Nature. 2006;442:282. doi: 10.1038/nature04969. [DOI] [PubMed] [Google Scholar]
- (3).Tans SJ, Verschueren ARM, Dekker C. Nature. 1998;393:49. [Google Scholar]
- (4).Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramirez JT, Yacaman MJ. Nanotechnology. 2005;16:2346. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- (5).Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat. Mater. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- (6).Nanomaterials Production 2002-2016: Production Volumes, Revenues and End User Market Demand. http://www.researchandmarkets.com/research/97chkk/nanomaterials_prod (accessed Sep 12, 2013)
- (7).Colvin VL. Nat. Biotechnol. 2003;21:1166. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- (8).Maynard AD, Kuempel ED. J. Nanopart. Res. 2005;7:587. [Google Scholar]
- (9).Song Y, Li X, Du X. Eur. Respir. J. 2009;34:559. doi: 10.1183/09031936.00178308. [DOI] [PubMed] [Google Scholar]
- (10).Gottschalk F, Nowack B. J. Environ. Monit. 2011;13:1145. doi: 10.1039/c0em00547a. [DOI] [PubMed] [Google Scholar]
- (11).Kendall M, Holgate S. Respirology. 2012;17:743. doi: 10.1111/j.1440-1843.2012.02171.x. [DOI] [PubMed] [Google Scholar]
- (12).Wiecinski PN, Metz KM, Mangham AN, Jacobson KH, Hamers RJ, Pedersen JA. Nanotoxicology. 2009;3:202. doi: 10.1080/17435390902859556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pietroiusti A. Nanoscale. 2012;4:1231. doi: 10.1039/c2nr11688j. [DOI] [PubMed] [Google Scholar]
- (14).Chao J.-b., Liu J.-f., Yu S.-j., Feng Y.-d., Tan Z.-q., Liu R, Yin Y.-g. Anal. Chem. 2011;83:6875. doi: 10.1021/ac201086a. [DOI] [PubMed] [Google Scholar]
- (15).Judy JD, Unrine JM, Bertsch PM. Environ. Sci. Technol. 2011;45:776. doi: 10.1021/es103031a. [DOI] [PubMed] [Google Scholar]
- (16).Werlin R, Priester JH, Mielke RE, Kramer S, Jackson S, Stoimenov PK, Stucky GD, Cherr GN, Orias E, Holden PA. Nat. Nanotechnol. 2011;6:65. doi: 10.1038/nnano.2010.251. [DOI] [PubMed] [Google Scholar]
- (17).De Jong WH, Borm PJA. Int. J. Nanomed. 2008;3:133. [Google Scholar]
- (18).Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5th Garland Science; New York: 2007. [Google Scholar]
- (19).Mu Q, Liu W, Xing Y, Zhou H, Li Z, Zhang Y, Ji L, Wang F, Si Z, Zhang B, Yan B. J. Phys. Chem. C. 2008;112:3300. [Google Scholar]
- (20).Yang J, Lee JY, Too HP, Chow GM. Biophys. Chem. 2006;120:87. doi: 10.1016/j.bpc.2005.10.011. [DOI] [PubMed] [Google Scholar]
- (21).Yi CQ, Liu DD, Fong CC, Zhang JC, Yang MS. ACS Nano. 2010;4:6439. doi: 10.1021/nn101373r. [DOI] [PubMed] [Google Scholar]
- (22).Puddu V, Perry CC. ACS Nano. 2012;6:6356. doi: 10.1021/nn301866q. [DOI] [PubMed] [Google Scholar]
- (23).Fischer NO, Verma A, Goodman CM, Simard JM, Rotello VM. J. Am. Chem. Soc. 2003;125:13387. doi: 10.1021/ja0352505. [DOI] [PubMed] [Google Scholar]
- (24).Liu YP, Meyer-Zaika W, Franzka S, Schmid G, Tsoli M, Kuhn H. Angew. Chem., Int. Ed. 2003;42:2853. doi: 10.1002/anie.200250235. [DOI] [PubMed] [Google Scholar]
- (25).Zhang Y, Tekobo S, Tu Y, Zhou QF, Jin XL, Dergunov SA, Pinkhassik E, Yan B. ACS Appl. Mater. Interfaces. 2012;4:4099. doi: 10.1021/am300840p. [DOI] [PubMed] [Google Scholar]
- (26).Sun EY, Josephson L, Kelly KA, Weissleder R. Bioconjugate Chem. 2005;17:109. doi: 10.1021/bc050290e. [DOI] [PubMed] [Google Scholar]
- (27).Zhou H, Mu Q, Gao N, Liu A, Xing Y, Gao S, Zhang Q, Qu G, Chen Y, Liu G, Zhang B, Yan B. Nano Lett. 2008;8:859. doi: 10.1021/nl0730155. [DOI] [PubMed] [Google Scholar]
- (28).Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat. Biotech. 2005;23:1418. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- (29).Fourches D, Pu DQY, Tropsha A. Comb. Chem. High Throughput Screening. 2011;14:217. doi: 10.2174/138620711794728743. [DOI] [PubMed] [Google Scholar]
- (30).Shaw SY, Westly EC, Pittet MJ, Subramanian A, Schreiber SL, Weissleder R. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7387. doi: 10.1073/pnas.0802878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Fourches D, Pu DQY, Tassa C, Weissleder R, Shaw SY, Mumper RJ, Tropsha A. ACS Nano. 2010;4:5703. doi: 10.1021/nn1013484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J. Am. Chem. Soc. 2010;132:5761. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- (33).Rao CNR, Biswas K. Annu. Rev. Anal. Chem. 2009;2:435. doi: 10.1146/annurev-anchem-060908-155236. [DOI] [PubMed] [Google Scholar]
- (34).Zimbone M, Calcagno L, Messina G, Baeri P, Compagnini G. Mater. Lett. 2011;65:2906. [Google Scholar]
- (35).Patil S, Sandberg A, Heckert E, Self W, Seal S. Biomaterials. 2007;28:4600. doi: 10.1016/j.biomaterials.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Elzey S, Tsai DH, Yu LL, Winchester MR, Kelley ME, Hackley VA. Anal. Bioanal. Chem. 2013;405:2279. doi: 10.1007/s00216-012-6617-z. [DOI] [PubMed] [Google Scholar]
- (37).Baer DR, Gaspar DJ, Nachimuthu P, Techane SD, Castner DG. Anal. Bioanal. Chem. 2010;396:983. doi: 10.1007/s00216-009-3360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zhou H, Li X, Lemoff A, Zhang B, Yan B. Analyst. 2010;135:1210. doi: 10.1039/c0an00066c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhou HY, Du FF, Li X, Zhang B, Li W, Yan B. J. Phys. Chem. C. 2008;112:19360. [Google Scholar]
- (40).Zhang B, Yan B. Anal. Bioanal. Chem. 2010;396:973. doi: 10.1007/s00216-009-2996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang Y, Yan B, Chen L. Chem. Rev. 2012;113:1391. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- (42).Kumari A, Yadav SK. Expert Opinion on Drug Delivery. 2011;8:141. doi: 10.1517/17425247.2011.547934. [DOI] [PubMed] [Google Scholar]
- (43).Zhao F, Zhao Y, Liu Y, Chang XL, Chen CY, Zhao YL. Small. 2011;7:1322. doi: 10.1002/smll.201100001. [DOI] [PubMed] [Google Scholar]
- (44).Treuel L, Jiang XE, Nienhaus GU. J. R. Soc., Interface. 2013;10 doi: 10.1098/rsif.2012.0939. art.no.20120939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Fadaee-Shohada MJ, Hirst RA, Rutman A, Roberts IS, O'Callaghan C, Andrew PW. PLoS One. 2010;5:e10450. doi: 10.1371/journal.pone.0010450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhao YN, Sun XX, Zhang GN, Trewyn BG, Slowing II, Lin VSY. ACS Nano. 2011;5:1366. doi: 10.1021/nn103077k. [DOI] [PubMed] [Google Scholar]
- (47).Gao NN, Zhang Q, Mu QX, Bai YH, Li LW, Zhou HY, Butch ER, Powell TB, Snyder SE, Jiang GB, Yan B. ACS Nano. 2011;5:4581. doi: 10.1021/nn200283g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Corazzari I, Gilardino A, Dalmazzo S, Fubini B, Lovisolo D. Toxicol. In Vitro. 2013;27:752. doi: 10.1016/j.tiv.2012.12.016. [DOI] [PubMed] [Google Scholar]
- (49).Seib FP, Jones GT, Rnjak-Kovacina J, Lin Y, Kaplan DL. Adv. Healthcare Mater. 2013 doi: 10.1002/adhm.201300034. Published online April 26, 2013. DOI: 10.1002/adhm.201300034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Gasser M, Rothen-Rutishauser B, Krug HF, Gehr P, Nelle M, Yan B, Wick P. J. Nanobiotechnol. 2010:8. doi: 10.1186/1477-3155-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kapralov AA, Feng WH, Amoscato AA, Yanamala N, Balasubramanian K, Winnica DE, Kisin ER, Kotchey GP, Gou PP, Sparvero LJ, Ray P, Mallampalli RK, Klein-Seetharaman J, Fadeel B, Star A, Shvedova AA, Kagan VE. ACS Nano. 2012;6:4147. doi: 10.1021/nn300626q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ge CC, Du JF, Zhao LN, Wang LM, Liu Y, Li DH, Yang YL, Zhou RH, Zhao YL, Chai ZF, Chen CY. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16968. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Prado-Gotor R, Grueso E. PCCP. 2011;13:1479. doi: 10.1039/c0cp00901f. [DOI] [PubMed] [Google Scholar]
- (54).Guo L, Bussche AV, Buechner M, Yan AH, Kane AB, Hurt RH. Small. 2008;4:721. doi: 10.1002/smll.200700754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Hellstrand E, Lynch I, Andersson A, Drakenberg T, Dahlback B, Dawson KA, Linse S, Cedervall T. FEBS J. 2009;276:3372. doi: 10.1111/j.1742-4658.2009.07062.x. [DOI] [PubMed] [Google Scholar]
- (56).Kumar P, Bohidar HB. Colloids Surf., A. 2010;361:13. [Google Scholar]
- (57).Bakshi MS, Zhao L, Smith R, Possmayer F, Petersen NO. Biophys. J. 2008;94:855. doi: 10.1529/biophysj.107.106971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Wang LY, Castranova V, Mishra A, Chen B, Mercer RR, Schwegler-Berry D, Rojanasakul Y. Part. Fibre Toxicol. 2010;7:31. doi: 10.1186/1743-8977-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Beck-Broichsitter M, Ruppert C, Schmehl T, Guenther A, Betz T, Bakowsky U, Seeger W, Kissel T, Gessler T. Nanomed. -Nanotechnol. Biol. Med. 2011;7:341. doi: 10.1016/j.nano.2010.10.007. [DOI] [PubMed] [Google Scholar]
- (60).Herzog E, Byrne HJ, Davoren M, Casey A, Duschl A, Oostingh GJ. Toxicol. Appl. Pharmacol. 2009;236:276. doi: 10.1016/j.taap.2009.02.007. [DOI] [PubMed] [Google Scholar]
- (61).Schleh C, Muhlfeld C, Pulskamp K, Schmiedl A, Nassimi M, Lauenstein HD, Braun A, Krug N, Erpenbeck VJ, Hohlfeld JM. Respiratory Research. 2009;10:90. doi: 10.1186/1465-9921-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Peetla C, Stine A, Labhasetwar V. Mol. Pharm. 2009;6:1264. doi: 10.1021/mp9000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Baoukina S, Monticelli L, Marrink SJ, Tieleman DP. Langmuir. 2007;23:12617. doi: 10.1021/la702286h. [DOI] [PubMed] [Google Scholar]
- (64).Peetla C, Labhasetwar V. Mol. Pharm. 2008;5:418. doi: 10.1021/mp700140a. [DOI] [PubMed] [Google Scholar]
- (65).Roiter Y, Ornatska M, Rammohan AR, Balakrishnan J, Heine DR, Minko S. Langmuir. 2009;25:6287. doi: 10.1021/la900119a. [DOI] [PubMed] [Google Scholar]
- (66).Roiter Y, Ornatska M, Rammohan AR, Balakrishnan J, Heine DR, Minko S. Nano Lett. 2008;8:941. doi: 10.1021/nl080080l. [DOI] [PubMed] [Google Scholar]
- (67).Mecke A, Majoros IJ, Patri AK, Baker JR, Holl MMB, Orr BG. Langmuir. 2005;21:10348. doi: 10.1021/la050629l. [DOI] [PubMed] [Google Scholar]
- (68).Tsogas I, Tsiourvas D, Nounesis G, Paleos CM. Langmuir. 2006;22:11322. doi: 10.1021/la0620861. [DOI] [PubMed] [Google Scholar]
- (69).Hartono D, Hody; Yang KL, Yung LYL. Biomaterials. 2010;31:3008. doi: 10.1016/j.biomaterials.2010.01.003. [DOI] [PubMed] [Google Scholar]
- (70).Hartono D, Qin WJ, Yang KL, Yung LYL. Biomaterials. 2009;30:843. doi: 10.1016/j.biomaterials.2008.10.037. [DOI] [PubMed] [Google Scholar]
- (71).Ionov M, Gardikis K, Wrobel D, Hatziantoniou S, Mourelatou H, Majoral JP, Klajnert B, Bryszewska M, Demetzos C. Colloids Surf., B. 2011;82:8. doi: 10.1016/j.colsurfb.2010.07.046. [DOI] [PubMed] [Google Scholar]
- (72).Bhattacharya S, Srivastava A. Langmuir. 2003;19:4439. [Google Scholar]
- (73).Wang B, Zhang LF, Bae SC, Granick S. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18171. doi: 10.1073/pnas.0807296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Zhang XF, Yang SH. Langmuir. 2011;27:2528. doi: 10.1021/la104449y. [DOI] [PubMed] [Google Scholar]
- (75).Harishchandra RK, Saleem M, Galla HJ. J. R. Soc., Interface. 2010;7:S15. doi: 10.1098/rsif.2009.0329.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Goertz MP, Goyal N, Bunker BC, Montano GA. J. Colloid Interface Sci. 2011;358:635. doi: 10.1016/j.jcis.2011.02.063. [DOI] [PubMed] [Google Scholar]
- (77).Mecke A, Lee DK, Ramamoorthy A, Orr BG, Holl MMB. Langmuir. 2005;21:8588. doi: 10.1021/la051800w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Hirano A, Uda K, Maeda Y, Akasaka T, Shiraki K. Langmuir. 2010;26:17256. doi: 10.1021/la103615b. [DOI] [PubMed] [Google Scholar]
- (79).Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Holl MMB. Nano Lett. 2008;8:420. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- (80).Ramachandran S, Merrill NE, Blick RH, van der Weide DW. Biosensors & Bioelectronics. 2005;20:2173. doi: 10.1016/j.bios.2004.08.042. [DOI] [PubMed] [Google Scholar]
- (81).Ramachandran S, Kumar GL, Blick RH, van der Weide DW. Appl. Phys. Lett. 2005:86. [Google Scholar]
- (82).Sachan AK, Harishchandra RK, Bantz C, Maskos M, Reichelt R, Galla HJ. ACS Nano. 2012;6:1677. doi: 10.1021/nn204657n. [DOI] [PubMed] [Google Scholar]
- (83).Peetla C, Labhasetwar V. Langmuir. 2009;25:2369. doi: 10.1021/la803361y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Hong SP, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi XY, Balogh L, Orr BG, Baker JR, Holl MMB. Bioconjugate Chem. 2004;15:774. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- (85).Hong SP, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Holl MMB. Bioconjugate Chem. 2006;17:728. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- (86).Peetla C, Rao KS, Labhasetwar V. Mol. Pharm. 2009;6:1311. doi: 10.1021/mp900011h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Li Y, Chen X, Gu N. J. Phys. Chem. B. 2008;112:16647. doi: 10.1021/jp8051906. [DOI] [PubMed] [Google Scholar]
- (88).Yang K, Ma YQ. Nat. Nanotechnol. 2010;5:579. doi: 10.1038/nnano.2010.141. [DOI] [PubMed] [Google Scholar]
- (89).Monticelli L, Salonen E, Ke PC, Vattulainen I. Soft Matter. 2009;5:4433. [Google Scholar]
- (90).Park J, Lu W. Physical Review E. 2009;80 doi: 10.1103/PhysRevE.80.021607. art.no.021607. [DOI] [PubMed] [Google Scholar]
- (91).Choe S, Chang R, Jeon J, Violi A. Biophys. J. 2008;95:4102. doi: 10.1529/biophysj.107.123976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Jusufi A, DeVane RH, Shinoda W, Klein ML. Soft Matter. 2011;7:1139. [Google Scholar]
- (93).Song B, Yuan HJ, Jameson CJ, Murad S. Mol. Phys. 2011;109:1511. [Google Scholar]
- (94).Fiedler SL, Violi A. Biophys. J. 2010;99:144. doi: 10.1016/j.bpj.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Ding HM, Ma YQ. Nanoscale. 2012;4:1116. doi: 10.1039/c1nr11425e. [DOI] [PubMed] [Google Scholar]
- (96).Ginzburg VV, Balijepailli S. Nano Lett. 2007;7:3716. doi: 10.1021/nl072053l. [DOI] [PubMed] [Google Scholar]
- (97).Kelly CV, Leroueil PR, Orr BG, Holl MMB, Andricioaei I. J. Phys. Chem. B. 2008;112:9346. doi: 10.1021/jp8013783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Qiao R, Roberts AP, Mount AS, Klaine SJ, Ke PC. Nano Lett. 2007;7:614. doi: 10.1021/nl062515f. [DOI] [PubMed] [Google Scholar]
- (99).Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Adv. Drug Del. Rev. 2009;61:428. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Lynch I, Dawson KA, Linse S. Sci. STKE. 2006;2006:pe14. doi: 10.1126/stke.3272006pe14. [DOI] [PubMed] [Google Scholar]
- (101).Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd Springer; Berlin, Germany: 2009. [Google Scholar]
- (102).Casanova D, Giaume D, Moreau M, Martin JL, Gacoin T, Boilot JP, Alexandrou A. J. Am. Chem. Soc. 2007;129:12592. doi: 10.1021/ja0731975. [DOI] [PubMed] [Google Scholar]
- (103).Rocker C, Potzl M, Zhang F, Parak WJ, Nienhaus GU. Nat. Nanotechnol. 2009;4:577. doi: 10.1038/nnano.2009.195. [DOI] [PubMed] [Google Scholar]
- (104).Calzolai L, Franchini F, Gilliland D, Rossi F. Nano Lett. 2010;10:3101. doi: 10.1021/nl101746v. [DOI] [PubMed] [Google Scholar]
- (105).Li N, Zeng S, He L, Zhong WW. Anal. Chem. 2011;83:6929. doi: 10.1021/ac201889j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Ipe BI, Shukla A, Lu H, Zou B, Rehage H, Niemeyer CM. ChemPhysChem. 2006;7:1112. doi: 10.1002/cphc.200500660. [DOI] [PubMed] [Google Scholar]
- (107).Rich RL, Myszka DG. Curr. Opin. Biotechnol. 2000;11:54. doi: 10.1016/s0958-1669(99)00054-3. [DOI] [PubMed] [Google Scholar]
- (108).Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2050. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Kaufman ED, Belyea J, Johnson MC, Nicholson ZM, Ricks JL, Shah PK, Bayless M, Pettersson T, Feldotö Z, Blomberg E, Claesson P, Franzen S. Langmuir. 2007;23:6053. doi: 10.1021/la063725a. [DOI] [PubMed] [Google Scholar]
- (110).Gerdon AE, Wright DW, Cliffel DE. Anal. Chem. 2004;77:304. doi: 10.1021/ac048953t. [DOI] [PubMed] [Google Scholar]
- (111).Shenderova OA, Zhirnov VV, Brenner DW. Crit. Rev. Solid State Mater. Sci. 2002;27:227. [Google Scholar]
- (112).Kroto HW. J. Chem. Soc., Faraday Trans. 1990;86:2465. [Google Scholar]
- (113).Geim AK, Novoselov KS. Nat. Mater. 2007;6:183. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- (114).Iijima S, Ichihashi T. Nature. 1993;363:603. [Google Scholar]
- (115).Nepal D, Geckeler KE. Small. 2006;2:406. doi: 10.1002/smll.200500351. [DOI] [PubMed] [Google Scholar]
- (116).Li XJ, Chen W, Zhan QW, Dai LM, Sowards L, Pender M, Naik RR. J. Phys. Chem. B. 2006;110:12621. doi: 10.1021/jp061518d. [DOI] [PubMed] [Google Scholar]
- (117).Bradley K, Briman M, Star A, Gruner G. Nano Lett. 2004;4:253. [Google Scholar]
- (118).Karajanagi SS, Vertegel AA, Kane RS, Dordick JS. Langmuir. 2004;20:11594. doi: 10.1021/la047994h. [DOI] [PubMed] [Google Scholar]
- (119).Han ZJ, Ostrikov K, Tan CM, Tay BK, Peel SAF. Nanotechnology. 2011:22. doi: 10.1088/0957-4484/22/29/295712. [DOI] [PubMed] [Google Scholar]
- (120).Matsuura K, Saito T, Okazaki T, Ohshima S, Yumura M, Iijima S. Chem. Phys. Lett. 2006;429:497. [Google Scholar]
- (121).Lin Y, Allard LF, Sun YP. J. Phys. Chem. B. 2004;108:3760. [Google Scholar]
- (122).Su ZD, Leung T, Honek JF. J. Phys. Chem. B. 2006;110:23623. doi: 10.1021/jp065837g. [DOI] [PubMed] [Google Scholar]
- (123).Balavoine F, Schultz P, Richard C, Mallouh V, Ebbesen TW, Mioskowski C. Angew. Chem., Int. Ed. 1999;38:1912. doi: 10.1002/(SICI)1521-3773(19990712)38:13/14<1912::AID-ANIE1912>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- (124).Ling WL, Biro A, Bally I, Tacnet P, Deniaud A, Doris E, Frachet P, Schoehn G, Pebay-Peyroula E, Arlaud GJ. ACS Nano. 2011;5:730. doi: 10.1021/nn102400w. [DOI] [PubMed] [Google Scholar]
- (125).Nepal D, Geckeler KE. Small. 2007;3:1259. doi: 10.1002/smll.200600511. [DOI] [PubMed] [Google Scholar]
- (126).Zorbas V, Ortiz-Acevedo A, Dalton AB, Yoshida MM, Dieckmann GR, Draper RK, Baughman RH, Jose-Yacaman M, Musselman IH. J. Am. Chem. Soc. 2004;126:7222. doi: 10.1021/ja049202b. [DOI] [PubMed] [Google Scholar]
- (127).Karajanagi SS, Yang HC, Asuri P, Sellitto E, Dordick JS, Kane RS. Langmuir. 2006;22:1392. doi: 10.1021/la0528201. [DOI] [PubMed] [Google Scholar]
- (128).Shen JW, Wu T, Wang Q, Kang Y. Biomaterials. 2008;29:3847. doi: 10.1016/j.biomaterials.2008.06.013. [DOI] [PubMed] [Google Scholar]
- (129).Fu ZM, Luo Y, Derreumaux P, Wei GH. Biophys. J. 2009;97:1795. doi: 10.1016/j.bpj.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Kang Y, Liu YC, Wang Q, Shen JW, Wu T, Guan WJ. Biomaterials. 2009;30:2807. doi: 10.1016/j.biomaterials.2009.01.024. [DOI] [PubMed] [Google Scholar]
- (131).Linse S, Cabaleiro-Lago C, Xue WF, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8691. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Benyamini H, Shulman-Peleg A, Wolfson HJ, Belgorodsky B, Fadeev L, Gozin M. Bioconjugate Chem. 2006;17:378. doi: 10.1021/bc050299g. [DOI] [PubMed] [Google Scholar]
- (133).Belgorodsky B, Fadeev L, Kolsenik J, Gozin M. ChemBioChem. 2006;7:1783. doi: 10.1002/cbic.200600237. [DOI] [PubMed] [Google Scholar]
- (134).Nakamura E, Isobe H. Acc. Chem. Res. 2003;36:807. doi: 10.1021/ar030027y. [DOI] [PubMed] [Google Scholar]
- (135).Chen BX, Wilson SR, Das M, Coughlin DJ, Erlanger BF. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10809. doi: 10.1073/pnas.95.18.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Zhu ZW, Schuster DI, Tuckerman ME. Biochemistry. 2003;42:1326. doi: 10.1021/bi020496s. [DOI] [PubMed] [Google Scholar]
- (137).Sijbesma R, Srdanov G, Wudl F, Castoro JA, Wilkins C, Friedman SH, Decamp DL, Kenyon GL. J. Am. Chem. Soc. 1993;115:6510. [Google Scholar]
- (138).Noon WH, Kong YF, Ma JP. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6466. doi: 10.1073/pnas.022532599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Braden BC, Goldbaum FA, Chen BX, Kirschner AN, Wilson SR, Erlanger BF. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12193. doi: 10.1073/pnas.210396197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Kotelnikova RA, Bogdanov GN, Frog EC, Kotelnikov AI, Shtolko VN, Romanova VS, Andreev SM, Kushch AA, Fedorova NE, Medzhidova AA, Miller GG. J. Nanopart. Res. 2003;5:561. [Google Scholar]
- (141).Sreejith S, Ma X, Zhao Y. J. Am. Chem. Soc. 2012;134:17346. doi: 10.1021/ja305352d. [DOI] [PubMed] [Google Scholar]
- (142).Liang LJ, Wang Q, Wu T, Shen JW, Kang Y. Chin. J. Chem. Phys. 2009;22:627. [Google Scholar]
- (143).Hu WB, Peng C, Lv M, Li XM, Zhang YJ, Chen N, Fan CH, Huang Q. ACS Nano. 2011;5:3693. doi: 10.1021/nn200021j. [DOI] [PubMed] [Google Scholar]
- (144).De M, Chou SS, Dravid VP. J. Am. Chem. Soc. 2011;133:17524. doi: 10.1021/ja208427j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Steinmuller-Nethl D, Kloss FR, Najam-U-Haq M, Rainer M, Larsson K, Linsmeier C, Koehler G, Fehrer C, Lepperdinger G, Liu X, Memmel N, Bertel E, Huck CW, Gassner R, Bonn G. Biomaterials. 2006;27:4547. doi: 10.1016/j.biomaterials.2006.04.036. [DOI] [PubMed] [Google Scholar]
- (146).Kong XL, Huang LCL, Hsu CM, Chen WH, Han CC, Chang HC. Anal. Chem. 2004;77:259. doi: 10.1021/ac048971a. [DOI] [PubMed] [Google Scholar]
- (147).Worrall JWE, Verma A, Yan HH, Rotello VM. Chem. Commun. 2006:2338. doi: 10.1039/b517421j. [DOI] [PubMed] [Google Scholar]
- (148).Jiang X, Jiang UG, Jin YD, Wang EK, Dong SJ. Biomacromolecules. 2005;6:46. doi: 10.1021/bm049744l. [DOI] [PubMed] [Google Scholar]
- (149).Chah S, Hammond MR, Zare RN. Chem. Biol. 2005;12:323. doi: 10.1016/j.chembiol.2005.01.013. [DOI] [PubMed] [Google Scholar]
- (150).Fischer NO, McIntosh CM, Simard JM, Rotello VM. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5018. doi: 10.1073/pnas.082644099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Brewer SH, Glomm WR, Johnson MC, Knag MK, Franzen S. Langmuir. 2005;21:9303. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- (152).Dominguez-Medina S, McDonough S, Swanglap P, Landes CF, Link S. Langmuir. 2012;28:9131. doi: 10.1021/la3005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Bhattacharya R, Mukherjee P, Xiong Z, Atala A, Soker S, Mukhopadhyay D. Nano Lett. 2004;4:2479. [Google Scholar]
- (154).Mukherjee P, Bhattacharya R, Wang P, Wang L, Basu S, Nagy JA, Atala A, Mukhopadhyay D, Soker S. Clin. Cancer. Res. 2005;11:3530. doi: 10.1158/1078-0432.CCR-04-2482. [DOI] [PubMed] [Google Scholar]
- (155).Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM. J. Am. Chem. Soc. 2004;126:739. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]
- (156).Verma A, Simard JM, Rotello VM. Langmuir. 2004;20:4178. doi: 10.1021/la036183v. [DOI] [PubMed] [Google Scholar]
- (157).Teichroeb JH, Forrest JA, Jones LW. Eur. Phys. J. E: Soft Matter Biol. Phys. 2008;26:411. doi: 10.1140/epje/i2007-10342-9. [DOI] [PubMed] [Google Scholar]
- (158).Pramanik S, Banerjee P, Sarkar A, Bhattacharya SC. J. Lumin. 2008;128:1969. [Google Scholar]
- (159).You CC, De M, Han G, Rotello VM. J. Am. Chem. Soc. 2005;127:12873. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- (160).Deng ZJ, Liang MT, Monteiro M, Toth I, Minchin RF. Nat. Nanotechnol. 2011;6:39. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- (161).Peng ZG, Hidajat K, Uddin MS. J. Colloid Interface Sci. 2004;271:277. doi: 10.1016/j.jcis.2003.12.022. [DOI] [PubMed] [Google Scholar]
- (162).Mahmoudi M, Shokrgozar MA, Sardari S, Moghadam MK, Vali H, Laurent S, Stroeve P. Nanoscale. 2011;3:1127. doi: 10.1039/c0nr00733a. [DOI] [PubMed] [Google Scholar]
- (163).Ishii D, Kinbara K, Ishida Y, Ishii N, Okochi M, Yohda M, Aida T. Nature. 2003;423:628. doi: 10.1038/nature01663. [DOI] [PubMed] [Google Scholar]
- (164).Xiao Q, Huang S, Qi ZD, Zhou B, He ZK, Liu Y. Biochim. Biophys. Acta, Proteins Proteomics. 2008;1784:1020. doi: 10.1016/j.bbapap.2008.03.018. [DOI] [PubMed] [Google Scholar]
- (165).Pompa PP, Chiuri R, Manna L, Pellegrino T, del Mercato LL, Parak WJ, Calabi F, Cingolani R, Rinaldi R. Chem. Phys. Lett. 2006;417:351. doi: 10.1063/1.2221693. [DOI] [PubMed] [Google Scholar]
- (166).Shen XC, Liou XY, Ye LP, Liang H, Wang ZY. J. Colloid Interface Sci. 2007;311:400. doi: 10.1016/j.jcis.2007.03.006. [DOI] [PubMed] [Google Scholar]
- (167).Wang Q, Kuo YC, Wang YW, Shin G, Ruengruglikit C, Huang QR. J. Phys. Chem. B. 2006;110:16860. doi: 10.1021/jp062279x. [DOI] [PubMed] [Google Scholar]
- (168).Larsericsdotter H, Oscarsson S, Buijs J. J. Colloid Interface Sci. 2001;237:98. doi: 10.1006/jcis.2001.7485. [DOI] [PubMed] [Google Scholar]
- (169).Karlsson M, Carlsson U. Biophys. J. 2005;88:3536. doi: 10.1529/biophysj.104.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Karlsson M, Martensson LG, Jonsson BH, Carlsson U. Langmuir. 2000;16:8470. [Google Scholar]
- (171).Lundqvist M, Sethson I, Jonsson B-H. Langmuir. 2005;21:5974. doi: 10.1021/la050569j. [DOI] [PubMed] [Google Scholar]
- (172).Vertegel AA, Siegel RW, Dordick JS. Langmuir. 2004;20:6800. doi: 10.1021/la0497200. [DOI] [PubMed] [Google Scholar]
- (173).Shang W, Nuffer JH, Dordick JS, Siegel RW. Nano Lett. 2007;7:1991. doi: 10.1021/nl070777r. [DOI] [PubMed] [Google Scholar]
- (174).Nygren P, Lundqvist M, Broo K, Jonsson B-H. Nano Lett. 2008;8:1844. doi: 10.1021/nl080386s. [DOI] [PubMed] [Google Scholar]
- (175).Lord MS, Cousins BG, Doherty PJ, Whitelock JM, Simmons A, Williams RL, Milthorpe BK. Biomaterials. 2006;27:4856. doi: 10.1016/j.biomaterials.2006.05.037. [DOI] [PubMed] [Google Scholar]
- (176).Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Nano Lett. 2007;7:914. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]
- (177).Delgado AD, Leonard M, Dellacherie E. Langmuir. 2001;17:4386. [Google Scholar]
- (178).Milani S, Bombelli FB, Pitek AS, Dawson KA, Radler J. ACS Nano. 2012;6:2532. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- (179).Prapainop K, Wentworth P. Eur. J. Pharm. Biopharm. 2011;77:353. doi: 10.1016/j.ejpb.2010.12.026. [DOI] [PubMed] [Google Scholar]
- (180).Gref R, Luck M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Muller RH. Colloids Surf., B. 2000;18:301. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- (181).Walkey CD, Olsen JB, Guo HB, Emili A, Chan WCW. J. Am. Chem. Soc. 2012;134:2139. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- (182).Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14265. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Cedervall T, Lynch I, Foy M, Berggad T, Donnelly SC, Cagney G, Linse S, Dawson KA. Angew. Chem., Int. Ed. 2007;46:5754. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- (184).Wiogo HTR, Lim M, Bulmus V, Yun J, Amal R. Langmuir. 2010;27:843. doi: 10.1021/la104278m. [DOI] [PubMed] [Google Scholar]
- (185).Mosqueira VCF, Legrand P, Gulik A, Bourdon O, Gref R, Labarre D, Barratt G. Biomaterials. 2001;22:2967. doi: 10.1016/s0142-9612(01)00043-6. [DOI] [PubMed] [Google Scholar]
- (186).Labarre D, Vauthier C, Chauvierre C, Petri B, Muller R, Chehimi MM. Biomaterials. 2005;26:5075. doi: 10.1016/j.biomaterials.2005.01.019. [DOI] [PubMed] [Google Scholar]
- (187).Mu QX, Li ZW, Li X, Mishra SR, Zhang B, Si ZK, Yang L, Jiang W, Yan B. J. Phys. Chem. C. 2009;113:5390. [Google Scholar]
- (188).Maiorano G, Sabella S, Sorce B, Brunetti V, Malvindi MA, Cingolani R, Pompa PP. ACS Nano. 2010;4:7481. doi: 10.1021/nn101557e. [DOI] [PubMed] [Google Scholar]
- (189).Lesniak A, Campbell A, Monopoli MP, Lynch I, Salvati A, Dawson KA. Biomaterials. 2010;31:9511. doi: 10.1016/j.biomaterials.2010.09.049. [DOI] [PubMed] [Google Scholar]
- (190).Jeng ES, Barone PW, Nelson JD, Strano MS. Small. 2007;3:1602. doi: 10.1002/smll.200700141. [DOI] [PubMed] [Google Scholar]
- (191).Chen H, Zou QC, Yu H, Peng M, Song GW, Zhang JZ, Chai SG, Zhang YH, Yan CE. Microchim. Acta. 2010;168:331. [Google Scholar]
- (192).Wang GL, Murray RW. Nano Lett. 2004;4:95. [Google Scholar]
- (193).Zheng M, Jagota A, Semke ED, Diner BA, McLean RS, Lustig SR, Richardson RE, Tassi NG. Nat. Mater. 2003;2:338. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- (194).Hughes ME, Brandin E, Golovchenko JA. Nano Lett. 2007;7:1191. doi: 10.1021/nl062906u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Li D, Li GP, Guo WW, Li PC, Wang EK, Wang J. Biomaterials. 2008;29:2776. doi: 10.1016/j.biomaterials.2008.03.007. [DOI] [PubMed] [Google Scholar]
- (196).Gearheart LA, Ploehn HJ, Murphy CJ. J. Phys. Chem. B. 2001;105:12609. [Google Scholar]
- (197).Li X, Peng YH, Qu XG. Nucleic Acids Res. 2006;34:3670. doi: 10.1093/nar/gkl513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Gigliotti B, Sakizzie B, Bethune DS, Shelby RM, Cha JN. Nano Lett. 2006;6:159. doi: 10.1021/nl0518775. [DOI] [PubMed] [Google Scholar]
- (199).Zheng M, Jagota A, Strano MS, Santos AP, Barone P, Chou SG, Diner BA, Dresselhaus MS, McLean RS, Onoa GB, Samsonidze GG, Semke ED, Usrey M, Walls DJ. Science. 2003;302:1545. doi: 10.1126/science.1091911. [DOI] [PubMed] [Google Scholar]
- (200).Rajendra J, Rodger A. Chem. -Eur. J. 2005;11:4841. doi: 10.1002/chem.200500093. [DOI] [PubMed] [Google Scholar]
- (201).Rajendra J, Baxendale M, Rap LGD, Rodger A. J. Am. Chem. Soc. 2004;126:11182. doi: 10.1021/ja048720j. [DOI] [PubMed] [Google Scholar]
- (202).Goodman CM, Chari NS, Han G, Hong R, Ghosh P, Rotello VM. Chem. Biol. Drug Des. 2006;67:297. doi: 10.1111/j.1747-0285.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- (203).Dong L, Li Y, Zhang Y, Chen X, Hu Z. Microchim. Acta. 2007;159:49. [Google Scholar]
- (204).Xiao J, Chen J, Ren F, Chen Y, Xu M. Microchim. Acta. 2007;159:287. [Google Scholar]
- (205).Zheng JH, Wu X, Wang MQ, Ran DH, Xu W, Yang JH. Talanta. 2008;74:526. doi: 10.1016/j.talanta.2007.06.014. [DOI] [PubMed] [Google Scholar]
- (206).Kim EY, Stanton J, Vega RA, Kunstman KJ, Mirkin CA, Wolinsky SM. Nucleic Acids Res. 2006;34:54. doi: 10.1093/nar/gkl147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Liu SQ, Xu JJ, Chen HY. Colloids Surf., B. 2004;36:155. doi: 10.1016/j.colsurfb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- (208).Li X, Peng YH, Ren JS, Qu XG. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19658. doi: 10.1073/pnas.0607245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Ren HL, Wang C, Zhang JL, Zhou XJ, Xu DF, Zheng J, Guo SW, Zhang JY. ACS Nano. 2010;4:7169. doi: 10.1021/nn101696r. [DOI] [PubMed] [Google Scholar]
- (210).Rahban M, Divsalar A, Saboury AA, Golestani A. J. Phys. Chem. C. 2010;114:5798. [Google Scholar]
- (211).Heller DA, Jeng ES, Yeung TK, Martinez BM, Moll AE, Gastala JB, Strano MS. Science. 2006;311:508. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- (212).Bhattacharyya SS, Paul S, De A, Das D, Samadder A, Boujedaini N, Khuda-Bukhsh AR. Toxicol. Appl. Pharmacol. 2011;253:270. doi: 10.1016/j.taap.2011.04.010. [DOI] [PubMed] [Google Scholar]
- (213).Dukovic G, Balaz M, Doak P, Berova ND, Zheng M, McLean RS, Brus LE. J. Am. Chem. Soc. 2006;128:9004. doi: 10.1021/ja062095w. [DOI] [PubMed] [Google Scholar]
- (214).Dovbeshko GI, Repnytska OP, Obraztsova ED, Shtogun YV. Chem. Phys. Lett. 2003;372:432. [Google Scholar]
- (215).Wu YR, Phillips JA, Liu HP, Yang RH, Tan WH. ACS Nano. 2008;2:2023. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Han G, Chari NS, Verma A, Hong R, Martin CT, Rotello VM. Bioconjugate Chem. 2005;16:1356. doi: 10.1021/bc050173j. [DOI] [PubMed] [Google Scholar]
- (217).Tokuyama H, Yamago S, Nakamura E, Shiraki T, Sugiura Y. J. Am. Chem. Soc. 1993;115:7918. [Google Scholar]
- (218).Zhao X, Johnson JK. J. Am. Chem. Soc. 2007;129:10438. doi: 10.1021/ja071844m. [DOI] [PubMed] [Google Scholar]
- (219).Gao HJ, Kong Y, Cui DX, Ozkan CS. Nano Lett. 2003;3:471. [Google Scholar]
- (220).Moulton SE, Minett AI, Murphy R, Ryan KP, McCarthy D, Coleman JN, Blau WJ, Wallace GG. Carbon. 2005;43:1879. [Google Scholar]
- (221).Varghese N, Mogera U, Govindaraj A, Das A, Maiti PK, Sood AK, Rao CNR. ChemPhyschem. 2009;10:206. doi: 10.1002/cphc.200800459. [DOI] [PubMed] [Google Scholar]
- (222).Liu M, Zhao HM, Chen S, Yu HT, Quan X. Chem. Commun. 2012;48:564. doi: 10.1039/c1cc16429e. [DOI] [PubMed] [Google Scholar]
- (223).Tang LH, Chang HX, Liu Y, Li JH. Adv. Funct. Mater. 2012;22:3083. [Google Scholar]
- (224).Zhang X, Servos MR, Liu JW. Langmuir. 2012;28:3896. doi: 10.1021/la205036p. [DOI] [PubMed] [Google Scholar]
- (225).Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal. Chem. 2006;78:8313. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Foley EA, Carter JD, Shan F, Guo T. Chem. Commun. 2005:3192. doi: 10.1039/b503425f. [DOI] [PubMed] [Google Scholar]
- (227).McIntosh CM, Esposito EA, Boal AK, Simard JM, Martin CT, Rotello VM. J. Am. Chem. Soc. 2001;123:7626. doi: 10.1021/ja015556g. [DOI] [PubMed] [Google Scholar]
- (228).Railsback JG, Singh A, Pearce RC, McKnight TE, Collazo R, Sitar Z, Yingling YG, Melechko AV. Adv. Mater. 2012;24:4261. doi: 10.1002/adma.201104891. [DOI] [PubMed] [Google Scholar]
- (229).Cao YWC, Jin RC, Mirkin CA. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- (230).Han G, Martin CT, Rotello VM. Chem. Biol. Drug Des. 2006;67:78. doi: 10.1111/j.1747-0285.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- (231).Chen P, Pan D, Fan CH, Chen JH, Huang K, Wang DF, Zhang HL, Li Y, Feng GY, Liang PJ, He L, Shi YY. Nat. Nanotechnol. 2011;6:639. doi: 10.1038/nnano.2011.141. [DOI] [PubMed] [Google Scholar]
- (232).Fu AH, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. J. Am. Chem. Soc. 2004;126:10832. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- (233).Zheng JW, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Nano Lett. 2006;6:1502. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Torimoto T, Yamashita M, Kuwabata S, Sakata T, Mori H, Yoneyama H. J. Phys. Chem. B. 1999;103:8799. [Google Scholar]
- (235).Zhu RR, Wang SL, Zhang R, Sun XY, Yao SD. Chin. J. Chem . 2007;25:958. [Google Scholar]
- (236).Ashikaga T, Wada M, Kobayashi H, Mori M, Katsumura Y, Fukui H, Kato S, Yamaguchi M, Takamatsu T. Mutat. Res.-Genet. Toxicol. Environ. Mutag. 2000;466:1. doi: 10.1016/s1383-5718(99)00228-4. [DOI] [PubMed] [Google Scholar]
- (237).Donaldson K, Beswick PH, Gilmour PS. Toxicol. Lett. 1996;88:293. doi: 10.1016/0378-4274(96)03752-6. [DOI] [PubMed] [Google Scholar]
- (238).Dunford R, Salinaro A, Cai LZ, Serpone N, Horikoshi S, Hidaka H, Knowland J. FEBS Lett. 1997;418:87. doi: 10.1016/s0014-5793(97)01356-2. [DOI] [PubMed] [Google Scholar]
- (239).Jose GP, Santra S, Mandal SK, Sengupta TK. J. Nanobiotechnol. 2011;9:9. doi: 10.1186/1477-3155-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Zinchenko AA, Luckel F, Yoshikawa K. Biophys. J. 2007;92:1318. doi: 10.1529/biophysj.106.094185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Acosta E. Curr. Opin. Colloid Interface Sci. 2009;14:3. [Google Scholar]
- (242).Velikov KP, Pelan E. Soft Matter. 2008;4:1964. [Google Scholar]
- (243).Shan CS, Yang HF, Song JF, Han DX, Ivaska A, Niu L. Anal. Chem. 2009;81:2378. doi: 10.1021/ac802193c. [DOI] [PubMed] [Google Scholar]
- (244).Casey A, Davoren M, Herzog E, Lyng FM, Byrne HJ, Chambers G. Carbon. 2007;45:34. [Google Scholar]
- (245).Shen WZ, Wang H, Guan RG, Li ZJ. Colloids Surf., A. 2008;331:263. [Google Scholar]
- (246).Casey A, Herzog E, Lyng FM, Byrne HJ, Chambers G, Davoren M. Toxicol. Lett. 2008;179:78. doi: 10.1016/j.toxlet.2008.04.006. [DOI] [PubMed] [Google Scholar]
- (247).Rajesh C, Majumder C, Mizuseki H, Kawazoe Y. J. Chem. Phys. 2009;130 doi: 10.1063/1.3079096. art.no.124911. [DOI] [PubMed] [Google Scholar]
- (248).Rafeeqi T, Kaul G. Adv. Sci. Lett. 2011;4:536. [Google Scholar]
- (249).Cazorla C. Thin Solid Films. 2010;518:6951. [Google Scholar]
- (250).Kloepfer JA, Mielke RE, Nadeau JL. Appl. Environ. Microbiol. 2005;71:2548. doi: 10.1128/AEM.71.5.2548-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Lam CW, James JT, McCluskey R, Hunter RL. Toxicol. Sci. 2004;77:126. doi: 10.1093/toxsci/kfg243. [DOI] [PubMed] [Google Scholar]
- (252).Lin DH, Xing BS. Environ. Sci. Technol. 2008;42:5580. doi: 10.1021/es800422x. [DOI] [PubMed] [Google Scholar]
- (253).Lu L, Sun RWY, Chen R, Hui CK, Ho CM, Luk JM, Lau GKK, Che CM. Antiviral Ther. 2008;13:253. [PubMed] [Google Scholar]
- (254).Navarro E, Baun A, Behra R, Hartmann NB, Filser J, Miao AJ, Quigg A, Santschi PH, Sigg L. Ecotoxicology. 2008;17:372. doi: 10.1007/s10646-008-0214-0. [DOI] [PubMed] [Google Scholar]
- (255).Shi XH, von dem Bussche A, Hurt RH, Kane AB, Gao HJ. Nat. Nanotechnol. 2011;6:714. doi: 10.1038/nnano.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (256).Jiang XE, Rocker C, Hafner M, Brandholt S, Dorlich RM, Nienhaus GU. ACS Nano. 2010;4:6787. doi: 10.1021/nn101277w. [DOI] [PubMed] [Google Scholar]
- (257).Goldstein JL, Anderson RGW, Brown MS. Nature. 1979;279:679. doi: 10.1038/279679a0. [DOI] [PubMed] [Google Scholar]
- (258).Krpetic Z, Porta F, Caneva E, Dal Santo V, Scari G. Langmuir. 2010;26:14799. doi: 10.1021/la102758f. [DOI] [PubMed] [Google Scholar]
- (259).Faklaris O, Joshi V, Irinopoulou T, Tauc P, Sennour M, Girard H, Gesset C, Arnault JC, Thorel A, Boudou JP, Curmi PA, Treussart F. ACS Nano. 2009;3:3955. doi: 10.1021/nn901014j. [DOI] [PubMed] [Google Scholar]
- (260).Schrand AM, Lin JB, Hens SC, Hussain SM. Nanoscale. 2011;3:435. doi: 10.1039/c0nr00408a. [DOI] [PubMed] [Google Scholar]
- (261).Singh S, Kumar A, Karakoti A, Seal S, Self WT. Mol. BioSyst. 2010;6:1813. doi: 10.1039/c0mb00014k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (262).Taylor U, Klein S, Petersen S, Kues W, Barcikowski S, Rath D. Cytometry Part A. 2010;77A:439. doi: 10.1002/cyto.a.20846. [DOI] [PubMed] [Google Scholar]
- (263).Geiser M, Rothen-Rutishauser B, Kapp N, Schurch S, Kreyling W, Schulz H, Semmler M, Hof VI, Heyder J, Gehr P. Environ. Health Perspect. 2005;113:1555. doi: 10.1289/ehp.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (264).Nativo P, Prior IA, Brust M. ACS Nano. 2008;2:1639. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- (265).Porter AE, Gass M, Bendall JS, Muller K, Goode A, Skepper JN, Midgley PA, Welland M. ACS Nano. 2009;3:1485. doi: 10.1021/nn900416z. [DOI] [PubMed] [Google Scholar]
- (266).Pantarotto D, Singh R, McCarthy D, Erhardt M, Briand JP, Prato M, Kostarelos K, Bianco A. Angew. Chem., Int. Ed. 2004;43:5242. doi: 10.1002/anie.200460437. [DOI] [PubMed] [Google Scholar]
- (267).Mu Q, Broughton DL, Yan B. Nano Lett. 2009;9:4370. doi: 10.1021/nl902647x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Salvati A, Aberg C, dos Santos T, Varela J, Pinto P, Lynch I, Dawson KA. Nanomed. -Nanotechnol. Biol. Med. 2011;7:818. doi: 10.1016/j.nano.2011.03.005. [DOI] [PubMed] [Google Scholar]
- (269).Bussy C, Cambedouzou J, Lanone S, Leccia E, Heresanu V, Pinault M, Mayne-I'Hermite M, Brun N, Mory C, Cotte M, Doucet J, Boczkowski J, Launoist P. Nano Lett. 2008;8:2659. doi: 10.1021/nl800914m. [DOI] [PubMed] [Google Scholar]
- (270).Qu M, Mehrmohammadi M, Emelianov S. Small. 2011;7:2858. doi: 10.1002/smll.201100960. [DOI] [PubMed] [Google Scholar]
- (271).Vercauteren D, Deschout H, Remaut K, Engbersen JFJ, Jones AT, Demeester J, De Smedt SC, Braeckmans K. ACS Nano. 2011;5:7874. doi: 10.1021/nn2020858. [DOI] [PubMed] [Google Scholar]
- (272).Chernenko T, Matthaus C, Milane L, Quintero L, Amiji M, Diem M. ACS Nano. 2009;3:3552. doi: 10.1021/nn9010973. [DOI] [PubMed] [Google Scholar]
- (273).Vasir JK, Labhasetwar V. Biomaterials. 2008;29:4244. doi: 10.1016/j.biomaterials.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (274).Shan YP, Hao XA, Shang X, Cai MJ, Jiang JG, Tang ZY, Wang HD. Chem. Commun. 2011;47:3377. doi: 10.1039/c1cc00040c. [DOI] [PubMed] [Google Scholar]
- (275).Shan YP, Ma SY, Nie LY, Shang X, Hao X, Tang ZY, Wang HD. Chem. Commun. 2011;47:8091. doi: 10.1039/c1cc11453k. [DOI] [PubMed] [Google Scholar]
- (276).Wilhelm C, Gazeau F, Roger J, Pons JN, Bacri JC. Langmuir. 2002;18:8148. [Google Scholar]
- (277).Raemy DO, Limbach LK, Rothen-Rutishauser B, Grass RN, Gehr P, Birbaum K, Brandenberger C, Gunther D, Stark WJ. Eur. J. Pharm. Biopharm. 2011;77:368. doi: 10.1016/j.ejpb.2010.11.017. [DOI] [PubMed] [Google Scholar]
- (278).Limbach LK, Li YC, Grass RN, Brunner TJ, Hintermann MA, Muller M, Gunther D, Stark WJ. Environ. Sci. Technol. 2005;39:9370. doi: 10.1021/es051043o. [DOI] [PubMed] [Google Scholar]
- (279).Cho EC, Xie JW, Wurm PA, Xia YN. Nano Lett. 2009;9:1080. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- (280).Jin H, Heller DA, Strano MS. Nano Lett. 2008;8:1577. doi: 10.1021/nl072969s. [DOI] [PubMed] [Google Scholar]
- (281).Yuan HY, Li J, Bao G, Zhang SL. Phys. Rev. Lett. 2010;105 doi: 10.1103/PhysRevLett.105.138101. art.no.138101. [DOI] [PubMed] [Google Scholar]
- (282).Zhang SL, Li J, Lykotrafitis G, Bao G, Suresh S. Adv. Mater. 2009;21:419. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (283).Kang B, Chang SQ, Dai YD, Yu DC, Chen D. Small. 2010;6:2362. doi: 10.1002/smll.201001260. [DOI] [PubMed] [Google Scholar]
- (284).Lunov O, Syrovets T, Loos C, Beil J, Delecher M, Tron K, Nienhaus GU, Musyanovych A, Mailander V, Landfester K, Simmet T. ACS Nano. 2011;5:1657. doi: 10.1021/nn2000756. [DOI] [PubMed] [Google Scholar]
- (285).Becker ML, Fagan JA, Gallant ND, Bauer BJ, Bajpai V, Hobbie EK, Lacerda SH, Migler KB, Jakupciak JP. Adv. Mater. 2007;19:939. [Google Scholar]
- (286).Zahr AS, Davis CA, Pishko MV. Langmuir. 2006;22:8178. doi: 10.1021/la060951b. [DOI] [PubMed] [Google Scholar]
- (287).Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (288).Mok H, Bae KH, Ahn CH, Park TG. Langmuir. 2009;25:1645. doi: 10.1021/la803542v. [DOI] [PubMed] [Google Scholar]
- (289).Babic M, Horak D, Jendelova P, Glogarova K, Herynek V, Trchova M, Likavcanova K, Lesny P, Pollert E, Hajek M, Sykova E. Bioconjugate Chem. 2009;20:283. doi: 10.1021/bc800373x. [DOI] [PubMed] [Google Scholar]
- (290).Yang H, Fung SY, Liu MY. Angew. Chem., Int. Ed. 2011;50:9643. doi: 10.1002/anie.201102911. [DOI] [PubMed] [Google Scholar]
- (291).Oh E, Delehanty JB, Sapsford KE, Susumu K, Goswami R, Blanco-Canosa JB, Dawson PE, Granek J, Shoff M, Zhang Q, Goering PL, Huston A, Medintz IL. ACS Nano. 2011;5:6434. doi: 10.1021/nn201624c. [DOI] [PubMed] [Google Scholar]
- (292).Asati A, Santra S, Kaittanis C, Perez JM. ACS Nano. 2010;4:5321. doi: 10.1021/nn100816s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (293).Chithrani BD, Ghazani AA, Chan WCW. Nano Lett. 2006;6:662. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- (294).Chithrani BD, Chan WCW. Nano Lett. 2007;7:1542. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- (295).Qiu Y, Liu Y, Wang LM, Xu LG, Bai R, Ji YL, Wu XC, Zhao YL, Li YF, Chen CY. Biomaterials. 2010;31:7606. doi: 10.1016/j.biomaterials.2010.06.051. [DOI] [PubMed] [Google Scholar]
- (296).Bartneck M, Keul HA, Singh S, Czaja K, Bornemann J, Bockstaller M, Moeller M, Zwadlo-Klarwasser G, Groll J. ACS Nano. 2010;4:3073. doi: 10.1021/nn100262h. [DOI] [PubMed] [Google Scholar]
- (297).Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11613. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (298).Liang M, Lin IC, Whittaker MR, Minchin RF, Monteiro MJ, Toth I. ACS Nano. 2010;4:403. doi: 10.1021/nn9011237. [DOI] [PubMed] [Google Scholar]
- (299).Verma A, Uzun O, Hu YH, Hu Y, Han HS, Watson N, Chen SL, Irvine DJ, Stellacci F. Nat. Mater. 2008;7:588. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (300).Prapainop K, Witter DP, Wentworth P. J. Am. Chem. Soc. 2012;134:4100. doi: 10.1021/ja300537u. [DOI] [PubMed] [Google Scholar]
- (301).Lin JQ, Zhang HW, Chen Z, Zheng YG. ACS Nano. 2010;4:5421. doi: 10.1021/nn1010792. [DOI] [PubMed] [Google Scholar]
- (302).Su GX, Zhou HY, Mu QX, Zhang Y, Li LW, Jiao PF, Jiang GB, Yan B. J. Phys. Chem. C. 2012;116:4993. [Google Scholar]
- (303).Mu Q, Su G, Li L, Gilbertson BO, Yu LH, Zhang Q, Sun Y-P, Yan B. ACS Appl. Mater. Interfaces. 2012;4:2259. doi: 10.1021/am300253c. [DOI] [PubMed] [Google Scholar]
- (304).Florez L, Herrmann C, Cramer JM, Hauser CP, Koynov K, Landfester K, Crespy D, Mailander V. Small. 2012;8:2222. doi: 10.1002/smll.201102002. [DOI] [PubMed] [Google Scholar]
- (305).Park J-H, von Maltzahn G, Zhang L, Derfus AM, Simberg D, Harris TJ, Ruoslahti E, Bhatia SN, Sailor MJ. Small. 2009;5:694. doi: 10.1002/smll.200801789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (306).Banquy X, Suarez F, Argaw A, Rabanel JM, Grutter P, Bouchard JF, Hildgen P, Giasson S. Soft Matter. 2009;5:3984. [Google Scholar]
- (307).Liu WJ, Zhou XY, Mao ZW, Yu DH, Wang B, Gao CY. Soft Matter. 2012;8:9235. [Google Scholar]
- (308).Cho EC, Zhang Q, Xia YN. Nat. Nanotechnol. 2011;6:385. doi: 10.1038/nnano.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (309).Safi M, Courtois J, Seigneuret M, Conjeaud H, Berret JF. Biomaterials. 2011;32:9353. doi: 10.1016/j.biomaterials.2011.08.048. [DOI] [PubMed] [Google Scholar]
- (310).Zhou R, Zhou HY, Xiong B, He Y, Yeung ES. J. Am. Chem. Soc. 2012;134:13404. doi: 10.1021/ja304119w. [DOI] [PubMed] [Google Scholar]
- (311).Chen JM, Hessler JA, Putchakayala K, Panama BK, Khan DP, Hong S, Mullen DG, DiMaggio SC, Som A, Tew GN, Lopatin AN, Baker JR, Holl MMB, Orr BG. J. Phys. Chem. B. 2009;113:11179. doi: 10.1021/jp9033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Arvizo RR, Miranda OR, Thompson MA, Pabelick CM, Bhattacharya R, Robertson JD, Rotello VM, Prakash YS, Mukherjee P. Nano Lett. 2010;10:2543. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (313).Slowing II, Wu CW, Vivero-Escoto JL, Lin VSY. Small. 2009;5:57. doi: 10.1002/smll.200800926. [DOI] [PubMed] [Google Scholar]
- (314).Sayes CM, Gobin AM, Ausman KD, Mendez J, West JL, Colvin VL. Biomaterials. 2005;26:7587. doi: 10.1016/j.biomaterials.2005.05.027. [DOI] [PubMed] [Google Scholar]
- (315).Doshi N, Mitragotri S. J. R. Soc., Interface. 2010;7:S403. doi: 10.1098/rsif.2010.0134.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (316).Tong L, Zhao Y, Huff TB, Hansen MN, Wei A, Cheng JX. Adv. Mater. 2007;19:3136. doi: 10.1002/adma.200701974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Zhang W, Kalive M, Capco DG, Chen YS. Nanotechnology. 2010;21 doi: 10.1088/0957-4484/21/35/355103. [DOI] [PubMed] [Google Scholar]
- (318).Lin YS, Haynes CL. J. Am. Chem. Soc. 2010;132:4834. doi: 10.1021/ja910846q. [DOI] [PubMed] [Google Scholar]
- (319).Lauger P. J. Membr. Biol. 1980;57:163. doi: 10.1007/BF01869585. [DOI] [PubMed] [Google Scholar]
- (320).Park KH, Chhowalla M, Iqbal Z, Sesti F. J. Biol. Chem. 2003;278:50212. doi: 10.1074/jbc.M310216200. [DOI] [PubMed] [Google Scholar]
- (321).Chhowalla M, Unalan HE, Wang YB, Iqbal Z, Park K, Sesti F. Nanotechnology. 2005;16:2982. [Google Scholar]
- (322).Xu HF, Bai J, Meng J, Hao W, Xu HY, Cao JM. Nanotechnology. 2009:20. doi: 10.1088/0957-4484/20/28/285102. [DOI] [PubMed] [Google Scholar]
- (323).Kraszewski S, Tarek M, Treptow W, Ramseyer C. ACS Nano. 2010;4:4158. doi: 10.1021/nn100723r. [DOI] [PubMed] [Google Scholar]
- (324).Jakubek LM, Marangoudakis S, Raingo J, Liu XY, Lipscombe D, Hurt RH. Biomaterials. 2009;30:6351. doi: 10.1016/j.biomaterials.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (325).Holt BD, Short PA, Rape AD, Wang YL, Islam MF, Dahl KN. ACS Nano. 2010;4:4872. doi: 10.1021/nn101151x. [DOI] [PubMed] [Google Scholar]
- (326).Tarantola M, Schneider D, Sunnick E, Adam H, Pierrat S, Rosman C, Breus V, Sonnichsen C, Basche T, Wegener J, Janshoff A. ACS Nano. 2009;3:213. doi: 10.1021/nn800721j. [DOI] [PubMed] [Google Scholar]
- (327).Gupta AK, Gupta M. Biomaterials. 2005;26:1565. doi: 10.1016/j.biomaterials.2004.05.022. [DOI] [PubMed] [Google Scholar]
- (328).Gupta AK, Gupta M, Yarwood SJ, Curtis ASG. J. Controlled Release. 2004;95:197. doi: 10.1016/j.jconrel.2003.11.006. [DOI] [PubMed] [Google Scholar]
- (329).Soenen SJH, Nuytten N, De Meyer SF, De Smedt SC, De Cuyper M. Small. 2010;6:832. doi: 10.1002/smll.200902084. [DOI] [PubMed] [Google Scholar]
- (330).Pan Y, Leifert A, Ruau D, Neuss S, Bornemann J, Schmid G, Brandau W, Simon U, Jahnen-Dechent W. Small. 2009;5:2067. doi: 10.1002/smll.200900466. [DOI] [PubMed] [Google Scholar]
- (331).Karatas OF, Sezgin E, Aydin O, Culha M. Colloids Surf., B. 2009;71:315. doi: 10.1016/j.colsurfb.2009.02.020. [DOI] [PubMed] [Google Scholar]
- (332).Paunesku T, Vogt S, Lai B, Maser J, Stojicevic N, Thurn KT, Osipo C, Liu H, Legnini D, Wang Z, Lee C, Woloschak GE. Nano Lett. 2007;7:596. doi: 10.1021/nl0624723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (333).Zhou FF, Xing D, Wu BY, Wu SN, Ou ZM, Chen WR. Nano Lett. 2010;10:1677. doi: 10.1021/nl100004m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (334).Salnikov V, Lukyanenko YO, Frederick CA, Lederer WJ, Lukyanenko V. Biophys. J. 2007;92:1058. doi: 10.1529/biophysj.106.094318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (335).Conroy J, Byrne SJ, Gun'ko YK, Rakovich YP, Donegan JF, Davies A, Kelleher D, Volkov Y. Small. 2008;4:2006. doi: 10.1002/smll.200800088. [DOI] [PubMed] [Google Scholar]
- (336).Chen FQ, Gerion D. Nano Lett. 2004;4:1827. [Google Scholar]
- (337).Zakhidov ST, Marshak TL, Malolina EA, Kulibin AY, Zelenina IA, Pavluchenkova SM, Rudoy VM, Dement'eva OV, Skuridin SG, Evdokimov YM. Biol. Membr. 2010;27:349. [Google Scholar]
- (338).Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R, Masuda T, Danev R, Nagayama K, Kogure K, Harashima H. Biomaterials. 2009;30:2940. doi: 10.1016/j.biomaterials.2009.02.009. [DOI] [PubMed] [Google Scholar]
- (339).Martindale JL, Holbrook NJ. J. Cell. Physiol. 2002;192:1. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- (340).Xia T, Kovochich M, Brant J, Hotze M, Sempf J, Oberley T, Sioutas C, Yeh JI, Wiesner MR, Nel AE. Nano Lett. 2006;6:1794. doi: 10.1021/nl061025k. [DOI] [PubMed] [Google Scholar]
- (341).Karakoti A, Singh S, Dowding JM, Seal S, Self WT. Chem. Soc. Rev. 2010;39:4422. doi: 10.1039/b919677n. [DOI] [PubMed] [Google Scholar]
- (342).Jia HY, Liu Y, Zhang XJ, Han L, Du LB, Tian Q, Xut YC. J. Am. Chem. Soc. 2009;131:40. doi: 10.1021/ja808033w. [DOI] [PubMed] [Google Scholar]
- (343).George S, Pokhrel S, Xia T, Gilbert B, Ji ZX, Schowalter M, Rosenauer A, Damoiseaux R, Bradley KA, Madler L, Nel AE. ACS Nano. 2010;4:15. doi: 10.1021/nn901503q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (344).Wang H, Joseph JA. Free Radical Biol. Med. 1999;27:612. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- (345).Hwang ET, Lee JH, Chae YJ, Kim YS, Kim BC, Sang BI, Gu MB. Small. 2008;4:746. doi: 10.1002/smll.200700954. [DOI] [PubMed] [Google Scholar]
- (346).De Berardis B, Civitelli G, Condello M, Lista P, Pozzi R, Arancia G, Meschini S. Toxicol. Appl. Pharmacol. 2010;246:116. doi: 10.1016/j.taap.2010.04.012. [DOI] [PubMed] [Google Scholar]
- (347).Lu X, Qian JC, Zhou HJ, Gan Q, Tang W, Lu JX, Yuan Y, Liu CS. Int. J. Nanomed. 2011;6:1889. doi: 10.2147/IJN.S24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (348).Berg JM, Ho S, Hwang W, Zebda R, Cummins K, Soriaga MP, Taylor R, Guo B, Sayes CM. Chem. Res. Toxicol. 2010;23:1874. doi: 10.1021/tx100307h. [DOI] [PubMed] [Google Scholar]
- (349).Burello E, Worth AP. Nanotoxicology. 2011;5:228. doi: 10.3109/17435390.2010.502980. [DOI] [PubMed] [Google Scholar]
- (350).Thakor AS, Paulmurugan R, Kempen P, Zavaleta C, Sinclair R, Massoud TF, Gambhir SS. Small. 2011;7:126. doi: 10.1002/smll.201001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (351).Wu J, Sun JA, Xue Y. Toxicol. Lett. 2010;199:269. doi: 10.1016/j.toxlet.2010.09.009. [DOI] [PubMed] [Google Scholar]
- (352).Park EJ, Yi J, Chung YH, Ryu DY, Choi J, Park K. Toxicol. Lett. 2008;180:222. doi: 10.1016/j.toxlet.2008.06.869. [DOI] [PubMed] [Google Scholar]
- (353).Fenoglio I, Greco G, Livraghi S, Fubini B. Chem. -Eur. J. 2009;15:4614. doi: 10.1002/chem.200802542. [DOI] [PubMed] [Google Scholar]
- (354).Foucaud L, Goulaouic S, Bennasroune A, Laval-Gilly P, Brown D, Stone V, Falla J. Toxicol. In Vitro. 2010;24:1512. doi: 10.1016/j.tiv.2010.07.012. [DOI] [PubMed] [Google Scholar]
- (355).Palomaki J, Valimaki E, Sund J, Vippola M, Clausen PA, Jensen KA, Savolainen K, Matikainen S, Alenius H. ACS Nano. 2011;5:6861. doi: 10.1021/nn200595c. [DOI] [PubMed] [Google Scholar]
- (356).Ambrosi A, Pumera M. Chem. -Eur. J. 2010;16:1786. doi: 10.1002/chem.200902534. [DOI] [PubMed] [Google Scholar]
- (357).Zhang YB, Xu Y, Li ZG, Chen T, Lantz SM, Howard PC, Paule MG, Slikker W, Watanabe F, Mustafa T, Biris AS, Ali SF. ACS Nano. 2011;5:7020. doi: 10.1021/nn2016259. [DOI] [PubMed] [Google Scholar]
- (358).Wan R, Mo YQ, Zhang X, Chien SF, Tollerud DJ, Zhang QW. Toxicol. Appl. Pharmacol. 2008;233:276. doi: 10.1016/j.taap.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (359).Thubagere A, Reinhard BM. ACS Nano. 2010;4:3611. doi: 10.1021/nn100389a. [DOI] [PubMed] [Google Scholar]
- (360).Ahamed M, Akhtar MJ, Siddiqui MA, Ahmad J, Musarrat J, Al-Khedhairy AA, AlSalhi MS, Alrokayan SA. Toxicology. 2011;283:101. doi: 10.1016/j.tox.2011.02.010. [DOI] [PubMed] [Google Scholar]
- (361).Piao MJ, Kang KA, Lee IK, Kim HS, Kim S, Choi JY, Choi J, Hyun JW. Toxicol. Lett. 2011;201:92. doi: 10.1016/j.toxlet.2010.12.010. [DOI] [PubMed] [Google Scholar]
- (362).Xia T, Kovochich M, Liong M, Madler L, Gilbert B, Shi HB, Yeh JI, Zink JI, Nel AE. ACS Nano. 2008;2:2121. doi: 10.1021/nn800511k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (363).Karlsson HL, Cronholm P, Gustafsson J, Moller L. Chem. Res. Toxicol. 2008;21:1726. doi: 10.1021/tx800064j. [DOI] [PubMed] [Google Scholar]
- (364).Gerloff K, Albrecht C, Boots AW, Forster I, Schins RPF. Nanotoxicology. 2009;3:355. doi: 10.3109/17435390.2010.525670. [DOI] [PubMed] [Google Scholar]
- (365).Pujalte I, Passagne I, Brouillaud B, Treguer M, Durand E, Ohayon-Courtes C, L'Azou B. Part. Fibre Toxicol. 2011:8. doi: 10.1186/1743-8977-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (366).Kocbek P, Teskac K, Kreft ME, Kristl J. Small. 2010;6:1908. doi: 10.1002/smll.201000032. [DOI] [PubMed] [Google Scholar]
- (367).Zhang HY, Ji ZX, Xia T, Meng H, Low-Kam C, Liu R, Pokhrel S, Lin SJ, Wang X, Liao YP, Wang MY, Li LJ, Rallo R, Damoiseaux R, Telesca D, Madler L, Cohen Y, Zink JI, Nel AE. ACS Nano. 2012;6:4349. doi: 10.1021/nn3010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (368).Braydich-Stolle LK, Schaeublin NM, Murdock RC, Jiang J, Biswas P, Schlager JJ, Hussain SM. J. Nanopart. Res. 2009;11:1361. [Google Scholar]
- (369).Liu W, Wu YA, Wang C, Li HC, Wang T, Liao CY, Cui L, Zhou QF, Yan B, Jiang GB. Nanotoxicology. 2010;4:319. doi: 10.3109/17435390.2010.483745. [DOI] [PubMed] [Google Scholar]
- (370).Hussain S, Boland S, Baeza-Squiban A, Hamel R, Thomassen LCJ, Martens JA, Billon-Galland MA, Fleury-Feith J, Moisan F, Pairon JC, Marano F. Toxicology. 2009;260:142. doi: 10.1016/j.tox.2009.04.001. [DOI] [PubMed] [Google Scholar]
- (371).Yu KO, Grabinski CM, Schrand AM, Murdock RC, Wang W, Gu BH, Schlager JJ, Hussain SM. J. Nanopart. Res. 2009;11:15. [Google Scholar]
- (372).Bhattacharjee S, de Haan LHJ, Evers NM, Jiang X, Marcelis ATM, Zuilhof H, Rietjens I, Alink GM. Part. Fibre Toxicol. 2010:7. doi: 10.1186/1743-8977-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (373).Yin H, Casey PS, McCall MJ, Fenech M. Langmuir. 2010;26:15399. doi: 10.1021/la101033n. [DOI] [PubMed] [Google Scholar]
- (374).Chompoosor A, Saha K, Ghosh PS, Macarthy DJ, Miranda OR, Zhu ZJ, Arcaro KF, Rotello VM. Small. 2010;6:2246. doi: 10.1002/smll.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (375).Guo B, Zebda R, Drake SJ, Sayes CM. Part. Fibre Toxicol. 2009;6:4. doi: 10.1186/1743-8977-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (376).Liu YX, Chen ZP, Wang JK. J. Nanopart. Res. 2011;13:199. [Google Scholar]
- (377).Gharbi N, Pressac M, Hadchouel M, Szwarc H, Wilson SR, Moussa F. Nano Lett. 2005;5:2578. doi: 10.1021/nl051866b. [DOI] [PubMed] [Google Scholar]
- (378).Hirst SM, Karakoti AS, Tyler RD, Sriranganathan N, Seal S, Reilly CM. Small. 2009;5:2848. doi: 10.1002/smll.200901048. [DOI] [PubMed] [Google Scholar]
- (379).Ali SS, Hardt JI, Quick KL, Kim-Han JS, Erlanger BF, Huang TT, Epstein CJ, Dugan LL. Free Radical Biol. Med. 2004;37:1191. doi: 10.1016/j.freeradbiomed.2004.07.002. [DOI] [PubMed] [Google Scholar]
- (380).Watts PCP, Fearon PK, Hsu WK, Billingham NC, Kroto HW, Walton DRM. J. Mater. Chem. 2003;13:491. [Google Scholar]
- (381).Celardo I, De Nicola M, Mandoli C, Pedersen JZ, Traversa E, Ghibelli L. ACS Nano. 2011;5:4537. doi: 10.1021/nn200126a. [DOI] [PubMed] [Google Scholar]
- (382).Hamasaki T, Kashiwagi T, Imada T, Nakamichi N, Aramaki S, Toh K, Morisawa S, Shimakoshi H, Hisaeda Y, Shirahata S. Langmuir. 2008;24:7354. doi: 10.1021/la704046f. [DOI] [PubMed] [Google Scholar]
- (383).Martin R, Menchon C, Apostolova N, Victor VM, Alvaro M, Herance JR, Garcia H. ACS Nano. 2010;4:6957. doi: 10.1021/nn1019412. [DOI] [PubMed] [Google Scholar]
- (384).Amin KA, Hassan MS, Awad ET, Hashem KS. Int. J. Nanomed. 2011;6:143. doi: 10.2147/IJN.S15308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (385).Fenoglio I, Tomatis M, Lison D, Muller J, Fonseca A, Nagy JB, Fubini B. Free Radical Biol. Med. 2006;40:1227. doi: 10.1016/j.freeradbiomed.2005.11.010. [DOI] [PubMed] [Google Scholar]
- (386).Zhang LB, Laug L, Munchgesang W, Pippel E, Gosele U, Brandsch M, Knez M. Nano Lett. 2010;10:219. doi: 10.1021/nl903313r. [DOI] [PubMed] [Google Scholar]
- (387).Kyriakis JM, Avruch J. Physiol. Rev. 2012;92:689. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- (388).Peuschel H, Sydlik U, Haendeler J, Buchner N, Stockmann D, Kroker M, Wirth R, Brock W, Unfried K. Biol. Chem. 2010;391:1327. doi: 10.1515/BC.2010.131. [DOI] [PubMed] [Google Scholar]
- (389).Liu X, Sun JA. Biomaterials. 2010;31:8198. doi: 10.1016/j.biomaterials.2010.07.069. [DOI] [PubMed] [Google Scholar]
- (390).Kim JA, Lee NH, Kim BH, Rhee WJ, Yoon S, Hyeon T, Park TH. Biomaterials. 2011;32:2871. doi: 10.1016/j.biomaterials.2011.01.019. [DOI] [PubMed] [Google Scholar]
- (391).Mohamed BM, Verma NK, Prina-Mello A, Williams Y, Davies AM, Bakos G, Tormey L, Edwards C, Hanrahan J, Salvati A, Lynch I, Dawson K, Kelleher D, Volkov Y. J. Nanobiotechnol. 2011;9:29. doi: 10.1186/1477-3155-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (392).Lao F, Chen L, Li W, Ge CC, Qu Y, Sun QM, Zhao YL, Han D, Chen CY. ACS Nano. 2009;3:3358. doi: 10.1021/nn900912n. [DOI] [PubMed] [Google Scholar]
- (393).Hayden MS, Ghosh S. Cell. 2008;132:344. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- (394).Nishanth RP, Jyotsna RG, Schlager JJ, Hussain SM, Reddanna P. Nanotoxicology. 2011;5:502. doi: 10.3109/17435390.2010.541604. [DOI] [PubMed] [Google Scholar]
- (395).Cui YL, Liu HT, Zhou M, Duan YM, Li N, Gong XL, Hu RP, Hong MM, Hong FS. J. Biomed. Mater. Res., Part A. 2011;96A:221. doi: 10.1002/jbm.a.32976. [DOI] [PubMed] [Google Scholar]
- (396).Li A, Qin LL, Zhu D, Zhu RR, Sun J, Wang SL. Biomaterials. 2010;31:748. doi: 10.1016/j.biomaterials.2009.09.095. [DOI] [PubMed] [Google Scholar]
- (397).Manna SK, Sarkar S, Barr J, Wise K, Barrera EV, Jejelowo O, Rice-Ficht AC, Ramesh GT. Nano Lett. 2005;5:1676. doi: 10.1021/nl0507966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (398).Chou CC, Hsiao HY, Hong QS, Chen CH, Peng YW, Chen HW, Yang PC. Nano Lett. 2008;8:437. doi: 10.1021/nl0723634. [DOI] [PubMed] [Google Scholar]
- (399).Petersen EJ, Nelson BC. Anal. Bioanal. Chem. 2010;398:613. doi: 10.1007/s00216-010-3881-7. [DOI] [PubMed] [Google Scholar]
- (400).AshaRani PV, Mun GLK, Hande MP, Valiyaveettil S. ACS Nano. 2009;3:279. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- (401).Xing Y, Xiong W, Zhu L, Osawa E, Hussin S, Dai LM. ACS Nano. 2011;5:2376. doi: 10.1021/nn200279k. [DOI] [PubMed] [Google Scholar]
- (402).Zhu L, Chang DW, Dai LM, Hong YL. Nano Lett. 2007;7:3592. doi: 10.1021/nl071303v. [DOI] [PubMed] [Google Scholar]
- (403).Ahamed M, Karns M, Goodson M, Rowe J, Hussain SM, Schlager JJ, Hong YL. Toxicol. Appl. Pharmacol. 2008;233:404. doi: 10.1016/j.taap.2008.09.015. [DOI] [PubMed] [Google Scholar]
- (404).Ljungman M. Environ. Mol. Mutag. 2010;51:879. doi: 10.1002/em.20597. [DOI] [PubMed] [Google Scholar]
- (405).Waite KA, Eng C. Nat. Rev. Genet. 2003;4:763. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- (406).Zhang Y, Mu Q, Zhou H, Vrijens K, Roussel MF, Jiang G, Yan B. Cell Death Dis. 2012;3:e308. doi: 10.1038/cddis.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (407).Liu DD, Yi CQ, Zhang DW, Zhang JC, Yang MS. ACS Nano. 2010;4:2185. doi: 10.1021/nn901479w. [DOI] [PubMed] [Google Scholar]
- (408).Mu QX, Du GQ, Chen TS, Zhang B, Yan B. ACS Nano. 2009;3:1139. doi: 10.1021/nn900252j. [DOI] [PubMed] [Google Scholar]
- (409).Li Y, Liu Y, Fu YJ, Wei TT, Le Guyader L, Gao G, Liu RS, Chang YZ, Chen CY. Biomaterials. 2012;33:402. doi: 10.1016/j.biomaterials.2011.09.091. [DOI] [PubMed] [Google Scholar]
- (410).Braydich-Stolle LK, Lucas B, Schrand A, Murdock RC, Lee T, Schlager JJ, Hussain SM, Hofmann MC. Toxicol. Sci. 2010;116:577. doi: 10.1093/toxsci/kfq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (411).Chen GY, Yang HJ, Lu CH, Chao YC, Hwang SM, Chen CL, Lo KW, Sung LY, Luo WY, Tuan HY, Hu YC. Biomaterials. 2012;33:6559. doi: 10.1016/j.biomaterials.2012.05.064. [DOI] [PubMed] [Google Scholar]
- (412).Zhang TT, Stilwell JL, Gerion D, Ding LH, Elboudwarej O, Cooke PA, Gray JW, Alivisatos AP, Chen FF. Nano Lett. 2006;6:800. doi: 10.1021/nl0603350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (413).Bouwmeester H, Poortman J, Peters RJ, Wijma E, Kramer E, Makama S, Puspitaninganindita K, Marvin HJP, Peijnenburg A, Hendriksen PJM. ACS Nano. 2011;5:4091. doi: 10.1021/nn2007145. [DOI] [PubMed] [Google Scholar]
- (414).Cui DX, Tian FR, Ozkan CS, Wang M, Gao HJ. Toxicol. Lett. 2005;155:73. doi: 10.1016/j.toxlet.2004.08.015. [DOI] [PubMed] [Google Scholar]
- (415).Liu YX, Chen ZP, Gu N, Wang JK. Toxicol. Lett. 2011;205:130. doi: 10.1016/j.toxlet.2011.05.1031. [DOI] [PubMed] [Google Scholar]
- (416).Ding LH, Stilwell J, Zhang TT, Elboudwarej O, Jiang HJ, Selegue JP, Cooke PA, Gray JW, Chen FQF. Nano Lett. 2005;5:2448. doi: 10.1021/nl051748o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (417).Fujita K, Horie M, Kato H, Endoh S, Suzuki M, Nakamura A, Miyauchi A, Yamamoto K, Kinugasa S, Nishio K, Yoshida Y, Iwahashi H, Nakanishi J. Toxicol. Lett. 2009;191:109. doi: 10.1016/j.toxlet.2009.08.011. [DOI] [PubMed] [Google Scholar]
- (418).Khan JA, Pillai B, Das TK, Singh Y, Maiti S. ChemBioChem. 2007;8:1237. doi: 10.1002/cbic.200700165. [DOI] [PubMed] [Google Scholar]
- (419).Hanagata N, Zhuang F, Connolly S, Li J, Ogawa N, Xu MS. ACS Nano. 2011;5:9326. doi: 10.1021/nn202966t. [DOI] [PubMed] [Google Scholar]
- (420).Tsai YY, Huang YH, Chao YL, Hu KY, Chin LT, Chou SH, Hour AL, Yao YD, Tu CS, Liang YJ, Tsai CY, Wu HY, Tan SW, Chen HM. ACS Nano. 2011;5:9354. doi: 10.1021/nn2027775. [DOI] [PubMed] [Google Scholar]
- (421).Ge Y, Bruno M, Wallace K, Winnik W, Prasad RY. Proteomics. 2011;11:2406. doi: 10.1002/pmic.201000741. [DOI] [PubMed] [Google Scholar]
- (422).Haniu H, Matsuda Y, Takeuchi K, Kim YA, Hayashi T, Endo M. Toxicol. Appl. Pharmacol. 2010;242:256. doi: 10.1016/j.taap.2009.10.015. [DOI] [PubMed] [Google Scholar]
- (423).Yuan JF, Gao HC, Sui JJ, Chen WN, Ching CB. Toxicol. In Vitro. 2011;25:1820. doi: 10.1016/j.tiv.2011.09.022. [DOI] [PubMed] [Google Scholar]
- (424).Lenz EM, Wilson ID. J. Proteome Res. 2007;6:443. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- (425).Kim S, Kim S, Lee S, Kwon B, Choi J, Hyun JW, Kim S. Bull. Korean Chem. Soc. 2011;32:2021. [Google Scholar]
- (426).Feng JH, Liu HL, Bhakoo KK, Lu LH, Chen Z. Biomaterials. 2011;32:6558. doi: 10.1016/j.biomaterials.2011.05.035. [DOI] [PubMed] [Google Scholar]
- (427).Feng JH, Zhao J, Hao FH, Chen C, Bhakoo K, Tang HR. J. Nanopart. Res. 2011;13:2049. [Google Scholar]
- (428).Wang Y, Chen L. Nanomed. -Nanotechnol. Biol. Med. 2011;7:385. [Google Scholar]
- (429).Wang Y, Chen L, Liu P. Chem. -Eur. J. 2012;18:5935. doi: 10.1002/chem.201103571. [DOI] [PubMed] [Google Scholar]
- (430).Dubois F, Mahler B, Dubertret B, Doris E, Mioskowski C. J. Am. Chem. Soc. 2007;129:482. doi: 10.1021/ja067742y. [DOI] [PubMed] [Google Scholar]
- (431).Song HT, Choi JS, Huh YM, Kim S, Jun YW, Suh JS, Cheon J. J. Am. Chem. Soc. 2005;127:9992. doi: 10.1021/ja051833y. [DOI] [PubMed] [Google Scholar]
- (432).Li N, Binder WH. J. Mater. Chem. 2011;21:16717. [Google Scholar]
- (433).Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, Keller S, Radler J, Natile G, Parak WJ. Nano Lett. 2004;4:703. [Google Scholar]
- (434).Gole A, Murphy CJ. Chem. Mater. 2005;17:1325. [Google Scholar]
- (435).Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdörster G, McGrath JL. Biomaterials. 2009;30:603. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- (436).Aubin-Tam M-E, Hamad-Schifferli K. Langmuir. 2005;21:12080. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- (437).You C-C, De M, Han G, Rotello VM. J. Am. Chem. Soc. 2005;127:12873. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- (438).Gessner A, Lieske A, Paulke BR, Muller RH. Eur. J. Pharm. Biopharm. 2002;54:165. doi: 10.1016/s0939-6411(02)00081-4. [DOI] [PubMed] [Google Scholar]
- (439).Su G, Zhou H, Mu Q, Zhang Y, Li L, Jiao P, Jiang G, Yan B. J. Phys. Chem. C. 2012;116:4993. [Google Scholar]
- (440).Cho EC, Xie J, Wurm PA, Xia Y. Nano Lett. 2009;9:1080. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- (441).Wilhelm C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F. Biomaterials. 2003;24:1001. doi: 10.1016/s0142-9612(02)00440-4. [DOI] [PubMed] [Google Scholar]
- (442).Mailaender V, Lorenz MR, Holzapfel V, Musyanovych A, Fuchs K, Wiesneth M, Walther P, Landfester K, Schrezenmeier H. Mol. Imag. Biol. 2008;10:138. doi: 10.1007/s11307-007-0130-3. [DOI] [PubMed] [Google Scholar]
- (443).Zhang LW, Monteiro-Riviere NA. Toxicol. Sci. 2009;110:138. doi: 10.1093/toxsci/kfp087. [DOI] [PubMed] [Google Scholar]
- (444).Fernandes CF, Godoy JR, Döring B, Cavalcanti MCO, Bergmann M, Petzinger E, Geyer J. Biochem. Biophys. Res. Commun. 2007;361:26. doi: 10.1016/j.bbrc.2007.06.160. [DOI] [PubMed] [Google Scholar]
- (445).Fuller JE, Zugates GT, Ferreira LS, Ow HS, Nguyen NN, Wiesner UB, Langer RS. Biomaterials. 2008;29:1526. doi: 10.1016/j.biomaterials.2007.11.025. [DOI] [PubMed] [Google Scholar]
- (446).Leroueil PR, Hong S, Mecke A, Baker JR, Jr., Orr BG, Holl MMB. Acc. Chem. Res. 2007;40:335. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (447).Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Banaszak Holl MM. Nano Lett. 2008;8:420. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- (448).Gessner A, Waicz R, Lieske A, Paulke BR, Mäder K, Müller RH. Int. J. Pharm. 2000;196:245. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- (449).Lorenz S, Hauser CP, Autenrieth B, Weiss CK, Landfester K, Mailaender V. Macromol. Biosci. 2010;10:1034. doi: 10.1002/mabi.201000099. [DOI] [PubMed] [Google Scholar]
- (450).Yang H, Fung S-Y, Liu M. Angew. Chem., Int. Ed. 2011;50:9643. doi: 10.1002/anie.201102911. [DOI] [PubMed] [Google Scholar]
- (451).Nel AE, Maedler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- (452).Richards D, Ivanisevic A. Chem. Soc. Rev. 2012;41:2052. doi: 10.1039/c1cs15252a. [DOI] [PubMed] [Google Scholar]
- (453).Pelaz B, Charron G, Pfeiffer C, Zhao Y, de la Fuente JM, Liang X-J, Parak WJ, del Pino P. Small. 2012 doi: 10.1002/smll.201201229. DOI: 10.1002/smll.201201229. [DOI] [PubMed] [Google Scholar]
- (454).Wang Y, Ye C, Wu L, Hu Y. J. Pharm. Biomed. Anal. 2010;53:235. doi: 10.1016/j.jpba.2010.02.028. [DOI] [PubMed] [Google Scholar]
- (455).Lovric J, Cho SJ, Winnik FM, Maysinger D. Chem. Biol. 2005;12:1227. doi: 10.1016/j.chembiol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- (456).Schrand AM, Rahman MF, Hussain SM, Schlager JJ, Smith DA, Syed AF. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2010;2:544. doi: 10.1002/wnan.103. [DOI] [PubMed] [Google Scholar]
- (457).Niidome T, Yamagata M, Okamoto Y, Akiyama Y, Takahashi H, Kawano T, Katayama Y, Niidome Y. J. Controlled Release. 2006;114:343. doi: 10.1016/j.jconrel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- (458).Lemon BI, Crooks RM. J. Am. Chem. Soc. 2000;122:12886. [Google Scholar]
- (459).Liu J. a., Li H, Wang W, Xu H, Yang X, Liang J, He Z. Small. 2006;2:999. doi: 10.1002/smll.200500421. [DOI] [PubMed] [Google Scholar]
- (460).Susumu K, Mei BC, Mattoussi H. Nat. Protoc. 2009;4:424. doi: 10.1038/nprot.2008.247. [DOI] [PubMed] [Google Scholar]
- (461).Tan SJ, Jana NR, Gao S, Patra PK, Ying JY. Chem. Mater. 2010;22:2239. [Google Scholar]
- (462).Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. Nat. Biotechnol. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- (463).Akiyoshi K, Sasaki Y, Sunamoto J. Bioconjugate Chem. 1999;10:321. doi: 10.1021/bc9801272. [DOI] [PubMed] [Google Scholar]
- (464).Yu WW, Chang E, Falkner JC, Zhang J, Al-Somali AM, Sayes CM, Johns J, Drezek R, Colvin VL. J. Am. Chem. Soc. 2007;129:2871. doi: 10.1021/ja067184n. [DOI] [PubMed] [Google Scholar]
- (465).Lu W, Senapati D, Wang S, Tovmachenko O, Singh AK, Yu H, Ray PC. Chem. Phys. Lett. 2010;487:92. doi: 10.1016/j.cplett.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (466).Chung Y-C, Chen IH, Chen C-J. Biomaterials. 2008;29:1807. doi: 10.1016/j.biomaterials.2007.12.032. [DOI] [PubMed] [Google Scholar]
- (467).Ge C, Du J, Zhao L, Wang L, Liu Y, Li D, Yang Y, Zhou R, Zhao Y, Chai Z, Chen C. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16968. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (468).Leonov AP, Zheng J, Clogston JD, Stern ST, Patri AK, Wei A. ACS Nano. 2008;2:2481. doi: 10.1021/nn800466c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (469).Hauck TS, Ghazani AA, Chan WCW. Small. 2008;4:153. doi: 10.1002/smll.200700217. [DOI] [PubMed] [Google Scholar]
- (470).Alkilany AM, Nagaria PK, Hexel CR, Shaw TJ, Murphy CJ, Wyatt MD. Small. 2009;5:701. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- (471).Aillon KL, Xie Y, El-Gendy N, Berkland CJ, Forrest ML. Adv. Drug Del. Rev. 2009;61:457. doi: 10.1016/j.addr.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (472).Barreto JA, O'Malley W, Kubeil M, Graham B, Stephan H, Spiccia L. Adv. Mater. 2011;23:H18. doi: 10.1002/adma.201100140. [DOI] [PubMed] [Google Scholar]
- (473).Petros RA, DeSimone JM. Nat. Rev. Drug Discovery. 2010;9:615. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- (474).Wang M, Thanou M. Pharmacol. Res. 2010;62:90. doi: 10.1016/j.phrs.2010.03.005. [DOI] [PubMed] [Google Scholar]
- (475).Xie J, Liu G, Eden HS, Ai H, Chen X. Acc. Chem. Res. 2011;44:883. doi: 10.1021/ar200044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (476).Sha MY, Xu H, Natan MJ, Cromer R. J. Am. Chem. Soc. 2008;130:17214. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (477).Cho H-S, Dong Z, Pauletti GM, Zhang J, Xu H, Gu H, Wang L, Ewing RC, Huth C, Wang F, Shi D. ACS Nano. 2010;4:5398. doi: 10.1021/nn101000e. [DOI] [PubMed] [Google Scholar]
- (478).Albanese A, Tang PS, Chan WCW. Yarmush ML, editor. Annu. Rev. Biomed. Eng., Vol 14. 2012;14:1. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- (479).Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanomedicine. 2011;6:715. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (480).Pasqui D, Golini L, Della Giovampaola C, Atrei A, Barbucci R. Biomacromolecules. 2011;12:1243. doi: 10.1021/bm101521e. [DOI] [PubMed] [Google Scholar]
- (481).Di Guglielmo C, De Lapuente J, Porredon C, Ramos-Lopez D, Sendra J, Borras M. J. Nanosci. Nanotechnol. 2012;12:6185. doi: 10.1166/jnn.2012.6430. [DOI] [PubMed] [Google Scholar]
- (482).Li J, Ni X, Leong KW. J. Biomed. Mater. Res., Part A. 2003;65A:196. doi: 10.1002/jbm.a.10444. [DOI] [PubMed] [Google Scholar]
- (483).Zhang J, Ma PX. Nano Today. 2010;5:337. doi: 10.1016/j.nantod.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (484).Zhang L, Chan JM, Gu FX, Rhee J-W, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. ACS Nano. 2008;2:1696. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (485).Romberg B, Hennink WE, Storm G. Pharm. Res. 2008;25:55. doi: 10.1007/s11095-007-9348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (486).Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Bioconjugate Chem. 2004;15:79. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- (487).Akiyama Y, Mori T, Katayama Y, Niidome T. J. Controlled Release. 2009;139:81. doi: 10.1016/j.jconrel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- (488).Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- (489).Gu L, Fang RH, Sailor MJ, Park J-H. ACS Nano. 2012;6:4947. doi: 10.1021/nn300456z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (490).Larson TA, Joshi PR, Sokolov K. ACS Nano. 2012;6:9182. doi: 10.1021/nn3035155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (491).van Schooneveld MM, Vucic E, Koole R, Zhou Y, Stocks J, Cormode DP, Tang CY, Gordon RE, Nicolay K, Meijerink A, Fayad ZA, Mulder WJM. Nano Lett. 2008;8:2517. doi: 10.1021/nl801596a. [DOI] [PubMed] [Google Scholar]
- (492).Lynn DM, Anderson DG, Putnam D, Langer R. J. Am. Chem. Soc. 2001;123:8155. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- (493).Akinc A, Lynn DM, Anderson DG, Langer R. J. Am. Chem. Soc. 2003;125:5316. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- (494).Anderson DG, Lynn DM, Langer R. Angew. Chem. Int. Ed. 2003;42:3153. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- (495).Anderson DG, Akinc A, Hossain N, Langer R. Mol. Ther. 2005;11:426. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- (496).Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Bioconjugate Chem. 2006;17:1162. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- (497).Zugates GT, Tedford NC, Zumbuehl A, Jhunjhunwala S, Kang CS, Griffith LG, Lauffenburger DA, Langer R, Anderson DG. Bioconjugate Chem. 2007;18:1887. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]
- (498).Green JJ, Zugates GT, Tedford NC, Huang YH, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Adv. Mater. 2007;19:2836. [Google Scholar]
- (499).Schellenberger EA, Reynolds F, Weissleder R, Josephson L. ChemBioChem. 2004;5:275. doi: 10.1002/cbic.200300713. [DOI] [PubMed] [Google Scholar]
- (500).Kelly KA, Shaw SY, Nahrendorf M, Kristoff K, Aikawa E, Schreiber SL, Clemons PA, Weissleder R. Integr. Biol. 2009;1:311. doi: 10.1039/b821775k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (501).Miranda OR, You C-C, Phillips R, Kim I-B, Ghosh PS, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2007;129:9856. doi: 10.1021/ja0737927. [DOI] [PubMed] [Google Scholar]
- (502).Samanta A, Maiti KK, Soh K-S, Liao X, Vendrell M, Dinish US, Yun S-W, Bhuvaneswari R, Kim H, Rautela S, Chung J, Olivo M, Chang Y-T. Angew. Chem. Int. Ed. 2011;50:6089. doi: 10.1002/anie.201007841. [DOI] [PubMed] [Google Scholar]
- (503).Zhang B, Xing Y, Li Z, Zhou H, Mu Q, Yan B. Nano Lett. 2009;9:2280. doi: 10.1021/nl900437n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (504).Gao N, Zhang Q, Mu Q, Bai Y, Li L, Zhou H, Butch ER, Powell TB, Snyder SE, Jiang G, Yan B. ACS Nano. 2011;5:4581. doi: 10.1021/nn200283g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (505).Zhou H, Jiao P, Yang L, Li X, Yan B. J. Am. Chem. Soc. 2011;133:680. doi: 10.1021/ja108527y. [DOI] [PubMed] [Google Scholar]
- (506).Zhang Y, Yan B. Chem. Res. Toxicol. 2012;25:1212. doi: 10.1021/tx300059m. [DOI] [PubMed] [Google Scholar]
- (507).Zhang Y, Mu Q, Zhou H, Vrijens K, Roussel MF, Jiang G, Yan B. Cell Death Dis. 2012;3:e308. doi: 10.1038/cddis.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (508).Tropsha A. Mol. Inf. 2010;29:476. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- (509).Tropsha A. Predictive Quantitative Structure-Activity Relationship Modeling. In: John BT, David JT, editors. Compr. Med. Chem. II. Elsevier; Oxford: 2007. pp. 149–165. [Google Scholar]
- (510).Todeschini R, Consonni V. Handbook of Molecular Descriptors. Wiley-VCH; Weinheim, Germany: 2000. [Google Scholar]
- (511).Dearden JC, Cronin MTD, Kaiser KLE. SAR QSAR Environ. Res. 2009;20:241. doi: 10.1080/10629360902949567. [DOI] [PubMed] [Google Scholar]
- (512).Stouch TR, Kenyon JR, Johnson SR, Chen XQ, Doweyko A, Li Y. J. Comput. Aided Mol. Des. 2003;17:83. doi: 10.1023/a:1025358319677. [DOI] [PubMed] [Google Scholar]
- (513).Tropsha A, Golbraikh A. Curr. Pharm. Des. 2007;13:3494. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- (514).Netzeva TI, Worth AP, Aldenberg T, Benigni R, Cronin MTD, Gramatica P, Jaworska JS, Kahn S, Klopman G, Marchant CA, Myatt G, Nikolova-Jeliazkova N, Patlewicz GY, Perkins R, Roberts DW, Schultz TW, Stanton DT, van de Sandt JJM, Tong WD, Veith G, Yang CH. ATLA, Altern. Lab. Anim. 2005;33:155. doi: 10.1177/026119290503300209. [DOI] [PubMed] [Google Scholar]
- (515).Irwin JJ, Shoichet BK. J. Chem. Inf. Model. 2005;45:177. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (516).2013. Dragon, Version 6.0; TALETE s.r.l.: Italy.
- (517).2012. MOE, Version 2012.10; Chemical Computing Group: Canada.
- (518).Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov'ev V, Hoonakker F, Tetko IV, Marcou G. Curr. Comput.-Aided Drug Des. 2008;4:191. [Google Scholar]
- (519).Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat. Biotechnol. 2005;23:1418. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- (520).Tasis D, Tagmatarchis N, Bianco A, Prato M. Chem. Rev. 2006;106:1105. doi: 10.1021/cr050569o. [DOI] [PubMed] [Google Scholar]
- (521).Menard-Moyon C, Kostarelos K, Prato M, Bianco A. Chem. Biol. 2010;17:107. doi: 10.1016/j.chembiol.2010.01.009. [DOI] [PubMed] [Google Scholar]
- (522).Prato M, Kostarelos K, Bianco A. Acc. Chem. Res. 2008;41:60. doi: 10.1021/ar700089b. [DOI] [PubMed] [Google Scholar]
- (523).Heister E, Lamprecht C, Neves V, Tilmaciu C, Datas L, Flahaut E, Soula B, Hinterdorfer P, Coley HM, Silva SRP, McFadden J. ACS Nano. 2010;4:2615. doi: 10.1021/nn100069k. [DOI] [PubMed] [Google Scholar]