Abstract
Plant regeneration via in vitro culture can induce genetic and epigenetic variation; however, the extent of such changes in triticale is not yet understood. In the present study, metAFLP, a variation of methylation-sensitive amplified fragment length polymorphism analysis, was used to investigate tissue culture-induced variation in triticale regenerants derived from four distinct genotypes using androgenesis and somatic embryogenesis. The metAFLP technique enabled identification of both sequence and DNA methylation pattern changes in a single experiment. Moreover, it was possible to quantify subtle effects such as sequence variation, demethylation, and de novo methylation, which affected 19, 5.5, 4.5 % of sites, respectively. Comparison of variation in different genotypes and with different in vitro regeneration approaches demonstrated that both the culture technique and genetic background of donor plants affected tissue culture-induced variation. The results showed that the metAFLP approach could be used for quantification of tissue culture-induced variation and provided direct evidence that in vitro plant regeneration could cause genetic and epigenetic variation.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-015-0368-0) contains supplementary material, which is available to authorized users.
Keywords: Androgenesis, Demethylation, De novo methylation, Doubled haploid, In vitro culture, Sequence variation, Somatic embryogenesis
Introduction
Triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) is a fertile amphiploid derived from chromosome doubling of hybrids between two genera Triticum and Secale. Hexaploid forms have 42 chromosomes: 28 from wheat and 14 from rye (Secale cereale L.). The wheat complement of primary synthetic forms contained A and B genomes of durum wheat (Triticum durum Desf.) or other tetraploid species; in the contemporary cultivars, classified as the secondary triticales, majority of wheat chromosomes have been introgressed from hexaploid common wheat (Triticum aestivum L.). Triticale was artificially created with the aim of combining the productivity of wheat with the hardiness of rye (McGoverin et al. 2011). Triticale has excellent tolerance to water-limitation and salinity stress and displays high mineral efficiency (Blum 2014). Triticale is currently used for food, animal feed, and biofuel production (Hills et al. 2007), and is suitable for erosion control and as a cover crop (Ramirez-Garcia et al. 2015). The expansion of triticale cultivation has increased the need for improved breeding methodologies. One such technique is in vitro culture plant regeneration, which can provide breeders with homozygous lines (doubled haploids; DHs) in a single generation. DHs can be used for hybrid breeding, which is one of the most promising avenues for triticale improvement (Oettler et al. 2005). However, use of tissue culture can result in tissue culture-induced variation (TCIV) in regenerants (Dennis et al. 1987; Kaeppler and Phillips 1993a, b; Olhoft and Phillips 1994; Kaeppler et al. 1998) or somaclonal variation in progeny of the in vitro regenerated plants (Lorz and Scowcroft 1983; Larkin et al. 1984; Barwale and Widholm 1987; Breiman et al. 1987; Zehr et al. 1987; Bernardi et al. 1999; Kirikovich et al. 2003). Plant uniformity in in vitro regenerants and their progeny is compromised as a result of DNA methylation changes, cytological aberrations, transposon activation, and genomic variation (Phillips et al. 1994; Bednarek et al. 2007; Guo et al. 2007; Li et al. 2007; Ngezahayo et al. 2009; Baránek et al. 2010; Dann and Wilson 2011; Stroud et al. 2013; Wang et al. 2013; Zhang et al. 2014). Genetic and epigenetic changes induced at the DNA level due to plant tissue culture manipulation have been analyzed using a range of molecular markers (Polanco and MaL 2002; Xu et al. 2004; Smykal et al. 2007). More recently, the development and refinement of the metAFLP approach (Bednarek et al. 2007) allowed simultaneous quantification of sequence changes and DNA methylation patterns. The metAFLP method employs two isoschizomers, KpnI and Acc65I, which differ in their sensitivity towards recognition site DNA methylation. KpnI is insensitive and Acc65I is sensitive to restriction site methylation. This results in methylation-dependent differences in the amplified fragment length polymorphism (AFLP profiles) produced by the two enzymes, whereas sequence variation could be revealed based on the KpnI/MseI platform. AFLP profiles produced by metAFLP can be used for the identification of TCIV events and calculation of their quantitative characteristics. This technique has been used in a number of species to date, such as Hordeum vulgare (Bednarek et al. 2007), Gentiana pannonica (Fiuk et al. 2010), Phyllostachys praecox (Lu et al. 2012), and Poa annua (Chwedorzewska and Bednarek 2012). The ability of the extended metAFLP approach to characterize sequence changes as well as DNA methylation pattern alterations was demonstrated on limited triticale materials (Machczyńska et al. 2014a). Recently, global DNA methylation changes in triticale were analyzed using an RP-HPLC approach (Machczyńska et al. 2014b). This showed that global DNA methylation decreased in regenerants relative to the donor plants, but then increased in regenerant progeny. While RP-HPLC can be used for the analysis of global DNA methylation (Mankessi et al. 2011; Teyssier et al. 2013), this approach is not adequate for estimating subtle effects such as de novo methylation, demethylation, and sequence mutations that occur during in vitro plant differentiation and dedifferentiation (Zhang et al. 2010). It is not clear to what extent the metAFLP approach can identify the methylation changes identified by RP-HPLC or whether comparable estimates of similar characteristics, such as genome methylation (GM) and global DNA methylation, are derived via the two approaches independently.
The aim of this study was to use metAFLP to quantify TCIV in several triticale donor-regenerant sets developed using different androgenesis and somatic embryogenesis processes. In addition, metAFLP and RP-HPLC methods of GM and global DNA methylation were compared.
Materials and methods
Plant material
Four genotypes of DH regenerants derived from isolated microspores of triticale (Oleszczuk et al. 2004) were extracted from randomly chosen plants of the partly heterogeneous cv. Bogo (as it originated as a double cross) and were cloned by partitioning plant clumps after tillering. These served as explants for plant production via androgenesis in shed microspore culture (M) and anther culture (A), and by somatic embryogenesis from immature zygotic embryo culture (E) (Machczyńska et al. 2014b). Cloned individuals were kept in a growth chamber at a photoperiod of 16/8 h day/night at 16/12 °C to allow tillering. The same procedure (tillering and partitioning) was repeated every 2 weeks. Chosen cloned individuals (later called ‘donors’) of the four genotypes, their DH androgenic regenerants and homozygous regenerants derived from E, constituted the four sets (Si, where i = successive genotypes from 1 to 4) (Table 1).
Table 1.
Triticosecale Wittmack cv. Bogo plant material used for metAFLP analysis
| Set | Donors | Regenerants | ||
|---|---|---|---|---|
| Si | DiCn | RA | RM | RE |
| S1 | 9 | 25 | 12 | 25 |
| S2 | 8 | 20 | 8 | 15 |
| S3 | 12 | 22 | 5 | 15 |
| S4 | 10 | 12 | 12 | 17 |
| Total | 39 | 79 | 37 | 72 |
Si represents four sets (i = 1–4); DiCn indicates the number of donors used for in vitro plant regeneration, where Di indicates donor DH genotype (i = 1–4) and Cn indicates cloned individuals; RA represents regenerants derived from anther cultures; RM represents regenerants derived from shed microspore cultures; and RE represents regenerants derived from immature zygotic embryo cultures
MetAFLP procedure
DNA was extracted from fresh leaves of donor plants and their regenerants at the same developmental stage (flag leaf emerging) using a DNeasy Plant Mini Kit (Qiagen). DNA samples were characterized spectrophotometrically following verification of integrity and purity on 1.2 % agarose gels with ethidium bromide staining.
MetAFLP was performed as described elsewhere (Bednarek et al. 2007). Following adapter ligation, pre-selection, and selective amplification steps, samples were digested with Acc65I/MseI and KpnI/MseI endonucleases pairs (37 °C for 3 h, followed by 70 °C at 15 min). The arrangement of adapter and primer sequences is presented in Online Resource 1. PCR products were separated on a 7 % polyacrylamide gel. The metAFLP profiles for the Acc65I/MseI and KpnI/MseI AFLP platforms were scored as ‘1’ (the presence of a band) and ‘0’ (absence), and arranged in a form of a binary juxtaposed matrix.
MetAFLP characteristics
Theoretically, sixteen four-digit binary codes were possible (Bednarek et al. 2007). The first and third positions of the binary code indicated the presence or absence of a marker in the AFLP profile of a donor plant (D) digested with Acc65I/MseI and KpnI/MseI, respectively. The second and fourth positions reflected the same situation but for the regenerant (R). Four-digit binary codes were grouped into various events reflecting the different genetic background of those events. Genetic background sequence, demethylation, de novo methylation, and complex events were distinguished using the binary code and these reflected total tissue culture-induced events. Sites with non-methylated and methylated status in D and R were also identified. Sequence (SE), demethylation (DME), de novo methylation (DNME), and complex (CE) events were converted into sequence (SV), demethylation (DMV), de novo methylation (DNMV), and complex (CV) variation percentages using previously described formulae (Machczyńska et al. 2014a). Complex variation consisted of SV, DMV, and DNMV. These types of variation were extracted from CV and added to SV, DMV, and DNMV as a correction. All types of variation taken together described total tissue culture-induced variation (TTCIV). The metAFLP approach allowed quantification of non-methylated (SNMS) and methylated (SMS) sites. SNMS sites were those that remained non-methylated in donors (D) and regenerants (R), as well as those that underwent demethylation in regenerants. Sites that were methylated in D and R plus those that were de novo methylated in R were classified as SMS. Sites affected by methylation (SAM) were calculated as the sum of DNMV and DMV. Finally, metAFLP was used to evaluate the percentage of global genome restriction sites that were methylated in regenerants (GM). GM was defined as the sum of DNME and SMS divided by the sum of DNME, DME, SMS, and SNMS multiplied by 100. Detailed information regarding the quantitative characteristics of the metAFLP approach are published elsewhere (Machczyńska et al. 2014a).
Global DNA methylation evaluation using RP-HPLC
Briefly, 6 µg of DNA from each sample was enzymatically hydrolyzed to nucleotides using P1 nuclease and then dephosphorylated with alkaline phosphatase. The reaction mixture was centrifuged at 12,000 rpm for 5 min and used for reversed-phase liquid chromatography (RP-HPLC) analysis (Machczyńska et al. 2014b). A Waters 625 LC Chromatography System connected to a Millennium 32v 4.0 data processing station was used for nucleoside separation. RP-HPLC analysis was based on the protocol described by Johnston et al. (2005). Two eluents were used (eluent A: 0.5 % methanol in 10 mMKH2PO4 (v/v), and eluent B: 10 % methanol in 10 mM KH2PO4) with a linear gradient of 10 min of 100 % A and 100 % B, and 15 min with 100 % B and 100 % A, with 5 min of total running time. The percentage of 5mdC (5-methyldeoxycytidine) was quantified as total 5mdC content divided by the sum of 5mdC and dC (deoxycytidine) multiplied by 100. Three analytical measurements were performed for each DNA sample.
Data analysis
Mean and standard deviation (SD) values were calculated for metAFLP characteristics irrespective of sets or the in vitro tissue culture plant regeneration approach used. MetAFLP mean characteristics were also independently evaluated for all genotype sets and regenerants derived via anther culture, shed microspore culture, and immature zygotic embryo culture.
UPGMA
PAST software was used to analyze the metAFLP profiles of cloned individuals by UPGMA using Jaccard’s coefficient with 1000 bootstraps to estimate the robustness of the branches (Hammer et al. 2001).
ANOVA
R CRAN software was used for one-way ANOVA analysis with Tukey’s test to evaluate the differences between datasets. To avoid data correlation, sets were analyzed using uncorrelated metAFLP characteristics. The overall differences between the four sets as well as differences related to the in vitro tissue culture regeneration approaches and in vitro-induced variation characteristics were regarded as significant at a probability level of p ≤ 0.05 and α = 0.01.
Pearson correlation
Pearson correlation analysis was conducted for GM of metAFLP and global DNA methylation of RP-HPLC regenerant data (Machczyńska et al. 2014b) using the SAS statistical package (SAS Institute Inc 2004).
Results
Uniformity of doubled haploid cloned individuals
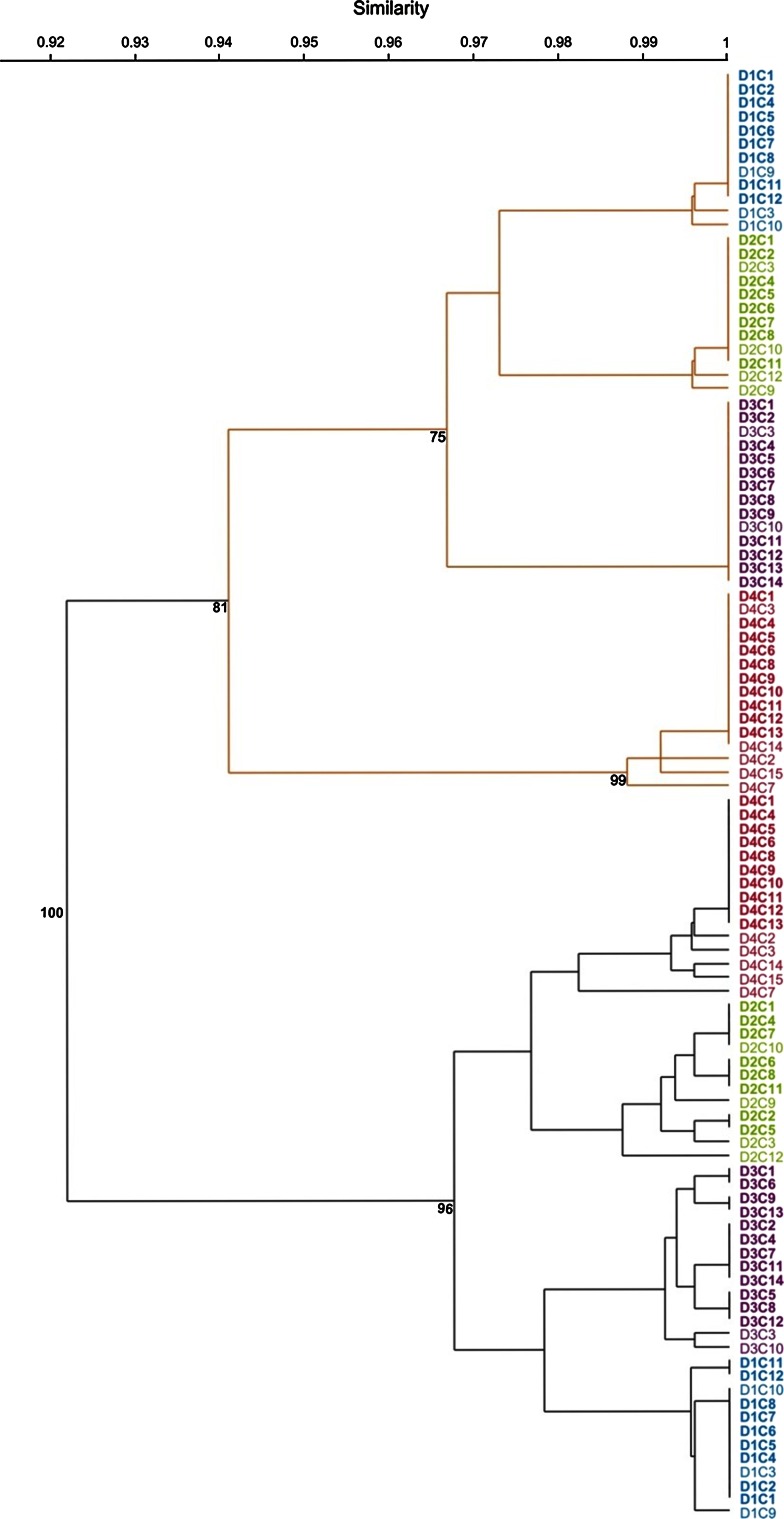
In total, 53 cloned individuals with no obvious morphological differences representing four distinct DH genotypes of cv. Bogo (DiCn, where Di represents donor DH genotype (i = 1–4) and Cn represents cloned individuals) were obtained. Cluster analysis based on 2720 metAFLP markers amplified with 14 selective primer pairs distinguished two separate data groups that reflected the Acc65I/MseI and KpnI/MseI digests (Fig. 1). Grouping based on Acc65I/MseI-derived markers placed the D1C and D3C individuals in one group and D2C and D4C individuals in a second cluster. The KpnI/MseI metAFLP platform grouped D1C and D2C together, while D3C and D4C formed two subclusters with D4C being most distinct from the others. The genetic distances between individuals of the same genotype were 0.012 and 0.018 for the KpnI/MseI and Acc65I/MseI platforms, respectively. In total, 9, 8, 12, and 10 genetically and epigenetically uniform cloned DH individuals (donors) representing four genotypes (D1, D2, D3, and D4) were produced and used for in vitro regeneration.
Fig. 1.
Clustering (UPGMA and Jaccard’s similarity index) of metAFLP data from 53 cloned individuals. Each individual is coded DiCn, where Di represents donor DH genotype (i = 1–4) and Cn represents cloned individuals. The lower (black) cluster reflects Acc65I/MseI data and the upper (orange) cluster reflects KpnI/MseI data. Cloned individuals of the same genotype are marked with the same color. Cloned individuals used for in vitro plant regeneration are shown in bold. Bootstrap values are indicated at the nodes
Molecular characteristics of regenerants derived from uniform donor plants
Donor plants were a source of explant tissue for regeneration of 79, 37, and 72 plants via A, M, and E methods, respectively (Table 1). Spontaneously doubled androgenesis-derived regenerants and regenerants derived via immature zygotic embryos exhibited no apparent morphological differences compared to the donors.
Amplification using 14 selective primer combinations was performed on DNA samples from donors and their regenerants from all four genotype sets. In total, 2720 bands were produced with an average of 49 products per primer combination. The highest number of amplified fragments was observed for the CpXpG AGA/M CAA and the lowest was for the CpG GCA/M CGC primer combinations. In total, there were 1429 polymorphic and 1291 monomorphic fragments in Acc65I/MseI, and 1172 and 1544 in KpnI/MseI, respectively (Online Resource 2). When all genotype sets were considered, a total of 1072 polymorphic and 935 monomorphic fragments were shared between the Acc65I/MseI and KpnI/MseI digests.
MetAFLP results
MetAFLP marker data were converted to four-digit binary codes. Codes (1111) related to non-methylated sites in donor and regenerant were most abundant (range 1865–12181, depending upon set and in vitro tissue culture regeneration method) (Online Resource 3). The less frequent events were those encoded as 0110 (range 22–95) and classified as demethylation and sequence events.
Calculation of the mean values of the metAFLP characteristics without separate consideration of the in vitro tissue culture regeneration methods or genotype sets used showed that 19.16, 5.48, 4.48, and 29.12 % of sites changed with respect to SV, DMV, DNMV, and TTCIV, respectively (Table 2). Up to 61.06 % of sites had non-methylated status in donors and regenerants (SNMS), and up to 4.33 % of sites were methylated (SMS). The percentage of SAM (10.59 %) was lower than the level of GM (12.19 %), as determined from methylation of the restriction sites and their vicinities.
Table 2.
Mean quantitative metAFLP characteristics for different genotype sets and in vitro tissue culture regeneration methods
| MetAFLP quantitative characteristics | Si | RA (%) | RM (%) | RE (%) | Mean (all regeneration approaches) (%) |
|---|---|---|---|---|---|
| Sequence variation (SV) | S1 | 11.02 ± 0.93 | 11.62 ± 1.62 | 11.49 ± 0.62 | 11.38 ± 1.18 |
| S2 | 19.73 ± 1.14 | 18.55 ± 1 | 19.88 ± 0.75 | 19.64 ± 1.04 | |
| S3 | 26.37 ± 0.76 | 25.76 ± 0.82 | 21.45 ± 0.82 | 24.54 ± 2.26 | |
| S4 | 20.87 ± 0.9 | 20.67 ± 0.92 | 21.56 ± 0.74 | 21.10 ± 0.9 | |
| Mean (all sets) | 19.5 ± 5.51 | 19.15 ± 5.19 | 18.6 ± 4.35 | 19.16 ± 5.21 | |
| Demethylation (DMV) | S1 | 4.5 ± 0.54 | 4.07 ± 0.6 | 4.88 ± 0.32 | 4.48 ± 0.6 |
| S2 | 5.67 ± 0.75 | 6.28 ± 0.3 | 6.22 ± 0.57 | 5.94 ± 0.64 | |
| S3 | 5.34 ± 0.36 | 5.35 ± 0.47 | 5.74 ± 0.36 | 5.48 ± 0.42 | |
| S4 | 6.12 ± 0.43 | 5.86 ± 0.66 | 6.12 ± 0.49 | 6.04 ± 0.53 | |
| Mean (all sets) | 5.4 ± 0.75 | 5.39 ± 0.99 | 5.74 ± 0.7 | 5.48 ± 0.86 | |
| De novo methylation (DNMV) | S1 | 2.84 ± 1.14 | 3.58 ± 0.99 | 3.04 ± 0.79 | 3.18 ± 1.02 |
| S2 | 5.48 ± 1.61 | 4.08 ± 1.1 | 4.63 ± 0.85 | 5.0 ± 1.35 | |
| S3 | 4.85 ± 1.02 | 5.06 ± 0.78 | 4.35 ± 0.61 | 4.71 ± 0.87 | |
| S4 | 5.29 ± 0.74 | 5.21 ± 1.05 | 4.71 ± 0.75 | 5.03 ± 0.86 | |
| Mean (all sets) | 4.61 ± 1.59 | 4.48 ± 1.16 | 4.18 ± 1.02 | 4.48 ± 1.29 | |
| Total tissue culture-induced variation (TTCIV) | S1 | 18.36 ± 1.43 | 19.28 ± 1.8 | 19.41 ± 0.91 | 19.02 ± 1.48 |
| S2 | 30.88 ± 0.89 | 28.92 ± 0.9 | 30.73 ± 0.84 | 30.59 ± 1.1 | |
| S3 | 36.55 ± 1.14 | 36.17 ± 0.83 | 31.55 ± 0.93 | 34.73 ± 2.36 | |
| S4 | 32.28 ± 1.1 | 31.74 ± 1.25 | 32.39 ± 1 | 32.17 ± 1.11 | |
| Mean (all sets) | 29.51 ± 6.84 | 29.02 ± 6.46 | 28.52 ± 5.55 | 29.12 ± 6.5 | |
| Genome methylation (GM) | S1 | 7.95 ± 1.84 | 9.21 ± 1.34 | 7.07 ± 1.11 | 8.14 ± 1.74 |
| S2 | 16.33 ± 2.03 | 14.80 ± 1.5 | 15.17 ± 1.83 | 15.73 ± 1.83 | |
| S3 | 10.71 ± 1.39 | 10.88 ± 0.86 | 10.54 ± 0.89 | 10.68 ± 1.13 | |
| S4 | 14.45 ± 1.09 | 14.89 ± 1.43 | 13.63 ± 1.01 | 14.24 ± 1.24 | |
| Mean (all sets) | 12.36 ± 2.58 | 12.44 ± 1.96 | 11.6 ± 2.55 | 12.19 ± 2.4 | |
| Sites affected by methylation (SAM) | S1 | 7.34 ± 1.07 | 7.66 ± 1.18 | 7.92 ± 0.85 | 7.64 ± 1.06 |
| S2 | 13.66 ± 1.3 | 12.56 ± 0.8 | 13.52 ± 0.75 | 13.48 ± 1.08 | |
| S3 | 10.19 ± 0.99 | 10.41 ± 0.78 | 10.09 ± 0.87 | 10.19 ± 0.96 | |
| S4 | 11.41 ± 0.76 | 11.07 ± 0.99 | 10.83 ± 0.87 | 11.07 ± 0.91 | |
| Mean (all sets) | 10.65 ± 2.54 | 10.42 ± 2.27 | 10.59 ± 2.12 | 10.59 ± 2.36 | |
| Sites with methylated status in donor and regenerant (SMS) | S1 | 3.94 ± 0.68 | 4.29 ± 0.67 | 2.97 ± 0.32 | 3.76 ± 0.8 |
| S2 | 4.42 ± 0.75 | 4.60 ± 0.6 | 4.03 ± 0.43 | 4.23 ± 0.6 | |
| S3 | 3.13 ± 0.42 | 3.13 ± 0.43 | 3.94 ± 0.49 | 3.42 ± 0.57 | |
| S4 | 5.93 ± 0.45 | 6.26 ± 0.55 | 5.71 ± 0.4 | 5.94 ± 0.5 | |
| Mean (all sets) | 4.35 ± 1.03 | 4.57 ± 1.09 | 4.16 ± 1.02 | 4.33 ± 1.08 | |
| Sites with non-methylated status in donor and regenerant (SNMS) | S1 | 70.8 ± 1.05 | 69.71 ± 1.29 | 70.56 ± 0.85 | 70.32 ± 1.17 |
| S2 | 58.65 ± 1.6 | 60.40 ± 0.6 | 59.46 ± 1.01 | 59.15 ± 1.43 | |
| S3 | 56.57 ± 0.92 | 56.92 ± 0.85 | 60.19 ± 0.53 | 57.91 ± 1.72 | |
| S4 | 56.88 ± 0.86 | 56.78 ± 1.04 | 56.93 ± 0.73 | 56.87 ± 0.85 | |
| Mean (all sets) | 60.72 ± 5.89 | 60.95 ± 5.54 | 61.78 ± 5.37 | 61.06 ± 5.7 |
Percentages of sites affected are shown. RA, anther culture-derived regenerants; RM, shed microspore-derived regenerants; RE, immature zygotic embryo-derived regenerants. Si (where i = 1–4) indicates genotype sets encompassing donor plants from four genotypes of DH of triticale cv. Bogo and their regenerants. Standard deviation is shown (±)
When regeneration methods (but not genotype sets) were considered, somatic embryogenesis-derived regeneration (RE) yielded the lowest values for SV, DNMV, and TTCIV as well as for GM and SMS. DMV and SAM values were lowest with the shed microspore method (RM). Values for all the quantitative characteristics were highest with anther-based regeneration (RA). When genotype sets were considered, values of SV, DMV, DNMV, TTCIV, GM, and SAM were generally lowest with the S1 set. Each set exhibited metAFLP values that varied with the different regeneration approaches (Table 2).
ANOVA analysis
Comparison of sets by means of mean value of all metAFLP characteristics
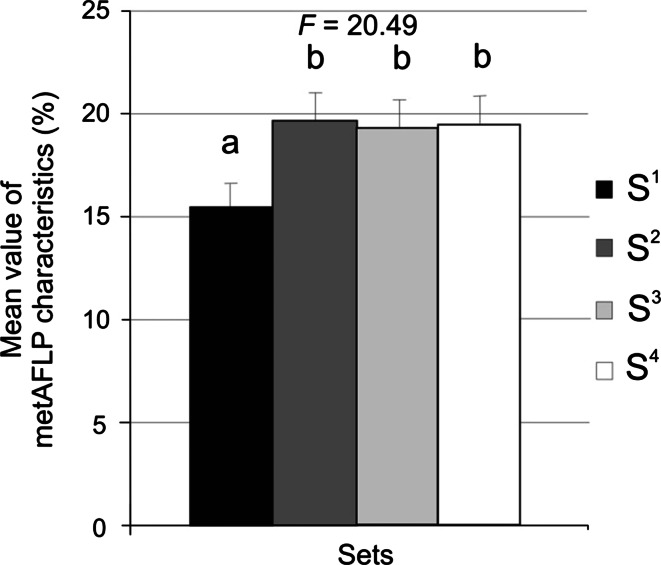
Genotype sets were compared, independent of the tissue culture plant regeneration method used, by examination of mean metAFLP values (for F statistics see Fig. 2). Tukey analysis grouped S2, S3, and S4 together, but S1 was distinct (Fig. 2).
Fig. 2.
Grouping of genotype sets (Tukey’s multiple range test at 5 % significance level) using mean values of all metAFLP characteristics with error bars. Statistically similar sets (Si, where i = 1–4) are indicated with the same letter. F indicates ANOVA statistics (p ≤ 0.05 and α = 0.01)
Comparison of sets by individual metAFLP characteristics
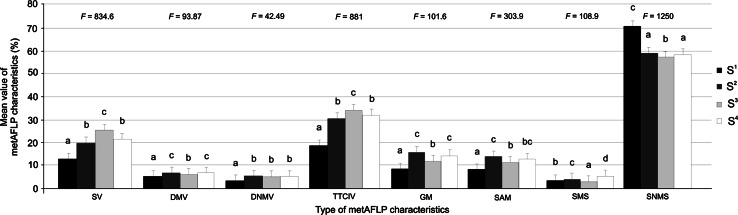
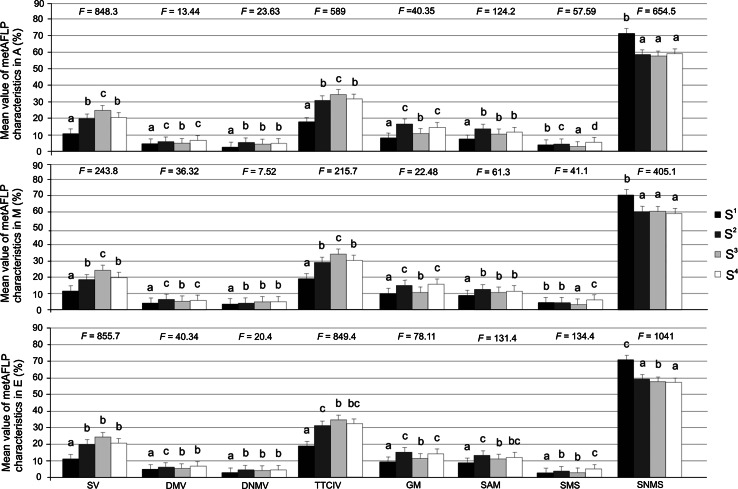
Most genotype sets differed with regard to their metAFLP characteristics (for F statistics see Fig. 3). Tukey analysis differentiated the sets into three broad groups: S1, S3, and S2 with S4. Specifically, S2 and S4 significantly differed only with respect to SMS and were not significantly different for SV, DMV, TTCIV, GM, and SNMS. For DNMV, S2, S3, and S4 formed a single group that was distinct from S1. For SAM, S1, S2, and S3 differed from each other, but S4 was similar to both S2 and S3 (Fig. 3).
Fig. 3.
Grouping of genotype sets (Tukey’s multiple range test at 5 % significance level) for each metAFLP characteristic with error bars. SV, sequence variation; DMV, demethylation variation; DNMV, de novo methylation variation; TTCIV, total tissue culture-induced variation; GM, genome methylation; SAM, sites affected by methylation; SMS, sites with methylated status in donor and regenerants; SNMS, sites with non-methylated status in donor and regenerants. For each characteristic, statistically similar sets (Si, where i = 1–4) are indicated with the same letter. F indicates ANOVA statistics (p ≤ 0.05 and α = 0.01)
Comparison of in vitro tissue culture regeneration methods
No statistically significant differences were observed between the different regeneration methods (RA, RM, and RE) when genotype sets and different metAFLP characteristics were considered together (ANOVA F = 0.145).
Comparison of genotype sets and different plant regeneration methods with overall metAFLP values
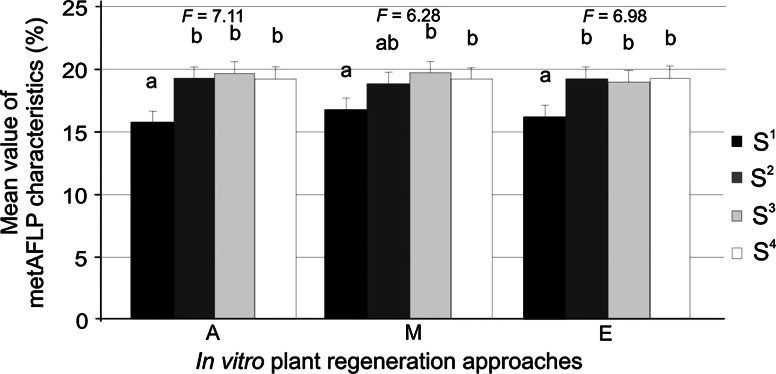
Overall mean metAFLP characteristics were compared for the different genotype sets and regeneration methods (ANOVA; see Fig. 4 for F statistics). The S1 genotype exhibited markedly lower values than the S2, S3, and S4 sets. No statistically significant differences were noted between the S2, S3, and S4 groups for any of the regeneration methods. For the shed microspore culture approach, S2 was similar to both S1 and S3/S4 (Fig. 4).
Fig. 4.
Grouping of genotype sets (Tukey’s multiple range test at 5 % significance level) for mean metAFLP characteristics with respect to plant regeneration method with error bars. A, anther culture-derived regeneration; M, shed microspore-derived regeneration; E, immature zygotic embryo-derived regeneration. For each regeneration approach, statistically similar genotype sets (Si, where i = 1–4) are indicated with the same letter. F indicates ANOVA statistics (p ≤ 0.05 and α = 0.01)
Comparison of genotype sets and different regeneration methods with individual metAFLP characteristics
Individual metAFLP characteristics were compared for the different genotype sets and regeneration methods (ANOVA; see Fig. 5 for F statistics). With the exception of SMS, patterns of similarity between the genotype sets were identical between the shed microspore and anther regeneration methods. For these regeneration approaches, no significant differences were observed between the S2 and S4 genotypes for SV, DMV, TTCIV, and GM, and there were no significant differences between S2, S3, and S4 for DNMV, SAM, and SNMS. Genomic set groupings were generally different for immature zygotic embryo-derived regeneration compared to the other two regeneration approaches.
Fig. 5.
Grouping of genotype sets (Tukey’s multiple range test at 5 % significance level) for individual metAFLP characteristics with respect to plant regeneration method with error bars. For each regeneration approach, statistically similar genotype sets (Si, where i = 1–4) are indicated with the same letter. F indicates ANOVA statistics (p ≤ 0.05 and α = 0.01). Abbreviations as in Figs. 3 and 4
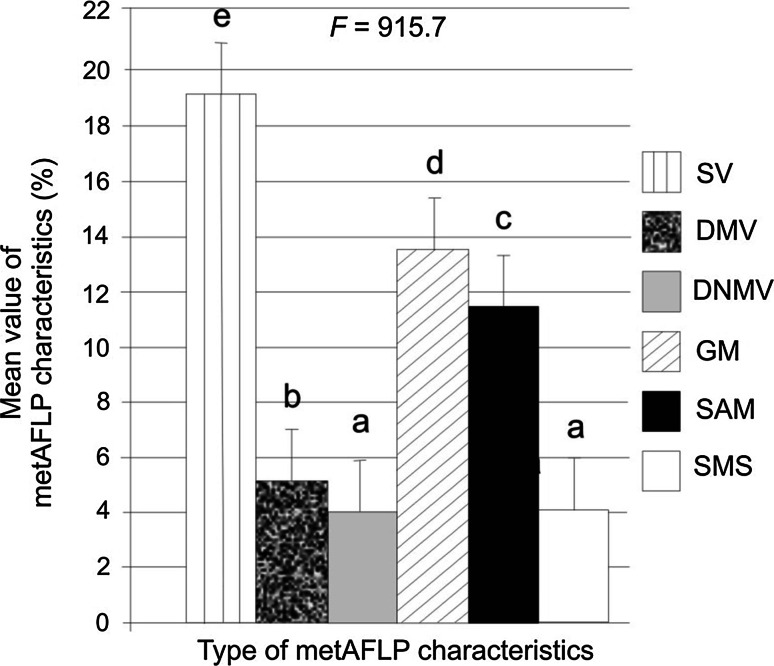
Comparison of metAFLP characteristics
Significant differences were observed between the different metAFLP characteristics with the exception of DNMV and SMS (Tukey’s test, Fig. 6).
Fig. 6.
Grouping of metAFLP characteristics (Tukey’s multiple range test at 5 % significance level) with error bars. Statistically similar characteristics are indicated by the same letter. F indicates ANOVA statistics (p ≤ 0.05 and α = 0.01). Abbreviations as in Fig. 3
Comparison of GM and global DNA methylation data from metAFLP and RP-HPLC analyses.
In our previous study, we observed a decrease in global DNA methylation of regenerants compared to donors irrespective of the in vitro tissue culture approach used for plant regeneration. RP-HPLC showed that global DNA methylation of donors and regenerants was 25.39 and 24.1 %, respectively (Machczyńska et al. 2014b). MetAFLP estimates of GM of the restriction sites in regenerants were twofold lower (Table 2) than RP-HPLC values. RP-HPLC estimated the decrease in global DNA methylation between donors and regenerants to be ~1.29 %. While GM values were lower using metAFLP, the decrease in methylation between donors and regenerants was also estimated as ~1 % (DNMV–DMV; Table 2).
RP-HPLC and metAFLP estimates of global DNA methylation and GM were compared using Pearson correlation analysis. When genotype sets and in vitro tissue culture regeneration approaches were not taken into consideration, limited correlation was observed (r = 0.15, p < 0.02). When sets were considered separately, a high level of correlation between RP-HPLC and metAFLP was seen for S1 but not for S2–S4 (Table 3).
Table 3.
Pearson correlation coefficients comparing genome methylation (GM) estimates by metAFLP and global DNA methylation by RP-HPLC in different genotype sets
| Set (Si) | Pearson correlation coefficient (r); p value |
|---|---|
| S1 | 0.38; p < 0.001 |
| S2 | −0.17; p < 0.27 |
| S3 | −0.06; p < 0.70 |
| S4 | 0.09; p < 0.57 |
Si represents genotype, where i = 1–4
Pearson correlation was also used to compare estimates of global DNA methylation by RP-HPLC and GM by metAFLP for the different in vitro plant regeneration approaches (Table 4). Significant correlations were observed for the anther- and embryo-derived, but not the shed microspore-derived, regenerants.
Table 4.
Pearson correlation comparing genome methylation estimates by metAFLP and global DNA methylation estimates by RP-HPLC for different plant regeneration approaches
| In vitro tissue culture regeneration approaches | Pearson correlation coefficient (r); p value |
|---|---|
| Anther culture | −0.28; p < 0.01 |
| Shed microspore culture | 0.15; p < 0.28 |
| Immature zygotic embryo culture | 0.28; p < 0.01 |
Discussion
Several studies have compared the molecular characteristics of tissue culture-derived regenerants and their donor plants (Bouman and Klerk 2001; Hossain et al. 2003; Bhatia et al. 2009). However, the plants used as a source of explants in these studies were not assessed for uniformity (Teyssier et al. 2013), and this may be of critical importance for the quantitative evaluation of TCIV (Bednarek et al. 2007; Machczyńska et al. 2014a). To address this, we used specific plant resources to examine TCIV in this study. Four randomly selected and genetically distinct DH genotypes derived from isolated microspores from the cv. Bogo triticale served as donor plants. DH donor plants were cloned by partitioning plant clumps after tillering. This gave sufficient numbers of explants to produce simultaneous androgenic and somatic regenerants for quantitative analysis. To ensure that cloned individuals were epigenetically and genetically identical, individuals were analyzed using the same primer combinations as for analysis of genotype sets. A lack of morphological and minor molecular differences between cloned individuals from the same donor plant were seen (0–1.8 % for the Acc65I/MseI metAFLP platform and 0–1.2 % for the KpnI/MseI digest), but these could be attributed to the metAFLP experimental error (Meudt and Clarke 2007). Although we did our best to pick leaves for DNA isolation at the same developmental stages, some differences among samples may have not been excluded. The differences observed with the Acc65I/MseI digest related to DNA methylation may reflect this. It cannot be excluded that variation between cloned individuals of the same genotype might be a consequence of genomic factors such as activity of mobile elements (Schwartz and Dennis 1986; Khan et al. 2013) or of DNA polymerase slippage (Alhani and Wilkinson 1998). Nevertheless, a sufficient number of cloned individuals were identified that had no detectable variation using the Acc65I/MseI and KpnI/MseI metAFLP platforms. Of 53 clones, 39 individuals representing four genotypes of cv. Bogo were used for plant regeneration via the A, M, and E approaches.
Recently, we described an extended metAFLP approach that facilitated the evaluation of numerous quantitative characteristics (Machczyńska et al. 2014a). The observed TCIV was higher than that found in barley (Bednarek et al. 2007) and, whereas methylation pattern changes were predominant in barley, most of the observed variation in preliminary triticale study was a result of sequence changes (Machczyńska et al. 2014a). The level of SV observed in the regenerants could be attributed to the activation of transposable elements that is thought to take place in parallel with genomic DNA demethylation (Brettell and Dennis 1991; Liu et al. 2004) in response to abiotic stresses (Kashkush et al. 2003). The number of SAM was comparable to the level of sequence alterations. This suggested that tissue culture prompted numerous sequence and DNA methylation changes in triticale, possibly reflecting previously observed genome instabilities (Lapitan et al. 1984; Bento et al. 2011) such as chromosome rearrangements (Oleszczuk et al. 2011). The epigenetic and genetic changes did not cause any readily apparent morphological consequences in the regenerants, suggesting that the changes may have occurred in genomic regions that did not affect morphological traits. Alternatively, changes in DNA methylation pattern and/or sequence mutations affecting essential traits might be rare, or regenerants with such changes might be eliminated during plant regeneration. It is possible that the number of regenerants analyzed may have been sufficient to detect epigenetic and genetic variation, but not to identify morphological mutants. However, changes in DNA methylation level caused phenotypic variation in other organisms, such as Linaria flower (Cubas et al. 1999) and Arabidopsis thaliana (Soppe et al. 2000), and the DNA methylation-derived changes were heritable (Kathiria et al. 2010). Thus, even though no phenotypic variation was observed in triticale regenerants, successive regenerant progenies may display morphological changes.
It is possible that donor plant genotype may influence TCIV. Our evaluation of four sample sets comprising distinct donor genotypes allowed examination of the role of genotype on TCIV. We previously demonstrated that donor genotype had an effect on TCIV in barley (Bednarek et al. 2007). However, in that case, it was possible that the observed “donor effect” was a statistical artifact of the Tukey–Kramer test which, when used to compare variable numbers of regenerants within a data set, compares sets in pairs and is capable of delivering a non-existent “genotype effect”. The genotype sets in the present study also contained different numbers of regenerants. The most different set was represented by the largest number of regenerants, which may favor the non-existent “genotype effect” explanation. The S1 set contained highly uniform regenerants, which resulted in lower metAFLP values compared to the other sets. Tukey’s tests for metAFLP characteristics distinguished between most of the sets, which favored the donor effect hypothesis.
Another factor that may contribute to TCIV is the method used for in vitro plant regeneration (Bairu et al. 2011). In triticale, DHs may be produced by androgenesis (Immonen and Robinson 2000; Oleszczuk et al. 2004; Würschum et al. 2012) or by artificial crossing with maize (Wȩdzony et al. 1998), wheat (Pratap et al. 2005), or pearl millet (Inagaki and Hash 1998). Artificial crossing necessitates the use of chromosome doubling agents that may generate additional variation (Liu et al. 2009; Wu et al. 2012; Wang et al. 2014). In androgenesis, spontaneous doubling rate in triticale varies from 0 to 50 % (Arzani and Darvey 2001; Ślusarkiewicz-Jarzina and Ponitka 2003; Oleszczuk et al. 2004; Lantos et al. 2014). Androgenesis is the most frequently adopted method for the evaluation of DH plants in cereals (Maluszyński et al. 2003) and was therefore used here. By contrast, with isolated microspore (Oleszczuk et al. 2004) or chromosome elimination methods (Powell et al. 1986), plant regeneration using anthers or immature zygotic embryos as sources of tissues can proceed via an intermediate callus stage (Kim et al. 2003; Seguí-Simarro and Nuez 2007). Numerous previous studies (Phillips et al. 1994; Young et al. 1999; Wang et al. 2013) suggested that the presence of the callus stage was the most probable source of TCIV. In our experiments, E and A regeneration involved an intermediate callus phase; however, no significant differences in TCIV were observed between regeneration approaches when genotype sets were analyzed together. These results were consistent with those for barley (Bednarek et al. 2007), where the levels of TCIV in androgenesis and immature zygotic embryo-derived regenerants were comparable. This is also consistent with studies using Freesia hybrida, in which sequence and methylation changes were similar for direct regeneration and indirect regeneration proceeding via the callus stage (Gao et al. 2010). By contrast, studies in barley revealed a higher level of DNA methylation in regenerants from anther culture than in those derived from Hordeum bulbosum (Devaux et al. 1993). When tissue culture methods were compared for each individual genotype in our dataset, differences in TCIV between regeneration approaches were observed. Observed differences were mostly attributable to the S1 set, which supported either the donor hypothesis or the statistically-derived “genotype effect”. However, individual metAFLP characteristics were more discriminative: some metAFLP types distinguished most of the sets, while others separated S1 only. Thus, it is possible that plant regeneration approaches influence TCIV but that the differences are a result of subtle effects (possibly random fluctuations) that can only be observed by comparison of different metAFLP characteristics.
Assuming that metAFLP can quantify GM through examination of the restriction sites recognized by the isoschizomers used in the approach and that RP-HPLC can be used to evaluate global DNA methylation, it might be expected that outcomes of the two methods would be correlated; however, the expected correlation was not apparent. MetAFLP uses changes at restriction sites recognized by Acc65I and KpnI isoschizomers as a proxy for whole-GM; however, it is possible that this may not reflect changes affecting the whole genome accurately. This suggests that the restriction sites used in metAFLP might not be randomly distributed across chromosomes. Genetic mapping with AFLP markers in rye showed that mapped markers were not evenly distributed across chromosomes but formed clusters of tightly linked markers (Bednarek et al. 2003). If this is the case with the restriction sites used here, then the weak correlation between the metAFLP and RP-HPLC estimates of GM and global DNA methylation suggests that metAFLP reflects differences within particular genomic regions rather than across the whole genome.
It was previously demonstrated that global DNA methylation, as estimated using RP-HPLC analysis, decreased in regenerants compared to donors irrespective of the regeneration approaches used (Machczyńska et al. 2014b). The decrease in DNA methylation was about 1.2 %, which is consistent with comparative methylation values from the metAFLP approach. However, RP-HPLC and metAFLP global DNA methylation and GM were poorly correlated for all genotype sets except S1, which exhibited relatively high correlation. The correlation for the S1 set may reflect either the larger number of individuals used for the analysis or the putative genotypic effects of the D1 genotype. Similar analyses of larger sets would be needed to discriminate between the two hypotheses.
The direction of DNA methylation alteration in triticale differed from that in barley (Bednarek et al. 2007). Presently, it is unclear whether these opposite directions are species- or even cultivar-specific. One may speculate, however, that, as a result of its more recent development and polyploidization, triticale or some of its forms might have a less stable genome than barley (Bento et al. 2008) and might therefore be more prone to genomic changes. However, it remains possible that the opposing directions of change may be attributable to the different genotypes. Cytological evidence suggests that some triticale forms are highly stable under tissue culture treatment, while others exhibit more instability (Oleszczuk et al. 2011). Additional complexity may underlie GM, as demonstrated by the different change directions observed in a single species, barley, using the methylation-sensitive amplified polymorphism (Li et al. 2007) and metAFLP (Bednarek et al. 2007) approaches. Whether these differences reflect genuine genotype effects or simply the differences in the genomic regions examined by the different methodological approaches remains to be investigated.
The metAFLP characteristics used for the evaluation of differences between genotype sets proved to discriminate sets and tissue culture approaches. The characteristics were also distinct from each other. This was consistent with data from barley (Bednarek et al. 2007) and suggested that the different metAFLP quantitative characteristics reflected different phenomena (sequence changes, alterations in site methylation patterns, etc.). If the different metAFP characteristics were not linked to biological phenomena, then they would most likely not have been statistically discriminated from one another. Taken together, our results demonstrate the value of the metAFLP approach for the examination of TCIV and show that such variation is linked to genetic background related to in vitro plant regeneration approaches.
Conclusions
Regeneration of triticale plants via in vitro tissue culture was error-prone and affected DNA sequence and methylation patterns, irrespective of the culture method used. One of the most frequently observed variation types was the alteration in sequence between donor and regenerant plants. Numerous changes to DNA methylation pattern were also observed in regenerants compared to donors. Observed changes initially appeared unlinked to donor genotype or the tissue regeneration approach used; however, differences became apparent when individual metAFLP characteristics were examined. Both metAFLP and RP-HPLC estimates of GM and global DNA methylation indicated a decrease in methylation in regenerants compared to donors; however, RP-HPLC methylation estimates were in most cases poorly correlated with estimates of methylation from metAFLP examination of isoschizomer restriction sites. In summary, metAFLP quantitative characteristics were useful for evaluation of TCIV, which appeared to be linked to genotype background and in vitro regeneration approaches.
Electronic supplementary material
Acknowledgments
For statistical assistance, the authors acknowledge Dr. Dariusz Robert Mańkowski from the Plant Breeding and Acclimatization Institute of the National Research Institute in Radzików, Poland.
References
- Alhani MC, Wilkinson MJ. Inter simple sequence repeat polymerase chain reaction for the detection of somaclonal variation. Plant Breed. 1998;117(6):573–575. doi: 10.1111/j.1439-0523.1998.tb02210.x. [DOI] [Google Scholar]
- Arzani A, Darvey N. The effect of colchicine on triticale anther-derived plants: microspore pre-treatment and haploid-plant treatment using a hydroponic recovery system. Euphytica. 2001;122(2):235–241. doi: 10.1023/A:1012966506030. [DOI] [Google Scholar]
- Bairu M, Aremu A, Van Staden J. Somaclonal variation in plants: causes and detection methods. Plant Growth Regul. 2011;63(2):147–173. doi: 10.1007/s10725-010-9554-x. [DOI] [Google Scholar]
- Baránek M, Křižan B, Ondrušíková E, Pidra M. DNA-methylation changes in grapevine somaclones following in vitro culture and thermotherapy. Plant Cell Tissue Organ Cult. 2010;101(1):11–22. doi: 10.1007/s11240-009-9656-1. [DOI] [Google Scholar]
- Barwale UB, Widholm JM. Somaclonal variation in plants regenerated from cultures of soybean. Plant Cell Rep. 1987;6:365–368. doi: 10.1007/BF00269562. [DOI] [PubMed] [Google Scholar]
- Bednarek PT, Lewandowska R, Golas T, Pasnik M. The chromosomal location of rye AFLP bands. Cell Mol Biol Lett. 2003;8(4):955–961. [PubMed] [Google Scholar]
- Bednarek PT, Orłowska R, Koebner RMD, Zimny J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.) BMC Plant Biol. 2007;7:10. doi: 10.1186/1471-2229-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento M, Pereira HS, Rocheta M, Gustafson P, Viegas W, Silva M. Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in Triticale. PLoS One. 2008;3(1):e1402. doi: 10.1371/journal.pone.0001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento M, Gustafson JP, Viegas W, Silva M. Size matters in Triticeae polyploids: larger genomes have higher remodeling. Genome. 2011;54(3):175–183. doi: 10.1139/G10-107. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Croy RRD, Natali L, Cavallini A, Durante M. Evidence for seed proteins somaclonal variation in the progenies of regenerated plants of Pisum sativum L. J Genet Breed. 1999;53(4):355–358. [Google Scholar]
- Bhatia R, Singh KP, Jhang T, Sharma TR. Assessment of clonal fidelity of micropropagated gerbera plants by ISSR markers. Sci Hortic. 2009;119(2):208–211. doi: 10.1016/j.scienta.2008.07.024. [DOI] [Google Scholar]
- Blum A. The abiotic stress response and adaptation of triticale: a review. Cereal Res Commun. 2014;42(3):359–375. doi: 10.1556/CRC.42.2014.3.1. [DOI] [Google Scholar]
- Bouman H, Klerk G-J. Measurement of the extent of somaclonal variation in begonia plants regenerated under various conditions: comparison of three assays. Theor Appl Genet. 2001;102:111–117. doi: 10.1007/s001220051625. [DOI] [Google Scholar]
- Breiman A, Rotem-Abarbanell D, Karp A, Shaskin H. Heritable somaclonal variation in wild barley (Hordeum spontaneum) Theor Appl Genet. 1987;74(1):104–112. doi: 10.1007/BF00290092. [DOI] [PubMed] [Google Scholar]
- Brettell RS, Dennis E. Reactivation of a silent Ac following tissue culture is associated with heritable alterations in its methylation pattern. Mol Gen Genet. 1991;229(3):365–372. doi: 10.1007/BF00267457. [DOI] [PubMed] [Google Scholar]
- Chwedorzewska KJ, Bednarek PT. Genetic and epigenetic variation in a cosmopolitan grass Poa annua from Antarctic and Polish populations. Pol Polar Res. 2012;33(1):63–80. [Google Scholar]
- Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401(6749):157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Dann A, Wilson C. Comparative assessment of genetic and epigenetic variation among regenerants of potato (Solanum tuberosum) derived from long-term nodal tissue-culture and cell selection. Plant Cell Rep. 2011;30(4):631–639. doi: 10.1007/s00299-010-0983-9. [DOI] [PubMed] [Google Scholar]
- Dennis ES, Brettell RIS, Peacock WJ. A tissue culture induced Adh1 null mutant of maize results from a single base change. Mol Gen Genet. 1987;210:181–183. doi: 10.1007/BF00337777. [DOI] [Google Scholar]
- Devaux P, Kilian A, Kleinhofs A. Anther culture and Hordeum bulbosum derived barley doubled haploids: mutations and methylation. Mol Gen Genet. 1993;241:674–679. doi: 10.1007/BF00279910. [DOI] [PubMed] [Google Scholar]
- Fiuk A, Bednarek P, Rybczyński J. Flow cytometry, HPLC-RP, and metAFLP analyses to assess genetic variability in somatic embryo-derived plantlets of Gentiana pannonica Scop. Plant Mol Biol Rep. 2010;28(3):413–420. doi: 10.1007/s11105-009-0167-3. [DOI] [Google Scholar]
- Gao X, Yang D, Cao D, Ao M, Sui X, Wang Q, Kimatu JN, Wang L. In vitro micropropagation of Freesia hybrida and the assessment of genetic and epigenetic stability in regenerated plantlets. J Plant Growth Regul. 2010;29(3):257–267. doi: 10.1007/s00344-009-9133-4. [DOI] [Google Scholar]
- Guo WL, Wu R, Zhang YF, Liu XM, Wang HY, Gong L, Zhang ZH, Liu B. Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsis lanceolata Benth. et Hook. f. Plant Cell Rep. 2007;26:1297–1307. doi: 10.1007/s00299-007-0320-0. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- Hills MJ, Hall LM, Messenger DF, Graf RJ, Beres BL, Eudes F. Evaluation of crossability between triticale (× Triticosecale Wittmack) and common wheat, durum wheat and rye. Environ Biosaf Res. 2007;6(4):249–257. doi: 10.1051/ebr:2007046. [DOI] [PubMed] [Google Scholar]
- Hossain AM, Konisho K, Mineo M, Kazuhiro N. Somaclonal variation of regenerated plants in chili pepper (Capsicum annuum L.) Euphytica. 2003;130(2):233–239. doi: 10.1023/A:1022856725794. [DOI] [Google Scholar]
- Immonen S, Robinson J. Stress treatments and ficoll for improving green plant regeneration in triticale anther culture. Plant Sci. 2000;150(1):77–84. doi: 10.1016/S0168-9452(99)00169-7. [DOI] [Google Scholar]
- Inagaki MN, Hash CT. Production of haploids in bread wheat, durum wheat and hexaploid triticale crossed with pearl millet. Plant Breed. 1998;117(5):485–487. doi: 10.1111/j.1439-0523.1998.tb01978.x. [DOI] [Google Scholar]
- Johnston JW, Harding K, Bremner DH, Souch G, Green J, Lynch PT, Grout B, Benson EE. HPLC analysis of plant DNA methylation: a study of critical methodological factors. Plant Physiol Biochem. 2005;43(9):844–853. doi: 10.1016/j.plaphy.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Kaeppler SM, Phillips RL. DNA methylation and tissue culture-induced variation in plants. Vitro Cell Dev Biol Plant. 1993;29(3):125–130. doi: 10.1007/BF02632283. [DOI] [Google Scholar]
- Kaeppler SM, Phillips RL. Tissue culture-induced DNA methylation variation in maize. Proc Natl Acad Sci USA. 1993;90(19):8773–8776. doi: 10.1073/pnas.90.19.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler SM, Phillips RL, Olhoft P. Molecular basis of herutable tissue culture: induced variation in plants. In: Jain SM, Brar DS, Ahloowalia BS, editors. Somaclonal variation and induced mutation in crop improvement. Dordrecht: Kluwer Academic Publishers; 1998. pp. 465–484. [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33(1):102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I. Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 2010;153(4):1859–1870. doi: 10.1104/pp.110.157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Enjalbert J, Marsollier A-C, Rousselet A, Goldringer I, Vitte C. Vernalization treatment induces site-specific DNA hypermethylation at the VERNALIZATION-A1 (VRN-A1) locus in hexaploid winter wheat. BMC Plant Biol. 2013;13(1):209. doi: 10.1186/1471-2229-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Oh S, Liu J. Control of direct and indirect somatic embryogenesis by exogenous growth regulators in immature zygotic embryo cultures of rose. Plant Cell Tissue Organ Cult. 2003;74(1):61–66. doi: 10.1023/A:1023355729046. [DOI] [Google Scholar]
- Kirikovich SS, Svirshchevskaya AM, Levites EV. Variation at isozyme loci in seed offspring of sugar beet gynogenetic lines. Sugar Tech. 2003;5(4):289–292. doi: 10.1007/BF02942487. [DOI] [Google Scholar]
- Lantos C, Bóna L, Boda K, Pauk J. Comparative analysis of in vitro anther- and isolated microspore culture in hexaploid Triticale (× Triticosecale Wittmack) for androgenic parameters. Euphytica. 2014;197(1):27–37. doi: 10.1007/s10681-013-1031-y. [DOI] [Google Scholar]
- Lapitan NL, Sears RG, Gill BS. Translocations and other karyotypic structural changes in wheat × rye hybrids regenerated from tissue culture. Theor Appl Genet. 1984;68(6):547–554. doi: 10.1007/BF00285012. [DOI] [PubMed] [Google Scholar]
- Larkin PJ, Ryan SA, Brettell RIS, Scowcroft WR. Heritable somaclonal variation in wheat. Theor Appl Genet. 1984;67(5):443–455. doi: 10.1007/BF00263410. [DOI] [PubMed] [Google Scholar]
- Li X, Yu X, Wang N, Feng Q, Dong Z, Liu L, Shen J, Liu B. Genetic and epigenetic instabilities induced by tissue culture in wild barley (Hordeum brevisubulatum (Trin.) Link) Plant Cell Tissue Organ Cult. 2007;90(2):153–168. doi: 10.1007/s11240-007-9224-5. [DOI] [Google Scholar]
- Liu ZL, Han FP, Tan M, Shan XH, Dong YZ, Wang XZ, Fedak G, Hao S, Liu B. Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theor Appl Genet. 2004;109(1):200–209. doi: 10.1007/s00122-004-1618-8. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Lin H, Mo XY, Long T, Zhang HY. Genetic variation in colchicine-treated regenerated plants of Eucalyptus globulus Labill. J Genet. 2009;88(3):345–348. doi: 10.1007/s12041-009-0051-9. [DOI] [PubMed] [Google Scholar]
- Lorz H, Scowcroft WR. Variability among plants and their progeny regenerated from protoplasts of Su/su heterozygotes of Nicotiana tabacum. Theor Appl Genet. 1983;66(1):67–75. doi: 10.1007/BF00281851. [DOI] [PubMed] [Google Scholar]
- Lu Y-Q, Wang D-M, Li H-Y, Jia Q, Wu Z, Lu W-F. A comparison on DNA methylation levels in bamboo at five developmental stages. J For Res. 2012;23(1):157–159. doi: 10.1007/s11676-012-0247-3. [DOI] [Google Scholar]
- Machczyńska J, Orłowska R, Mańkowski DR, Zimny J, Bednarek P. DNA methylation changes in triticale due to in vitro culture plant regeneration and consecutive reproduction. Plant Cell Tissue Organ Cult. 2014;119(2):289–299. doi: 10.1007/s11240-014-0533-1. [DOI] [Google Scholar]
- Machczyńska J, Orłowska R, Zimny J, Bednarek P. Extended metAFLP approach in studies of tissue culture induced variation (TCIV) in triticale. Mol Breed. 2014;34(3):845–854. doi: 10.1007/s11032-014-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszyński M, Kasha KJ, Forster BP, Szarejko I. Doubled haploid production in crop plants: a manual. Dordrecht: Kluwer Academic Publishers; 2003. Published doubled haploid protocols in plant species; pp. 309–335. [Google Scholar]
- Mankessi F, Saya AR, Favreau B, Doulbeau S, Conéjéro G, Lartaud M, Verdeil J-L, Monteuuis O. Variations of DNA methylation in Eucalyptus urophylla × Eucalyptus grandis shoot tips and apical meristems of different physiological ages. Physiol Plant. 2011;143(2):178–187. doi: 10.1111/j.1399-3054.2011.01491.x. [DOI] [PubMed] [Google Scholar]
- McGoverin CM, Snyders F, Muller N, Botes W, Fox G, Manley M. A review of triticale uses and the effect of growth environment on grain quality. J Sci Food Agric. 2011;91(7):1155–1165. doi: 10.1002/jsfa.4338. [DOI] [PubMed] [Google Scholar]
- Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;12(3):106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Ngezahayo F, Xu C, Wang H, Jiang L, Pang J, Liu B. Tissue culture-induced transpositional activity of mPing is correlated with cytosine methylation in rice. BMC Plant Biol. 2009 doi: 10.1186/1471-2229-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettler G, Tams SH, Utz HF, Bauer E, Melchinger AE. Prospects for hybrid breeding in winter triticale—I: heterosis and combining ability for agronomic traits in European elite germplasm. Crop Sci. 2005;45(4):1476–1482. doi: 10.2135/cropsci2004.0462. [DOI] [Google Scholar]
- Oleszczuk S, Sowa S, Zimny J. Direct embryogenesis and green plant regeneration from isolated microspores of hexaploid triticale (× Triticosecale Wittmack) cv. Bogo. Plant Cell Rep. 2004;22(12):885–893. doi: 10.1007/s00299-004-0796-9. [DOI] [PubMed] [Google Scholar]
- Oleszczuk S, Rabiza-Swider J, Zimny J, Lukaszewski A. Aneuploidy among androgenic progeny of hexaploid triticale (× Triticosecale Wittmack) Plant Cell Rep. 2011;30(4):575–586. doi: 10.1007/s00299-010-0971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olhoft PM, Phillips RL (1994) DNA methylation and tissue culture-induced variation in mayze. Conference material (Plant Genome II Conference, San Diego, CA, January, 1994)
- Phillips RL, Kaeppler SM, Olhoft P. Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci USA. 1994;91(12):5222–5226. doi: 10.1073/pnas.91.12.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco C, ML Ruiz. AFLP analysis of somaclonal variation in Arabidopsis thaliana regenerated plants. Plant Sci. 2002;162(5):817–824. doi: 10.1016/S0168-9452(02)00029-8. [DOI] [Google Scholar]
- Powell W, Caligari PDS, Dunwell JM. Field performance of lines derived from haploid and diploid tissues of Hordeum vulgare. Theor Appl Genet. 1986;72(4):458–465. doi: 10.1007/BF00289527. [DOI] [PubMed] [Google Scholar]
- Pratap A, Sethi GS, Chaudhary HK. Relative efficiency of different Gramineae genera for haploid induction in triticale and triticale × wheat hybrids through the chromosome elimination technique. Plant Breed. 2005;124(2):147–153. doi: 10.1111/j.1439-0523.2004.01059.x. [DOI] [Google Scholar]
- Ramirez-Garcia J, Gabriel JL, Alonso-Ayuso M, Quemada M. Quantitative characterization of five cover crop species. J Agr Sci. 2015;153:1174–1185. doi: 10.1017/S0021859614000811. [DOI] [Google Scholar]
- SAS Institute Inc . SAS/STAT 9.1 user’s guide. Cary: SAS Publishing, SAS Institute Inc.; 2004. [Google Scholar]
- Schwartz D, Dennis ES. Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol Gen Genet. 1986;205:476–482. doi: 10.1007/BF00338085. [DOI] [Google Scholar]
- Seguí-Simarro JM, Nuez F. Embryogenesis induction, callogenesis, and plant regeneration by in vitro culture of tomato isolated microspores and whole anthers. J Exp Bot. 2007;58(5):1119–1132. doi: 10.1093/jxb/erl271. [DOI] [PubMed] [Google Scholar]
- Ślusarkiewicz-Jarzina A, Ponitka A. Efficient production of spontaneous and induced doubled haploid triticale plants derived from anther culture. Cereal Res Commun. 2003;31(3–4):289–296. [Google Scholar]
- Smykal P, Valledor L, Rodriguez R, Griga M. Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.) Plant Cell Rep. 2007;26(11):1985–1998. doi: 10.1007/s00299-007-0413-9. [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6(4):791–802. doi: 10.1016/S1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Stroud H, Ding B, Simon SA, Feng S, Bellizzi M, Pellegrini M, Wang G-L, Meyers BC, Jacobsen SE. Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife. 2013 doi: 10.7554/eLife.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C, Maury S, Beaufour M, Grondin C, Delaunay A, Le Metté C, Ader K, Cadene M, Label P, Lelu-Walter M-A. In search of markers for somatic embryo maturation in hybrid larch (Larix × eurolepis): global DNA methylation and proteomic analyses. Physiol Plant. 2013 doi: 10.1111/ppl.12081. [DOI] [PubMed] [Google Scholar]
- Wang X, Wu R, Lin X, Bai Y, Song C, Yu X, Xu C, Zhao N, Dong Y, Liu B. Tissue culture-induced genetic and epigenetic alterations in rice pure-lines, F1 hybrids and polyploids. BMC Plant Biol. 2013;13(1):77. doi: 10.1186/1471-2229-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dong B, Jiang J, Fang W, Guan Z, Liao Y, Chen S, Chen F. Characterization of in vitro haploid and doubled haploid Chrysanthemum morifolium plants via unfertilized ovule culture for phenotypical traits and DNA methylation pattern. Front Plant Sci. 2014 doi: 10.3389/fpls.2014.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wȩdzony M, Marcińska I, Ponitka A, ŚLusarkiewicz-Jarzina A, Woźna J. Production of doubled haploids in triticale (× Triticosecale Wittm.) by means of crosses with maize (Zea mays L.) using picloram and dicamba. Plant Breed. 1998;117(3):211–215. doi: 10.1111/j.1439-0523.1998.tb01928.x. [DOI] [Google Scholar]
- Wu J-H, Ferguson AR, Murray BG, Jia Y, Datson PM, Zhang J. Induced polyploidy dramatically increases the size and alters the shape of fruit in Actinidia chinensis. Ann Bot. 2012;109(1):169–179. doi: 10.1093/aob/mcr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Tucker MR, Reif JC, Maurer HP. Improved efficiency of doubled haploid generation in hexaploid triticale by in vitro chromosome doubling. BMC Plant Biol. 2012;12:109. doi: 10.1186/1471-2229-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Li X, Korban S. DNA-methylation alterations and exchanges during in vitro cellular differentiation in rose (Rosa hybrida L.) Theor Appl Genet. 2004;109(5):899–910. doi: 10.1007/s00122-004-1717-6. [DOI] [PubMed] [Google Scholar]
- Young WP, Schupp JM, Keim P. DNA methylation and AFLP marker distribution in the soybean genome. Theor Appl Genet. 1999;99:785–790. doi: 10.1007/s001220051297. [DOI] [Google Scholar]
- Zehr BE, Williams ME, Duncan RD, WIdholm JM. Somaclonal variation among the progeny of plants regenerated from callus cultures of seven inbred lines of maize. Can J Bot. 1987;61:491–499. doi: 10.1139/b87-061. [DOI] [Google Scholar]
- Zhang M, Wang H, Dong Z, Qi B, Xu K, Liu B. Tissue culture-induced variation at simple sequence repeats in sorghum (Sorghum bicolor L.) is genotype-dependent and associated with down-regulated expression of a mismatch repair gene, MLH3. Plant Cell Rep. 2010;29(1):51–59. doi: 10.1007/s00299-009-0797-9. [DOI] [PubMed] [Google Scholar]
- Zhang D, Wang Z, Wang N, Gao Y, Liu Y, Wu Y, Bai Y, Zhang Z, Lin X, Dong Y, Ou X, Xu C, Liu B. Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS One. 2014;9(5):e96879. doi: 10.1371/journal.pone.0096879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.