Abstract
Metals such as arsenic, cadmium, beryllium, and nickel are known human carcinogens; however, other transition metals, such as tungsten (W), remain relatively uninvestigated with regard to their potential carcinogenic activity. Tungsten production for industrial and military applications has almost doubled over the past decade and continues to increase. Here, for the first time, we demonstrate tungsten’s ability to induce carcinogenic related endpoints including cell transformation, increased migration, xenograft growth in nude mice, and the activation of multiple cancer related pathways in transformed clones as determined by RNA seq. Human bronchial epithelial cell line (Beas-2B) exposed to tungsten developed carcinogenic properties. In a soft agar assay, tungsten-treated cells formed more colonies than controls and the tungsten-transformed clones formed tumors in nude mice. RNA-sequencing data revealed that the tungsten-transformed clones altered the expression of many cancer-associated genes when compared to control clones. Genes involved in lung cancer, leukemia, and general cancer genes were deregulated by tungsten. Taken together, our data shows the carcinogenic potential of tungsten. Further tests are needed, including in vivo and human studies, in order to validate tungsten as a carcinogen to humans.
Keywords: Tungsten, Cancer, Beas-2B, RNA-Seq, In Vitro, Nude Mice
Introduction
Tungsten is used in many industrial and military functions because of its distinct physical properties, such as the exceptional hardness of tungsten carbide. Due to tungsten’s high melting point it has become crucial in a broad range of industrial activities. Tungsten is found in electronics, light bulb filaments, cemented tungsten carbide grinding wheels, carbide tipped tools and armaments. According to a report from the EPA, tungsten enters our environment through ore processing, alloy fabrication, tungsten carbide production and use, as well as during municipal waste combustion [4]. Tungsten production is increasing, in 2011 there were 72,000 tons produced while in 2002 there were only 40,000 tons produced [37].
Federal facilities have detected dissolved tungsten in groundwater in areas where small munitions ranges were located [13]. Additionally, the U.S. Army made tungsten/nylon projectiles from tungsten powder. The coatings of the tungsten/nylon projectiles form oxides, which further oxidized to become water-soluble. High levels of tungsten were found in soil pore-water beneath bullet collection areas up to 400 mg/L at depths up to 65 cm. Concentrations of 400 mg/L equate to ~2 mM tungsten. Tungsten was measured at concentrations up to 560 μg/L in down gradient monitoring wells, which equates to ~3 μM [4].
As a result, environmental exposures to tungsten via food, water and soil have become of significant concern. Tungsten in ground water can accumulate in plants used for food by both humans and other species [1]. There has been recent discussion concerning tungsten as an emerging chemical toxicant of environmental health concerns. In vitro and animal studies point to tungsten toxicity leading to pulmonary inflammation and the development of cancer [38].
The association between metal exposure and cancer is supported by various molecular and epidemiological studies. Cadmium, chromium (VI), arsenic, and nickel are IARC class I human carcinogens and other metals such as vanadium that are not established as IARC carcinogens have demonstrated tumorigenic tendencies [3,6]. Tungsten is a transition metal in the same block as many of the carcinogenic metals on the periodic table and holds potential to induce cancer-associated effects. A small, underwhelming number of studies have been conducted to investigate the possibility of tungsten as a carcinogen. An epidemiological investigation by Wild et al. revealed that hard-metal plant workers co-exposed to tungsten carbide and cobalt displayed an increased risk to lung cancer compared to a control group [40].
Tungsten is popular in industry due to its remarkable robustness; the free element has the highest melting point of all the elements. Tungsten inert gas welding is a process in which fusion is produced by heating with an arc established between a non-consumable tungsten electrode and a base metal. Workers exposed to welding fumes have higher incidences of impaired lung function, chronic obstructive lung diseases, asthma, and lung cancer [26]. Historically, the heaviest metal exposures occur in the workplace or in environmental settings in close proximity to industrial sources [17]. Given the large amount of evidence characterizing metals as carcinogens and the wide use of tungsten in industrial settings, there is a need for basic research to investigate the possibility of tungsten as a carcinogen.
Methods
Cell Culture
Immortalized human bronchial epithelial cells (Beas-2B; #CRL-9609, ATCC, Manassas, VA) were adapted to serum growth after their purchase and have been carefully maintained. Beas-2B were cultured in 1 x Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 100 μg mL−1 Pen Strep (GIBCO, Grand Island, NY). The cells were maintained in 10 cm2 polystyrene tissue culture dishes in an incubator at 37°C with 5% CO2. Media was changed every 4 days as well as passaged using 0.05% trypsin-EDTA (GIBCO) as described previously [10]. Cells were split and seeded at the same concentration, 3 × 105, in the presence of Na2WO4, (Sigma-Aldrich) dissolved in distilled water, concentrations ranging from 50μM to 250 μM. After each week they were placed into soft agar, to temporally investigate tungsten induced transformed the cells.
Colony Transformation
After 6 weeks of treatment, the Beas-2B cells with or without treatment of Na2WO4 were tested for anchorage-independent growth. A bottom layer of 0.5% [w/v] agar (BD Biosciences, San Diego, CA) and a top layer of 0.35% agar was placed in 6-well non-treated, polystyrene, plate, as described in Sato and Kan’s standard protocol, except for 5000 cells were seeded in each well of a 6-well plate in triplicates [34]. The cells were incubated for one month in soft agar without tungsten treatment at 37°C, 95% air /5% CO2. Individual transformed colonies were picked from the agar and grown into a monolayer. RNA from colonies was collected in TRIzol Reagent (Invitrogen) for RNA Seq. To observe how many transformed colonies there were per each dose, wells were stained overnight with INT-BCIP solution (Roche, New York, NY) in 0.1 Tris/0.05 M MgCl2/0.1 M NaCl.
Migration Assay
Five tungsten-transformed clones, five control clones that formed spontaneously without any treatment and five parental Beas-2B cells were seeded at a density of 1.5 × 105 on each reservoir in Ibidi Culture-Insert (Ibidi, Munich, Germany), the silicon insert allows for a uniformed wound of 500 μM between the seeded cells. Cells were cultured as described, until they reached confluency (48 hours), then the insert was removed. The cells were washed with PBS to remove floating cells and DMEM was replaced on the cells. Images of the same field of view were taken using a Nikon camera on a Nikon TMS-F microscope (Duesseldorf, Germany). Images were taken immediately, eight-hours later and then at 20-hours to watch the migration of cells over the scratch.
In vivo tumorigenesis
Twelve, six-week old female athymic nude mice obtained from the National Cancer Institute (NCI, Frederick) were subcutaneously injected in the left or right flank with either control (Beas-2B that spontaneously grew in agar without any treatment) or Na2WO4 transformed cells. All mice had access to food and water ad libitum and were handled in accordance with NIH and Institutional Animal Care and Use Committee (IACUC) guidelines. Each mouse was injected in one location on each of the left and right flanks with 0.1cc of 5 million cells of either control or tungsten-transformed cells. Each mouse is injected on one side with control and the other with tungsten-transformed cells to ensure the same biological environment. Each cell line was subcutaneously injected into three mice, in triplicates.
Sequencing reads mapping and preprocessing
Alterations in gene expression were studied by comparing tungsten-transformed cells to control clones (vide supra). All raw sequencing reads were mapped to the human genome (GRCh37/hg19) by using Bowtie aligner (0.12.9) with v2 and m1 parameters. The mapped reads were subsequently sorted and filtered by removing the PCR duplicates with samtools (0.1.19) before further analysis.
RNA Sequencing
To obtain significant differential expressed genes, the tungsten-transformed colonies were compared to control colonies. RNA isolation is conducted by standard protocol of TRIzol Reagent (Invitrogen). The RNA was quantified using NanoDrop ND-1000 flurospectrometer (Thermo). The reads after PCR duplicates removal from data preprocessing were then assigned to Ensemble gene model (Homo_sapiens.GRCh37.71.gtf) with HTseq (0.6.1.p.1) [28, 33]. For the statistical analysis, DESeq2 R/Bioconductor package was used [24]. The raw reads counts were modeled and normalized by following a negative binomial distribution as previously described [24]. The common dispersion and statistical significance for genes cross sample groups were estimated and calculated using a general linear model [31, 33]. To obtain adjusted p value for each gene, the FDR method for multiple hypothesis test has been applied to those genes that have the summed reads counts per million (CPM) of all samples greater than 5. The heatmap was generated using heatmap.2 function from gplots package in R (version 3.1.0). In the heatmap, rows represent genes and columns represent the comparison of control clones vs. tungsten clones. Green color indicates genes that are down regulated in control clones or tungsten clones and red color indicates the genes that are up regulated in control clones or tungsten clones.
Results
Tungsten demonstrated positive results in a battery of tests evaluating carcinogenesis
The treated cells were tested for anchorage independent growth. The result is a colony of cells that can grow independent of checkpoints that usually limits the ability of these cells to form colonies in agar. However, there was a low level of sporadic colony growth in the untreated Beas-2B as compared to the chronically treated cells; nonetheless, there were statically significantly fewer clones from untreated cells compared to clones derived from cells treated with Na2WO4. The results clearly display that Na2WO4 augments anchorage independent growth at all dosages (figures 1 and 2).
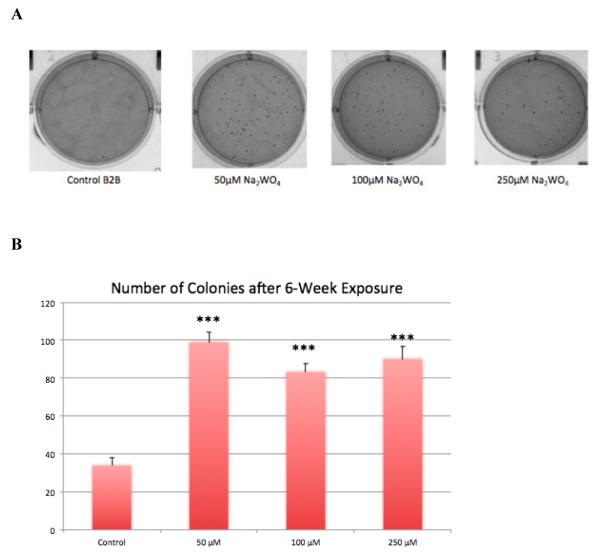
Figure 1. Transformation Assay.
After 6 weeks of chronic exposure to Na2WO4, Beas-2B cells were placed into soft agar. (A) Representative picture of colony formation for each dose showing that W-treated cells formed more transformed colonies than the control. (B) The control cells transformed at an average of 33 colonies per well. At the lowest dose, 50 μM, there was an average of 99 colonies per well. The higher doses, 100 and 250 μM, grew an average of 83 and 89 colonies per well, respectively. W-treated cells show a statistically significantly higher number of transformed colonies than control (p<0.001). *** p-value < 0.001 compared to control
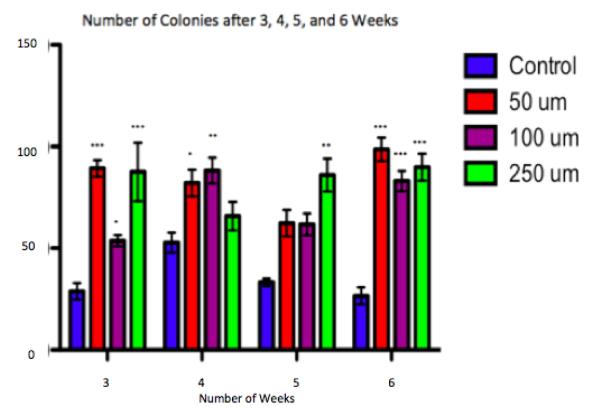
Figure 2. Time Series of Transformation.
After three, four, five and six weeks of chronic exposure to sodium tungstate, Beas-2B cells were placed into soft agar in order to evaluate the temporal induction of anchorage independent growth. Untreated controls grew significantly fewer colonies than W-treated cells at each time point. At each of the four time points, colony formation seemed to be variable amongst the doses. This figure shows that after three weeks of chronic treatment, W-treated cells became transformed at each dose. * p-value<0.05, ** p-value<0.01, *** p-value<0.001 compared to control
After six weeks, the soft agar was stained with INT/BCIP and then photographed. Figure 1A represents plates of cell growth in soft agar at Na2WO4 exposure concentrations from 0-250 μM. As depicted in the wells of figure 1A, all tungsten treated cells exhibited increased anchorage-independent growth in soft agar. Ten clones from control and W treated cells were isolated from the soft agar, trypsinized and then grown into confluent monolayers. Thus, each sub-cloned cell line comprised a cell population that originated from a single cell. Surviving clones were used to assay migration employing the scratch test assay, tumor formation in nude mice, as well as gene expression analysis with RNA Sequencing.
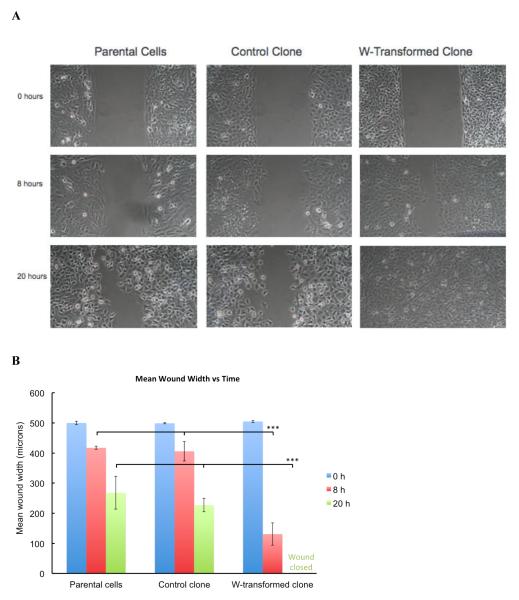
The cell scratch assay assessed cell migration and evaluates wound healing in vivo. The results of the scratch test showed that Na2WO4 transformed clones were able to heal the wound 20 hours after the scratch was made (figure 3). W-transformed clones were compared to spontaneous transformed control clones, which were unable to heal the wound within the same 20 hours timeframe.
Figure 3. Scratch Test.
500 micron simulated wounds were created in confluent monolayers. (A) Representative pictures are taken in time intervals to capture the migration of the cells showing that the W-transformed clones migrated more quickly than control clone or parental Beas-2B cells. Images were taken at 100X magnification at the same field of view. (B) W-transformed clone cells reduced mean wound width more quickly than either the control clone or parental cells. Wounds were statistically significantly smaller 8 and 20 hours after wound creation in the W-transformed clone as compared to control clone or parental cells (p<0.001). *** p<0.001 compared to control
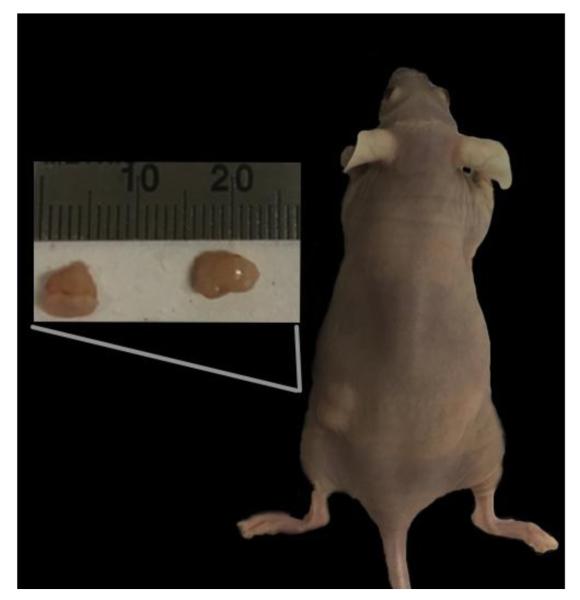
A month after subcutaneous injection into 12 female athymic nude mice, 100% of the tungsten-transformed cells formed visible tumors, while none of the spontaneous transformed control cells formed tumors. The mice were sacrificed after 5 months and the tumors were dissected and frozen for further testing (figure 4).
Figure 4. Tungsten-transformed clones grew tumors in vivo.
Twelve, six-week old female athymic nude mice were subcutaneously injected in the left flank with Na2WO4 transformed cells or the right flank with sporadic growth control cells. Each mouse is injected on one side with control and the other with tungsten-transformed cells to ensure the same biological environment. This figure shows a representative mouse that has formed tumors on the left side where they were injected with W-transformed cells, unlike the sporadic growth control cells.
RNA-sequencing of tungsten-transformed clones revealed key pathways and genes involved in carcinogenesis
Overall changes in gene expression due to tungsten exposure
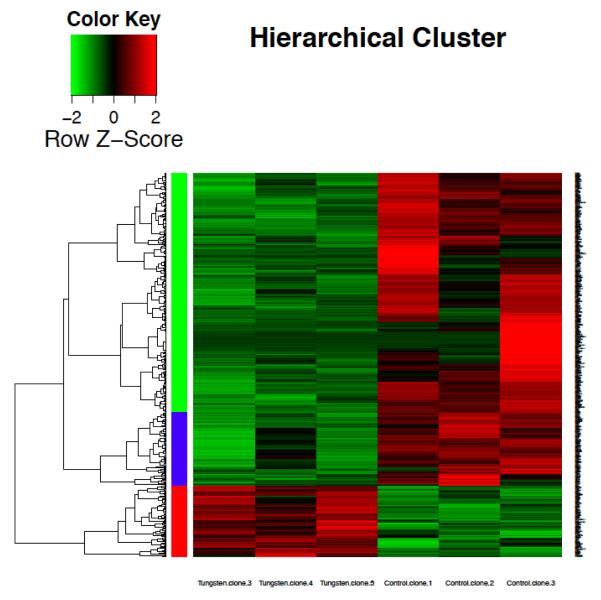
Additional analysis of W-transformed clones and control clones displayed differences in gene expression. Through RNA sequencing, it was revealed that 16,448 genes were altered. Of those genes, 535 passed FDR less than 0.05. Figure 5A shows a heatmap that provides an overall view of the change in gene expression between the tungsten clones and spontaneous clones. Figure 5A illustrates the changes in gene expression of the tungsten-transformed clones compared to the control.
Figure 5. Gene expression profiles of W-transformed clones.
(A) Heat Map. Hierarchical cluster analysis of significantly differentially expressed genes in W-control clones compared to clones that spontaneously grew in soft agar (control). The bar relates the color code to the expression value of normalized counts with DESeq2 package.
In order to investigate the specific gene expression and pathway changes induced by tungsten, we uploaded the gene list into the Ingenuity Pathway Analysis (IPA). IPA revealed that the top diseases according to p-value were “Cancer (423 affected molecules)”, “Inflammatory Response (104 affected molecules)”, “Organismal Injury and Abnormalities (263 affected molecules)”, and “Tumor Morphology (74 affected molecules)”. Some of the top canonical pathways included, “Role of Tissue Factor in Cancer”, “Molecular Mechanisms of Cancer”, “mTOR signaling”, “Chronic Myeloid Leukemia Signaling”, “p53 Signaling”, and “PI3K/Akt signaling”. All of the top canonical pathways are involved in cancer and their dysregulation induced by tungsten further points towards the notion that tungsten is a carcinogen.
Genes involved in Cancer
After investigating many of the altered genes from the gene list, it was evident that many of the genes were involved in certain types of cancers including lung cancer, leukemia, as well as, many genes common to all cancers. Genes involved in lung cancer included, NQO1 (NAD(P)H dehydrogenase, quinone 1) (up-regulated); CRYAB (crystallin, alpha B) (up-regulated); S100A4 (S100 calcium binding protein A4) (up-regulated); SPK2 (S-phase kinase-associated protein 2) (up-regulated); and HTATIP2 (HIV-1 Tat interactive protein 2) (up-regulated). Leukemia-associated genes seemed to predominate altered cancer genes. Some of the dysregulated leukemia genes include, CD74 (major histocompatibility complex, class II invariant chain) (up-regulated); CTGF (connective tissue growth factor) (down-regulated); HOXB5 (homeobox B5) (down-regulated); MST4 (serine/threonine protein kinase 26) (up-regulated); CSF3 (colony stimulating factor 3) (up-regulated); along with several other leukemia-associated genes. Many of the altered genes were found to be common in many types of cancers. These genes include CDNK1A (cyclin-dependent kinase inhibitor 1A) (down-regulated) and TGF1B (transforming growth factor, beta 1) (down-regulated). Among others cancer-associated genes that were dysregulated include, NFKB, AKT2, TGFB2, and IGFBP2.
Discussion
Previously, arsenic, chromium, nickel and vanadium have been studied for their ability to induce transformation of Beas-2B cells [12, 35]. This study explores the carcinogenic potential of tungsten, a metal that has not been evaluated in metal-induced carcinogenesis investigations. Here, for the first time, we demonstrate the carcinogenic properties of tungsten. We evaluated the metal’s tumorigenicity by performing a battery of tests that investigate carcinogenic endpoints. This study shows that chronic exposure to Na2WO4 induces anchorage independent growth, altered migration ability, as well as, formation of tumors in nude athymic female mice. By demonstrating the carcinogenic potential of tungsten both in vitro and in vivo testing models, we further establish the hypothesis that tungsten is carcinogenic.
RNA-sequencing revealed major gene expression changes between the 3 tungsten-transformed clones and the 3 control clones that grew spontaneously in soft agar. Given that this work was performed in a lung cell line, we used IPA to investigate if there were any altered genes involved in lung carcinogenesis. We found several genes involved in respiratory tract cancers that were altered by tungsten and the direction of their regulation, either up or down, supported their involvement in cancer. NQO1, which is an NADPH dehydrogenase, was up-regulated during tungsten-induced cell transformation. Several studies have reported the dysregulation of NQO1 in lung cancer [5, 19, 39]. While Iskander et al. demonstrated that NQO1 knockout mice displayed an increased risk of lymphoma [19], we found the gene to be up-regulated after tungsten exposure. Both up- and down-regulation of NQO1 protein holds potential to induce carcinogenesis. For example, NQO1 plays a cytoprotective role after exposure to toxicants and it prevents degradation of the tumor suppressor p53; however, NQO1 is known to activate lung carcinogens [39]. It is likely that cell type plays a large role in whether or not the dysregulation of the NQO1 gene will facilitate carcinogenesis. Bey et al. reports that upregulation of NQO1 facilitates the growth of non-small-cell lung cancers [5], which is in line with our data showing that up-regulation of NQO1 will facilitate lung cancers.
Tungsten exposure increased the expression of CRYAB. The protein product of CRYAB acts similar to a chaperone protein but holds its target protein in large aggregates. A recent investigation by Qin et al. reports that overexpression of the CRYAB gene promotes the progression of non-small cell lung cancer and patients displaying up-regulation of both the mRNA and protein levels of CRYAB have a poor prognosis [32].
Tungsten increased the expression of S100A4 gene and dysfunction in this gene has been reported to increase the incidence of lung carcinoma. The protein product of this gene binds calcium and is involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation [29]. SPK2 gene encodes an F-box protein, which is part of an ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box). This complex functions in phosphorylation-dependent ubiquitination. Spk2 protein plays an intricate role in S-phase of the cell cycle and is an essential element of the cyclin A-CDK2 kinase. Tungsten upregulated the expression of SPK2 and increased levels of Spk2 protein have been reported to enhance the growth of small cell lung cancers [42]. The protein product of the HTATIP2 gene is called Tip30 and acts as a serine/threonine kinase that is involved in the regulation of major cell signaling players such as Myc, Vegf, and Akt. Tungsten increased the expression of HTATIP2 and there is an association between up-regulation of HTATIP2 expression and cancer [21].
Along with respiratory tract cancers, genes involved in leukemia were also dysregulated during tungsten induced cell transformation. CD74 (major histocompatibility complex, class II) was up-regulated by tungsten. Patients with Chronic lymphocytic leukemia (CLL) display an increased expression of CD74. High expression of CD74 was associated with advanced stages of CLL. A tungsten-induced up-regulation of CD74 may contribute to leukemia development and provides for a link between the known association of tungsten and leukemia [7]. CTGF was down-regulated by tungsten. Over-expression of CTGF is seen in lymphoblastic leukemia and targeting the CTGF protein has given positive results in combatting the disease. Although, tungsten induced a down-regulation of this gene, it points in the direction that tungsten affects genes involved in leukemia. Had a B-cell line been used for the study or a different dose of tungsten, the gene expression of CTGF may be affected differently [25].
HOXB5, homeobox 5, was one of the down-regulated genes altered by tungsten exposure. Increased expression of this gene was associated with a distinct biologic subset of acute myeloid leukemia (AML) and expression has been identified in acute promyelocytic leukemia [36]. Although the W-induced expression of HOXB5 is down, one study demonstrates that expression is different among different types of leukemia [23]. HOXB5 gene expression can be epigenetically regulated. A ChIP experiment demonstrated binding of the polycomb repressive complex members: EZH2 and SUZ12 to the HOXB5 gene. EZH2 and SUZ12 are histone methyltransferases for different lysines on H3 [8]. Tungsten exposure also altered the expression of MST4 (serine/threonine protein kinase 26), another gene involved in leukemia. Up-regulation of this gene by tungsten may facilitate carcinogenesis given that the protein product of the MST4 gene is involved in the differentiation of NB4 cells, an acute promyelocytic leukemia cell line and expression is also found in Jurkat cells, acute T-cell leukemia cell line [16, 22]. Tungsten also up-regulated the expression of CSF3 (colony stimulating factor 3). Up-regulation of this protein increases proliferation of HL-60 cells, which are a promyelocytic leukemia cell line [41]. Presence of CSF3 activates two STAT isoforms (transcription factors involved in cell growth) that induce differentiation of HL-60 cells [9].
Along with altering the expression of many genes specific to certain types of cancers, tungsten affected several genes that play a general role in carcinogenesis and are involved in many cancer types. Tungsten-transformed clones displayed a down-regulation in the CDNK1A gene (cyclin-dependent kinase inhibitor 1A), which encodes a key protein involved in the cell cycle. This gene encodes a potent cyclin-dependent kinase inhibitor called p21. P21 acts as a regulator of cell cycle progression at G1 by binding to and inhibiting the activity of cyclin-CDK2 complexes. CDNK1A was decreased due to tungsten exposure and a decrease in CDNK1A expression has been associated with many cancers. Reduced levels of CDNK1A have been shown by several studies to facilitate the growth of Hct116 cells, a colorectal cancer cell line [2, 15]. Down-regulation of CDNK1A increases the growth of ALVA31 cells, a prostatic carcinoma cell line [27]. Nakagawa et al. found that down-regulation of CDNK1A by siRNA resulted in increased proliferation of H358 cells, a lung adenocarcinoma cell line, due to increased progression of the cell cycle at G1 [30].
Another gene often dysregulated in cancer is TGF1B (transforming growth factor, beta 1). TGF1B encodes a cytokine that regulates proliferation, differentiation, adhesion, migration, and other functions in many cell types. Many cells have TGFB receptors, and the protein positively and negatively regulates many other growth factors. This gene is frequently up-regulated in tumor cells. Tungsten exposure decreased TGF1B expression and a number of investigations have reported on reduced TGF1B levels in carcinogenesis. Several studies have demonstrated that presence of TGF1B protein decreases cellular proliferation in lymphoma cell lines, such as Mac-2A cells, a cutaneous T-cell lymphoma line, and DB cells, a B-cell lymphoma line [11, 20]. Turley et al. found that repressing TGF1B mRNA levels by siRNA increased proliferation of HL60 cells, a leukemia cell line [37]. Presence of TGFB protein decreased the growth of UMSCC6 cells, a head and neck cancer cell line [14].
Conclusion
Metals are largely associated with inducing toxic and carcinogenic effects. For the first time, we present evidence characterizing the cytotoxicity and carcinogenicity of tungsten, an element that has been rarely investigated in metal toxicology. Several tests evaluating carcinogenic endpoints were conducted to assess tungsten’s carcinogenic potential. Tungsten-treated cells formed colonies in soft agar and the clones formed tumors in nude mice. The tungsten-transformed clones demonstrated migration ability by displaying a positive result in the scratch test. Along with affirmative results from this battery of tests measuring carcinogenic parameters, RNA extracted from tungsten-transformed clones revealed that many cancer-associated genes were dysregulated following tungsten induced cell transformation. These genes included a few genes common to many cancers such as CDNK1A and TGFB1. Several genes involved in lung cancer and leukemia also demonstrated altered expression levels.
Our data showcases the carcinogenic properties of tungsten and points to the notion that tungsten is a carcinogen. Further studies are needed in order to establish this metal as a carcinogen including animal bioassays, mechanistic studies, and epidemiological investigations. Chronic, low-dose exposures in animals are needed in order to represent exposures relevant to humans.
Highlights.
Tungsten (W) induces cell transformation and increases migration in vitro.
W increases xenograft growth in nude mice.
W altered the expression of cancer-related genes such as those involved in leukemia.
Some of the dysregulated leukemia genes include, CD74, CTGF, MST4, and HOXB5.
For the first time, data is presented that demonstrates tungsten’s carcinogenic potential.
Table.
Gene definitions and functions.
| Gene | Up or down regulated |
Function | Citations |
|---|---|---|---|
|
NQO1 NAD(P)H dehydrogenase, quinone 1 |
Up | This gene encodes a cytoplasmic 2- electron reductase. It functions to reduce quinones to hydroquinones. Dysregulation of NQO1 expression is reported in lung cancer. |
Bey et al., 2007; Iskander et al., 2008; |
|
CRYAB crystallin, alpha B |
Up |
CRYAB protein product holds its target protein in large aggregates. Overexpression of the CRYAB gene promotes the progression of non-small cell lung cancer. |
Qin et al., 2014 |
|
S100A4 S100 calcium binding protein A4 |
Up | This protein is involved in cell cycle progression, differentiation, and other cellular processes. Dysfunction of S100A4 gene has been reported to increase the incidence of lung carcinoma. |
Naaman et al., 2004 |
|
SPK2 S-phase kinase-associated protein 2 |
Up | Spk2 protein plays an intricate role in S- phase of the cell cycle and is an essential element of the cyclin A-CDK2 kinase. Increased levels of Spk2 enhances the growth of small cell lung cancers. |
Yokoi et al., 2002 |
|
CD74 Major histocompatibility complex, class II |
Up | Patients with Chronic lymphocytic leukemia (CLL) display an increased expression of CD74. High expression of CD74 was associated with advanced stages of CLL. |
Butrym et al., 2013 |
|
HOXB5 Homeobox 5 |
Down | The encoded protein is involved in gut and lung development. This gene has been associated with acute myeloid leukemia (AML). |
Liu et al., 2011; Cai et al., 2012 |
|
MST4 Serine/threonine protein kinase 26 |
Up | Up-regulation of this gene is involved in the differentiation of NB4 cells, an acute promyelocytic leukemia cell line, and expression is also found in Jurkat cells, an acute T-cell leukemia cell line |
Hattori et al., 2007; Lin et al., 2001 |
|
CSF3 Colony stimulating factor 3 |
Up | Up-regulation of this protein increases proliferation of HL-60 cells, which are a promyelocytic leukemia cell line. |
Yamaguchi et al., 1999 |
|
CDNK1A cyclin-dependent kinase inhibitor 1A |
Down | This gene encodes p21, which acts as a regulator of cell cycle progression at G1 by binding to and inhibiting the activity of cyclin-CDK2 complexes. Its role in carcinogenesis may be due to its influence on apoptosis following caspase activation. |
Hata et al., 2005; Nakagawa et al., 2005 |
|
TGF1B transforming growth factor, beta 1 |
Down |
TGF1B encodes a cytokine that regulates proliferation, differentiation, adhesion, migration, and other functions in many cell types. Presence of Tgf1b protein decreases cellular proliferation in lymphoma cell lines, Mac-2A and DB cells. Repressing TGF1B mRNA levels by siRNA increased proliferation of HL60 cells, a leukemia cell line. |
Chen et al., 2007; Turley et al., 1996; Cohen et al., 2009 |
Acknowledgements
This work was supported by National Institutes of Health, National Institute of Environmental Health Sciences Grants R01ES023174 P30ES000260 and R01ES022935
We also acknowledge financial support from Regione Autonoma Sardegna L.R.7/2007, project CRP 26712.
Footnotes
Conflict of Interest Statemtent
None of the authors involved in the manuscript have a conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adamakis IDS, et al. Tungsten toxicity in plants. Plants. 2012;1(2):82–89. doi: 10.3390/plants1020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer SY, et al. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci USA. 1998;95(12):6791–6. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arita A, et al. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1(3):222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Association of State and Territorial Solid Waste Management Officials Tungsten Issues Paper. 2011. ASTSWMO.
- 5.Bey EA, et al. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc Natl Acad Sci USA. 2007;104(28):11832–7. doi: 10.1073/pnas.0702176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brocato J, et al. Basic mechanics of DNA methylation and the unique landscape of the DNA methylome in metal-induced carcinogenesis. Crit Rev Toxicol. 2013;43(6):493–514. doi: 10.3109/10408444.2013.794769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butrym A, et al. High CD74 expression correlates with ZAP70 expression in B cell chronic lymphocytic leukemia patients. Med Oncol. 2013;30(2):560. doi: 10.1007/s12032-013-0560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai L, et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell. 2013;49(3):571–82. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty A, et al. Granulocyte colony-stimulating factor activation of Stat3 alpha and Stat3 beta in immature normal and leukemic human myeloid cells. Blood. 1996;88(7):2442–9. [PubMed] [Google Scholar]
- 10.Chen H, et al. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis. 2010;31(12):2136–2144. doi: 10.1093/carcin/bgq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, et al. Resistance to TGF-beta 1 correlates with aberrant expression of TGF-beta receptor II in human B-cell lymphoma cell lines. Blood. 2007;109(12):5301–7. doi: 10.1182/blood-2006-06-032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy HA, et al. Gene expression changes in human lung cells exposed to arsenic, chromium, nickel or vanadium indicate the first steps in cancer. Metallomics. 2012;4:784–793. doi: 10.1039/c2mt20074k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clausen JL, et al. Fate and Transport of Tungsten at Camp Edwards Small Arms Ranges. ERDC TR-07-05. Strategic Environmental Research and Development Program. U.S. Army Corps of Engineers Engineer Research and Development Center. 2007.
- 14.Cohen J, et al. Attenuated transforming growth factor beta signaling promotes nuclear factor-kappaB activation in head and neck cancer. Cancer Res. 2009;69(8):3415–24. doi: 10.1158/0008-5472.CAN-08-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hata T, et al. Role of p21waf1/cip1 in effects of oxaliplatin in colorectal cancer cells. Mol Cancer Ther. 2005;(10):1585–94. doi: 10.1158/1535-7163.MCT-05-0011. [DOI] [PubMed] [Google Scholar]
- 16.Hattori H, et al. RNAi screen identifies UBE2D3 as a mediator of all-trans retinoic acid-induced cell growth arrest in human acute promyelocytic NB4 cells. Blood. 2007;110(2):640–50. doi: 10.1182/blood-2006-11-059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes RB. The carcinogenicity of metals in humans. Cancer Causes Control. 1997;8(3):371–385. doi: 10.1023/a:1018457305212. [DOI] [PubMed] [Google Scholar]
- 18.Huang Z, et al. Tumor suppressor Alpha B-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene. 2011;31(32):3709–20. doi: 10.1038/onc.2011.529. [DOI] [PubMed] [Google Scholar]
- 19.Iskander K, et al. Disruption of NAD(P)H:quinone oxidoreductase 1 gene in mice leads to radiation-induced myeloproliferative disease. Cancer Res. 2008;68(19):7915–22. doi: 10.1158/0008-5472.CAN-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knaus PI, et al. A dominant inhibitory mutant of the type II transforming growth factor beta receptor in the malignant progression of a cutaneous T-cell lymphoma. Mol Cell Biol. 1996;16(7):3480–9. doi: 10.1128/mcb.16.7.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumtepe Y, et al. High serum HTATIP2/TIP30 level in serous ovarian cancer as prognostic or diagnostic marker. European Journal of Med Research. 2013;18(18) doi: 10.1186/2047-783X-18-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin JL, et al. MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene. 2001;20(45):6559–69. doi: 10.1038/sj.onc.1204818. [DOI] [PubMed] [Google Scholar]
- 23.Liu HC, et al. Expression of HOXB genes is significantly different in acute myeloid leukemia with a partial tandem duplication of MLL vs. a MLL translocation: a cross-laboratory study. Cancer Genet. 2011;204(5):252–9. doi: 10.1016/j.cancergen.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu H, et al. Targeting connective tissue growth factor (CTGF) in acute lymphoblastic leukemia preclinical models: anti-CTGF monoclonal antibody attenuates leukemia growth. Ann Hematol. 2014;93(3):485–492. doi: 10.1007/s00277-013-1939-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meo SA, et al. Health hazards of welding fumes. Saudi Med J. 2003;24(11):1176–1182. [PubMed] [Google Scholar]
- 27.Moffatt KA, et al. Growth inhibitory effects of 1alpha, 25-dihydroxyvitamin D(3) are mediated by increased levels of p21 in the prostatic carcinoma cell line ALVA-31. Cancer Res. 2001;61(19):7122–9. [PubMed] [Google Scholar]
- 28.Morgan M, et al. ShortRead: A Bioconductor Package for Input, Quality Assessment and Exploration of High-Throughput Sequence Data. Bioinformatics. 2009;25(19):2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naaman C, et al. Cancer predisposition in mice deficient for the metastasis-associated Mts1(S100A4) gene. Oncogene. 2004;23(20):3670–80. doi: 10.1038/sj.onc.1207420. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa M, et al. Phorbol ester-induced G1 phase arrest selectively mediated by protein kinase Cdelta-dependent induction of p21. J Biol Chem. 2005;280(40):33926–34. doi: 10.1074/jbc.M505748200. [DOI] [PubMed] [Google Scholar]
- 31.Oshlack A, et al. From RNA-seq Reads to Differential Expression Results. Genome Biology. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin H, et al. Elevated expression of CRYAB predicts unfavorable prognosis in non-small cell lung cancer. Med Oncol. 2014;8(142) doi: 10.1007/s12032-014-0142-1. [DOI] [PubMed] [Google Scholar]
- 33.Robinson MD, et al. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato JD, et al. Media for Culture of Mammalian Cells Unit 1.2. Current Protocols in Cell Biology. Core Publication. 1998 doi: 10.1002/0471143030.cb0102s00. 1.2.7. [DOI] [PubMed] [Google Scholar]
- 35.Sun H, et al. Comparison of gene expression profiles in chromate transformed BEAS-2B cells. PLoS One. 2011;6:e17982. doi: 10.1371/journal.pone.0017982. 10.1371/journal.pone.0017982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson A, et al. Global down-regulation of HOX gene expression in PML-RARalpha + acute promyelocytic leukemia identified by small-array real-time PCR. Blood. 2003;101(4):1558–65. doi: 10.1182/blood.V101.4.1558. [DOI] [PubMed] [Google Scholar]
- 37.Turley JM, et al. Transforming growth factor beta 1 functions in monocytic differentiation of hematopoietic cells through autocrine and paracrine mechanisms. Cell Growth Differ. 1996;7(11):1535–44. [PubMed] [Google Scholar]
- 38.Tyrrell J, Galloway TS, Abo-Zaid G, Melzer D, Depledge MH, Osborne NJ. High Urinary Tungsten Concentration Is Associated with Stroke in the National Health and Nutrition Examination Survey 1999–2010. PLOS One. 2013 doi: 10.1371/journal.pone.0077546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiencke JK, et al. Lung cancer in Mexican-Americans and African-Americans is associated with the wild-type genotype of the NAD(P)H: quinone oxidoreductase polymorphism. Cancer Epidemiol Biomarkers Prev. 1997;6(2):87–92. [PubMed] [Google Scholar]
- 40.Wild P, et al. Lung cancer and exposure to metals: the epidemiological evidence. Methods Mol Biol. 2009;472:139–167. doi: 10.1007/978-1-60327-492-0_6. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi T, et al. The role of STAT3 in granulocyte colony-stimulating factor-induced enhancement of neutrophilic differentiation of Me2SO-treated HL-60 cells. GM-CSF inhibits the nuclear translocation of tyrosine-phosphorylated STA T3. J Biol Chem. 1999;274(22):15575–81. doi: 10.1074/jbc.274.22.15575. [DOI] [PubMed] [Google Scholar]
- 42.Yokoi S, et al. A novel target gene, SKP2, within the 5p13 amplicon that is frequently detected in small cell lung cancers. Am J Pathol. 2002;161(1):207–16. doi: 10.1016/S0002-9440(10)64172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]