Abstract
Taxonomic differentiation among morphologically identical Ascaris species is a debatable scientific issue in the context of Ascariasis epidemiology. To explain the disease epidemiology and also the taxonomic position of different Ascaris species, genome information of infecting strains from endemic areas throughout the world is certainly crucial. Ascaris population from human has been genetically characterized based on the widely used genetic marker, internal transcribed spacer1 (ITS1). Along with previously reported and prevalent genotype G1, 8 new sequence variants of ITS1 have been identified. Genotype G1 was significantly present among female patients aged between 10 to 15 years. Intragenic linkage disequilibrium (LD) analysis at target locus within our study population has identified an incomplete LD value with potential recombination events. A separate cluster of Indian isolates with high bootstrap value indicate their distinct phylogenetic position in comparison to the global Ascaris population. Genetic shuffling through recombination could be a possible reason for high population diversity and frequent emergence of new sequence variants, identified in present and other previous studies. This study explores the genetic organization of Indian Ascaris population for the first time which certainly includes some fundamental information on the molecular epidemiology of Ascariasis.
Keywords: Ascariasis, Ascaris sp, Genetic diversity, Phylogeny, Internal transcribed spacer 1 (ITS1), Sequence typing
1. Introduction
Human Ascariasis caused by gastrointestinal nematode Ascaris lumbricoides (L) is one of the major Soil Transmitted Helminthiases (STHs).The disease has been included in World Health Organization (WHO) list of Neglected Tropical Diseases (NTD), infecting more than one billion people [1]. Transmission is normally through the ingestion of infective Ascaris sp. egg in sewage contaminated soil and vegetables. Majority of infections are asymptomatic, while some chronic infection develops symptoms like abdominal pain, nausea, lung inflammation, anemia, stunted growth, diminished physical fitness etc. [1]. This has a certain impact on socio-economic development of low-income countries [2]. Like human, pigs are also infected with closely related species of A. lumbricoides (L), Ascaris suum Goeze [3]. Taxonomic separation between A. lumbricoides and A. suum represents a debatable scientific issue in the context of Ascariasis epidemiology due to the absence of distinguishing morphological characteristics among them [4]. Proper identification and genetic characterization of infecting strains from endemic areas throughout the world are certainly important to explain the disease epidemiology and also the taxonomic status of two Ascaris species. Several molecular epidemiological investigation based on polymorphic markers like-internal transcribed spacer 1 (ITS1), mitochondrial cytochrome c oxidase subunit 1 (cox1), NADH dehydrogenase subunit 1 (nad1) and microsatellite markers have been proposed to explain the origin of the two ascarid taxa in their respective hosts and their taxonomic status [5–8]. However, non-repetitive genomic regions have been preferred over repetitive regions as genotyping marker for their high genetic stability and evolutionary significance. Single nucleotide substitution occurred just once in the phylogenetic history of a species, unlikely to mutate again to either a novel or ancestral genotype [9]. Using the non-repetitive marker ITS1, 5 Ascaris genotypes (G1-G5) in human and 3 Ascaris genotypes (G1-G3) in pig have been identified. G1 frequently infects human, while G3 is predominant in pigs. The other three has been detected in lower frequencies in their respective hosts [5]. Recently, a study from Brazil based on ITS1 marker reported a new Ascaris genotype G6 in human [10]. However, no such information regarding genetic pattern and diversity of Ascaris population from India are available still date. Hence, the present study was designed to generate an idea about genetic patterns and diversity of Indian Ascaris population and also to determine their phylogenetic relation with the global Ascaris population. The result revealed a considerable amount of polymorphism within our Ascaris population. Along with the previously reported and widely distributed genotype G1 [5,10], 8 new sequence variants of ITS1 (IND1-IND8) have been identified. Since, Ascaris sp. multiply through sexual reproduction and genetic recombination during meiosis is a natural phenomenon [11], Efforts were also made to determine whether this population diversity are associated with genetic shuffling. Intragenic linkage disequilibrium (LD) among our study population was evaluated at ITS1 locus to identify potential recombination events within them. Moreover, any significant association of Ascaris genotypes with patient's age and sex was also evaluated.
2. Material and methods
2.1. Sample collection and detection of Ascaris sp.:
A total of 35 Ascaris isolates from human were included in our study. Fecal samples were collected from people of “low socio-economic community of Kolkata” through an on-going field project, studying the parasite burden of those communities. Poor hygiene, sanitation and malnutrition were common in those communities [12]. The ethical clearance for this study has been provided by NICED IEC (i.e. National Institute of Cholera and Enteric Diseases Institutional Ethical Committee). Informed consents have been obtained from the patients (in case of children consents have been obtained from their parents). The parasite's eggs within the fecal were primarily detected by microscopy [13]. DNA was isolated directly from microscopy positive fecal samples using STOOL DNA Minikit (QIAGEN, USA) as per manufacturer's protocol.
2.2. Polymerase chain reaction (PCR) amplification and DNA sequencing
Partial amplification of target gene (ITS1) was performed using gene specific primer pairs (Table 1). In all cases the PCR reaction was performed in 50 μl reaction volume containing approximately 0.4 μg and 0.1 μg of template DNA for primary and nested PCR respectively, 10 pM of each primer, 2.5 mM MgCl2, 1 μg of Bovine Serum Albumin (SIGMA, USA), 200 μM dNTP and 2.5 U of Taq DNA polymerase (Bioline, USA) with the reaction parameters as initial denaturation for 5 min or 4 min (Primary and Nested respectively) at 94 °C. This was followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 65 °C or 60 °C (Primary and Nested respectively) for 30 s, extension at 72 °C for 30 s. This was again followed by the final extension for 10 min at 72 °C. The amplified PCR products were then separated by electrophoresis on 1.5% agarose gels (SIGMA, USA) according to their size. PCR products of expected sizes were extracted from gels and purified (ROCHE, Germany). The Purified PCR products were then sequenced directly with specific primers (marked with a in Table 1) using the ‘BigDye Terminator V3.1 cycle sequencing kit’ (APPLIED BIOSYSTEMS, USA) as per the manufacturer's protocol. The labeled DNA fragments were further purified by sodium acetate and ethanol precipitation. The sequencing was carried out in an ABI 310 PRISM Automated Genetic Analyzer. Accuracy of DNA sequencing data has been confirmed by sequencing in both directions and also by repetition of DNA sequencing with a new PCR product for all study isolates.
Table 1.
List of gene specific primers, used in the study.
| Gene name | PCR round | Primer name | Primer sequence (5′ to 3′) |
|---|---|---|---|
| Internal transcribed spacer 1 (ITS1) | Primary | ITS F 1 | CGAGCAGAAAAAAAAAAGTCTCC |
| ITS R1 | GGAATGAACCCGATGGCGCAAT | ||
| Secondary | ITS F 2a | CGAGCAGAAAAAAAAAAAAGTCTCC | |
| ITS R 2a | GCTGCGTTCTTCATCGAT |
Gene specific primer pairs used for sequencing of amplified PCR products.
2.3. Analysis of sequence polymorphisms
ITS1 sequences of our study isolates were aligned with all previously published sequences of corresponding loci (downloaded from NCBI GenBank database, accession numbers have been provided in Table 2) using ClustalW multiple alignment program of MEGA version 4 software [14]. Nucleotide position of each single nucleotide polymorphism (SNP) within the target loci was identified from the aligned sequences. The nucleotide positions of SNPs within the target loci were relative to the reference sequence of G1 genotype (GenBank accession number AJ554036) (Table 2). Variable sequences of our target loci (in respect to the reference sequence) were submitted to NCBI GenBank database with accession numbers JN176638 – JN176674.
Table 2.
List of polymorphic sites identified within internal transcribed spacer 1 (ITS1) region among various Ascaris isolates from India and Worldwide.
| Sample ID | GenBank accession number | Genotypes/haplotypes | Country | Host | Nucleotide variation at alignment positiona | References |
|---|---|---|---|---|---|---|
| WP01 | AJ554036 | G1b | Bac/Brd/Jae/Chf | Hug/Ph | 120T, 122T, 123T, 124T, 125T, 127T, 128--i, 129 --i, 130G, 131C, 132G,133G, 134A, 135C, 139T, 142A, 148 --i, 150T, 151T, 155A, 156T, 157--i, 162T, 167A, 168A, 169G, 172T, 173G, 176T, 181T, 183T, 203G, 205C, 206G, 207C, 218T, 229T, 231C, 233T, 238A, 248T | [5,10,21] |
| WP40 | AJ554037 | G2b | Chf | Hug/Ph | 128--i/T, 133G/Sj | [5] |
| WP17 | AJ554038 | G3b | Jae/Chf | Hug/Ph | 128--i/T, 133G/C, 248T/A | [5,21] |
| WP03 | AJ554039 | G4b | Chf | Hug | 173G/Rk | [5] |
| WP37 | AJ554040 | G5b | Chf | Hug | 133G/Sj, 248T/Wl | [5] |
| DL04 | EF153621 | G6b | Brd | Hug | 120T/--i | [10,17] |
| AJ000895H | AJ000895 | Alm | Aun | Hug | 205C/Sj | [7] |
| AJ000896P | AJ000896 | Aso | UKp/Deq | Ph | 128--i/T, 129--i/T, 133G/C, 205C/Sj, 248T/A | [7] |
| DL02Ec9 | GQ339794 | H1b | Brd | Hug | 203G/A | [17] |
| DL04Ec1 | EU635686 | H2b | Brd | Hug | 124T/--i, 127T/--i, 156T/C, 231C/T | [17] |
| DL04Ec2 | EU635687 | H3b | Brd | Hug | 127T/--i, 183T/A | [17] |
| DL04Ec3 | EU635688 | H4b | Brd | Hug | 156T/C, 127T/--i | [17] |
| DL13Ec2 | GQ339795 | H5b | Brd | Hug | 167A/G, 120T/--i | [17] |
| DL13Ec4 | GQ339796 | H6b | Brd | Hug | 150T/C | [17] |
| DL15Ec1 | GQ339797 | H7b | Brd | Hug | 120T/--i, 229T/C | [17] |
| DL16Ec1 | GQ339798 | H8b | Brd | Hug | 120T/C, 233T/C | [17] |
| DL16Ec3 | GQ339799 | H9b | Brd | Hug | 124T/--i, 218T/C | [17] |
| DL17Ec2 | EU635694 | H10b | Brd | Hug | 127T/--i, 238A/G | [17] |
| DL17Ec14 | EU635695 | H11b | Brd | Hug | 127T/--i, 130G/T, 155A/G, 156T/C, 231C/T | [17] |
| 041-1Ec4 | GQ339800 | H12b | Brd | Hug | 120T/--i, 229T/A, 248T/C | [17] |
| 104-5Ec1 | GQ339801 | H13b | Brd | Hug | 124T/C | [17] |
| I158 | JN176638 | G1b | INDr | Hug | Is | [*]t |
| I305 | JN176639 | G1b | INDr | Hug | Is | [*]t |
| I300 | JN176640 | G1b | INDr | Hug | Is | [*]t |
| I152 | JN176641 | G1b | INDr | Hug | Is | [*]t |
| I6 | JN176642 | G1b | INDr | Hug | Is | [*]t |
| I172 | JN176643 | G1b | INDr | Hug | Is | [*]t |
| I149 | JN176644 | G1b | INDr | Hug | Is | [*]t |
| I31 | JN176645 | G1b | INDr | Hug | Is | [*]t |
| I203 | JN176646 | G1b | INDr | Hug | Is | [*]t |
| I170 | JN176647 | G1b | INDr | Hug | Is | [*]t |
| I449 | JN176648 | G1b | INDr | Hug | Is | [*]t |
| I450 | JN176649 | G1b | INDr | Hug | Is | [*]t |
| I2878 | JN176655 | G1b | INDr | Hug | Is | [*]t |
| I198 | JN176658 | G1b | INDr | Hug | Is | [*]t |
| I189 | JN176659 | G1b | INDr | Hug | Is | [*]t |
| I33 | JN176660 | G1b | INDr | Hug | Is | [*]t |
| I2212 | JN176663 | G1b | INDr | Hug | Is | [*]t |
| I2946 | JN176664 | G1b | INDr | Hug | Is | [*]t |
| I236 | JN176665 | G1b | INDr | Hug | Is | [*]t |
| I2910 | JN176666 | G1b | INDr | Hug | Is | [*]t |
| I17 | JN176667 | G1b | INDr | Hug | Is | [*]t |
| I2864 | JN176668 | G1b | INDr | Hug | Is | [*]t |
| I2838 | JN176669 | G1b | INDr | Hug | Is | [*]t |
| I2942 | JN176670 | G1b | INDr | Hug | Is | [*]t |
| I611 | JN176672 | G1b | INDr | Hug | Is | [*]t |
| I75 | JN176673 | G1b | INDr | Hug | Is | [*]t |
| I161 | JN176674 | G1b | INDr | Hug | Is | [*]t |
| I451 | JN176651 | IND1u | INDr | Hug | 142A/C, 148--i/G, 157--i/T | [*]t |
| I2990 | JN176653 | IND2u | INDr | Hug | 130G/A, 131C/G, 173G/C | [*]t |
| I2788 | JN176654 | IND3u | INDr | Hug | 122T/A | [*]t |
| I288 | JN176656 | IND4u | INDr | Hug | 162T/--i | [*]t |
| I196 | JN176657 | IND5u | INDr | Hug | 120T/A | [*]t |
| I2905 | JN176661 | IND6u | INDr | Hug | 122T/A, 123T/A, 124T/A, 125T/A, 131C/G, 132G/A, 134A/G, 135C/A, 151T/G, 168A/T, 169G/T, 172T/A, 181T/G | [*]t |
| I567 | JN176662 | IND7u | INDr | Hug | 134A/G | [*]t |
| I722 | JN176671 | IND8u | INDr | Hug | 139T/--i | [*]t |
Previously reported Ascaris genotypes (G1– G6)/haplotypes (H1– H13).
Ba: Bangladesh.
Br: Brazil.
Ja: Japan.
Ch: China.
Hu: human.
P: pig.
--: nucleotide deletion.
S: nucleotide G or C.
R: nucleotide A or G.
W: nucleotide A or T.
Al: A. lumbricoides (without nomenclature).
Au: Australia.
As: A. suum (without nomenclature).
UK: United Kingdom.
De: Denmark.
IND: India.
I: similar to reference sequence G1.
[*]: identified in this present study.
New sequence variants of ITS1, identified in our study.
Intragenic LD and number of recombination events at ITS1 locus among our study population were also assessed by using DnaSP version 5.10.01 (www.ub.es/dnasp/) software.
2.4. Statistical analysis
Associations of Ascaris genotypes with patient's age and sex have been evaluated by Epi-Info version 3.5.4 software [15].
2.5. Phylogenetic analysis
Phylogenetic trees were constructed from the previously aligned ITS1 sequences by MEGA version 4 software [14]. Two individual methods [i.e. Neighbor-Joining (NJ) and Maximum parsimony (MP)] were used to confirm the topology of the tree. In both cases, widely distributed and most prevalent Ascaris genotype G1 (Genbank ID AJ554036) was considered as an out-group. The bootstrap values were also analyzed to estimate confidence intervals. Genetic distance analysis among our study isolates was also performed using MEGA version 4 software [14].
3. Results
Among our 35 study isolates, majority (27) corresponded to previously reported and widely distributed genotype G1 [5,10]. Along with genotype G1, 8 new sequence variants of ITS1 have also been identified. These new sequence variants have assigned alphanumerical codes beginning with letter ‘IND’ to indicate their Indian origin (i.e. IND1-IND8) (Table 2).
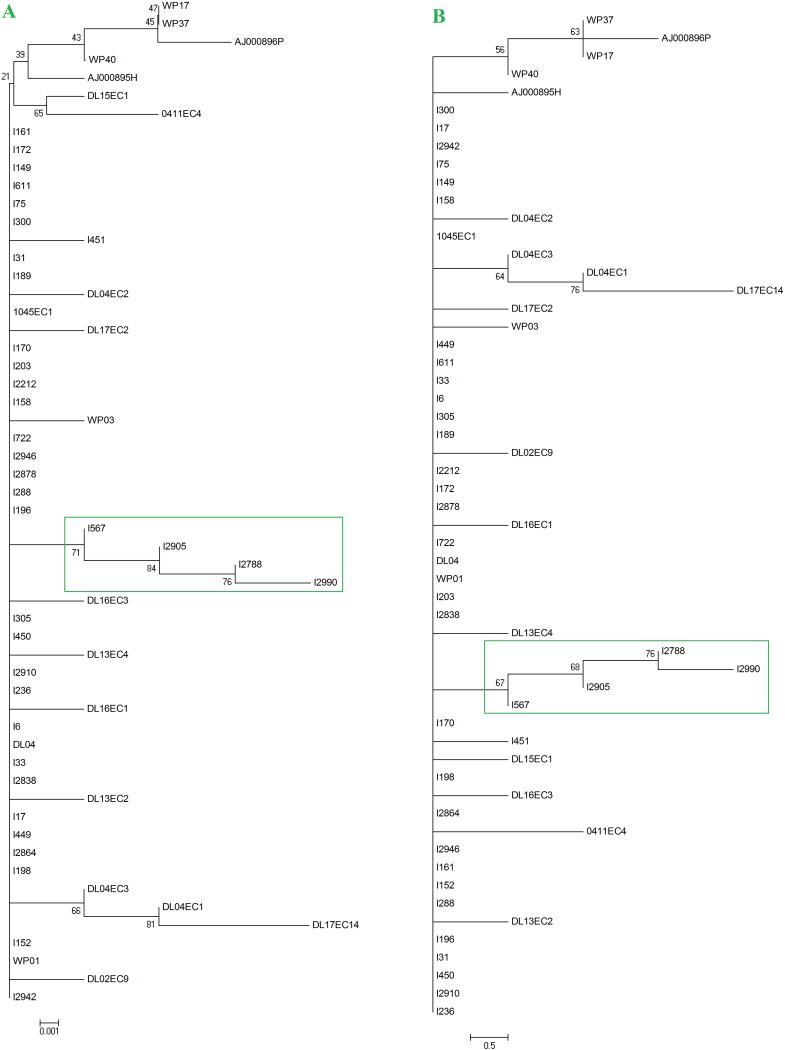
Sequence comparison of our study isolates with the global Ascaris population has revealed 17 new SNPs, which were present in 8 newly identified ITS1 sequences from our Indian Ascaris population (i.e. IND1-IND8) (Table 2). Phylogenetic comparison of our study isolates with the global Ascaris population using two individual methods (i.e. NJ and MP) generates trees with similar topology. In both cases, few of our Indian isolates (with newly identified ITS1 sequences) formed distinct cluster with high bootstrap value (marked with green color in both trees), which may indicates their distinct phylogenetic position in respect to global Ascaris population (Fig. 1). We have also performed genetic distance analysis among our study isolates. The result has been provided in data 1. Intragenic LD between pairs of polymorphic sites at ITS1 locus of our study population was also evaluated to identify potential recombination events within them. Among 171 pairwise comparisons, 92 were significant by Chi-square test and 89 were significant after Bonferroni correction (Table 3). An incomplete LD value (|D′| Y = 0.9818 + 0.1974X, where Y is the LD value and X is the nucleotide distance in kilobases) was also detected (Table 3). Moreover, intragenic recombination analysis at ITS1 locus of our study isolates has identified 2 potential recombination events within our study population (Table 3). Moreover, any significant association of Ascaris genotypes with patient's age and sex was also studied, which revealed that G1 genotype was significantly present among female patients (co-efficient value = 0.815, p value = 0.000002) aged between 10 to 15 years (co-efficient value = 0.690, p value = 0.000105). The age and sex information of the patients, included in our study has been provided in Table 4.
Fig. 1.
Phylogenetic comparison of our study isolates with the global Ascaris population: Internal transcribed spacer 1 (ITS1) sequences of our study isolates were aligned with ITS1 sequences from the global Ascaris population using ClustalW multiple alignment program of MEGA version 4 software. Two separate phylogenetic trees were generated from this alignment using two individual methods- [A] Neighbor-Joining (NJ) and [B] Maximum parsimony (MP). In both cases, widely distributed and most prevalent Ascaris genotype G1 (Genbank ID AJ554036) was considered as out-group. The bootstrap values were also analyzed to estimate confidence intervals. Phylogenetic comparison using these two individual methods (i.e. NJ and MP) generates trees with similar topology. In both cases, few of our Indian isolates (with newly identified ITS1 sequences) formed distinct cluster with high bootstrap value (marked with green color in both trees), which may indicates their distinct phylogenetic position in respect to global Ascaris population.
Table 3.
Intragenic linkage disequilibrium (LD) and recombination analysis at internal transcribed spacer 1 (ITS1) region of our study isolates.
| Population | Number of samples analyzed | Number of polymorphic sites analyzed | Number of pairwise comparisons | Number of significant pairwise comparisons a | Intragenic LD(|D′|) b | Rm c |
|---|---|---|---|---|---|---|
| Indian | 35 | 21 | 171 | 92(89) | Y = 0.9818 + 0.1974X | 2 |
Number of significant pairwise comparisons by Chi square test (after Bonferroni correction).
Intragenic linkage disequilibrium (LD), where Y is the LD value and X is the nucleotide distance in kilobases.
Minimum number of intragenic recombination events.
Table 4.
Age and sex information of patients, included in the study.
| Sample ID | Patient information |
GenBank accession number | Genotypes/haplotypes | |
|---|---|---|---|---|
| Age (Y:M) | Sex (M/F) | |||
| I158 | 10.2 | M | JN176638 | G1a |
| I305 | 12.6 | F | JN176639 | G1a |
| I300 | 10.7 | F | JN176640 | G1a |
| I152 | 14.11 | F | JN176641 | G1a |
| I6 | 10.7 | F | JN176642 | G1a |
| I172 | 12.5 | F | JN176643 | G1a |
| I149 | 12.1 | F | JN176644 | G1a |
| I31 | 15.0 | F | JN176645 | G1a |
| I203 | 12.6 | F | JN176646 | G1a |
| I170 | 11.8 | M | JN176647 | G1a |
| I449 | 11.7 | F | JN176648 | G1a |
| I450 | 13.9 | F | JN176649 | G1a |
| I2878 | 15.10 | F | JN176655 | G1a |
| I198 | 27.1 | F | JN176658 | G1a |
| I189 | 14.5 | M | JN176659 | G1a |
| I33 | 12.6 | F | JN176660 | G1a |
| I2212 | 26.9 | M | JN176663 | G1a |
| I2946 | 9.7 | M | JN176664 | G1a |
| I236 | 14.4 | F | JN176665 | G1a |
| I2910 | 10.9 | F | JN176666 | G1a |
| I17 | 10.1 | F | JN176667 | G1a |
| I2864 | 11.3 | F | JN176668 | G1a |
| I2838 | 13.2 | F | JN176669 | G1a |
| I2942 | 12.8 | F | JN176670 | G1a |
| I611 | 10.8 | F | JN176672 | G1a |
| I75 | 15.4 | F | JN176673 | G1a |
| I161 | 14.3 | F | JN176674 | G1a |
| I451 | 8.0 | F | JN176651 | IND1b |
| I2990 | 6.5 | M | JN176653 | IND2b |
| I2788 | 9.7 | M | JN176654 | IND3b |
| I288 | 12.7 | F | JN176656 | IND4b |
| I196 | 9.2 | M | JN176657 | IND5b |
| I2905 | 7.6 | M | JN176661 | IND6b |
| I567 | 37.5 | F | JN176662 | IND7b |
| I722 | 23.5 | F | JN176671 | IND8b |
Previously reported Ascaris genoype G1, identified in our study.
New sequence variants of target locus (i.e. ITS1), identified in our study.
4. Discussion
A. lumbricoides and A. suum are two of the world's most common soil transmitted nematode and together cause serious health and socio-economic problems. Ascariasis has been considered as Neglected Tropical Diseases (NTD) by WHO, since it is highly prevalent in poor urban and rural areas and has a certain impact on patient's health, physical fitness and productivity [1,2]. Morphological similarity of these two nematodes entails ongoing uncertainty concerning their taxonomic status and argues for the need to explore deeper into their molecular epidemiology [4]. A recent surveillance study among school children from south India revealed a highest prevalence of Ascaris species among all STHs infections. Co-infection with other STHs has also been reported [16]. Even though few surveillance studies on Ascaris infection have been conducted in India, diagnosis of this parasite was solely based on microscopy. Differentiation between Ascaris species certainly cannot be confirmed by microscopy but require detailed molecular epidemiological study based on genetic markers. In the present study, Ascaris population from human has been genetically characterized based on widely used genetic marker ITS1.
Sequence analysis of our study isolates has identified G1 as a dominant genotype. As much as 27 among 35 study isolates were corresponding to this widely distributed genotype. This result corroborates with previous report from China, where genotype G1 was dominant among human and G3 among pig [5]. Our study has also identified 8 new sequence variants of ITS1 (IND1-IND8) within our Indian Ascaris population. Similar finding was previously reported by Leles et.al from Brazil. They have also identified 13 new Ascaris haplotypes (H1–H13) from human [17]. Sequence comparison of our study isolates with previously reported Ascaris sequences has identified 17 new SNPs within our study isolates. Moreover, all of these SNPs were present within 8 newly identified sequence variants of ITS1 (IND1–IND8), which indicates their distinct genetic organization. This finding was further well supported by the observation of phylogenetic analysis. All the previously reported Ascaris sequences were retrieved from NCBI database and Ascaris sequences from our study isolates were phylogenetically compared with them. Phylogenetic analysis revealed an interesting scenario. Few of our Indian isolates (with new variations of ITS1 sequences) formed a separate cluster with high bootstrap value, indicating their distinct phylogenetic position in respect to the global Ascaris population. Moreover, Intragenic LD analysis between pairs of polymorphic sites at ITS1 locus has identified an incomplete LD value with two potential recombination events within our study population. This finding was quite compatible with a previous report by Li et.al. They have identified a similar type of observation (presence of intragenic LD value and recombination events) in gp60 locus of another enteric parasite, Cryptosporidium homonis [18]. Since, Ascaris sp. multiply through sexual reproduction [11], genetic recombination during meiosis could be a natural phenomenon. Furthermore, a recent study has identified the molecular evidence of polyandry in A.suum. Single female of A. suum can mate with multiple males, which can also increase the chance of genetic variations [19]. Such high possibilities of genetic shuffling could be associated with increasing population diversity in a restricted geographic region [5,7,10,17] and frequent emergence of new sequence variants, identified in present as well as in previous studies [17]. Attempts were also made to determine whether any statistically significant association exists between the identified Ascaris genotypes and patient's age and sex. Genotype G1 was found to be significantly present among female patients (co-efficient value = 0.815, p value = 0.000002) aged between 10 to 15 years (co-efficient value = 0.690, p value = 0.000105). This finding was quite congruous with a previous report by Anuar et.al [20]. They have reported that Ascariasis was significantly related to patients aged < 15 years and earning low household income.
Since, Ascariasis is one of the major Soil Transmitted Helminthiases (STHs) and has been declared as Neglected Tropical Diseases (NTD) by WHO, genome information of its infecting strains from different parts of the world is certainly crucial to investigate the disease epidemiology. This study explores the genetic organization of Indian Ascaris population for the first time; it will certainly include some fundamental information on the molecular epidemiology of Ascariasis.
Acknowledgments
This study was jointly supported by the grants from National Institute of Infectious Diseases, Japan, Okayama University Program of Founding Research Centre for Emerging and Re-emerging Infectious Disease (OUP 2-5), and Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors also acknowledge to the patients, provide their fecal samples for the study. Authors would like to thank Mrs. Debarati Ganguly for her immense help regarding proof-reading of this manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.csbj.2015.08.006.
Contributor Information
Koushik Das, Email: koushikdas55@gmail.com.
Punam Chowdhury, Email: punam.bt07@gmail.com.
Sandipan Ganguly, Email: sandipanganguly@gmail.com.
Appendix A. Supplementary data
Genetic distance analysis among our study isolates using MEGA version 4 software.
References
- 1.Dold C., Holland C.V. Ascaris and ascariasis. Microbes Infect. 2011;13:632–637. doi: 10.1016/j.micinf.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 2.De Silva N.R., Brooker S., Hotez P.J., Montresor A., Engels D. Soil-transmitted helminth infections: updating the global picture. Trends Parasitol. 2003;19:547–551. doi: 10.1016/j.pt.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Nejsum P., Jr Parker E.D., Frydenberg J., Roepstorff A., Boes J. Ascariasis is a zoonosis in Denmark. J Clin Microbiol. 2005;43:1142–1148. doi: 10.1128/JCM.43.3.1142-1148.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavallero S., Snabel. V., Pacella F., Perrone V., D'Amelio S. Phylogeographical studies of Ascaris spp. based on ribosomal and mitochondrial DNA sequences. PLoS Negl Trop Dis. 2013;7(4):2170. doi: 10.1371/journal.pntd.0002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng W., Yuan K., Zhou X., Hu M., EL-Osta Y.G. Molecular epidemiological investigation of Ascaris genotypes in China based on single-strand conformation polymorphism analysis of ribosomal DNA. Electrophoresis. 2003;24:2308–2315. doi: 10.1002/elps.200305455. [DOI] [PubMed] [Google Scholar]
- 6.Peng W., Yuan K., Hu M., Zhou X., Gasser R.B. Mutation scanning-coupled analysis of haplotypic variability in mitochondrial DNA regions reveals low gene flow between human and porcine Ascaris in endemic regions of China. Electrophoresis. 2005;26:4317–4326. doi: 10.1002/elps.200500276. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X., Chilton N.B., Jacobs E., Boes J., Gasser R.B. Characterization of Ascaris from human and pig hosts by nuclear ribosomal DNA sequences. Int J Parasitol. 1999;29:469–478. doi: 10.1016/s0020-7519(98)00226-4. [DOI] [PubMed] [Google Scholar]
- 8.Criscione C.D., Anderson J.D., Sudimack D., Peng W., Jha B. Disentangling hybridization and host colonization in parasitic roundworms of humans and pigs. Proc Biol Sci. 2007;274:2669–2677. doi: 10.1098/rspb.2007.0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharya D., Haque R., Singh U. Coding and noncoding genomic regions of Entamoeba histolytica have significantly different rates of sequence polymorphisms: implications for epidemiological studies. J Clin Microbiol. 2005;43:4815–4819. doi: 10.1128/JCM.43.9.4815-4819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leles D., Araújo A., Vicente A.C.P., Iñiguez A.M. Molecular diagnosis of ascariasis from human feces and description of a new Ascaris sp. genotype in Brazil. Vet Parasitol. 2009;163:167–170. doi: 10.1016/j.vetpar.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein P. Spermatogenesis and spermiogenesis in Ascaris lumbricoides Var. suum. J Morphol. 1977;154(3):317–337. doi: 10.1002/jmor.1051540302. [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee A.K., Chowdhury P., Bhattacharya M.K., Ghosh M., Rajendran K. BMC research notes. Vol. 2. 2009. Hospital-based surveillance of enteric parasites in Kolkata; p. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee A.K., Chowdhury P., Das K., Raj D., Karmakar S. India: a survey report. 2 (3) 2013. Helminth burden among school going children of southern Bengal; pp. 189–191. (G.J.B.A.H.S) [Google Scholar]
- 14.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 15.Dean A.G., Arner T.G., Sunki G.G., Friedman R., Lantiga M. Centers for Disease Control and Prevention; Atlanta, Georgia, USA: 2007. Epi InfoTM, a database and statistic program for public health professional. [Google Scholar]
- 16.Ragunathan L., Kalivaradhan S.K., Ramadass S., Nagaraj M., Ramesh K. Helminthic infections in school children in Puducherry, South India. J Microbiol Immunol Infect. 2010;43(3):228–232. doi: 10.1016/S1684-1182(10)60036-9. [DOI] [PubMed] [Google Scholar]
- 17.Leles D., Araújo A., Vicente A.C.P., Iñiguez A.M. ITS1 intra-individual variability of Ascaris isolates from Brazil. Parasitol Int. 2010;59:93–96. doi: 10.1016/j.parint.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Li N., Xiao L., Cama V.A., Ortega Y., Gilman R.H. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis. 2013;19:1573–1582. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C., Yuan K., Tang X., Hu N., Peng W. Molecular genetic evidence for polyandry in Ascaris suum. Parasitol Res. 2011;108(3):703–708. doi: 10.1007/s00436-010-2116-3. [DOI] [PubMed] [Google Scholar]
- 20.Anuar T.S., Md Salleh F., Moktar N. Soil-transmitted Helminth infections and associated risk factors in three Orang Asli tribes in Peninsular Malaysia. Sci Rep. 2014;4:4101. doi: 10.1038/srep04101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishiwata K., Shinohara A., Yagi K., Horii Y., Tsuchiya K. Identification of tissue-embedded ascarid larvae by ribosomal DNA sequencing. Parasitol Res. 2004;92:50–52. doi: 10.1007/s00436-003-1010-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic distance analysis among our study isolates using MEGA version 4 software.