Abstract
Nonalcoholic fatty-liver disease (NAFLD) prevalence is increasing worldwide, with the affected US population estimated near 30%. Diet is a recognized risk factor in the NAFLD spectrum, which includes non-alcoholic steatohepatitis (NASH) and fibrosis. Low hepatic copper (Cu) was recently linked to clinical NAFLD/NASH severity. Simple sugar consumption including sucrose and fructose is implicated in NAFLD, while consumption of these macronutrients also decrease liver Cu levels. Though dietary sugar and low Cu are implicated in NAFLD, transcript-level responses that connect diet and pathology are not established. We have developed a mature rat model of NAFLD induced by dietary Cu deficiency, human-relevant high sucrose intake (30% w/w), or both factors in combination. Compared to the control diet with adequate Cu and 10% (w/w) sucrose, rats fed either high sucrose or low Cu diets had increased hepatic expression of genes involved in inflammation and fibrogenesis, including hepatic stellate cell activation, while the combination of diet factors also increased ATP citrate lyase (Acly) and fatty-acid synthase (Fasn) gene transcription (Fold change >2, p <0.02). Low dietary Cu decreased hepatic and serum Cu (p ≤0.05), promoted lipid peroxidation, and induced NAFLD-like histopathology, while the combined factors also induced fasting hepatic insulin resistance and liver damage. Neither low Cu nor 30% sucrose in the diet led to enhanced weight gain. Taken together, transcript profiles, histological and biochemical data indicate that low Cu and high sucrose promote hepatic gene expression and physiological responses associated with NAFLD and NASH, even in the absence of obesity or severe steatosis.
Keywords: liver, copper, inflammation, fibrosis, steatosis, non-alcoholic steatohepatitis
1. INTRODUCTION
Obesity and its related health disorders are increasing worldwide, particularly in developed countries such as the United States [1–4]. Metabolic Syndrome (MetS) has been used to describe the multi-factorial series of disorders often associated with the co-morbidity of obesity. Factors associated with obesity include insulin resistance, hyperglycemia, cardiovascular disease, altered lipid metabolism, and a complex cross-talk between metabolic and inflammatory pathways [5]. Non-alcoholic fatty liver disease (NAFLD), the hepatic manifestation of MetS, is likewise becoming more prevalent, it is the most frequently diagnosed liver disease in Western countries as well as a health disparity for some minority groups [6]. It is currently estimated that in developed countries, 25–30% of adults [7–10] and 3–10% of children [11] have NAFLD. The emergence of pediatric NAFLD may be particularly alarming as the diagnosis can include progression to cirrhosis [12]. NAFLD includes a spectrum of liver pathology ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), fibrosis and cirrhosis [13]. First identified in 1980s, the cause of the disease is still unknown [14]. Recent studies have suggested that progression to NASH may increase mortality risk [15], while fibrosis severity is a critical diagnostic tool for chronic liver disease [16,17] and indicative of long-term liver complications and enhanced mortality risk [18,19]. The molecular mechanisms driving NAFLD progression to NASH have remained elusive, though NAFLD patients presenting with any degree of systemic inflammation and any degree of fibrosis are thought to be at highest risk for NASH [15].
This complex systemic manifestation describes an integrative metabolic response during NAFLD. In this context, the dietary status of the micronutrient copper (Cu) has a fundamental influence on lipid metabolism, though underlying mechanisms of influence are unclear (for review see [20]). Currently, the American or Western diet may be low in essential dietary nutrients such as Cu [21]. Cu deficiency induces NAFLD symptoms in rodent models [22–25], and retrospective analysis of NAFLD patients revealed decreased hepatic Cu levels that correlated with NAFLD/NASH progression [22,23]. Additionally, NAFLD patients with low Cu displayed increased levels of hepatic iron [22]. Biopsies from NAFLD patients with hepatic iron overload also revealed an association with increased oxidative stress and NASH [26]. In vivo rodent models of dietary Cu deficiency also displayed increased hepatic iron content, [22] as well as steatosis and insulin resistance [23]. Intestine-specific genetic inactivation of high-affinity Cu import also resulted in Fe accumulation in the liver in Kupffer cells, linking intestinal Cu transport and altered hepatic Fe metabolism [27].
In addition to Cu, another nutrient that affects NAFLD is dietary sugar, including fructose and sucrose. High fructose consumption, potentially from high-fructose corn syrup (HFCS) in beverages, is implicated as a factor driving the metabolic dysregulation underlying MetS. HFCS includes fructose in roughly equal proportion to glucose, similar to the 1:1 ratio in sucrose, and that both sugar sources have equivalent metabolic effects despite recent public focus on fructose [28]. Consumption of fructose in the U.S. has increased over the past 30 years, possibly by as much as 25% [29], via food sources such as fruit juice and HFCS in soft drinks [30,31]. Recent estimates place the mean fructose consumption among Americans at 10% of dietary intake and as high as 15% in up to one-fourth of adolescents [29,30]. Approximately 10% of adults consume as many as 25% of their daily calories from added sugar [32]. Importantly, an indirect role of fructose in oxidative stress may be occurring via the down-regulation of the Ctr1 Cu importer, as indicated in a weanling rat model of NAFLD [24], whereby low Cu reduces antioxidant capacity by limiting Cu-Zn superoxide dismutase activity and promoting hepatic iron accumulation [33].
Two recent studies have assessed the combination of high fructose diets (30% or 3% fructose added to a standard purified diet that already included 50% sucrose) with CuD diets in weanling rats [24,25]. Interestingly, high fructose intake exacerbated both Cu deficiency and hepatic iron overload, caused increased oxidative stress, and decreased antioxidant defenses. These experimental diets, however, with the experimental AIN76A standard purified diet formulation of 50% sucrose (and thus 25% fructose) diverge greatly from human dietary patterns. Though the existing data support links between dietary sugars, Cu status, and NAFLD, the molecular mechanisms by which Cu deficiency and dietary sugars interact to both induce and aggravate NAFLD/NASH disease progression is unclear.
Thus, while a low Cu diet is sufficient to induce its deficiency and to initiate lipogenesis with subsequent NAFLD symptoms, low hepatic Cu may also be involved in aggravating inflammation to promote progression of NAFLD to NASH, especially when coupled to excessive fructose consumption. The goal of the present study is to test the hypothesis that the liver transcriptomic response to sub-optimal Cu nutrition and Western diet-relevant dietary sugar/fructose intake, separately and in combination, can reveal gene expression pathways by which the diet factors promote steatosis and NAFLD spectrum symptoms in a mature rodent model. In our study, analysis of either low Cu or 30% sucrose in the diet identified differentially expressed genes that involved in inflammatory and fibrogenesis responses, while the combination of both factors also caused up-regulation of fatty-acid synthesis genes despite fewer overall transcript changes compared to the impact caused by either the low Cu or high sucrose diet. Moreover, observation of differentially expressed genes corresponding to a pro-fibrotic state, coupled with the development of insulin resistance in rats fed the combined Cu deficient/30% sucrose diet, indicate that low dietary Cu and sucrose consumptions are previously unrecognized, independent, and synergistic risk factors contributing to the progression of NAFLD.
2. METHODS
2.1 Animal Husbandry and Tissue Collection
Animal experiments and husbandry were reviewed approved by the University of Alaska Anchorage (UAA) Institutional Animal Care and Use Committee and performed in accordance with US Public Health Service Policy as documented by The Guide for the Care and Use of Laboratory Animals 8th Edition [34]. Mature (6 months old) male Wistar Rats that had been allowed ad libitum access to Mazuri Rodent Diet (PMI Nutrition, St. Louis) were used in the study. Twenty-four rats were randomly assigned to one of four groups and fed for 12 weeks with diets based on the Purified AIN76A formulation, modified for target sucrose and Cu content as indicated in Table 1 (Custom Animal Diets, Bangor, NJ). Sucrose and Cu content in diets were as follows: CuD/30%- Cu deficient (<0.3 mg Cu/kg)/30% sucrose, CuA/30%- Cu adequate (125 mg/kg)/30% sucrose, CuD/10%- <0.3 mg/kg Cu/10% sucrose, and D) CuA (125 mg/kg Cu)/10% sucrose (control). Starch and dextrin were used to equalize carbohydrates. Cu levels were selected consistent with deficient and ‘normal’ Cu diets used by Aigner et al. [23]. Cu content was verified by inductively coupled plasma mass spectrometry (ICP-MS). All animals were kept on a 12:12 light cycle with food and water supplied ad libitum. Rats were fasted for 6–8 hours prior to euthanization by CO2 asphyxiation after 12 weeks during the active (dark) period. Serum was isolated from whole blood and frozen at −80° C. Livers were perfused with saline via the hepatic portal vein prior to excision, and lobes were separated and stored (frozen at −80°C, or fixed with 4% formaldehyde (Fisher Chemical) in PBS (Sigma) for 1 week followed by storage in PBS with 0.02% sodium azide).
Table 1.
Experimental diet composition by mass (g/kg indicated).
| CuD/30% sucrose | CuA/30% sucrose | CuD/10% sucrose | CuA/10% sucrose | |
|---|---|---|---|---|
| Casein | 149.10 | 149.10 | 149.10 | 149.10 |
| Sucrose | 310.90 | 310.60 | 100.00 | 100.00 |
| Maltose Dextrin | 0.00 | 0.00 | 155.90 | 155.90 |
| Corn Starch | 410.00 | 410.00 | 465.00 | 464.70 |
| Cellulose | 40.00 | 40.00 | 40.00 | 40.00 |
| Salt mix, 76, Cu-def | 35.00 | 35.00 | 35.00 | 35.00 |
| Vit. Mix, 76A | 10.00 | 10.00 | 10.00 | 10.00 |
| Dl-Methionine | 3.00 | 3.00 | 3.00 | 3.00 |
| Choline Bitartrate | 2.00 | 2.00 | 2.00 | 2.00 |
| Cu carbonate | 0.00 | 0.22 | 0.00 | 0.22 |
| Corn Oil | 40.00 | 40.00 | 40.00 | 40.00 |
| FD&C red | 0.00 | 0.08 | 0.00 | 0.08 |
| Total mass | 1000.00 | 1000.00 | 1000.00 | 1000.00 |
2.2. Gene Expression
Hepatic RNA from was isolated from flash-frozen liver tissue using the NucleoSpin RNA II kit (Machery-Nagel). Hepatic transcript profiles (n = 6 per diet treatment) were analyzed by individual Agilent Sureprint G3 Rat 8X 60K platform according to standard protocols through the Institute for Systems Biology (University of Washington, Seattle, WA). Features were extracted with Agilent Feature Extraction and imported into GeneSpring (v12.6, Agilent Technologies, Inc.). Data was normalized by upper quartile rank (also called percentile normalization). Transcript expression levels that changed as a result of the experimental diets compared to control were identified by a fold change (FC) greater than 2.0 and moderated T-test (P < 0.02) with Benjamini-Hochberg correction for multiple comparisons as this order of tests is reported to be both statistically stringent while maintaining biological relevance when compared to either FC or T-test alone [35]. Differentially expressed transcripts and normalized expression values are provided in Supplementary Tables S1–3. Transcript expression levels that met these criteria were separated into lists of those unique to each diet and those shared between diets, and were analyzed in terms of metabolic networks using the Ingenuity Pathways Analysis (IPA, Ingenuity® Systems) software. Top Canonical Pathways and Top Tox Functions in IPA were used to examine metabolic pathways and biological functions that are impacted as suggested by altered transcript expression levels. Select transcripts of interest from pathways represented by IPA were verified in silico by comparison to existing literature describing NAFLD/NASH, as well as qRT-PCR (primers are listed in Supp. Table S4). The WebGestalt platform [36] was used to search for enriched gene ontology (GO) terms in the differentially expressed gene sets from the different comparisons, using as a reference list the set of rat genes represented in the microarray used. GO terms with Benjami-Hochberg corrected p-values < 0.05 were selected as enriched [37].
Raw qPCR amplification data was analyzed by LinRegPCR (linregpcr.nl/[38]). Relative quantification used the comparative ΔΔCt method with actin-beta (ACTB) and beta-2 macroglobulin (B2m) as reference transcripts [39], expressed per gene as fold change (2−ΔΔCt). Quantitative RT-PCR profiles of inflammatory transcripts were performed by SA Biosciences RT2 Cytokines and Chemokines Rat PCR array.
2.3. Gene nomenclature
Gene symbols referring specifically to expression in the rat model are consistent with the Rat Genome Database (http://rgd.mcw.edu/) and are matched with human orthologs defined by the HUGO Gene Nomenclature Committee (www.genenames.org). Gene symbols discussing expression in multiple species or human cells/tissue follow the HUGO nomenclature. Accession numbers are included in Table 2.
Table 2.
IPA identified pathways with differentially expressed transcripts indicated for each test diet when compared to the control diet. Each asterisk in the table indicates that a specific transcript was differentially expressed (up-regulated) compared to the control diet. A down-pointing arrow indicates a decrease in transcript level compared to the control.
| Accession | Symbol | Name | Significant expression change in: | ||
|---|---|---|---|---|---|
| CuA/30% | CuD/10% | CuD/30% | |||
| TNF family | |||||
| NM_012675 | Tnf | Tumor necrosis factor (alpha) | * | ||
| NM_212507 | Ltb | Lymphotoxin beta a.k.a. TNFc | * | ||
| NM_001109112 | Tnfsf13b | B-cell activating factor | * | * | |
| NM_145765 | Tnfsf15 | Vascular endothelial growth inhibitor | * | * | * |
| NM_019135 | Tnfrsf8 | a.k.a. CD30 | * | ||
| NM_001025773 | Tnfrsf9 | a.k.a. CD137 | * | * | |
| NM_001108207 | Tnfrsf21 | Death receptor 6 | * | * | |
| T Helper Cell Cytokines/Receptors | |||||
| NM_201270 | Il-4 | Interleukin-4 (from Th2 cells) | * | * | |
| NM_053828 | Il-13 | Interleukin-13 (from Th2 cells) | * | ||
| NM_001191988 | Il-22 | Interleukin-22 (from Th17 cells) | * | ||
| NM_013163 | Il-2ra | Interleukin-2 receptor alpha (ligand is from Th1 cells) | * | * | * |
| NM_001191937 | IL-17rd | Interleukin-17 receptor d (ligand is from Th17 cells) | * | ||
| NM_001012469 | Il-21r | Interleukin-21 receptor (ligands from Th2, Th17 cells) | * | * | |
| NM_001191869 | Il-22ra1 | Interleukin-22 receptor alpha 1 (ligand is from Th17 cells) | * | * | |
| Chemokines | |||||
| NM_031530 | Ccl2 | C-C motif ligand 2 | * | ||
| NM_013025 | Ccl3 | C-C motif ligand 3 | * | ||
| NM_001007612 | Ccl7 | C-C motif ligand 7 | * | ||
| NM_030845 | Cxcl1 | C-X-C motif ligand 1 | * | ||
| NM_053647 | Cxcl2 | C-X-C motif ligand 2 | * | ||
| NM_001017496 | Cxcl13 | C-X-C motif ligand 13 |

|
||
| NM_017183 | Cxcr2 | C-X-C motif receptor 2 | * | * | |
| Extravasation and Infiltration | |||||
| NM_001011889 | Cldn9 | Claudin 9 | * | ||
| NM_053457 | Cldn11 | Claudin 11 | * | ||
| NM_001107112 | Cldn17 | Claudin 17 | * | ||
| NM_001008514 | Cldn19 | Claudin 19 | * | ||
| NM_001109394 | Cldn20 | Claudin 20 | * | ||
| ENSRNOT00000015940 | Itga2 | Integrin, alpha 2 | * | ||
| NM_138879 | Sele | Selectin E | * | * | * |
| NM_019177 | Sell | Selectin L | * | ||
| NM_013114 | Selp | Selectin P | * | ||
| Ecm Remodeling | |||||
| ENSRNOT00000026719 | Col6a3 | Procollagen, Type VI, alpha 3 | * | * | |
| NM_212528 | Col11a2 | Collagen, Type XI, alpha 2 | * | ||
| HS6ST2 | Hs6st2 | Heparan sulfate 6-O-sulfotransferase 2 | * | * | |
| XM_344461 | Hs6st3 | Heparan sulfate 6-O-sulfotransferase 3 | * | * | |
| NM_001108237 | Lama1 | Laminin, alpha 1 | * | * | * |
| NM_001105925 | Mmp17 | Matrix mettalopeptidase 17 | * | ||
| XM_002742434 | Mmp25 | Matrix mettalopeptidase 25 | * | * | |
| NM_031641 | Sult4a1 | Sulfotransferase family 4A, member 1 | * | * | |
| NM_001109393 | Timp4 | Tissue inhibitor of MMP4 | * | * | |
| Other Inflammatory | |||||
| NM_001029901 | Csf1r | Colony stimulating factor 1 receptor | * | * | |
| NM_133555 | Csf2rb | Colony stimulating factor 2 receptor beta | * | * | |
| NM_001106685 | Csf3r | Colony stimulating factor 3 receptor | * | * | * |
| NM_053953 | Il-1r2 | Interleukin-1 receptor 2 | * | ||
| NM_001106418 | Il-7r | Interleukin-7 receptor | * | ||
| NM_001191750 | Il-12rb2 | Interleukin-12 receptor beta 2 | * | * | |
| NM_017208 | Lbp | Lipopolysaccharide binding protein | * | ||
| NM_001006961 | Osm | Oncostatin M | * | ||
| NM_001005384 | Osmr | Oncostatin M receptor | * | * | |
| Hedgehog Signaling Pathway - EMT Of Hscs | |||||
| NM_001191910 | Gli1 | Gli family zinc finger 1 | * | * | * |
| NM_001107169 | Gli2 | Gli family zinc finger 2 | * | * | |
| NM_080405 | Gli3 | Gli family zinc finger 3 | * |

|
|
| Fibroblast Proliferation And Differentiation | |||||
| NM_053809 | Fgf4 | Fibroblast growth factor 4 | * | * | |
| NM_022211 | Fgf5 | Fibroblast growth factor 5 | * | * | * |
| NM_130816 | Fgf11 | Fibroblast growth factor 11 | * | * | * |
| NM_130751 | Fgf22 | Fibroblast growth factor 22 | * | ||
| NM_130754 | Fgf23 | Fibroblast growth factor 23 | * | * | |
| NM_031131 | Tgfb2 | Transforming growth factor beta 2 | * | * | |
| NM_021586 | Ltbp2 | Latent transforming growth factor beta binding protein | * | * | |
| Markers Of HSC Activation | |||||
| NM_017009 | Gfap | Glial fibrillary acidic protein | * | ||
| NM_012604 | Myh3 | Myosin (II), heavy chain 3, skeletal muscle, embryonic | * | * | * |
| NM_001106017 | Myl7 | Myosin light chain 7, regulatory | * | * | * |
| XM_001078857 | Myh13 | Myosin (II), heavy chain 13, skeletal | * | * | |
| NM_001100690 | Myh14 | Myosin (II), heavy chain 14, non-muscle | * | ||
| NM_145879 | Socs1 | Suppressor of cytokine signaling 1 | * | * | |
| Metabolism And Mets-Related | |||||
| NM_012544 | Ace | Angiotensin I-converting enzyme | * | * | |
| NM_016987 | Acly | ATP-citrate lyase | * | ||
| NM_001004085 | Crat | Carnitine O-acetyltransferase |

|
||
| NM_031241 | Cyp8b1 | Cytochrome P450, family 8, subfamily b, polypeptide 1 |

|

|
|
| NM_017332 | Fasn | Fatty acid synthase | * | ||
| NM_023964 | Gapdhs | Glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | * | * | * |
| NM_021669 | Ghrl | Ghrelin/obestatin prepropeptide | * | * | |
| NM_001109615 | Gys1 | Glycogen synthase 1, muscle | * | * | * |
| NM_001192008 | Idi2 | Isopentenyl-diphosphate delta isomerase 2 | * | * | |
| NM_130741 | Lcn2 | Lipocalin 2 | * | ||
| NM_012859 | Lipe | Lipase, hormone sensitive | * | ||
| NM_017341 | Lipf | Gastric lipase | * | ||
| NM_053994 | Pdha2 | Pyruvate dehydrogenase (lipoamide) alpha 2 | * | ||
| NM_057135 | Pfkfb3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | * | * | * |
| NM_013161 | Pnlip | Pancreatic lipase | * | * | |
| NM_017136 | Sqle | Squalene epoxidase | * | ||
| NM_001107846 | Steap2 | Six transmembrane epithelial antigen of the prostate 2 | * | * | * |
2.4. Histology
Paraffin-embedded or frozen liver samples were sectioned and stained with hematoxylin and eosin (H&E) and Oil Red O (ORO) stains. Whole-slide images of each ORO and H&E tissue section were captured by Olympus NanoZoomer Digital Pathology Microscope. ORO images were captured near hepatic portal triads using NDP software at 200x magnification and divided into quadrants. Quadrants were coded with a random numeral to eliminate bias during evaluation, and NIH ImageJ was used to attain digital image analysis (DIA) of % stained area. H&E images were used to determine presence histological characteristics including lymphocyte infiltration, potential Mallory-Denk bodies, and ballooning hepatocytes features [2,40]. Coherent Anti-Stokes Raman Scattering Spectroscopy (CARS) was performed by Dr. T. Le (Desert Research Institute), as described in [41].
2.5. Biochemical Analyses
Concentrations of serum glucose and serum insulin were determined using a glucose fluorometric assay kit (Cayman Chemical) and rat insulin enzyme immunoassay kit (SPI-bio), respectively. To assess insulin resistance, a homeostatic model assessment of insulin resistance (HOMA-IR) modified specifically for Wistar rats [42], was used as a mathematical model defining insulin resistance based on serum glucose and insulin concentration. Serum alanine transaminase level (ALT) was measured using a colorimetric assay kit (Cayman Chemical). Serum triglycerides were quantitated with the EnzyChrom Triglyceride Assay Kit from BioAssay Systems. Malondialdehyde (MDA) concentrations in serum and hepatic tissue homogenates were determined using a TBARS assay (Cayman Chemical). Free fatty acid concentrations were determined using fluorometric assays for oleic acid (Cayman Chemical) and palmitic acid (BioAssay Systems).
2.6. Elemental Analysis
Approximately 30 mg of hepatic tissue was dehydrated for 24 hours at 37°C, after which tissue samples and serum were analyzed for transition metal content using inductively-coupled plasma mass spectrometry (ICP-MS) by the Elemental Analysis Core at the Oregon Health & Science University in Portland, OR.
2.7 Statistical Analysis
Elemental, biochemical, and quantified histological data have been expressed as means with error bars representing standard error of the mean (± SEM.). Statistical analyses of biochemical, histological, and elemental data were carried out using the GraphPad Prism software and included either Student’s t-test when comparing only two groups, or single-factor ANOVA followed by Dunnett’s or Šídák’s multiple comparison test where appropriate [43]. ROUT outlier analysis [44] was used to eliminate outliers (two outliers in liver Cu were eliminated).
3. RESULTS
3.1. Low dietary Cu and high sucrose induce transcript responses in signaling, inflammation, and fibrosis pathways in a mature rat model of NAFLD
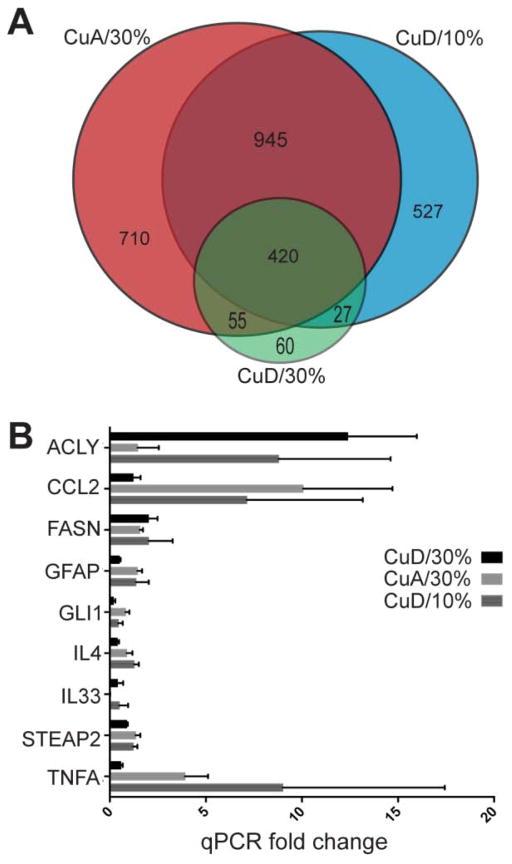
Intersection of transcripts whose abundance was significantly altered as a result of each diet is showed in Figure 1A. A extensive overlap (n = 420) in differentially expressed genes was observed between all the studied diet treatments, This number covers more than 70% of the transcriptional changes in the low Cu (CuD) combined with 30% sucrose, indicating that the transcriptional response under this diet scenario is principally covered by each of the conditions for itself, where the particular common changes can be classified as a local response, basically confined to 60 genes. The preponderance (more than 90%) of differentially expressed transcripts in each Venn diagram section were up-regulated (Supp. Table S5) The observation that nearly all changed transcripts are up-regulated has also been observed in human transcriptomic study of NAFLD, with ten times as many up-regulated compared to down-regulated transcript levels [45].
Figure 1. Relationships of transcript profiles and quantitative RT-PCR analysis of selected transcripts.
A. Venn diagram indicating overlap of transcripts and numbers in categories for CuD/30%, CuA/30% and CuD/10% compared to control CuA/10% diet. B. Mean fold change (2−ΔΔCt) for CuD/30%, CuA/30% and CuD/10% compared to CuA/10% with SEM is shown (n=3 per treatment).
The differentially expressed transcripts for each diet were analyzed by gene set enrichment analysis, which analyzes whether a priori-defined “sets” of biologically related genes are differentially represented when comparing two gene lists [46] and gene ontologies (GO) enrichment using a similar approach for defined sets of ontologies. Enriched processes identified across treatments in the GO analysis are characterized as corresponding to genes involved in signal transduction processes, particularly G-protein coupled receptor signaling, as well as sodium and potassium transport activity. (Supp. Fig. S6–S8). The CuD/30% diet also indicated response to Cu ion levels as well as monooxygenase activity and arachidonic acid epoxygenase activity (Supp. Fig. S8).
To identify pathways with potential roles in NAFLD and associated with genes that are differentially expressed in response to the diet treatments, lists of differentially expressed transcripts in each diet were compared to transcript expression patterns in the control (CuA/10% sucrose), and analyzed with IPA’s Core Analysis and Canonical Pathway functions. Transcripts of interest in each diet were selected from IPA-represented pathways using comparisons to existing literature on NAFLD progression. Of these, selected up-regulated relevant transcripts from each category were verified by qRT-PCR (Fig. 1B). Transcripts mapped to each IPA pathway are listed in Table 2 along with indication of differential expression levels for each diet. Specific nutrient-transcription responses with potential links to NAFLD and progression to NASH are described below.
3.2. Molecular profiles indicate immune cell infiltration in liver due to dietary CuD or 30% sucrose
Single-variable diets (CuD/10% sucrose and CuA/30% sucrose) were most significantly represented in IPA’s Top Canonical Pathway function by differentially expressed genes related to immune cell adhesion and diapedesis pathways, sharing the up-regulation of transcripts for claudins and selectins as well as transcripts involved in extracellular matrix (ECM) tissue remodeling (Table 2). The CuD/10% diet resulted uniquely in up-regulation of transcripts coding for the collagen receptor alpha 2 integrin (Itga2). These results suggest that the hepatic response to Cu deficiency or sucrose challenge in the diet may include tissue remodeling and infiltration of immune cells.
3.3. Inflammation, fibrosis and proliferation transcripts are up-regulated by 30% sucrose and low Cu
Consistent with immune cell infiltration and extravasation, Top Canonical Pathways analysis revealed an increase in inflammatory transcripts involved in the activation and proliferation of immune cells. Both single-variable diets resulted in up-regulation of transcripts corresponding to members of the tumor necrosis factor (TNF) superfamily, interleukins, and colony-stimulating factor receptors, while the CuA/30% sucrose diet uniquely increased several transcripts in each category such as Tnfα as well as C-C motif chemokines Ccl2 and Ccl3. Both CuA/30% and CuD/10% diets resulted in the up-regulation of Il4 gene expression, while CuD/10% alone up-regulated Il13. An additional T-helper cell profile, the Th17 profile, was represented in both CuA/30% and CuD/10% by up-regulation of genes coding for receptors of Il21 and Il22 and, in the CuA/30% diet alone, for the receptor for Il17. Additional inflammation-related transcripts were up-regulated in the single-factor CuA/30% and CuD/10% diet treatments.
Significant up-regulation of gene transcripts related to the inflammatory response in the absence of severe steatosis was an unexpected finding in this study. Therefore, a qPCR profile of inflammatory transcripts across diets was used to confirm the microarray-based observations. While both the CuD/10% diet and the CuA/30% resulted in altered expression of transcripts related to inflammation, the CuA/30% diet initiated the greatest patterns of altered transcript expression levels overall, most notably those related to the TNF and interleukin (IL) families (Fig. 2). Additionally, the gene coding for Il33 was significantly up-regulated (Fold Change > 1.5 and p < 0.05, n= 3) in the CuD/10% diet. The CuD/30% diet, in contrast, had fewer up-regulated inflammation-related transcripts compared to the CuA/10% sucrose control diet, supporting the observation of a negative synergism in gene expression patterns for the CuD/30% sucrose diet, where the combination of factors has less of an effect than single factors (i.e. CuD or 30% sucrose alone), The single-variable diets as well as the CuD/30% diet resulted in up-regulation of hepatic inflammatory transcripts compared to control, although the inflammatory profiles and relative magnitude of gene expression response were different across diets.
Figure 2. CuD or high sucrose initiates transcript expression associated with inflammation.
Cytokines and chemokines qPCR array showing the number of transcripts with fold change >1.5 up or down compared to CuA/10% diet.
Perhaps the most unexpected pattern of gene expression changes involved the observed increased in transcripts related to fibrosis progression. All tested diets shared increased levels of some of the fibrosis-relevant transcripts indicative of hepatic stellate cell (HSC) activation such as FGFs 5 and 11 and myosins Myh3 and Myl7). The CuD/10% diet alone resulted in the up-regulation of the genes coding for Fgf22 and the glial fibrillary acidic protein (Gfap), another possible marker for the earliest stages of HSC activation during inflammation. This also suggests a specific HSC response to Cu deficiency that may exacerbate or accelerate NAFLD progression. In addition to up-regulation of the Gfap transcript in the CuD/10% diet, an increase in transcript levels coding for the suppressor of cytokine signaling 1 (Socs1), which is also associated with HSC activation, was observed as a result of both CuA/30% and CuD/10% diets.
The two single-variable diets CuA/30% and CuD/10% resulted in transcript level increases for the transforming growth factor beta, isoform 2 (Tgfb2) and its associated binding protein (Ltbp2), as well as for sulfotransferases involved in the ECM sulfation of glycosaminoglycans related to Tgfb activation. Of the Hedgehog pathway effectors, Gli1 transcript levels were increased in all three treatment diets, while a Gli2 transcript increase was shared by single variable diets, and Gli3 transcript was up-regulated in the CuA/30% diet alone). The combined CuD/30% sucrose diet, however, induced significant down-regulation of the Gli3 transcript compared to the control CuA/10% diet.
3.4. Dietary sucrose and CuD alter transcripts involved in lipid metabolism and MetS
Differentially expressed transcripts indicate that a Cu deficiency (CuD) and 30% sucrose diet significantly affects hepatic metabolism. While the CuD/30% diet resulted in the fewest gene expression changes, the top metabolic pathway uniquely affected by this diet involved Acetyl-CoA Biosynthesis III (from Citrate). The up-regulation of genes coding for ATP citrate lyase (Acly) and fatty acid synthase (FASN) as a result of the CuD/30% diet (Fig. 1B, Table 2) indicate a switch from carbohydrate to lipid metabolism via use of excess citrate. All three treatment diets shared an increase in glycogen synthase 1 (Gys1) transcripts as well as in transcripts involved in cellular citrate increase, such as an isoform of glyceraldehyde-3-phosphate dehydrogenase (Gapdh), and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (Pfkfb3). Transcripts related to triglyceride (TG) breakdown in the CuA/30% and CuD/10% diets included pancreatic lipase (Pnlip), while CuD/10% alone up-regulated hormone sensitive lipase (Lipe) and CuA/30% sucrose alone increased gastric lipase (LIPF). Mevalonate pathway transcripts related to cholesterol synthesis were also up-regulated, including increases in isopentenyl-diphosphate delta isomerase 2 (Idi2) transcripts in both CuA/30% and CuD/10% diets, while the transcript coding for squalene epoxidase (Sqle) was increased by the CuA/30% diet. Other transcripts associated with MetS included increased gene expression of the angiotensin-1 converting enzyme (Ace) in CuA/30% and CuD/10% diets. Correspondingly in the CuA/30% diet, we observed increased lipocalin 2 (Lcn2), and decreased cytochrome p450 family 8 subfamily B1 (Cyp8b1) gene expression in both CuA/30% and CuD/30% diets, while all three diets led to increased transcript levels of laminin alpha 1 (Lama1) and the six-transmembrane epithelial antigen of the prostate 2 (Steap2) metalloreductase. Thus, either dietary Cu deficiency or high sucrose diet induces a distinct transcript profile that is consistent with steatosis, NAFLD and NASH, while combined, the two dietary factors appear to favor a switch to lipogenic metabolism, including Acly and Fasn.
3.5. Diets with 30% sucrose or low Cu promote histopathology consistent with NAFLD and NASH and disrupt metal balance
Histological analysis indicated that rats fed with CuD or 30% sucrose displayed at least some histological characteristics of NAFLD compared to rat fed on the CuA/10% sucrose control diet (Fig. 3A). The CuD/30% sucrose diet induced the most severe pathology, where ballooned hepatocytes and structures consistent with Mallory-Denk bodies were observed (Fig. 3B), along with some potential inflammatory cell infiltration of the liver tissue indicated by areas with numerous small, dark-stained cells. Though there is some expected heterogeneity in pathology, close examination of liver tissue from the CuD/30%-fed animals also reveals features suggestive of immune cell infiltration in addition to apparent mild steatosis (not shown).
Figure 3. Dietary CuD influences hepatic pathology and steatosis.
A) Representative ORO and H&E images of hepatic tissue with indicated diet (20X objective). CuD diets evidenced significant increases in lipid staining. B) White arrowheads indicate possible Mallory-Denk bodies (20X objective, digital zoom 2X). C) Digital image analysis of ORO stained lipid accumulation by % area, evaluated by ANOVA followed by Dunnett’s multiple comparison test. Data (columns) represent means +SEM (n=6). D) CARS imaging confirms lipid accumulation in 30%/CuD liver compared to 30%/CuA liver.
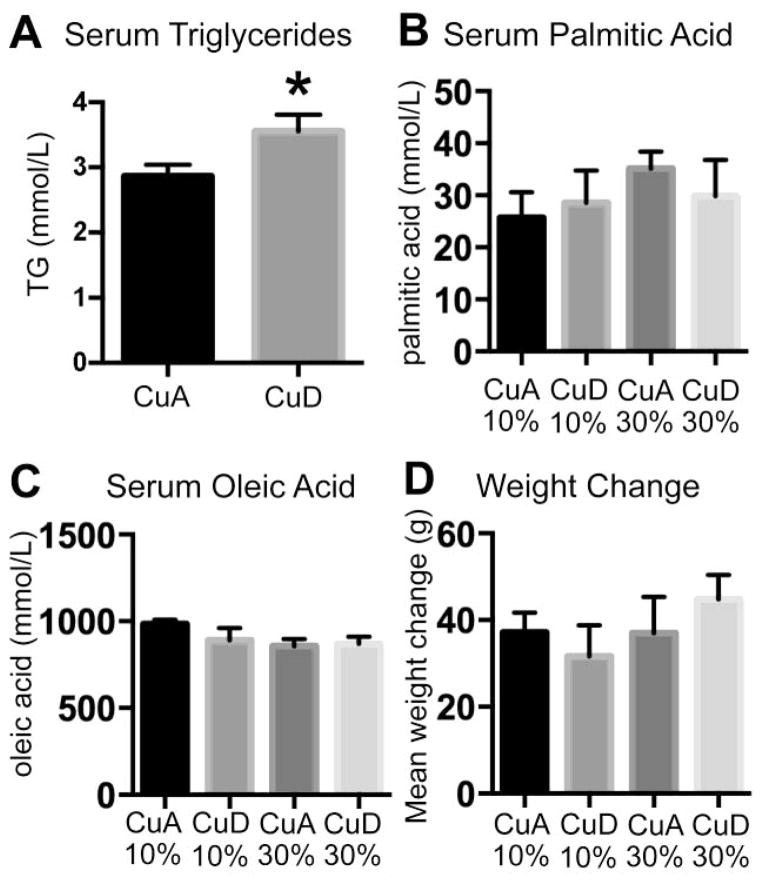
Oil red O (ORO) lipid staining of hepatic sections followed by digital image analysis (DIA) was used in this case to quantitatively analyze hepatic lipid accumulation. This method has previously been shown to be the most accurate histological method to determine hepatic lipid accumulation [47]. CuD/10% and CuD/30% diets significantly increased the percentage of stained area in rat hepatic tissue (Figure 3C). Induction of steatosis by CuD was confirmed by quantitation of lipids using Coherent Anti-Stokes Raman Scattering Spectroscopy (CARS) liver tissue sections from separate animals fed CuA/30% and CuD/30% diets (Fig. 3D). These data indicate that CuD diets are sufficient to induce modest steatosis in mature rats by 12 weeks regardless of sucrose content, while a combination of both Cu deficiency and sucrose dietary factors induces more characteristic histological features of NAFLD progression. No significant differences were observed in serum TG concentration across diets. However, when grouped by Cu content, serum TG levels were modestly but significantly higher in rats fed CuD diets (Figure 4A). No significant differences (P > 0.05) in two tested fatty acids (FA), palmitic acid and oleic acid, were observed (Fig. 4B–C). Thus, Cu deficiency slightly increased fasting circulation of TG by 12 weeks without a concurrent increase in fasting serum free fatty acid levels. No significant weight difference existed between rats fed experimental vs. control diets (Fig. 4D), indicating that the molecular responses to low dietary Cu and high sucrose may act independently of obesity-induced changes commonly observed with other NAFLD models. The histological analysis of rats fed the defined diets indicated that both Cu limitation and sucrose consumption, similar to the Western diet, alter physiological functions promoting NAFLD or NASH conditions. These results support the hypothesis that nutrient-induced changes in hepatic function are driven by specific transcriptional responses, such as hedgehog and FGFs, which promote fibrosis, and the differential expressions of gene transcripts related to lipid metabolism and altered lipid synthesis. Histologic features and lipid accumulation were most pronounced in the combined Cu deficient and 30% sucrose diet, where differentially expressed transcripts involved predominantly genes belonging to primary metabolism.
Figure 4. Dietary CuD alters circulating triglyceride levels, but not fatty acids or weight gain.
A. Serum triglyceride concentration by dietary Cu content, 10% and 30% sucrose are grouped. B. Serum palmitic acid concentration. C. Serum oleic acid concentration. D. Weight change after 12 weeks ad lib feeding purified diets.
Elemental analysis of serum and liver tissue indicated that serum and hepatic Cu levels were within the expected ranges for rats fed Cu-adequate diets [48,49], while the Cu-deficient diets significantly reduced hepatic and serum Cu levels (Fig. 5A, 5B). An increase in liver Fe was observed in CuD fed rats, with no statistically significant change in serum Fe at either sucrose level (Figure 5C, 5D). The data also suggested an influence of sucrose on Cu levels. However, these differences were not statistically significant when correcting for multiple comparisons with a 95% confidence interval in the experiments. These results confirm that Cu influences Fe status, which is consistent with other studies in rats as well as observations in NAFLD patients with low serum and liver Cu. These data thus indicate that hepatic Fe retention resulting in serum/liver partitioning of metals occurs when dietary Cu is low, similar to genetic inactivation of intestinal Cu uptake [27].
Figure 5. CuD and sucrose influence Cu and Fe status.
Mean + SEM (n=5–6) for hepatic Cu (A) and Fe (B) and serum Cu (C) and Fe (D). Significant differences between treatments are indicated by asterisks (P ≤0.05).
3.6. Moderate sucrose consumption and low dietary Cu promote biochemical responses consistent with MetS and lipid peroxidation
Since diets low in Cu, high in sucrose, or both promoted specific transcriptional responses consistent with NAFLD and MetS, as well as in some steatohepatitis, we tested the hypothesis that specific gene expression alterations would result in biochemical changes consistent with NAFLD and MetS.
Insulin resistance is considered a key indicator of MetS and may be observed even in the absence of obesity. While there were no significant differences in serum glucose across diets (Fig. 6A), significant hyperinsulinemia was observed in rats fed the CuD/30% diet (Fig. 6B). A modified version of HOMA-IR verified specifically for Wistar rats [42] was applied for the serum glucose and insulin data from all diets. As only the CuD/30% diet showed significant hyperinsulinemia, the HOMA-IR values for rats fed the CuD/30% diet were compared to the control diet [43]. The CuD/30% diet significantly increased the HOMA-IR value (Padj = 0.0262) above that of the control, placing it within the established range for insulin resistance in Wistar rats (Figure 6C). Thus, the combination of high dietary sucrose and low Cu synergistically contribute to the development of insulin resistance as a result of hyperinsulinemia.
Figure 6. Individual and synergistic effects of CuD and sucrose on insulin resistance and oxidative stress.
A) Serum glucose; B) serum insulin; C) modified HOMA-IR evaluating insulin resistance: dotted and dashed lines indicate validated HOMA-IR values for normal and insulin-resistant Wistar rats [42]. D–E) Levels of D) hepatic and E) serum malondialdehyde (MDA) as assessed by TBARS assay. F) Serum ALT level. Mean + S.E.M. (n=5–6) is indicated in A–F. Asterisks indicated significance of data vs. control: *P<0.05, **P< 0.01.
Hepatic and serum malondialdehyde (MDA) levels were investigated as an indirect measure of lipid peroxidation/oxidative stress, together with analysis of ALT level as an indicator of liver damage. While hepatic MDA levels were significantly increased in rats fed CuD/10% and CuD/30% diets (Fig. 6D), only a mild but relatively insignificant increase was found with CuA/30%, suggesting that Cu limitation may increase hepatic oxidative stress. Serum MDA and ALT concentrations were found to be significantly higher in the CuD/30% diet vs. the CuA/10% control diet (Fig. 6E–F). These results indicate that the CuD/10% diet is sufficient to cause increased oxidative stress and that dietary CuD concurrent with 30% dietary sucrose induces oxidative stress and mild liver damage.
4. DISCUSSION
4.1. Cu and sucrose nutrition influence specific gene expression consistent with NAFLD spectrum
The primary conceptual advance of this study is that the common dietary factors, moderate sucrose consumption and sub-optimal dietary Cu, induce specific inflammation and fibrosis transcriptional responses in the liver, even in the absence of obesity or severe steatosis. These inflammatory and fibrosis responses are accompanied by altered lipid metabolism, while Cu deficiency appears to drive hepatic lipid accumulation. Our data also suggest that sucrose consumption consistent with the Western diet disrupts Cu homeostasis. This indicates that diet composition, independent of quantity, may be a critical factor in NAFLD and progression to NASH. More broadly, our results indicate that dietary Cu content, in addition to sucrose (or fructose), may be general effectors of inflammatory responses consistent with NAFLD disease progression, possibly influencing broader pathologies in MetS.
Notably, the transcriptomic profiles and biochemical indication of NASH in our mature rat model occurred without the dramatic physiological changes that accompany diet treatments in weanling rats (where Cu deficiency may have a much greater effect due to developmental demands), and without the extreme levels of fructose feeding and/or obesity in many Westernized Diet models (reviewed in [50]). Without significant weight gain and with only modest steatosis, and a lack of increased serum FFA levels (all of which associated with NAFLD and obesity models, but not necessarily of NASH), we observed changes both at the transcriptome and physiological levels that are characteristic of NASH pathology. For example, in our study we identified up-regulation of gene transcripts that are associated with NASH and MetS and have been documented in both animal and clinical studies including angiotensin I converting enzyme (ACE) in the promotion of HSC activation [2,51] and association with MetS [5,52–54]. Similarly, up-regulation of LAMA1 has been correlated with NASH and cardiovascular disease in clinical settings [52], while LCN2 and STEAP2 have been associated with altered iron metabolism and systemic inflammatory status [55–58]. CYP8B1, whose transcript expression is down-regulated in the CuA/30% and CuD/30% diets, is involved in bile acid synthesis. Its gene transcript has also been found to be expressed at lower levels in clinical transcriptomic studies of NASH [59].
While much research to date has focused on the causative role that nutrient factors in the Western Diet play in the development of hepatic steatosis, it has been suggested that inflammation can precede steatosis in some cases [60]. Induction of transcript expressions leading to inflammation and fibrosis, despite only mild steatosis, is one of the most surprising results of this study. Our nutrient associated transcript expression profiles indicate that HSC activation occurs together with activation of ECM remodeler proteins, FGFs, and Tgfβ2, as well as myosin isoforms that are related to increase in contractility/migration of activated HSCs. Other studies in both rats and humans indicated that increased expression of GFAP may serve as an early marker for HSC activation during Hepatitis C infection, and can be used as a predictor for fibrosis progression, though its expression decreases as fibrosis progresses [61,62]. Additionally, SOCS1 may promote HSC resistance to elimination by natural killer cells [63]. Specifically analyzing hepatic tissue, the inflammation-induced activation of the hepatic stellate cells (HSCs) may occur via the Hedgehog signaling pathway and GLI transcription factor induced epithelial-to-mesenchymal transition (EMT), causing HSCs to behave as collagen-depositing myofibroblasts [64,65]. Although it has not yet been determined whether the TGFB2 isoform influences hepatic fibrosis via HSC activation, this protein has been shown to play a causal role in models of renal fibrosis [66], and is implicated in the induction of EMT in tumor cells in vitro [67].
Although the extent of differential transcript expression observed in response to the CuD/30% sucrose diet was limited compared to the single-factor diets, its transcriptomic profile shared several patterns of up-regulated pro-fibrotic transcripts with the single-variable diets, including differential gene expression of GLI1, FGFs 5 and 11, and myosin isoforms MYH3 and MYL7. However, rats fed the CuD/30% diet did undergo physiological and transcriptional changes indicative of NAFLD and possibly NASH, including significant increases in serum ALT consistent with liver damage and insulin resistance, both of which are strongly associated with NAFLD to NASH progression [15,68].
4.2. Cu and dietary sucrose as modifiers of metabolic pathways
In contrast to inflammation and fibrosis, patterns of primary metabolic pathway alterations seemed similar across diets. Dietary CuD and sucrose both independently and in combination resulted in the up-regulation of GAPDH and PFKFB3 gene transcripts, with both enzymes being involved in citrate accumulation. These suggested a potential increase in the rate of glycolysis as a result of CuD diets with a resulting increase in acetyl-CoA production. The CuD diets seemed to result in the altered expression of gene transcripts whose gene products would further increase mitochondrial acetyl-CoA levels, including pyruvate dehydrogenase (PDHA2) in the CuD/10% diet, while down-regulation of carnitine O-acetyltransferase (CRAT) in the CuD/30% diet could attenuate the normal decrease of acetyl-CoA during O-carnitine production. Increased acetyl-CoA (as a result of varied processes) can proceed to a mitochondrial buildup of citrate and the resulting shuttling of excess citrate out of the mitochondria into cytosol, whereby it is converted back into acetyl-CoA by action of ATP citrate lyase (ACLY). This process serves as a metabolic connection between carbohydrate and lipid metabolism when production of fatty acids by fatty acid synthase (FASN) is needed, as well as through action of the mevalonate pathway as an entry into cholesterol synthesis [69–71]. This increase in fatty acid synthesis may not necessarily manifest itself in an increase of circulating FFAs. Rather, FFAs synthesized in the liver may be converted to TG and accumulate (causing steatosis, as observed with our low Cu diets) or be secreted as VLDL [72,73]. Notably, the combined CuD/30% diet uniquely up-regulated both ACLY and FASN, indicating that CuD and sucrose each contribute to the buildup of citrate and that the two diets act synergistically to lead to an increase in transcript levels of genes coding for enzymes associated with fatty acid synthesis. The observed increase in transcripts coding for lipases, commonly expressed by Kupffer cells [74], suggests TG breakdown within the hepatic tissue and potential contribution to increased FFA flux from hepatic tissue into serum over time. Therefore, the up-regulation of the ACLY and FASN transcripts in the liver suggests a mechanism for steatosis and hepatic lipotoxicity independent of adipose lipolysis.
4.3. Conclusions
In conclusion, our nutrigenomics approach in the mature rat model allowed for unique insight into how specific factors in the Western diet can influence gene expression to promote steatosis and NAFLD spectrum conditions. Specifically, the approach allowed us to identify and specify changes in gene expression observed in response to the experimental diets, specifically in lipid metabolism and fibrosis, which provide molecular-level connections between diet and NAFLD-like pathology. This model may provide additional insight into the pathogenic mechanisms behind NAFLD when compared to high-fat diet models, as the increasing prevalence of NAFLD in Western cultures parallels the increase in carbohydrate and simple sugar consumption, whereas caloric contribution of fats has declined [75,76]. This is the first study to our knowledge that examines differential gene expression responses to Western diet relevant sugar consumption and Cu deficiency in a mature rodent model. Results from our study confirm that inadequate dietary Cu can significantly influence hepatic lipid metabolism gene expression, a response that manifests as hepatic lipid accumulation. Further, we found that inflammation and fibrosis gene expression is promoted by inadequate Cu and high sugar, even without severe steatosis described in the lipotoxicity model of NAFLD progression.
Supplementary Material
Highlights.
Low dietary Cu and human-relevant sucrose consumption promote inflammation and fibrosis gene expression changes consistent with NAFLD in a mature rat model
Low dietary Cu promotes steatosis as well as gene expression changes in lipid synthesis
Cu deficiency and sucrose consumption promote NAFLD-like pathology in mature rats, even in the absence of obesity or severe steatosis
Acknowledgments
The authors thank Eric Murphy and Christa Eussen for assistance with animal care to develop the mature rat model, Christine Couturier for assistance with qPCR data analysis, Valérie Copié, Lei Yu and Mario Kratz for critical reading and discussion of the manuscript, and Kristopher Short and Michael Walleri for lab assistance.
Grant support: This work was supported by P20RR016466-11 (Sub-Project ID: 7979 to JLB), 8P20GM103395-12 (JLB Project), UAA Innovate (JLB), 1S10RR025512-01 to MR, and the University of Washington Diabetes Research Center P30 DK017047.
Abbreviations
- ALT
alanine transaminase
- CARS
Coherent Anti-Stokes Raman Scattering
- Cu
copper
- Fe
iron
- CuA
copper adequate
- CuD
copper deficient
- ECM
extracellular matrix
- EMT
epithelial-to-mesenchymal transition
- FC
fold change
- FFA
free fatty-acid
- FGF
fibroblast growth factor
- H&E
hematoxylin and eosin
- HFCS
high-fructose corn syrup
- HOMA-IR
homeostatic model assessment of insulin resistance
- HSC
hepatic stellate cell
- ICP-MS
inductively-coupled mass spectrometry
- IL
interleukin
- IPA
Ingenuity Pathways Analysis
- MDA
malondialdehyde
- MetS
metabolic syndrome
- NAFLD
non-alcoholic fatty-liver disease
- NASH
non-alcoholic steatohepatitis
- ORO
oil red O
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative reverse transcription-PCR
- TBARS
Thiobarbituric Acid Reactive Substances
- TG
triglyceride
- TNF
tumor necrosis factor
Footnotes
Disclosures: The authors have no conflicts of interest to disclose.
Transcript profiling: Transcript profile data is deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE58875.
Author contributions: ST: acquisition of data; analysis and interpretation of data; drafting of the manuscript, statistical analysis; MD: acquisition of data, analysis of data; MR: analysis and interpretation of data, critical revision of manuscript; MPC: analysis and interpretation of data; ML: analysis and interpretation of data, critical revision of manuscript; JLB: acquisition of data; analysis and interpretation of data; drafting of the manuscript, statistical analysis, study concept and design.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McArdle Ma, Finucane OM, Connaughton RM, McMorrow AM, Roche HM. Mechanisms of obesity-induced inflammation and insulin resistance: insights into the emerging role of nutritional strategies. Front Endocrinol (Lausanne) 2013;4:52. doi: 10.3389/fendo.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres DM, Harrison Sa. Diagnosis and Therapy of Nonalcoholic Steatohepatitis. Gastroenterology. 2008;134:1682–98. doi: 10.1053/j.gastro.2008.02.077. [DOI] [PubMed] [Google Scholar]
- 3.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity in the United States, 2009–2010. NCHS Data Brief. 2012:1–8. [PubMed] [Google Scholar]
- 4.Baker RG, Hayden MS, Ghosh S. NF-kB, Inflammation, and Metabolic Disease. Cell Metab. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckel RH, Alberti KGMM, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–28. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 6.Weston SR, Leyden W, Murphy R, Bass NM, Bell BP, Manos MM, et al. Racial and ethnic distribution of nonalcoholic fatty liver in persons with newly diagnosed chronic liver disease. Hepatology. 2005;41:372–9. doi: 10.1002/hep.20554. [DOI] [PubMed] [Google Scholar]
- 7.Attar BM, Van Thiel DH. Current concepts and management approaches in nonalcoholic fatty liver disease. Scientific World Journal. 2013:1–10. doi: 10.1155/2013/481893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik A, Košir R, Rozman D. Genomic aspects of NAFLD pathogenesis. Genomics. 2013;102:84–95. doi: 10.1016/j.ygeno.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Lazo M, Hernaez R. Non-alcoholic fatty liver disease and mortality among US adults: prospective cohort study. BMJ. 2011;343:1–9. doi: 10.1136/bmj.d6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trauner M, Arrese M, Wagner M. Fatty liver and lipotoxicity. Biochim Biophys Acta. 2010;1801:299–310. doi: 10.1016/j.bbalip.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Nobili V, Siotto M, Bedogni G, Ravà L, Pietrobattista A, Panera N, et al. Levels of serum ceruloplasmin associate with pediatric nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2013;56:370–5. doi: 10.1097/MPG.0b013e31827aced4. [DOI] [PubMed] [Google Scholar]
- 12.Roberts EA. Pediatric nonalcoholic fatty liver disease (NAFLD): A “growing” problem? J Hepatol. 2007;46:1133–42. doi: 10.1016/j.jhep.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Cohen-Naftaly M, Friedman SL. Current status of novel antifibrotic therapies in patients with chronic liver disease. Therap Adv Gastroenterol. 2011;4:391–417. doi: 10.1177/1756283X11413002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim JS, Mietus-Snyder M, Valente A, Schwarz J-M, Lustig RH. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nat Rev Gastroenterol Hepatol. 2010;7:251–64. doi: 10.1038/nrgastro.2010.41. [DOI] [PubMed] [Google Scholar]
- 15.Pais R, Charlotte F, Fedchuk L, Bedossa P, Lebray P, Poynard T, et al. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J Hepatol. 2013;59:550–6. doi: 10.1016/j.jhep.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 16.Paradis V, Bedossa P. Definition and natural history of metabolic steatosis: histology and cellular aspects. Diabetes Metab. 2008;34:638–42. doi: 10.1016/S1262-3636(08)74598-1. [DOI] [PubMed] [Google Scholar]
- 17.Standish Ra, Cholongitas E, Dhillon A, Burroughs aK, Dhillon aP. An appraisal of the histopathological assessment of liver fibrosis. Gut. 2006;55:569–78. doi: 10.1136/gut.2005.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angulo P, Bugianesi E, Bjornsson ES, Charatcharoenwitthaya P, Mills PR, Barrera F, et al. Simple Noninvasive Systems Predict Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2013;145:782–9. doi: 10.1053/j.gastro.2013.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–46. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 20.Burkhead JL, Lutsenko S. The Role of Copper as a Modifier of Lipid Metabolism. In: Baez Rodrigo Valenzuela., editor. Lipid Metabolism. InTech; Rijeka: 2013. [Google Scholar]
- 21.Collins JF, Klevay LM. Copper 1,2. Adv Nutr. 2011;2:520–2. doi: 10.3945/an.111.001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aigner E, Theurl I, Haufe H, Seifert M, Hohla F, Scharinger L, et al. Copper availability contributes to iron perturbations in human nonalcoholic fatty liver disease. Gastroenterology. 2008;135:680–8. doi: 10.1053/j.gastro.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Aigner E, Strasser M, Haufe H, Sonnweber T, Hohla F, Stadlmayr A, et al. A role for low hepatic copper concentrations in nonalcoholic Fatty liver disease. Am J Gastroenterol. 2010;105:1978–85. doi: 10.1038/ajg.2010.170. [DOI] [PubMed] [Google Scholar]
- 24.Song M, Schuschke Da, Zhou Z, Chen T, Pierce WM, Wang R, et al. High fructose feeding induces copper deficiency in Sprague-Dawley rats: a novel mechanism for obesity related fatty liver. J Hepatol. 2012;56:433–40. doi: 10.1016/j.jhep.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song M, Schuschke Da, Zhou Z, Chen T, Shi X, Zhang J, et al. Modest fructose beverage intake causes liver injury and fat accumulation in marginal copper deficient rats. Obesity (Silver Spring) 2013;000:1–7. doi: 10.1002/oby.20380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maliken BD, Nelson JE, Klintworth HM, Beauchamp M, Yeh MM, Kowdley KV. Hepatic reticuloendothelial system cell iron deposition is associated with increased apoptosis in nonalcoholic fatty liver disease. Hepatology. 2013;57:1806–13. doi: 10.1002/hep.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nose Y, Kim BE, Thiele DJ. Ctr1 drives intestinal copper absorption and is essential for growth, iron metabolism, and neonatal cardiac function. Cell Metab. 2006;4:235–44. doi: 10.1016/j.cmet.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z, Lowndes J, Rippe J. High-fructose corn syrup and sucrose have equivalent effects on energy-regulating hormones at normal human consumption levels. Nutr Res. 2013;33:1043–52. doi: 10.1016/j.nutres.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Keim NL, Havel PJ. Fructose - Absorption and Metabolism. In: Caballero B, editor. Encycl Hum Nutr. 3. Vol. 2. 2013. pp. 361–5. [Google Scholar]
- 30.Vos M, Kimmons, Gillespie C, Welsh J, Michels Blanck H. Dietary fructose consumption among US children and adults: The third national health and nutrition examination survey (NHANES III) Medscape J Med. 2008;10:160. [PMC free article] [PubMed] [Google Scholar]
- 31.Abid A, Taha O, Nseir W, Farah R, Grosovski M, Assy N. Soft drink consumption is associated with fatty liver disease independent of metabolic syndrome. J Hepatol. 2009;51:918–24. doi: 10.1016/j.jhep.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Zhang Z, Gregg EW, Flanders WD, Merritt R, Hu FB. Added sugar intake and cardiovascular diseases mortality among US adults. JAMA Intern Med. 2014;174:516–24. doi: 10.1001/jamainternmed.2013.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yilmaz Y. Review article: fructose in non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2012;35:1135–44. doi: 10.1111/j.1365-2036.2012.05080.x. [DOI] [PubMed] [Google Scholar]
- 34.Committee. Guide for the Care and Use of Laboratory Animals: Eighth Edition. Guid Care Use Lab Anim. 2011:118. [Google Scholar]
- 35.McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–71. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt439. web server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den hoff MJB, et al. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Yeh MM, Brunt EM. Pathology of fatty liver: differential diagnosis of non-alcoholic fatty liver disease. Diagnostic Histopathol. 2008;14:586–97. [Google Scholar]
- 41.Le TT, Ziemba A, Urasaki Y, Brotman S, Pizzorno G. Label-free Evaluation of Hepatic Microvesicular Steatosis with Multimodal Coherent Anti-Stokes Raman Scattering Microscopy. PLoS One. 2012:7. doi: 10.1371/journal.pone.0051092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cacho J, Sevillano J, de Castro J, Herrera E, Ramos MP. Validation of simple indexes to assess insulin sensitivity during pregnancy in Wistar and Sprague-Dawley rats. Am J Physiol Endocrinol Metab. 2008;295:E1269–76. doi: 10.1152/ajpendo.90207.2008. [DOI] [PubMed] [Google Scholar]
- 43.Jaccard J, Becker MA, Wood G. Pairwise multiple comparison procedures: A review. Psychol Bull. 1984;96:589–96. [Google Scholar]
- 44.Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiappini F, Barrier A, Saffroy R, Domart M-C, Dagues N, Azoulay D, et al. Exploration of global gene expression in human liver steatosis by high-density oligonucleotide microarray. Lab Invest. 2006;86:154–65. doi: 10.1038/labinvest.3700374. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette Ma, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levene AP, Kudo H, Armstrong MJ, Thursz MR, Gedroyc WM, Anstee QM, et al. Quantifying hepatic steatosis - more than meets the eye. Histopathology. 2012;60:971–81. doi: 10.1111/j.1365-2559.2012.04193.x. [DOI] [PubMed] [Google Scholar]
- 48.Ogra Y, Suzuki KT. Targeting of tetrathiomolybdate on the copper accumulating in the liver of LEC rats. J Inorg Biochem. 1998;70:49–55. doi: 10.1016/s0162-0134(98)00012-9. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki KT, Ogra Y, Ohmichi M. Molybdenum and copper kinetics after tetrathiomolybdate injection in LEC rats: specific role of serum albumin. J Trace Elem Med Biol. 1995;9:170–5. doi: 10.1016/S0946-672X(11)80043-X. [DOI] [PubMed] [Google Scholar]
- 50.Kanuri G, Bergheim I. In Vitro and in Vivo Models of Non-Alcoholic Fatty Liver Disease (NAFLD) Int J Mol Sci. 2013;14:11963–80. doi: 10.3390/ijms140611963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sookoian S, Gianotti TF, Rosselli MS, Burgueño AL, Castaño GO, Pirola CJ. Liver transcriptional profile of atherosclerosis-related genes in human nonalcoholic fatty liver disease. Atherosclerosis. 2011;218:378–85. doi: 10.1016/j.atherosclerosis.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 53.Ferder L, Ferder MD, Inserra F. The role of high-fructose corn syrup in metabolic syndrome and hypertension. Curr Hypertens Rep. 2010;12:105–12. doi: 10.1007/s11906-010-0097-3. [DOI] [PubMed] [Google Scholar]
- 54.Aronne LJ, Nelinson DS, Lillo JL. Obesity as a disease state: a new paradigm for diagnosis and treatment. Clin Cornerstone. 2009;9:9–25. doi: 10.1016/s1098-3597(09)80002-1. discussion 26–9. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Lam KSL, Kraegen EW, Sweeney G, Zhang J, Tso AWK, et al. Lipocalin-2 is an inflammatory marker closely associated with obesity, insulin resistance, and hyperglycemia in humans. Clin Chem. 2007;53:34–41. doi: 10.1373/clinchem.2006.075614. [DOI] [PubMed] [Google Scholar]
- 56.Xu G, Ahn J, Chang S, Eguchi M, Ogier A, Han S, et al. Lipocalin-2 induces cardiomyocyte apoptosis by increasing intracellular iron accumulation. J Biol Chem. 2012;287:4808–17. doi: 10.1074/jbc.M111.275719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dittrich aM, Meyer Ha, Hamelmann E. The role of lipocalins in airway disease. Clin Exp Allergy. 2012;43:503–11. doi: 10.1111/cea.12025. [DOI] [PubMed] [Google Scholar]
- 58.Catalán V, Gómez-Ambrosi J, Rodríguez A, Ramírez B, Rotellar F, Valentí V, et al. Six-transmembrane epithelial antigen of prostate 4 and neutrophil gelatinase-associated lipocalin expression in visceral adipose tissue is related to iron status and inflammation in human obesity. Eur J Nutr. 2013;52:1587–95. doi: 10.1007/s00394-012-0464-8. [DOI] [PubMed] [Google Scholar]
- 59.Lake AD, Novak P, Shipkova P, Aranibar N, Robertson D, Reily MD, et al. Decreased hepatotoxic bile acid composition and altered synthesis in progressive human nonalcoholic fatty liver disease. Toxicol Appl Pharmacol. 2013;268:132–40. doi: 10.1016/j.taap.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–46. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 61.Zakaria S, Youssef M, Moussa M, Akl M, El-Ahwany E, El-Raziky M, et al. Value of α-smooth muscle actin and glial fibrillary acidic protein in predicting early hepatic fibrosis in chronic hepatitis C virus infection. Arch Med Sci. 2010;6:356–65. doi: 10.5114/aoms.2010.14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carotti S, Morini S, Corradini SG, Burza MA, Molinaro A, Carpino G, et al. Glial Fibrillary Acidic Protein as an Early Marker of Hepatic Stellate Cell Activation in Chronic and Posttransplant Recurrent Hepatitis C. Liver Transplant. 2008;14:806–14. doi: 10.1002/lt.21436. [DOI] [PubMed] [Google Scholar]
- 63.Yi H-S, Jeong W-I. Interaction of hepatic stellate cells with diverse types of immune cells: Foe or friend? J Gastroenterol Hepatol. 2013;28 (Suppl 1):99–104. doi: 10.1111/jgh.12017. [DOI] [PubMed] [Google Scholar]
- 64.Xie G, Karaca G, Swiderska-Syn M, Michelotti Ga, Krüger L, Chen Y, et al. Cross-talk between notch and hedgehog regulates hepatic stellate cell fate. 2013 doi: 10.1002/hep.26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Omenetti A, Porrello A, Jung Y, Yang L, Popov Y, Choi SS, et al. Report Information from ProQuest. J Clin Invest. 2008;118:3331–42. doi: 10.1172/JCI35875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu L, Border Wa, Huang Y, Noble Na. TGF-beta isoforms in renal fibrogenesis. Kidney Int. 2003;64:844–56. doi: 10.1046/j.1523-1755.2003.00162.x. [DOI] [PubMed] [Google Scholar]
- 67.Kimura C, Hayashi M, Mizuno Y, Oike M. Endothelium-dependent epithelial-mesenchymal transition of tumor cells: exclusive roles of transforming growth factor β1 and β2. Biochim Biophys Acta. 2013;1830:4470–81. doi: 10.1016/j.bbagen.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Sreekumar R, Rosado B, Rasmussen D, Charlton M. Hepatic gene expression in histologically progressive nonalcoholic steatohepatitis. Hepatology. 2003;38:244–51. doi: 10.1053/jhep.2003.50290. [DOI] [PubMed] [Google Scholar]
- 69.Menendez Ja, Vazquez-Martin A, Ortega FJ, Fernandez-Real JM. Fatty acid synthase: association with insulin resistance, type 2 diabetes, and cancer. Clin Chem. 2009;55:425–38. doi: 10.1373/clinchem.2008.115352. [DOI] [PubMed] [Google Scholar]
- 70.Lanaspa Ma, Sanchez-Lozada LG, Choi Y-J, Cicerchi C, Kanbay M, Roncal-Jimenez Ca, et al. Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress: potential role in fructose-dependent and -independent fatty liver. J Biol Chem. 2012;287:40732–44. doi: 10.1074/jbc.M112.399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson D, Cox MM. Lehninger Principles of Biochemistry. 4. 2004. [Google Scholar]
- 72.Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48:434–41. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest. 2008;118:829–38. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuntz E, Kuntz H-D. Hepatology. 3. 2008. [Google Scholar]
- 75.Chun OK, Chung CE, Wang Y, Padgitt A, Song WO. Changes in intakes of total and added sugar and their contribution to energy intake in the U. S Nutrients. 2010;2:834–54. doi: 10.3390/nu2080834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prevention C for DC. Trends in intake of energy and macronutrients - United States, 1971–2000. Morb Mortal Wkly Rep. 2004;53:80–2. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.