Abstract
A robust, quantitative ultraperformance liquid chromatography ion trap multistage scanning mass spectrometric (UPLC/MS3) method was established to characterize and measure five deoxyguanosine (dG) adducts formed by reaction of the chemotherapeutic nitrogen mustard (NM) bis-(2-chloroethyl)ethylamine with calf thymus (CT) DNA. In addition to the known N7-guanine (NM-G) adduct and its crosslink (G-NM-G), the ring-opened formamidopyrimidine (FapyG) mono-adduct (NM-FapyG) and cross-links in which one (FapyG-NM-G) or both (FapyG-NM-FapyG) guanines underwent ring-opening to FapyG units were identified. Authentic standards of all adducts were synthesized and characterized by NMR and mass spectrometry. These adducts were quantified in CT DNA treated with NM (1 μM) as their deglycosylated bases. A two-stage neutral thermal hydrolysis was developed to mitigate the artifactual formation of ring-opened FapyG adducts involving hydrolysis of the cationic adduct at 37 °C, followed by hydrolysis of the FapyG adducts at 95 °C. The limit of quantification values ranged between 0.3 and 1.6 adducts per 107 DNA bases, when the equivalent of 5 μg DNA hydrolysate was assayed on column. The principal adduct formed was the G-NM-G cross-link, followed by the NM-G mono-adduct; the FapyG-NM-FapyG adduct was at the limit of detection. The NM-FapyG adducts formed in CT DNA at a level of ~20% that of the NM-G adduct. NM-FapyG has not been previously quanitified and the FapyG-NM-G and FapyG-NM-FapyG adducts have not be previously characterized. Our validated analytical method was then applied to measure DNA adduct formation in the MDA-MB-231 mammary tumor cell line exposed to NM (100 μM) for 24 h. The major adduct formed was NM-G (970 adducts per 107 bases), followed by G-NM-G (240 adducts per 107 bases) and NM-FapyG (180 adducts per 107 bases), and lastly the FapyG-NM-G cross-link adduct (6.0 adducts per 107 bases). These lesions are expected to contribute to the NM-mediated toxicity and genotoxicity in vivo.
Keywords: Nitrogen mustards, DNA adducts, mass spectrometry, cross-links, formamidopyrimidines
Introduction
Nitrogen mustards are a class of bifunctional chemotherapeutic agents that were among the first agents to show effectiveness against non-Hodgkin lymphoma in mice and early human clinical trials.1-3 Mechlorethamine (1949), chlorambucil (1954), cyclophosphamide (1959), uracil mustard (1962) and melphalan (1964) were among the first drugs approved by the FDA for clinical use (Figure 1). They are still used as part of combination drug therapies for the treatment of cancer and autoimmune disease. The mechanism of action of nitrogen mustards is through alkylation of DNA bases. Secondary malignancies, in particular acute myeloid leukemia, are often observed in patients treated with nitrogen mustards, and this side effect is a limiting factor in their clinical use.4, 5
Figure 1.
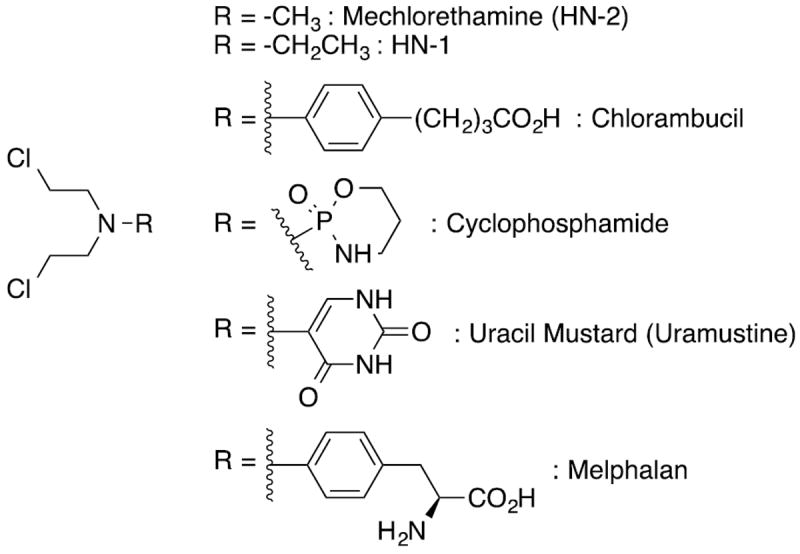
Early chemotherapeutic nitrogen mustard.
The chemical mechanism of DNA alkylation involves an initial intramolecular SN2 reaction to form an aziridinium ion intermediate, which is the DNA alkylating species.2, 6 As a bis-electrophile, the initial adduct can form a second aziridinium ion intermediate, which can undergo solvolysis to the corresponding alcohol affording a mono-adduct, react with nucleophilic sidechains of proteins (e.g., cysteine) to form DNA-protein cross-links,7 or react with another DNA base to generate DNA inter- and intrastrand cross-links.8-10 This is shown in Scheme 1 for the alkylation at the N7-position of deoxyguanosine (dG).
Scheme 1.
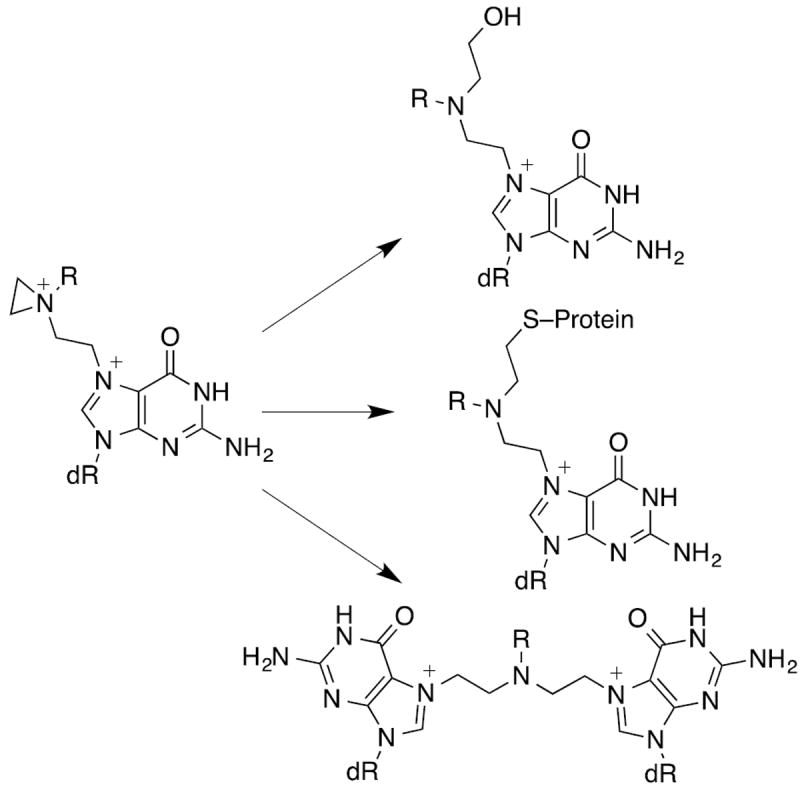
Interstrand DNA cross-links are regarded as highly cytotoxic lesions and although they generally represent only a small percentage of the total adduct burden, they are believed to play a disproportionately high role in the mechanism of action of nitrogen mustards and other DNA cross-linking agents.11 Interestingly, nitrogen mustards were shown to from interstrand cross-links in a 5’-GNC-3’ rather than 5’-GC-3’ sequence.12, 13
Numerous studies have examined the reaction of nitrogen mustards with nucleosides and single and double stranded DNA. The major adducts are from reaction at N7-dG and N3-dA with minor reaction at N3-dC, N1-dA and O6-dG.8, 14-21 In addition, cross-links have been characterized between N7-positions of dG, N3-positions of dA, and between the N7-position dG and N3-position of dA.9, 10 Cationic N7-dG adducts can also undergo a secondary reaction involving the addition of hydroxide to C8 followed by imidazole ring-opening to form an N5-substituted formamidopyrimidine adduct (Fapy-dG) in which the N5-substituent is derived from the alkylating agent. High pH is usually required to induce Fapy-dG formation for most alkylating agents. Mehta reported that alkylation of dG with a phosphoramide mustard (R= -PO2NH2) produced an unusually unstable adduct.22 The decomposition products, which formed at pH 7.4, were ascribed to imidazole ring opening based on their UV spectra. Chetsanga subsequently showed that reacting the phosphoramide mustard with DNA produced an adduct that was a substrate for Escherichia coli formamidopyrimidine glycosylase (FPG),23 suggesting the formation of a Fapy-dG adduct from the phosphoramide mustard. Hemminki observed FapyG adducts from the reaction of nor-nitrogen mustard (R= -H), mechlorethamine, chlorambucil and aziridine.15, 24-26 Interestingly, cultured human cells that overexpressed FPG were up to 100-fold less sensitive to thioTEPA or aziridine,27-29 10-fold less sensitive to 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU), and 2-fold less sensitive to the nitrogen mustard mafosfamide.30, 31 These results support a role for the N5-substituted Fapy-dG adducts in the cytotoxic mechanism of these agents.
The Fapy-dG adduct in which the formyl nitrogen is unsubstituted is a product of oxidative DNA damage and has been shown to be mutagenic in simian kidney (COS-7) and human embryonic kidney cell culture.32, 33 N5-Substituted Fapy-dG adducts derived from aflatoxin B1 epoxide (AFB1-Fapy-dG) and methylating agents (MeFapy-dG) have been reported to be persistent lesions in rodents.34, 35 The AFB1-Fapy-dG and MeFapy-dG adducts have also been shown to be mutagenic in COS-7 cells.36, 37 Given the importance of nitrogen mustards in cancer chemotherapy, it is surprising that N5-nitrogen mustard Fapy-dG adducts have received little attention. Recently, the Fapy-dG adduct derived from bis-(2-chloroethyl)ethylamine has been site-specifically incorporated into oligonucleotides by solid-phase methods and shown to be a substrate for FPG and E. coli Endonuclease IV.38
We report here the development of an analytical method to measure nitrogen mustard Fapy-dG adducts (NM-FapyG) from the reaction of bis-(2-chloroethyl)-ethylamine (NM) with calf thymus (CT) DNA by ion-trap multistage scan mass spectrometry. Further, we hypothesized the previously uncharacterized nitrogen mustard cross-links in which one (FapyG-NM-G) or both (FapyG-NM-FapyG) guanines undergo imidazole ring-opening to FapyG units should also be reaction products. The three Fapy adducts along with the N7-G mono-adduct (NM-G) and N7-cross-link (G-NM-G) were detected as their deglycosylated bases (Figure 2) and quantitated by the stable isotope dilution method,39 following neutral thermal hydrolysis and solid phase extraction (SPE).
Figure 2.
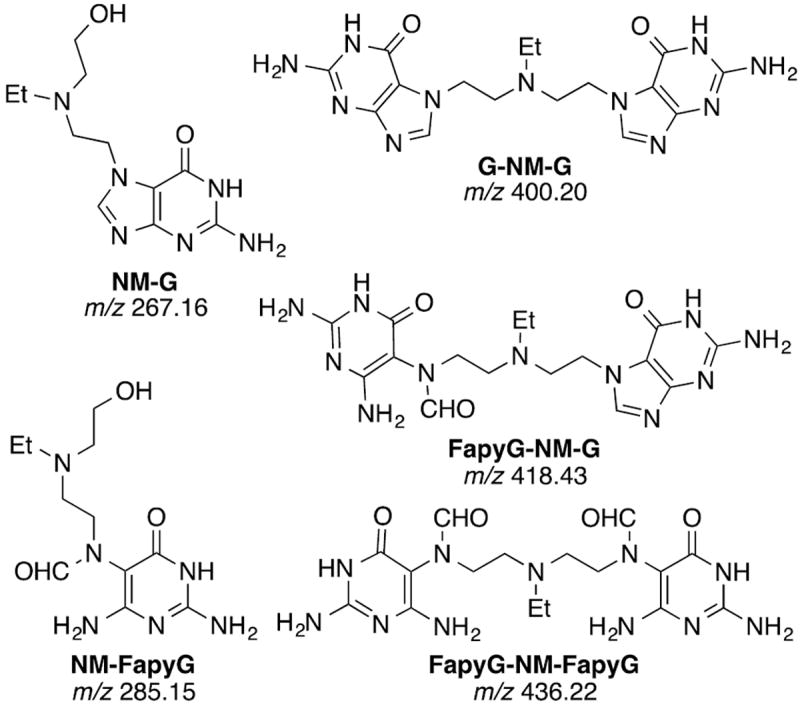
Guanine adducts of a bis-(2-chloroethyl)ethylamine
Experimental Procedures
Synthesis of standards
Authentic standards of NM-G, G-NM-G, NM-FapyG, FapyG-NM-G and FapyG-NM-FapyG were synthesized from dG and NM and described in the Supporting Information. The standards were characterized by NMR and MS and their UV extinction coefficients determined. Isotopically labeled NM-G, G-NM-G, FapyG-NM-G and FapyG-NM-FapyG were synthesized from dG and [2H5]-ethyl-NM; labeled NM-FapyG was synthesized from [15N5]-dG and NM.
Reaction of CT DNA with NM
CT DNA (50 μg) in 0.5 mL of 50 mM potassium phosphate buffer (pH 7.4) was reacted with NM (1 μM) at 37 °C for 14 h. The unreacted NM and decomposition products were removed by solvent extraction with ethyl acetate (3 × 500 μL). A solution of NaCl (5 M, 60 μL) was added, and the DNA was precipitated by addition of chilled ethanol (1 mL), followed by centrifugation The DNA pellets were washed with 70%, followed by 100% ethanol, then dried under vacuum for 10 min.
Neutral thermal hydrolysis of CT DNA and recovery of the NM-adducts
CT DNA (50 μg/mL) was dissolved in potassium phosphate (50 mM, pH 7.0,) and subjected to neutral thermal hydrolysis at 37 °C, 70 °C, or 95 °C for variable incubation times. Stable, isotopically labeled internal standards were added prior to hydrolysis at a level of 350 pg per 50 μg DNA; this level of spiking with the internal standards corresponds to 53 – 86 adducts per 107 bases. The hydrolysis was terminated by placing the solutions on ice and the residual DNA was removed by size exclusion filtration employing Pall 10 kDa molecular weight cut off filter and centrifugation at 5000 g for 20 min. The filters were washed with 2% HCO2H in H2O (300 μL), and the combined filtrates containing the adducts were evaporated to dryness by vacuum centrifugation at 65 °C.
The NM adducts were enriched by SPE using Sola SCX cartridges (Thermo Scientific, Bellafonte, PA) The SPE cartridge was pre-conditioned with 5% NH3 in CH3OH (1 mL) followed by 0.1 N HCl (1 mL). DNA hydrolysates were resuspended in 0.1 N HCl (1 mL) and loaded on to the SCX resin. The cartridges were sequentially washed with 0.1 N HCl; 2% HCO2H; 2% HCO2H in CH3OH; and H2O (1 mL each). The NM adducts were eluted from the SPE with a solvent mixture containing 25% H2O/ 70% CH3OH / 5% NH3 (1 mL). Samples were evaporated to dryness by vacuum centrifugation at 65 °C. The extracts were dissolved in 2 mM ammonium formate containing 5% CH3CN (50 μL) immediately prior to UPLC/MS3.
Method validation of NM-DNA adducts
The within-day and between-day reproducibility were assessed by measuring NM adducts from CT DNA treated with NM (1 μM) for 14 h. The DNA was subjected to neutral thermal hydrolysis for 4 h at 95 °C as described above. Calibration curves were constructed in a DNA matrix (50 μg), which underwent neutral thermal hydrolysis and SPE prior to addition of the adduct calibrants. The calibration sample set was comprised of triplicates at nine concentrations: 0, 0.2 to 6 pg μL-1 of unlabeled adduct standards for NM-G, NM-FapyG, FapyG-NM-G, FapyG-NM-FapyG, and 0, 0.2 to 30 pg μL-1 for G-NM-G, with 7 pg μL-1 isotopically labeled internal standards. A 5 μL sample volume out of 50 μL was injected on the column.
Treatment of NM-modified CT DNA with FPG
NM-modified CT DNA (17 μg) was diluted in sodium phosphate buffer (50 mM, pH 7.0, 200 μL) containing MgCl2 (10 mM) and DTT (1 mM).40, 41 Stable, isotopically labeled internal standards (117 pg, a level of internal standard corresponding between 53 to 86 adducts per 107 bases) were added, followed by the addition of FPG (40 units). The incubation was conducted at 37 °C and the released adducts were measured at time intervals of 7, 21, 30 and 44 h. The reaction was terminated by adding 2 volumes of chilled ethanol followed by sample storage at -20 °C for 1 h. The supernatant containing the released NM adducts was vacuum centrifuged to dryness and processed by SPE as described above. Additional FPG (40 units) was added to the remainder of the reaction at each time interval.
Treatment of MDA-MB-231 Cells with NM
The human breast cancer cell line MDA-MB-231 was obtained from American Type Culture Collection (Manassas, VA). Cells were cultured in Leibovitz’s L-15 medium (SIGMA. St Louis, MO) with a seeding density of 2 million in a T75 TPP® flask (SIGMA, St Louis, MO) supplemented with 10% Fetal Bovine Serum (FBS) and 1% Penicillin/Streptomycin (50 U/mL, 50 μg/mL) (SIGMA, St Louis, MO) at 37 °C without CO2. Cells were grown for 24 h and then treated with 100 μM NM (Mittenwald, Germany) in PBS. The culture media was 12 mL and the carcinogen volume was kept at less than 0.1% of the total media volume. The NM treatment was performed in duplicate along with a PBS (Corning Life Sciences, Corning, NY) serving as the negative control. The NM treatment was performed for 24 h. Two independent experiments were performed.
DNA isolation
After 24 h of treatment, the media was aspirated and the dead cells were removed and the adhered cells were washed twice with PBS and collected by incubating with 0.5% Trypsin-EDTA (GIBCO, Grand Island, NY) at 37 °C for 5 minutes. The trypsin was neutralized with equal volume of the culture media and centrifuged to pellet the cells. The cells were then resuspended in PBS (10 mL) and pelleted again. This washing was done twice more. Cell toxicity of the NM-treated cells versus PBS negative control cells was assessed by the Trypan blue exclusion assay using a Countess Automated Cell Counter (Invitrogen, Life Technologies, Carlsbad, CA). Cells were frozen at -80 °C before processed for DNA extraction.
The cells were homogenized and treated with RNase A (13 μL of 10 mg/mL) and RNase T1 (1.5 μL of stock 200KU/mL) (SIGMA, St Louis, MO) for 30 min at 37 °C, followed by addition of proteinase K (20 μL of 20 mg/mL) (Novagen- Merck, Darmstadt, Germany) and 30 μL of 10% SDS (Alfa Aesar, Ward Hill, MA) for 1 h at 37 °C. DNA was then extracted using DNeasy Blood & Tissue kit (Qiagen, Redwood City, CA) following manufacturer’s instructions with minor modifications. Neutral thermal hydrolysis of the NM-modified MDA-MB-231 Cell DNA was done for 4 h at 95 °C, or 72 h at 37 °C, followed by 4 h hydrolysis at 95 °C. The amount of DNA assayed ranged between 3.7 and 6.0 μg, with internal standards added at a level ranging between 240 and 360 adducts per 107 bases.
UPLC/MS3 measurements
UPLC/MS3 measurements were performed on a LTQ Velos Pro MS (Thermo Scientific, San Jose, CA), operating in the positive ESI mode. The MS was interfaced with an Advance CaptiveSpray™ source from Michrom Bioresource Inc. (Auburn, CA) and a NanoAcquity UPLC system (Waters Corp., Milford, MA). All DNA adducts were analyzed in a single scan event at the MS3 scan stage employing the MS parameters described in Table 1. Typical MS tuning parameters were as follows: capillary temperature, 270 °C; source spray voltage, 1.3 kV; S-lens RF level, 69%; no source fragmentation. Helium, set at 1 millitorr, was used as the collision and damping gas in the ion trap. There was no sheath or auxiliary gas. The reconstructed ion chromatograms were obtained using the m/z ions reported at the MS3 scan stage and employed for quantitative measurements.
Table 1.
ESI/MS3 parameters for NM-G adducts. The activation time for all analyses was 10 ms.
| MSn | Adduct | m/z | Isolation width (m/z) | CE | Activation Q | Ions monitored at MS3 scan stage |
|---|---|---|---|---|---|---|
|
| ||||||
| MS2 | NM-G | 267.2 | 3 | 35 | 0.25 | |
| MS3 | 116.1 | 2 | 40 | 0.35 | 72.1, 98.1 | |
|
| ||||||
| MS2 | [2H5]-NM-G | 272.2 | 3 | 35 | 0.25 | |
| MS3 | 121.1 | 2 | 40 | 0.35 | 77.1, 103.1 | |
|
| ||||||
| MS2 | NM-FapyG | 285.2 | 3 | 35 | 0.35 | |
| MS3 | 196.1 | 2 | 35 | 0.35 | 168.1 | |
|
| ||||||
| MS2 | [5N15]-NM-FapyG | 290.2 | 3 | 35 | 0.35 | |
| MS3 | 201.1 | 2 | 35 | 0.35 | 173.1 | |
|
| ||||||
| MS2 | G-NM-G | 400.2 | 2 | 35 | 0.30 | |
| MS3 | 249.1 | 2 | 32 | 0.32 | 178.1, 234.2 | |
|
| ||||||
| MS2 | [2H5]-G-NM-G | 405.2 | 4 | 35 | 0.30 | |
| MS3 | 254.2 | 2 | 32 | 0.32 | 178.1, 239.2 | |
|
| ||||||
| MS2 | FapyG-NM-G | 418.2 | 3 | 32 | 0.30 | |
| MS3 | 267.1 | 3 | 30 | 0.30 | 196.1 | |
|
| ||||||
| MS2 | [2H5]FapyG-NM-G | 423.3 | 4 | 32 | 0.30 | |
| MS3 | 272.2 | 2 | 30 | 0.30 | 196.1 | |
|
| ||||||
| MS2 | FapyG-NM-FapyG | 436.2 | 3 | 35 | 0.30 | |
| MS3 | 223.1 | 2 | 40 | 0.30 | 164.1 | |
The DNA adducts were separated on an Inetrsil C18 ODS-4 column (0.3 × 250 mm, 5 μm particle size) GL Sciences, Torrance, CA). Mobile phase A contained 2 mM ammonium formate and 5% CH3CN in H2O, and mobile phase B contained 2 mM ammonium formate and 5% H2O in CH3CN. A linear gradient was employed for separation of the DNA adducts. The solvent was held at 95:5 A/B for 1 min, and then increased to 5:95 A/B at 11 min, and held at this solvent composition for 2 min. The solvent composition was re- equilibrated to starting conditions over 2 min, followed by a 7 min equilibration, at a flow rate of 5 μL/min. The column was heated at 65 °C, to optimize the peak shape and chromatography. All connecting tubing employed was PEEKsil™ (50 μm inner diameter) because the NM G adducts strongly interacted with PEEK tubing, resulting in very broad peaks.
Statistical methods
The t-test or non-linear regression comparisons of curve fits were performed using GraphPad Prism (v. 6) for Windows, GraphPad Prism Software (La Jolla, CA). A P value <0.05 was considered statistically significant.
Results
Synthesis of Standards
Standards for the five adducts of interest (Figure 2) were synthesized from the reaction of dG with NM in trifluoroethanol as described in the Supporting Information. Isotopically labeled standards were synthesized using [2H5]-ethyl-NM. The isotopic label was lost at the MS2 scan stage for NM-FapyG; thus the labeled NM-FapyG was prepared from [15N5]-dG. The product ion spectra of the NM-G, G-NM-G, NM-FapyG, FapyG-NM-G, and FapyG-NM-FapyG adducts and isotopically labeled internal standards at the MS2 and MS3 scan stages with the proposed principal product ions are shown in Figures S16-S20 of the Supporting Information.
Calibration Curves and Quantitation of Monomer and Dimer Adducts of NM adducts
The linear regressions of the calibration curves were based on the ratio of the peak area of the unlabeled adduct to that of labeled internal standard plotted against the amount of unlabeled adduct versus the internal standard. Data were fitted to a straight line using ordinary least-squares with equal weightings (Figure S21 of the Supporting Information). For all five adducts, the linearity of the calibration curve was shown by the slope, the goodness-of-fit linear regression values, r2 > 0.99, over the range 0.1 to 30 pg or 150 pg of adduct.42 Based upon recommended guidelines for data acquisition and quality evaluation in environmental chemistry, the values of the limit of detection (LOD) and limit of quantitation (LOQ) for analytes were set at 3σ and 10σ standard deviation units above the background level signal of an uncontaminated matrix.43 The LOQ values ranged from 0.3 to 1.6 adducts per 107 DNA bases, when the equivalent of 5 μg DNA hydrolysate was assayed on column.
Development of an Adduct Enrichment Protocol
All measurements were performed with CT DNA (50 μg/mL) in potassium phosphate buffer (100 mM, pH 7.0) modified with NM (1 μM). Cationic N7-dG adducts are thermally labile. The FapyG lesion has been removed from DNA by hot piperidine treatment.44 We therefore decided to liberate the adducts of interest through a two-step process in which the NM-G and G-NM-G adducts would be removed by neutral thermal hydrolysis (pH 7.0, 95 °C) followed by hot piperidine treatment (0.2 M, 90°C, 1 h) to liberate the Fapy adducts. This protocol removed the DNA adducts of interest as their modified nucleobases while leaving the bulk unmodified DNA intact, which was removed by size exclusion filtration with a 10 kDa molecular weight cut off filter. Treatment of the NM-modified DNA with just hot piperidine induced artifactual NM-FapyG formation. In addition, the residual piperidine recovered following SPE, adversely affected the chromatography and signal of response for all adducts. We subsequently determined that prolonged hydrolysis of DNA at 95 °C in neutral phosphate buffer also caused hydrolysis of the Fapy adducts, thus obviating the need for the piperidine treatment.
The employment of SPE resins without polyethylene frits was essential for isolation of the modified nucleo- bases and optimal sensitivity by ion trap MS. We observed that solid phase resins containing polyethylene frits resulted in very strong ion suppression effects for all NM-G adducts, similar to the ion suppression effects previously seen with DNA adducts of heterocyclic aromatic amines enriched by SPE.45 A representative chromatogram of the adducts recovered from NM-modified CT DNA following neutral thermal hydrolysis (pH 7.0, 95 °C, 4 h) is shown in Figure 3.
Figure 3.
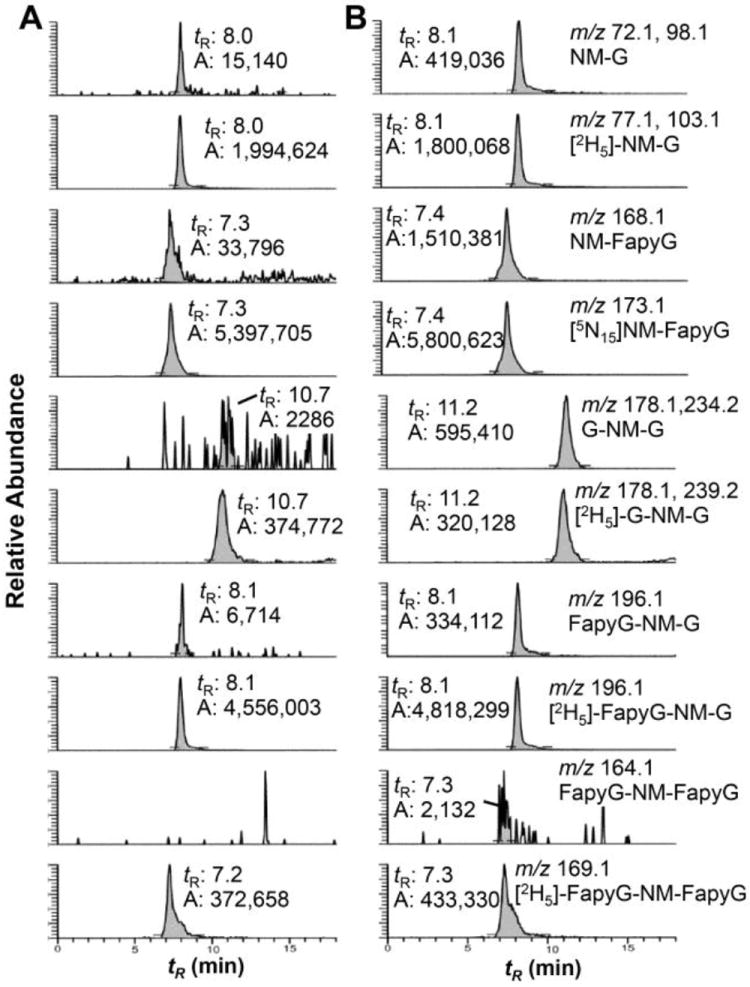
Neutral thermal hydrolysis of the NM-modified CT DNA for 4 h at 95 °C. (A) Untreated CT DNA and (B) DNA modified with NM (1 μM). Internal standards were added at a level ranging between 53 and 87 adducts per 107 bases. The retention time (tR) and area of peak integration (A) are reported.
Method Validation
The within-day and between-day reproducibility were determined for the reaction of CT DNA with NM, following neutral thermal hydrolysis at 95 °C for 4 h. Four independent replicate measurements were determined for the five adducts. The studies were repeated on 3 different days, and the data are summarized in Table 2. The within-day and between-day data precision, reported as the % coefficient of variation (% CV), are ≤10 % for the NM-G, NM-FapyG, FapyG-NM-G and G-NM-G adducts; formation of the FapyG-NM-FapyG adduct was below the LOQ value.
Table 2.
Performance of the Method: CT DNA Modified with NM (1μM) With-day and Between-day Reproducibility.a,b
| Adduct/107 bases | Day 1 (pH7) | Day 2 (pH7) | Day 3 (pH7) | CV (%) within-day | CV (%) between-day | |
|---|---|---|---|---|---|---|
|
| ||||||
| NM-G | Mean | 20.7 | 19.4 | 19.9 | ||
| SD | 1.7 | 0.2 | 1.3 | |||
| RSD (%) | 8.3 | 1.1 | 6.7 | 6.4 | 6.2 | |
|
| ||||||
| G-NM-G | Mean | 132 | 118 | 127 | ||
| SD | 14.1 | 1.3 | 9.7 | |||
| RSD (%) | 10.6 | 1.1 | 7.5 | 8.2 | 9.1 | |
|
| ||||||
| NM-FapyG | Mean | 19.9 | 18.8 | 19.2 | ||
| SD | 1.1 | 0.2 | 1.1 | |||
| RSD (%) | 5.7 | 1.0 | 5.4 | 4.8 | 5.2 | |
|
| ||||||
| FapyG-NM-G | Mean | 3.8 | 3.7 | 3.7 | ||
| SD | 0.2 | 0.1 | 0.2 | |||
| RSD (%) | 6.7 | 1.3 | 7.4 | 5.9 | 5.5 | |
CT DNA (50 μg/mL in potassium phosphate (50 mM, pH 7.0) was subjected to neutral thermal hydrolysis at 95 °C for 4 h.
FapyG-NM-FapyG was below the LOQ value (0.3 adduct per 107 bases) and not reported.
Time Course of recovery of NM adducts from CT DNA as a function of temperature and time, and artifactual formation of NM-FapyG adducts
The hydrolysis and recovery of NM CT DNA adducts was conducted at 70 and 95 °C as a function of time (Figure 4). The hydrolysis of the cationic NM-G and G-NM-G adducts was rapid at 95 °C, and the recovery of adducts reached a maximum level after ~0.5 h, whereas the NM-FapyG and FapyG-NM-G adducts were hydrolyzed more slowly, reaching their maximum level between 2 and 4 h. The FapyG-NM-FapyG adduct was observed at low levels, approaching the limit of detection (LOD). The cationic NM-G and G-NM-G adducts were released more slowly at 70 °C, reaching their maximum levels between 2 and 4 h. The levels of NM-FapyG and FapyG-NM-G continued to slowly increase, reaching levels of 2.5 and 1.2 adducts per 107 bases, respectively, over this 8 h time course.
Figure 4.
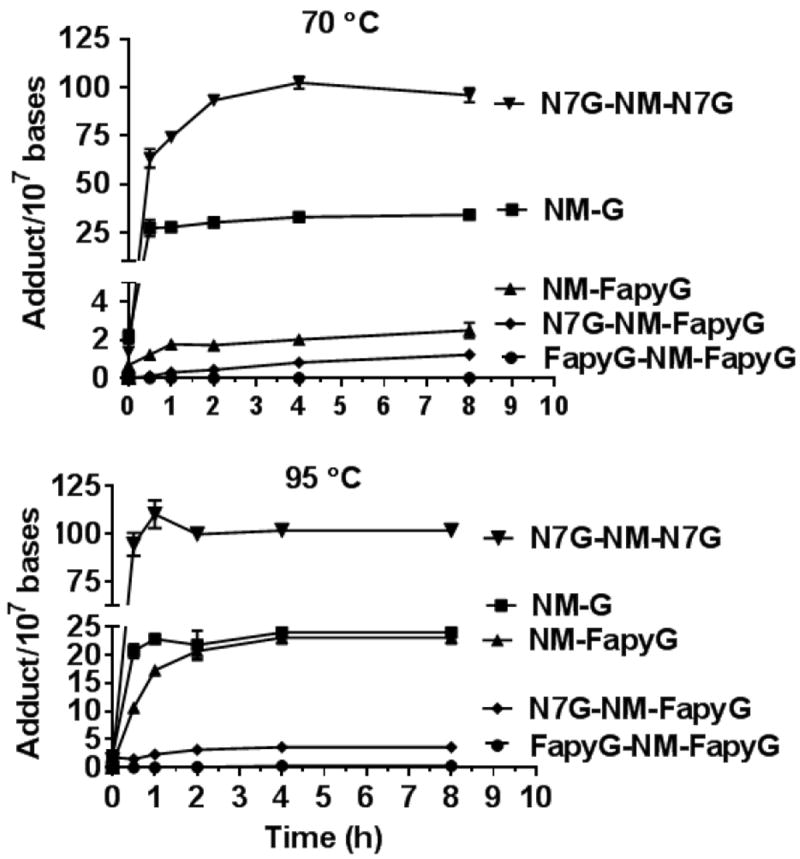
Time course of neutral thermal hydrolysis and recovery of NM-G adducts as a function of temperature (70 or 95°C) and time.
Once again, the FapyG-NM-FapyG level approached the LOD. The level of NM-FapyG was ~9-fold greater when the hydrolysis was conducted at 95 °C compared to 70 °C (23.1 ± 1.8 vs 2.5 ± 0.3 adducts per 107 DNA bases, t-test, p < 0.001, Mean ± SEM); the level of FapyG-NM-G was ~3-fold greater at 95 °C (3.6 ± 0.3 vs 1.2 ± 0.1 adducts per 107 DNA bases, t-test, p < 0.001); and the amount of NM-G decreased by ~0.3 fold when hydrolysis was done at 95 °C compared to hydrolysis at 70 °C (24.0 ± 1.6 vs 34.1 ± 1.9, p < 0.002). There are significant differences between the levels of NM-G, NM-FapyG, FapyG-NM-G adducts obtained by the two-step hydrolysis conditions (Figure 4). These kinetic data signify that a portion of the NM-G adducts undergo ring-opening to form NM-FapyG at 95 °C.
Since the cationic NM-G and NM-G-NM adducts undergo hydrolysis at elevated temperatures more rapidly than the ring-opened NM-FapyG adducts, we explored the selective depurination of the cationic adducts from DNA under mild hydrolysis at 37 °C while retaining the more stable NM-FapyG adducts in the DNA backbone.46 The time course of neutral thermal hydrolysis of NM-modified CT DNA was conducted over time up to 72 h, followed by precipitation of DNA and recovery of depurinated NM-G and G-NM-G adducts in the supernatant. The DNA was subjected to further hydrolysis at 95 °C to recover the three NM FapyG adducts.
The time course of hydrolysis is summarized in Figure 5A and 5B. The level of NM-G reached a plateau at ~32 h with greater than 95% of the NM-G released from the CT DNA. The hydrolysis of the G-NM-G adduct was less rapid than the mono-adduct, and ~15% of the adduct still remained bound to the DNA after 72 h. The ensuing hydrolysis of the partially depurinated DNA at 95 °C for 4 h showed that the remainder of G-NM-G was recovered at the elevated temperature (Figure 5B). The ring-opened NM Fapy adducts were relatively stable towards hydrolysis of DNA at 37 °C with <10% of the adducts recovered in the supernatant during the 72 h time course. The remaining ~ 90% of the NM-FapyG and FapyG-NM-G fractions were recovered from the DNA by the subsequent hydrolysis at 95 °C. The total amount of NM-FapyG and FapyG-NM-G recovered by sequential hydrolysis of NM-modified DNA at 37 °C followed by 95 °C were 6.9 ± 0.7 and 3.4 ± 0.6 adducts per 107 DNA bases, respectively. We conclude that neutral thermal hydrolysis at 37 °C for 72 h to remove cationic adducts, followed by hydrolysis of thermally stable ring-opened NM FapyG adducts at 95 °C greatly minimizes artifactual formation the NM FapyG adducts.
Figure 5.
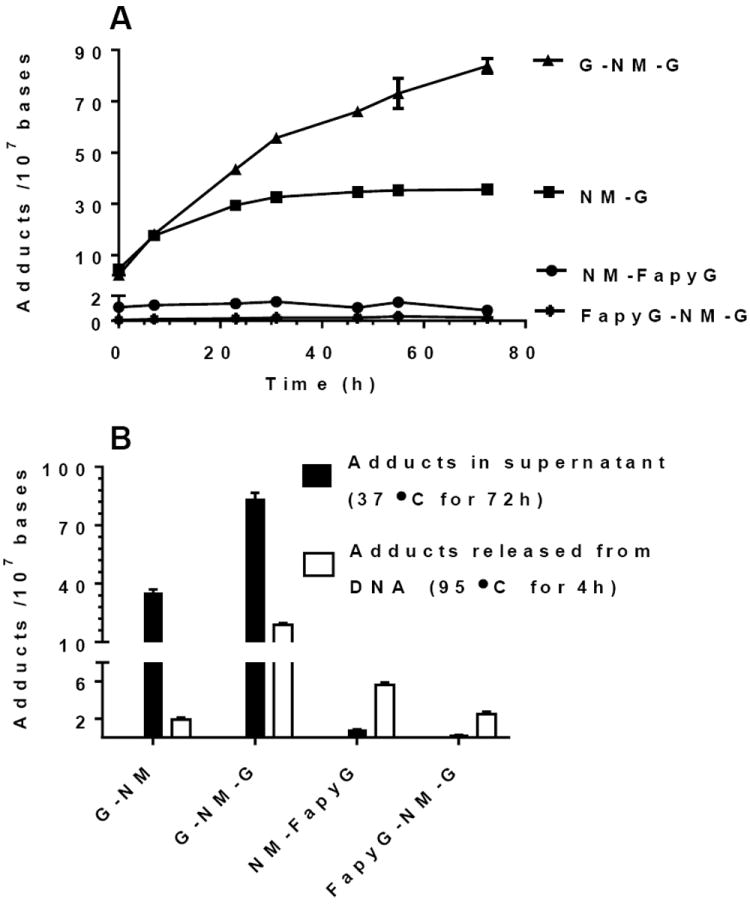
(A) Time course of the neutral thermal hydrolysis and recovery of the NM-G adducts in the supernatant as a function of time at 37 °C, (B) followed by neutral thermal hydrolysis of remaining NM-modified DNA at 95 °C for 4 h. The level of FapyG-NM-FapyG was below the limit of quantification (LOQ).
Enrichment of NM-FapyG adducts by FPG excision of NM CT-DNA
The NM-FapyG adduct is a substrate for FPG when placed in a 24-mer duplex.38 We examined the use of FPG to analyze the NM-FapyG in NM-modified CT DNA (Figure 6). The level of NM-FapyG and FapyG-NM-G based on sequential neutral thermal hydrolysis were, respectively, 6.9 ± 0.7 and 3.4 ± 0.6 adduct per adducts per 107 DNA bases versus 1 NM-FapyG adduct per 48 DNA bases in the 24-mer duplex.38 Despite the >50,000-fold dilution of NM-FapyG in CT DNA, FPG excised NM-FapyG from the CT DNA, albeit much less efficiently.
Figure 6.
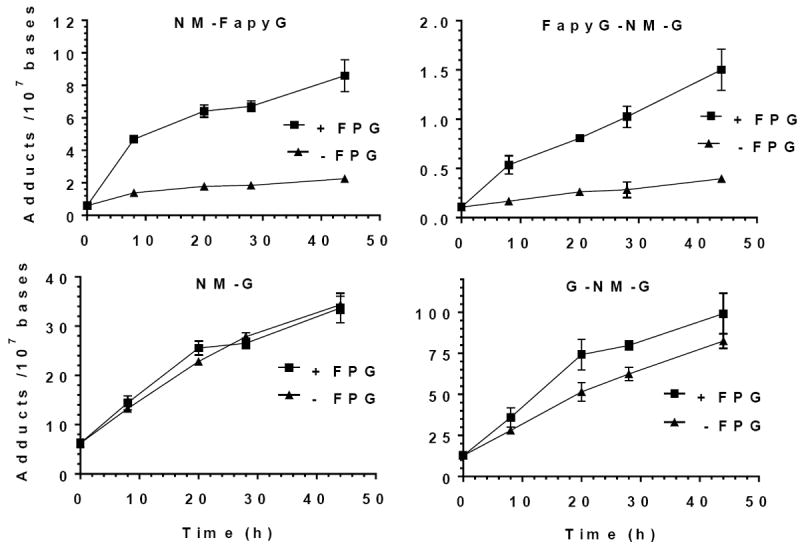
FPG Assay with CT DNA Modified with NM (1 μM)
The amount of NM-FapyG recovered by FPG from the treated CT DNA after 72 h was ~4-fold greater than the level of NM-FapyG due to background hydrolysis without FPG. The recovery of the NM G lesions, with and without FPG, versus time was assessed by non-linear regression analysis of adduct and showed significant differences for NM-FapyG and FapyG-NM-G adducts (p < 0.003). The levels of NM-FapyG and FapyG-NM-G recovered by FPG after 72 h were, respectively 8.6 ± 1.0 and 1.5 ± 0.2 adducts per 107 DNA bases. These adducts levels are similar to those recovered by sequential neutral thermal hydrolysis at 37 °C for 72 h followed by at 95 °C for 4 h (Figure 5A and B). In contrast, the recovery of the NM-G 72 h with FPG was nearly identical to that of the control without FPG (p = 0.78). Interestingly, the recovery of the G-NM-G adduct modestly increased in the presence of FPG over control (–FPG) (p = 0.01).
The recoveries of NM CT DNA adducts as a function of the different hydrolysis conditions are summarized in Table 3. The recoveries of cationic adducts, and NM-FapyG by the two stage neutral thermal hydrolysis at 37 °C followed by 95 °C are in excellent agreement to the level of adducts recovered by the FPG. In contrast, the amount of FapyG-NM-G recovered by treatment with FPG is about half the level recovered by two-stage neutral thermal hydrolysis. The FapyG-NM-G appears to be a poorer substrate for FPG than NM-FapyG, and the recovery of FapyG-NM-G did not go to completion at 44 h.
Table 3.
Mean Levels of NM Adduct for CT DNA as a Function of Neutral Thermal Hydrolysis Conditions a
| 95 °C (4 h) | 37 °C (72 h) adducts in supernatant | 95 °C (4 h) after 37 °C (72 h) | Sum of adducts 37 °C (72 h) and 95 °C (4 h) | FPG | |
|---|---|---|---|---|---|
| NM-G | 24.0 ± 1.6 | 33.8 ± 2.0 | 2.2 ± 0.4 | 36.0 ± 1.9 | 33.7 ± 3.0 |
| G-NM-G | 102 ± 3.0 | 75.7 ± 8.7 | 23.0 ± 4.8 | 98.7 ± 7.7 | 99.2 ± 12.3 |
| NM-FapyG | 23.1 ± 1.8 | 0.8 ± 0.1 | 6.1 ± 0.7 | 6.9 ± 0.7 | 8.6 ± 1.0 |
| FapyG-NM-G | 3.6 ± 0.3 | 0.3 ± 0.1 | 3.1 ± 0.6 | 3.4 ± 0.6 | 1.5 ± 0.2 |
3 independent measurements (Mean ± SD)
NM-DNA Adduct Formation in MB-231 Cells
Formation of the NM adduct was measured in MDA-MB-231 cells treated with NM (100 μM) for 24 h. DNA was hydrolyzed by neutral thermal hydrolysis (95 °C for 4 h, or 37 °C for 72 h, followed by hydrolysis of remaining NM adducts bound to DNA at 95 °C for 4 h). The results of NM DNA adduct formation are summarized in Figure 7, and chromatograms of DNA adducts of the untreated cells and NM treated cells as a function of hydrolysis conditions are shown in Supporting Figures S22-S23.
Figure 7.
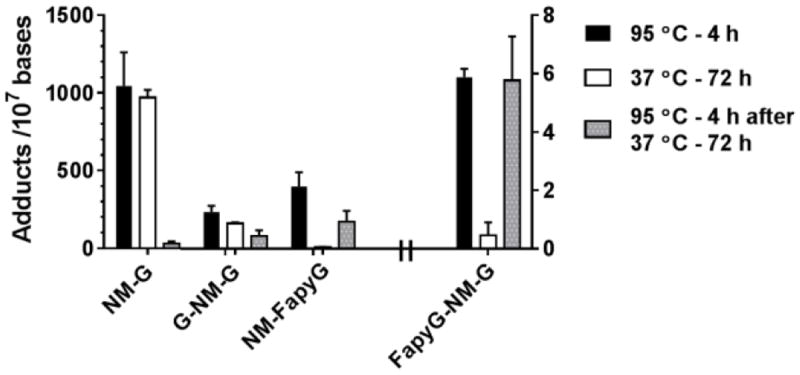
NM DNA adducts in MDA-MB-231 cells treated with NM (100 μM) for 24 h. Isolated DNA was hydrolyzed as described in Fig. 5 (N = 4 and reported as the average and SD of 2 independent experiments performed in duplicate). Levels of FapyG-NM-G are reported with the right Y scale, all other adducts are reported with the left Y scale
Similar to the data observed with CT DNA modified with NM, greater than 95% of the cationic NM-G adduct was depurinated from DNA over 72 h at 37 °C with little adduct retained to the DNA backbone. In contrast, only a trace of NM-FapyG was recovered in the supernatant following hydrolysis at 37 °C for 72 h with greater than 95% of this adduct being retained in the DNA backbone; NM-FapyG was recovered in the ensuing hydrolysis of DNA at 95 °C. The levels of NM-FapyG are about 20 – 25% of the level of NM-G formation, which is similar to the relative amounts observed in CT DNA treated with NM (1 μM). The G-NM-G cross-link was the major adduct formed with CT DNA, but the adduct levels were ~25% of the level of NM-G at this single time point in cellular DNA. The FapyG-NM-G cross-link was detected at a level of 6 adducts per 107 bases, or < 1% of the level of NM-G. Further studies are required to investigate the levels of NM DNA adducts formed as a function of dose and rate of adduct removal to understand relative levels of NM adduct formation and their persistence in cells and in vivo. These results demonstrate that both NM cross-links and FapyG adducts are formed at appreciable levels in nuclear DNA of MB-231 cells.
Discussion
The chemistry and reactivity of nitrogen8-10, 18, 20, 24, 47-56 and sulfur44, 57-64 mustards with nucleosides and DNA in vitro is well studied. There are also numerous studies that examine the mustard derived DNA adducts from treated cell culture and animals,46,48,50,54,56,59,64,65 including white blood cells of patients on chemotherapeutic regimens that include a nitrogen mustard.53, 57 A variety of methods have been used to identify these adducts including 32P-postlabeling,47, 58 co-chromatography by HPLC with authentic standards,8, 9, 24, 48, 57 and mass spectrometry.18, 20, 49-52, 56, 59, 64 Recently, tandem mass spectrometry with stable isotope dilution has been employed to identify and quantitate some of these DNA adducts.53, 54, 61-63 Collectively, the studies indicate that these electrophiles react predominantly at the N7 atom of dG, although N3-dA modification can also be significant. 66 The toxicity of mustard agents has been attributed to interstrand DNA cross-links. Interstrand cross-links inhibit DNA replication and transcription by preventing strand separation and thus, selectively induce toxicity and cell death of rapidly proliferating tumor cells over quiescent cells.11 The cationic N7-dG mono-adducts are generally believed to be benign since N7 does not participate in Watson-Crick base pairing; N7-adducts can undergo depurination and the cellular consequence of N7-dG alkylation is often attributed to the apurinic (AP) site or less abundant adducts that arise for reaction at other position, most notably O6-dG and N3-dA.67-69 However, some cationic N7-dG adducts can undergo ring-opening to produce stable N5-substituted Fapy-dG adducts.66 These lesions may persist in vivo, and contribute to the long term genotoxicity of alkylating agents.34, 35, 70 N5-Substituted FapyG adducts have not been extensively studied, unlike the unsubstituted counterpart derived from oxidative damage.71, 72
The scission of 8,9-bond of the purine ring of guanine to form a ring-opened Fapy adduct was first reported by Hems, who identified 2,6-diamino-4-hydroxy-5-formamidopyrmidine (FapyG) in which the formyl nitrogen is unsubstituted, as a major product formed after treatment of guanosine with ionizing radiation.73 Fapy-dG has been identified in oxidized DNA74 and also in liver of fish exposed to toxic chemicals.75 However, reports on the formation of other ring-opened N5-substituted FapyG adducts of genotoxic carcinogens in vivo are few. The major adduct of aflatoxin B1 (AFB1) is the cationic N7-G adduct formed by reaction of dG with AFB1 epoxide.34 A portion of N7-AFB1-G undergoes ring-opening to form the formamidopyrimidine derivative, AFB1–FapyG. This adduct persists in vivo and over time becomes the predominant AFB1-lesion in rodents.34 HPLC with radioactive detection was originally used to measure AFB1-N7-G and AFB1-FapyG from the DNA of livers of rodents treated with [3H]-AFB1. Both adducts were recently characterized by LC/MS2.76 One study reported the occurrence of N5-methyl-formamidopyrimidine (MeFapyG) formation in hepatic DNA of rats treated with radiolabeled N,N-dimethylnitrosamine (DMN), 1,2-dimethylhydrazine, or N-methyl-nitrosourea. The identity of the adduct as MeFapyG was based on HPLC co-chromatography of a synthetic standard with the radiolabeled adduct obtained from rat hepatic DNA.35, 77 However, a related study could not detect the MeFapyG lesion from the DNA of rodents treated with DMN.78 It was also reported that the C8-dG adduct of the carcinogenic aromatic amines 2-naphthylamine (2-NA) gave a related ring-opened product that was found to be a persistent lesion in urothelium but not in the liver of dogs given 2-NA.70
Previous studies of nitrogen and sulfur mustard adducts from their reaction with DNA in vitro, in cultured cells, or in vivo have concentrated on characterizing the N7-G and N3-A mono adducts and their cross-links. One study reported the identification of bis-N7-guanine cross-links in white blood cells of cancer patients receiving cyclophosphamide therapy,53 and sulfur mustard (bis-(2-chloroethyl)-sulfide, SM) also forms bis[2-(guanin-7-yl)ethyl]sulfide cross-links in tissues of rodents, and urine of rabbits exposed to SM.62, 63 The potential formation of ring-opened FapyG adducts was not addressed in those studies. Hoes et al identified the products from the reaction of melphalan with CT DNA by mass spectrometry.49 After the modification reaction and enzymatic hydrolysis, a product had a mass consistent with a 5’-p-(melphalan-FapyG)-p-C-3’ dinucleotide. Masses consistent with a melphalan-Fapy-dA adduct has also been reported.65, 79 However, no further characterization of these products was reported.
The labile N7-G and N3-A nitrogen and sulfur mustard adducts and their cross-links are often thermally hydolyzed from DNA under neutral8-10, 51, 57, 61 or acidic9, 10, 24, 46, 48, 50, 52, 53, 59 conditions. Alternatively, enzymatic digestion to the nucleosides has also been employed.18, 20, 47, 49, 54, 58, 65 and is necessary for DNA modifications that do not increase base lability (e.g., O6-dG adducts). Fapy nucleosides can exist as a number of slowly interconverting isomeric constituents such as α- and β-anomers, furnanose and hexanose forms of the deoxyribose, geometric isomers of the formamide group, and possible atroisomers.80 As a result, Fapy nucleoside adducts often exhibit poor chromatographic properties, which diminish the sensitivity of detection. The advantage of neutral thermal hydrolysis is that the labile adducts are selectively hydrolyzed while leaving the unmodified DNA in tact, allowing for its facile removal by precipitation and/or size exclusion filtration.
The data show the cationic NM-G and G-NM-G adducts can be converted to the NM-FapyG and Fapy-G-NM-G adducts at elevated temperature and neutral pH. The artifactual FapyG formation was mitigated by hydrolysis of cationic NM-G and G-NM-G adducts present in DNA under physiological conditions (pH 7.0, 37 °C, 72 h) prior to high temperature neutral thermal hydrolysis of the ring-opened FapyG adducts.
Our preliminary data show that NM forms high levels of DNA adducts in MDA-MB-231 cells. The relative amount of NM-FapyG to NM-G was comparable to that observed in vitro with CT DNA modified with NM. In contrast, the relative amount of G-NM-G to total NM DNA adducts was considerably lower in cells than in CT DNA. The lower amounts of G-NM-G formed in cells may be attributed to differences in reactivity of the NM in the nuclear genome where the DNA is packaged with histones versus commercial DNA, which has nearly all of protein removed during DNA isolation. Additionally, the cellular packaging of DNA is likely to prevent non-specific and inter-helix cross-linking, which may occur in the reaction of DNA in solution. We have also shown that the FapyG-NM-G cross-link is formed in cells and may contribute to the cytotoxicity of NM along with NM-G-NM.
Previously reported comparative quantitative measurements of cationic NM-G and its cross-linked adducts by MS are limited. Ganesan and Keating reported that the treatment of rat ovarian cells with phosphoramide mustard (6 μM) resulted in the formation of N-(2-hydroxyethyl)-N-[2-(N7-guaninyl)ethyl]-amine (NOR-G-OH) and the cross-linked adduct N,N-bis[2-(N7-guaninyl)-ethyl]amine (G-NOR-G) at levels of several adducts per 105 DNA bases.56 Wang et al reported that the SM (100 μM) also forms the structurally related N7-[2-[(2-hydroxyethyl)thio]ethyl]-guanine (N7-HETEG) and bis[2-(guanin-7-yl)ethyl]-sulfide (Bis-G), among other adducts, as the biomarkers for DNA damage in rodents and human keratinocytes, hepatocytes and lung fibroblasts.64 Very high levels N7-HETEG and G-SM-G cross-link (> 4 adducts per 105 bases) were formed in all cell lines. The t1/2 values of N7-HETEG and its cross-link showed no significant differences among the liver, lungs, and skin-derived cells. The relative amount of cross-link to N7-HETEG was >20% in all cell lines.
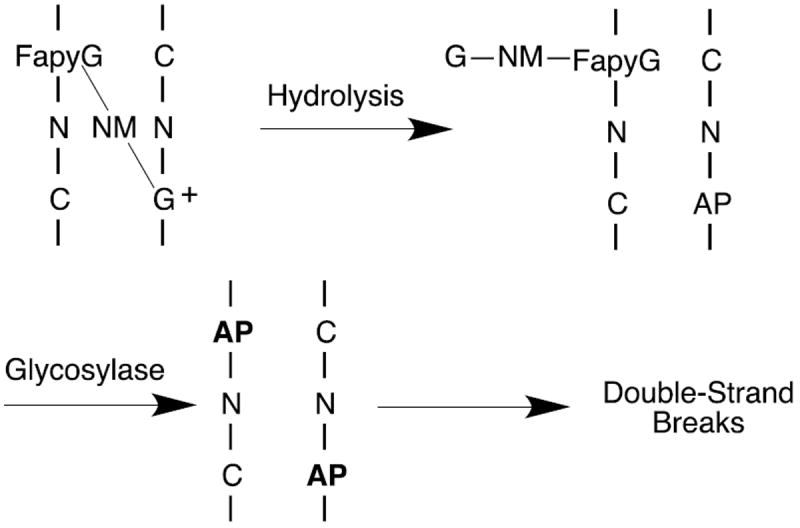
The NM-FapyG adduct was shown to be a good substrate for FPG.38 As expected, the NM-FapyG adduct was released when the NM-modified CT DNA was treated with FPG. The NM-FapyG level increased over the course of 72 h to 8.6 ± 1.0 adducts per 107 DNA bases, which compares favorably to the amount of adduct recovered under neutral thermal hydrolysis conditions. An unexpected result was that the level of the FapyG-NM-G cross-link adduct also increased over time to 1.5 ± 0.2 adducts per 107 bases after 72 h, indicating that this adduct is a substrate for FPG. The hydrolytic mechanism of glycosylases involves flipping the modified base out of the DNA helix and into the enzyme active site, which is likely to be inhibited by a cross-link. We therefore propose that the substrate for FPG is the Fapy-dG-NM-G mono-adduct where the cationic guanine has depurinated rather than the intact cross-link. In the case of interstrand cross-links, depurination of the cationic G followed by excision of the FapyG portion by a DNA glycosylase (e.g., NEIL1 or OGG1) could lead to highly cytotoxic double stand breaks (Scheme 2). The over-expression of FPG in human cultured cell was shown to have a modest (2-fold) protective effect against mafosfamide.30 While removal of the mafosfamide-FapyG mono-adduct is expected to be protective, repair of the much less abundant FapyG-mafosfamide-G adduct from hemi-depurinated interstrand cross-link would lead to double strand breaks and likely to enhance cyctotoxicity. Similarly, depurination of the bis-cationic N7-G cross-link followed by base excision repair of the hemi-depurinated G-NM-G adduct (e.g, N-methylpurine glycosylase, MPG) would also lead to double strand breaks. This is related to the observation that overexpression of MPG sensitized cells towards methylating agents.81 Defects or inhibition of homologous recombination has been shown to sensitize cells to mustard agents.82-84 An agent that could catalyze strand scission of an AP site would also be expected to enhance the therapeutic index of mustards and other DNA alkylating agents.85
Scheme 2.
Conclusions
We have developed an analytical protocol that allows simultaneous detection and quanititation of five dG adducts of a nitrogen mustard. The three FapyG adducts have not been fully characterized or previously quantitated, and two cross-links (FapyG-NM-G and FapyG-NM-FapyG) were previously unknown. The level of FapyG-NM-FapyG formed was observed at the LOD and its formation in cells is likely to be well below the LOD. NM-FapyG and FapyG-NM-G were observed in cultured MDA-MB-231 cells treated with NM suggesting that the contribution of these lesions to NM genotoxicity should be considered. With our validated analytical methodology, we will explore the relative contribution of ring-opened FapyG adducts of NM and their persistence in cell lines and experimental animals models. Such studies may provide further understanding about the contribution N5-substituted Fapy-dG lesions in the carcinogenicity of DNA alkylating agents and secondary tumor development from chemotherapeutic alkylating agents.
Supplementary Material
Synthesis and characterization (1H, 13C NMR, UV and HRMS) of synthetic standards. MS2 and MS3 scan stage of the labeled and unlabeled standards and proposed principal product ion assignments, and UPLC/MS3 analsyis of NM DNA adducts formed in MDA-MB-231 cells.
Acknowledgments
This work was supported by NIH grants P01 CA160032 (R.J.T and C.J.R), Center grants P30 CA77598 (R.J.T.), P30 CA068485 (C.J.R), and P30 ES00267 (C.J.R.).
Funding Sources
This work was supported by NIH grants P01 CA160032 (R.J.T and C.J.R), and Center grants P30 ES00267 (C.J.R.) and P30 CA068485 (C.J.R), Cancer Center Support grant no. CA-77598 (R.J.T.)
ABBREVIATIONS
- NM
bis-(2-chloroethyl)ethylamine
- CT DNA
calf thymus DNA
- Fapy
formamidopyrimidine
- FPG
formamidopyrimidine DNA glycosylase
- LOD
level of detection
- LOQ
level of quantitation
- CV
coefficient of variation
- SM
bis-(2-chloroethyl)sulfide
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information.
This material is available free of charge via the Internet at http://pubs.acs.org.”
The authors declare no competing financial interest.
References
- 1.Chabner BA, Roberts TG. Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 2.Emadi A, Jones RJ, Brodsky RA. Cyclophosphamide and cancer: Golden anniversary. Nat Rev Clin Oncol. 2009;6:638–647. doi: 10.1038/nrclinonc.2009.146. [DOI] [PubMed] [Google Scholar]
- 3.DeVita VT, Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68:8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- 4.Henne T, Schmähl D. Occurrence of second primary malignancies in man — a second look. Cancer Treat Rev. 1985;12:77–94. doi: 10.1016/0305-7372(85)90001-5. [DOI] [PubMed] [Google Scholar]
- 5.Povirk LF, Shuker DE. DNA damage and mutagenesis induced by nitrogen mustards. Mutat Res. 1994;318:205–226. doi: 10.1016/0165-1110(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 6.Rajski SR, Williams DM. DNA cross-linking agents as antitumor drugs. Chem Rev. 1998;98:2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 7.Michaelson-Richie ED, Ming X, Codreanu SG, Loeber RL, Liebler DC, Campbell C, Tretyakova NY. Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2011;10:2785–2796. doi: 10.1021/pr200042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osborne MR, Wilman DE, Lawley PD. Alkylation of DNA by the nitrogen mustard bis(2-chloroethyl)methylamine. Chem Res Toxicol. 1995;8:316–320. doi: 10.1021/tx00044a018. [DOI] [PubMed] [Google Scholar]
- 9.Osborne MR, Lawley PD. Alkylation of DNA by melphalan with special reference to adenine derivatives and adenine-guanine cross-linking. Chem Biol Interact. 1993;89:49–60. doi: 10.1016/0009-2797(93)03197-3. [DOI] [PubMed] [Google Scholar]
- 10.Balcome S, Park S, Quirk Dorr D, Hafner L, Phillips L, Tretyakova NY. Adenine-containing DNA-DNA cross-links of antitumor nitrogen mustards. Chem Res Toxicol. 2004;17:950–962. doi: 10.1021/tx0499463. [DOI] [PubMed] [Google Scholar]
- 11.Schärer OD. DNA interstrand crosslinks: Natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 12.Ojwang JO, Grueneberg DA, Loechler EL. Synthesis of a duplex oligonucleotide containing a nitrogen mustard interstrand DNA-DNA cross-link. Cancer Res. 1989;49:6529–6537. [PubMed] [Google Scholar]
- 13.Millard JT, Raucher S, Hopkins PB. Mechlorethamine cross-links deoxyguanosine residues at 5’-GNC sequences in duplex DNA fragments. J Am Chem Soc. 1990;112:2459–2460. [Google Scholar]
- 14.Brookes P, Lawley PD. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem J. 1961;80:496–503. doi: 10.1042/bj0800496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallama S, Hemminki K. Alkylation of guanosine by phosphoramide mustard, chloromethine hydrochloride and chlorambucil. Acta Pharmacol Toxicol. 1984;54:214–220. doi: 10.1111/j.1600-0773.1984.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 16.Kallama S, Hemminki K. Stabilities of 7-alkylguanosines and 7-deoxyguanosines formed by phosphoramide mustard and nitrogen mustard. Chem Biol Interact. 1986;57:85–96. doi: 10.1016/0009-2797(86)90051-7. [DOI] [PubMed] [Google Scholar]
- 17.Florea-Wang D, Haapala E, Mattinen J, Hakala K, Vilpo J, Hovinen J. Reactions of N,N-bis(2-chloroethyl)-p-aminophenylbutyric acid (chlorambucil) with 2’-deoxyadenosine. Chem Res Toxicol. 2003;16:403–408. doi: 10.1021/tx0256735. [DOI] [PubMed] [Google Scholar]
- 18.Florea-Wang D, Pawlowicz AJ, Sinkkonen J, Kronberg L, Vilpo J, Hovinen J. Reactions of 4-[bis(2-chloroethyl)amino]benzenebutanoic acid (Chlorambucil) with DNA. Chem Biodiversity. 2009;6:1002–1013. doi: 10.1002/cbdv.200800327. [DOI] [PubMed] [Google Scholar]
- 19.Haapala E, Hakala K, Jokipelto E, Vilpo J, Hovinen J. Reactions of N,N-bis(2-chloroethyl)-p-aminophenylbutyric acid (chlorambucil) with 2’-deoxyguanosine. Chem Res Toxicol. 2001;14:988–995. doi: 10.1021/tx000249u. [DOI] [PubMed] [Google Scholar]
- 20.Mohamed D, Mowaka S, Thomale J, Linscheid M. Chlorambucil-adducts in DNA analyzed at the oligonucleotide level using HPLC-ESI MS. Chem Res Toxicol. 2009;22:1435–1446. doi: 10.1021/tx900123r. [DOI] [PubMed] [Google Scholar]
- 21.Rojsitthisak P, Jongaroonngamsang N, Romero RM, Haworth IS. HPLC-UV, MALDI-TOF-MS and ESI-MS/MS analysis of the mechlorethamine DNA crosslink at a cytosine-cytosine mismatch pair. PLoS ONE. 2011;6:e20745. doi: 10.1371/journal.pone.0020745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta JR, Przybylski M, Ludlum DB. Alkylation of guanosine and deoxyguanosine by phosphoramide mustard. Cancer Res. 1980;40:4183–4186. [PubMed] [Google Scholar]
- 23.Chetsanga CJ, Polidori G, Mainwaring M. Analysis and excision of ring-opened phosphoramide mustard-deoxyguanine adducts in DNA. Cancer Res. 1982;42:2616–2621. [PubMed] [Google Scholar]
- 24.Hemminki K. DNA-binding products of nornitrogen mustard, a metabolite of cyclophosphamide. Chem Biol Interact. 1987;61:75–88. doi: 10.1016/0009-2797(87)90020-2. [DOI] [PubMed] [Google Scholar]
- 25.Hemminki K. Reactions of ethyleneimine with guanosine and deoxyguanosine. Chem Biol Interact. 1984;48:249–260. doi: 10.1016/0009-2797(84)90138-8. [DOI] [PubMed] [Google Scholar]
- 26.Hemminki K, Peltonen K, Vodicka P. Depurination from DNA of 7-methylguanine, 7-(2-aminoethyl)-guanine and ring-opened 7-methylguanines. Chem Biol Interact. 1989;70:289–303. doi: 10.1016/0009-2797(89)90051-3. [DOI] [PubMed] [Google Scholar]
- 27.Cussac C, Laval F. Reduction of the toxicity and mutagenicity of aziridine in mammalian cells harboring the Escherichia coli fpg gene. Nucleic Acids Res. 1996;24:1742–1746. doi: 10.1093/nar/24.9.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill R, Cussac C, Souhami R, Laval F. Increased resistance to N,N′,N″-triethylenethiophosphoramide (Thiotepa) in cells expressing the Escherichia coli formamidopyrimidine-DNA glycosylase. Cancer Res. 1996;56:3721–3724. [PubMed] [Google Scholar]
- 29.Kobune M, Xu Y, Baum C, Kelley MR, Williams DA. Retrovirus-mediated expression of the base excision repair proteins, formamidopyrimidine DNA glycosylase or human oxoguanine DNA glycosylase, protects hematopoietic cells from N, N′, N″-triethylenethiophosphoramide (thioTEPA)-induced toxicity in vitro and in vivo. Cancer Res. 2001;61:5116–5125. [PubMed] [Google Scholar]
- 30.Xu Y, Hansen WK, Rosenquist TA, Williams DA, Limp-Foster M, Kelley MR. Protection of mammalian cells against chemotherapeutic agents thiotepa, 1,3-N, N’-bis(2-chloroethyl)-N-nitrosourea, and mafosfamide using the DNA base excision repair genes Fpg and α-hOgg1: Implications for protective gene therapy applications. J Pharmacol Exp Ther. 2001;296:825–831. [PubMed] [Google Scholar]
- 31.He Y-H, Xu Y, Kobune M, Wu M, Kelley MR, Martin WJ. Escherichia coli FPG and human OGG1 reduce DNA damage and cytotoxicity by BCNU in human lung cells. Am J Physiol Lung Cell Mol Physiol. 2002;282:L50–55. doi: 10.1152/ajplung.00316.2001. [DOI] [PubMed] [Google Scholar]
- 32.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: Comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pande P, Haraguchi K, Jiang YL, Greenberg MM, Basu AK. Unlike catalyzing error-free bypass of 8-oxodGuo, DNA polymerase λ Is responsible for a significant part of Fapy·dG-Induced G → T mutations in human cells. Biochemistry. 2015;54:1859–1862. doi: 10.1021/acs.biochem.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Essigmann JM, Croy RG, Bennett RA, Wogan GN. Metabolic activation of aflatoxin B1: Patterns of DNA adduct formation, removal, and excretion in relation to carcinogenesis. Drug Metab Rev. 1982;13:581–602. doi: 10.3109/03602538209011088. [DOI] [PubMed] [Google Scholar]
- 35.Kadlubar FF, Beranek DT, Weis CC, Evans FE, Cox R, Irving CC. Characterization of the purine ring-opened 7-methylguanine and its persistence in rat bladder epithelial DNA after treatment with the carcinogen N-methylnitrosourea. Carcinogenesis. 1984;5:587–592. doi: 10.1093/carcin/5.5.587. [DOI] [PubMed] [Google Scholar]
- 36.Lin YC, Makarova AV, Burgers PM, Stone MP, Lloyd RS. Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis. 2014;357 doi: 10.1093/carcin/bgu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Earley LF, Minko IG, Christov PP, Rizzo CJ, Lloyd RS. Mutagenic spectra arising from replication bypass of the 2,6-diamino-4-hydroxy-N5-methyl formamidopyrimidine adduct in primate cells. Chem Res Toxicol. 2013;26:1108–1114. doi: 10.1021/tx4001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christov PP, Son KJ, Rizzo CJ. Synthesis and characterization of oligonucleotides containing a nitrogen mustard formamidopyrimidine monoadduct of deoxyguanosine. Chem Res Toxicol. 2014;27:1610–1618. doi: 10.1021/tx5002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tretyakova N, Goggin M, Sangaraju D, Janis G. Quantitation of DNA adducts by stable isotope dilution mass spectrometry. Chem Res Toxicol. 2012;25:2007–2035. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen EJ, Reipa V, Watson SS, Stanley DL, Rabb SA, Nelson BC. DNA damaging potential of photoactivated P25 titanium dioxide nanoparticles. Chem Res Toxicol. 2014;27:1877–1888. doi: 10.1021/tx500340v. [DOI] [PubMed] [Google Scholar]
- 41.Atha DH, Wang H, Petersen EJ, Cleveland D, Holbrook RD, Jaruga P, Dizdaroglu M, Xing B, Nelson BC. Copper oxide nanoparticle mediated DNA damage in terrestrial plant models. Environ Sci Technol. 2012;46:1819–1827. doi: 10.1021/es202660k. [DOI] [PubMed] [Google Scholar]
- 42.Magnusson B, Örnemark U. Eurachem Guide: The Fitness for Purpose of Analytical Methods - A Laboratory Guide to Method Validation and Related Topics. 2. 2014. Available from http://www.eurachem.org.
- 43.MacDougall D, Amore FJ, Cox GV, Crosby DG, Estes FL, Freeman DH, Gibbs WE, Gordon GE, Keith LH, Lal J, Langner RR, McClelland NI, Phillips WF, Pojasek RB, Sievers RE. Guidelines for data acquistion and data quality evaluation in environmental chemistry. Anal Chem. 1980;52:2242–2249. [Google Scholar]
- 44.Bauer GB, Povirk LF. Specificity and kinetics of interstrand and intrastrand bifunctional alkylation by nitrogen mustards at a G-G-C sequence. Nucleic Acids Res. 1997;25:1211–1218. doi: 10.1093/nar/25.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodenough AK, Schut HAJ, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2’-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemminki K. Binding of metabolites of cyclophosphamide to DNA in a rat liver microsomal system and in vivo in mice. Cancer Res. 1985;45:4237–4243. [PubMed] [Google Scholar]
- 47.Zhou GH, Teicher BA, Frei E. Postlabeling detection of DNA adducts of antitumor alkylating agents. Cancer Chemother Pharmacol. 1996;38:71–80. doi: 10.1007/s002800050450. [DOI] [PubMed] [Google Scholar]
- 48.Benson AJ, Martin CN, Garner RC. N-(2-Hydroxyethyl)-N-[2-(7-guaninyl)ethyl]amine, the putative major DNA adduct of cyclophosphamide in vitro and in vivo in the rat. Biochem Pharmacol. 1988;37:2979–2985. doi: 10.1016/0006-2952(88)90285-7. [DOI] [PubMed] [Google Scholar]
- 49.Hoes I, Lemière F, Van Dongen W, Vanhoutte K, Esmans EL, Van Bockstaele D, Berneman Z, Deforce D, Van den Eeckhout EG. Analysis of melphalan adducts of 2’-deoxynucleotides in calf thymus DNA hydrolysates by capillary high-performance liquid chromatography-electrospray tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 1999;736:43–59. doi: 10.1016/s0378-4347(99)00422-3. [DOI] [PubMed] [Google Scholar]
- 50.Mirkes PE, Brown NA, Kajbaf M, Lamb JH, Farmer PB, Naylor S. Identification of cyclophosphamide-DNA adducts in rat embryos exposed in vitro to 4-hydroperoxycyclophosphamide. Chem Res Toxicol. 1992;5:382–385. doi: 10.1021/tx00027a010. [DOI] [PubMed] [Google Scholar]
- 51.Sperry ML, Skanchy D, Marino MT. High-performance liquid chromatographic determination of N-[2-(hydroxyethyl)-N-(2-(7-guaninyl)ethyl)]methylamine, a reaction product between nitrogen mustard and DNA and its application to biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 1998;716:187–193. doi: 10.1016/s0378-4347(98)00296-5. [DOI] [PubMed] [Google Scholar]
- 52.Cushnir JR, Naylor S, Lamb JH, Farmer PB, Brown NA, Mirkes PE. Identification of phosphoramide mustard/DNA adducts using tandem mass spectrometry. Rapid Commun Mass Spectrom. 1990;4:410–414. doi: 10.1002/rcm.1290041014. [DOI] [PubMed] [Google Scholar]
- 53.Malayappan B, Johnson L, Nie B, Panchal D, Matter B, Jacobson P, Tretyakova NY. Quantitative high-performance liquid chromatography-electrospray ionization tandem mass spectrometry analysis of bis-N7-guanine DNA-DNA cross-links in white blood cells of cancer patients receiving cyclophosphamide therapy. Anal Chem. 2010 doi: 10.1021/ac902923s. [DOI] [PubMed] [Google Scholar]
- 54.Van den Driessche B, Esmans EL, Van der Linden A, van Dongen W, Schaerlaken E, Lemière F, Witters E, Berneman Z. First results of a quantitative study of DNA adducts of melphalan in the rat by isotope dilution mass spectrometry using capillary liquid chromatography coupled to electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:1999–2004. doi: 10.1002/rcm.2018. [DOI] [PubMed] [Google Scholar]
- 55.Osborne MR, Lawley PD, Crofton-Sleigh C, Warren W. Products from alkylation of DNA in cells by melphalan: Human soft tissue sarcoma cell line RD and Escherichia coli WP2. Chem Biol Interact. 1995;97:287–296. doi: 10.1016/0009-2797(95)03623-t. [DOI] [PubMed] [Google Scholar]
- 56.Ganesan S, Keating AF. Phosphoramide mustard exposure induces DNA adduct formation and the DNA damage repair response in rat ovarian granulosa cells. Toxicol Appl Pharmacol. 2015;282:252–258. doi: 10.1016/j.taap.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fidder A, Moes GW, Scheffer AG, van der Schans GP, Baan RA, de Jong LP, Benschop HP. Synthesis, characterization, and quantitation of the major adducts formed between sulfur mustard and DNA of calf thymus and human blood. Chem Res Toxicol. 1994;7:199–204. doi: 10.1021/tx00038a013. [DOI] [PubMed] [Google Scholar]
- 58.Niu T-q, Matijasevic Z, Austin-Ritchie P, Stering A, Ludlum DB. A 32P-postlabeling method for the detection of adducts in the DNA of human fibroblasts exposed to sulfur mustard. Chem Biol Interact. 1996;100:77–84. doi: 10.1016/s0009-2797(96)03690-3. [DOI] [PubMed] [Google Scholar]
- 59.Ludlum DB, Austin-Ritchie P, Hagopian M, Niu T-Q, Yu D. Detection of sulfur mustard-induced DNA modifications. Chem Biol Interact. 1994;91:39–49. doi: 10.1016/0009-2797(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 60.Fidder A, Noort D, de Jong LPA, Benschop HP, Hulst AG. N7-(2-Hydroxyethylthioethyl)-guanine: A novel urinary metabolite following exposure to sulphur mustard. Arch Toxicol. 1996;70:854–855. doi: 10.1007/s002040050350. [DOI] [PubMed] [Google Scholar]
- 61.Yue L, Wei Y, Chen J, Shi H, Liu Q, Zhang Y, He J, Guo L, Zhang T, Xie J, Peng S. Abundance of four sulfur mustard-DNA adducts ex vivo and in vivo revealed by simultaneous quantification in stable isotope dilution-ultrahigh performance liquid chromatography-tandem mass spectrometry. Chem Res Toxicol. 2014;27:490–500. doi: 10.1021/tx4003403. [DOI] [PubMed] [Google Scholar]
- 62.Yue L, Zhang Y, Chen J, Zhao Z, Liu Q, Wu R, Guo L, He J, Zhao J, Xie J, Peng S. Distribution of DNA adducts and corresponding tissue damage of Sprague-Dawley rats with percutaneous exposure to sulfur mustard. Chem Res Toxicol. 2015;28:532–540. doi: 10.1021/tx5004886. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Yue L, Nie Z, Chen J, Guo L, Wu B, Feng J, Liu Q, Xie J. Simultaneous determination of four sulfur mustard-DNA adducts in rabbit urine after dermal exposure by isotope-dilution liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;961:29–35. doi: 10.1016/j.jchromb.2014.04.050. [DOI] [PubMed] [Google Scholar]
- 64.Wang P, Zhang Y, Chen J, Guo L, Xu B, Wang L, Xu H, Xie J. Analysis of Different Fates of DNA Adducts in Adipocytes Post-sulfur Mustard Exposure in Vitro and in Vivo Using a Simultaneous UPLC-MS/MS Quantification Method. Chem Res Toxicol. 2015;28:1224–1233. doi: 10.1021/acs.chemrestox.5b00055. [DOI] [PubMed] [Google Scholar]
- 65.Edler M, Jakubowski N, Linscheid M. Quantitative determination of melphalan DNA adducts using HPLC-inductively coupled mass spectrometry. J Mass Spectrom. 2006;41:507–516. doi: 10.1002/jms.1009. [DOI] [PubMed] [Google Scholar]
- 66.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 67.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. The formation and biological significance of N7-guanine adducts. Mutat Res. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bignami M, O’Driscoll M, Aquilina G, Karran P. Unmasking a killer: DNA O6-methylguanine and the cytotoxicity of methylating agents. Mutat Res. 2000;462:71–82. doi: 10.1016/s1383-5742(00)00016-8. [DOI] [PubMed] [Google Scholar]
- 69.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kadlubar FF, Anson JF, Dooley KL, Beland FA. Formation of urothelial and hepatic DNA adducts from carcinogen 2-naphthylamine. Carcinogenesis. 1981;2:467–470. doi: 10.1093/carcin/2.5.467. [DOI] [PubMed] [Google Scholar]
- 71.Greenberg MM. The formamidopyrimidines: Purine lesions formed in competition with 8-oxopurines from oxidative stress. Acc Chem Res. 2012;45:588–597. doi: 10.1021/ar2002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dizdaroglu M, Kirkal iG, Jaruga P. Formamidopyrimidines in DNA: Mechanisms of formation, repair, and biological effects. Free Radic Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Hems G. Effect of ionizing radiation on aqueous solutions of guanylic acid and guanosine. Nature. 1958;181:1721–1722. doi: 10.1038/1811721a0. [DOI] [PubMed] [Google Scholar]
- 74.Karakaya A, Jaruga P, Bohr VA, Grollman AP, Dizdaroglu M. Kinetics of excision of purine lesions from DNA by Escherichia coli Fpg protein. Nucleic Acids Res. 1997;25:474–479. doi: 10.1093/nar/25.3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malins DC, Gunselman SJ. Fourier-transform infrared spectroscopy and gas chromatography-mass spectrometry reveal a remarkable degree of structural damage in the DNA of wild fish exposed to toxic chemicals. Proc Natl Acad Sci U S A. 1994;91:13038–13041. doi: 10.1073/pnas.91.26.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chawanthayatham S, Thiantanawat A, Egner PA, Groopman JD, Wogan GN, Croy RG, Essigmann JM. Prenatal exposure of mice to the human liver carcinogen aflatoxin B1 reveals a critical window of susceptibility to genetic change. Int J Cancer. 2015;136:1254–1262. doi: 10.1002/ijc.29102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beranek DT, Weis CC, Evans FE, Chetsanga CJ, Kadlubar FF. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983;110:625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- 78.Den Engelse L, Menkveld GJ, De Brij RJ, Tates AD. Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis. 1986;7:393–403. doi: 10.1093/carcin/7.3.393. [DOI] [PubMed] [Google Scholar]
- 79.Hoes I, Van Dongena W, Lemiere F, Esmansa EL, Van Bockstaeleb D, Berneman ZN. Comparison between capillary and nano liquid chromatography– electrospray mass spectrometry for the analysis of minor DNA– melphalan adducts. J Chromatogr B Analyt Technol Biomed Life Sci. 2000;748:197–212. doi: 10.1016/s0378-4347(00)00400-x. [DOI] [PubMed] [Google Scholar]
- 80.Brown KL, Deng JZ, Iyer RS, Iyer LG, Voehler MW, Stone MP, Harris CM, Harris TM. Unraveling the aflatoxin-FAPY conundrum: Structural basis for differential replicative processing of isomeric forms of the formamidopyrimidine-type DNA adduct of aflatoxin B1. J Am Chem Soc. 2006;128:15188–15199. doi: 10.1021/ja063781y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rinne ML, He Y-H, Pachkowski BF, Nakamura J, Kelley MR. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Res. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amrein L, Loignon M, Goulet A-C, Dunn M, Jean-Claude B, Aloyz R, Panasci L. Chlorambucil cytotoxicity in malignant B lymphocytes is synergistically increased by 2-(morpholin-4-yl)-benzo[h]chomen-4-one (NU7026)-mediated inhibition of DNA double-strand break repair via inhibition of DNA-dependent protein kinase. J Pharmacol Exp Ther. 2004;321:848–855. doi: 10.1124/jpet.106.118356. [DOI] [PubMed] [Google Scholar]
- 83.Jowsey PA, Williams FM, Blain PG. The role of homologous recombination in the cellular response to sulphur mustard. Toxicol Lett. 2010;197:12–18. doi: 10.1016/j.toxlet.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 84.De Silva I, McHugh P, Clingen P, Hartley J. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7890. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fkyerat A, Demeunynck M, Constant JF, Michon P, Lhomme J. A new class of artificial nucleases that recognize and cleave apurinic sites in DNA with great selectivity and efficiency. J Am Chem Soc. 1993;115:9952–9959. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synthesis and characterization (1H, 13C NMR, UV and HRMS) of synthetic standards. MS2 and MS3 scan stage of the labeled and unlabeled standards and proposed principal product ion assignments, and UPLC/MS3 analsyis of NM DNA adducts formed in MDA-MB-231 cells.


