Abstract
Advances in genome engineering technologies have made the precise control over genome sequence and regulation possible across a variety of disciplines. These tools can expand our understanding of fundamental biological processes and create new opportunities for therapeutic designs. The rapid evolution of these methods has also catalyzed a new era of genomics that includes multiple approaches to functionally characterize and manipulate the regulation of genomic information. Here, we review the recent advances of the most widely adopted genome engineering platforms and their application to functional genomics. This includes engineered zinc finger proteins, TALEs/TALENs, and the CRISPR/Cas9 system as nucleases for genome editing, transcription factors for epigenome editing, and other emerging applications. We also present current and potential future applications of these tools, as well as their current limitations and areas for future advances.
Genomic research has the potential to dramatically improve medicine, agriculture, biotechnology, and our fundamental understanding of living systems. Recent advances have generated extensive annotation of genomic and epigenomic regulatory modules within chromatin (Bernstein et al. 2010; The ENCODE Project Consortium 2012; Roadmap Epigenomics Consortium et al. 2015), as well as an understanding of genomic topological architecture (Dekker et al. 2013; Pombo and Dillon 2015). However, the roles of these numerous genes, regulatory elements, epigenetic marks, and topological domains in determining overall cell function remain incompletely understood. The recent development of genome engineering technologies has enabled precise interrogation of the function of these genomic features and their causal role in gene regulation. Additionally, these tools are facilitating the translation of this genomic information into tangible benefits for biotechnology, agriculture, and human therapeutics. In this Perspective, we discuss the recent advances to the most commonly used genome engineering technologies, including synthetic zinc finger (ZF) proteins, transcription activator-like effectors (TALEs), and CRISPR/Cas9 targeting systems, and their application in a new era of functional genomics.
Genome engineering technologies
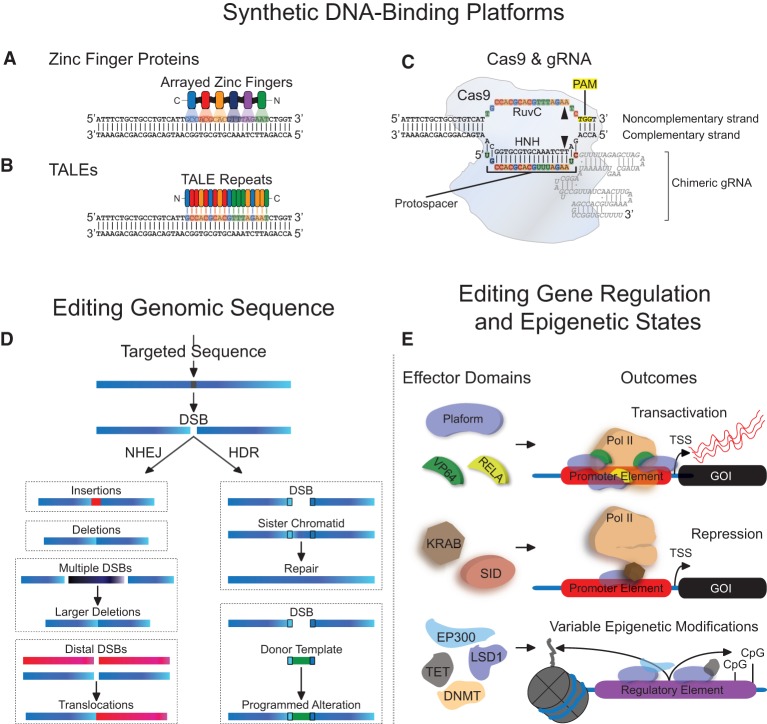
Cys2-His2 ZF domains are naturally occurring protein motifs which typically recognize three base pairs within the major groove of DNA (Pavletich and Pabo 1991; Wolfe et al. 2000). These modular ZF domains can be arrayed such that synthetic ZF DNA-binding proteins (DBPs) target a specific series of DNA triplets at unique genomic addresses (Fig. 1A; Liu et al. 1997; Gersbach et al. 2014). TALE proteins are components of plant pathogens that bind host DNA to facilitate virulence (Kay et al. 2007; Romer et al. 2007). TALEs consist of repeated DNA-binding domains containing repeat variable diresidues (RVDs), each of which recognizes a single nucleotide in target DNA (Boch et al. 2009; Moscou and Bogdanove 2009). Similar to ZFs, individual TALE RVDs can be linked in series to localize TALEs to target loci (Fig. 1B; Christian et al. 2010; Morbitzer et al. 2010; Cermak et al. 2011; Miller et al. 2011; Zhang et al. 2011). Clustered regularly interspaced short palindromic repeat (CRISPR) arrays and CRISPR-associated (Cas) proteins are components of bacterial and archaeal adaptive immune systems (Barrangou et al. 2007; Makarova et al. 2011). Unlike ZFs and TALEs, in which protein moieties dictate DNA recognition, CRISPR/Cas systems utilize RNA-mediated Watson-Crick bonding for recognition of nucleic acids.
Figure 1.
Zinc finger, TALE, and Cas9-gRNA platforms for editing genomic sequence and regulatory states. Individual zinc finger domains (A) and TALE repeats (B) that recognize unique triplets or single base pairs, respectively, can be arrayed in engineered proteins to target specific genomic sequences. (C) Cas9 in complex with a chimeric guide RNA (gRNA) can recognize a specific genomic address through complementarity between the protospacer segment of the gRNA and target DNA. The formation of this complex is dependent upon the presence of a protospacer adjacent motif (PAM). The RuvC and HNH nuclease domains of Cas9 cleave genomic DNA that matches the protospacer (i.e., the noncomplementary strand) and genomic DNA with complementarity to the protospacer (i.e., the complementary strand), respectively (indicated by black triangles). (D) Zinc fingers and TALEs fused to nuclease domains or Cas9 in complex with a gRNA can cleave targeted sequences to generate double-strand breaks (DSBs). DSB resolution through nonhomologous end joining (NHEJ) or homology-directed repair (HDR) can lead to various alterations in genomic sequence. (E) Zinc finger, TALE, or deactivated nuclease-null Cas9 (dCas9) platforms can also be fused to diverse effector domains to modify endogenous gene regulation and epigenetic states: (TSS) transcription start site; (GOI) gene of interest.
Prokaryotes harboring type II CRISPR/Cas systems transcribe CRISPR-RNAs (crRNAs) that hybridize with trans-activating crRNAs (tracrRNAs) that complex with the Cas9 nuclease (Brouns et al. 2008; Deltcheva et al. 2011; Jinek et al. 2012; Doudna and Charpentier 2014). A single crRNA-tracrRNA chimera, known as a guide RNA (gRNA), can be designed for simplified use in engineered systems (Jinek et al. 2012). The gRNA binds to and directs the Cas9 protein to DNA through regions of crRNA complementarity (termed “protospacer” sequences). A stringent prerequisite to protospacer hybridization is the presence of a protospacer adjacent motif (PAM) in the target DNA, which flanks the region of protospacer complementarity (Fig. 1C; Mojica et al. 2009; Anders et al. 2014; Sternberg et al. 2014; Kleinstiver et al. 2015). Interactions between the PAM-proximal “seed” nucleotides in the target site and the complementary gRNA sequence are also critical drivers of Cas9 targeting. The orthogonality of various prokaryotic Cas9 proteins with differing PAM requirements can be exploited for multiplex genome engineering efforts (Esvelt et al. 2013; Ran et al. 2015).
Engineered targeting of eukaryotic genomes with ZFs, TALEs, and type II CRISPR/Cas systems has established these technologies as useful resources for present and future genome engineering endeavors. Although each of these systems has been successfully incorporated into diverse genome engineering strategies, they each have unique benefits and limitations that depend upon the particular application (Gaj et al. 2013; Carroll 2014). Other genome engineering technologies, such as meganucleases and their fusion to TALE proteins (Boissel et al. 2014; Stoddard 2014) have also been successfully applied to gene editing in eukaryotes, but have been less widely adopted in the context of functional genomics due to the complexity involved in engineering meganucleases targeted to new sequences.
Editing genome sequences with programmable nucleases
Gene targeting based on homologous recombination can introduce exogenous DNA at genomic loci (Smithies et al. 1985; Thomas et al. 1986). The efficiency of this method is dramatically enhanced in the presence of double-strand breaks (DSBs) (Rouet et al. 1994). Cells generally use two distinct pathways to resolve DSBs (Chapman et al. 2012): homology-directed repair (HDR) and nonhomologous end joining (NHEJ) (Fig. 1D). DSB resolution through NHEJ occurs by direct ligation of DSB ends or through microhomology on DSB termini (Lieber 2010). This error-prone process results in small insertions or deletions (indels) at endogenous loci. The generation of two DSBs flanking a genomic region can also lead to NHEJ-mediated chromosomal deletions (Lee et al. 2010; Carlson et al. 2012; Kim et al. 2013; Essletzbichler et al. 2014) and inversions (Carlson et al. 2012; Lee et al. 2012; Xiao et al. 2013). Programmable nucleases can also be used to create NHEJ-mediated translocations in vivo (Brunet et al. 2009; Maddalo et al. 2014). HDR uses regions of homologous DNA on sister chromatids or exogenous DNA to repair DSBs. In contrast to NHEJ, HDR is a high-fidelity process leading to largely error-free correction at DSB sites. HDR can lead to specified incorporation of sequences that are rationally designed into donor DNA templates. In many instances, this is advantageous over NHEJ, in which DSB resolution is unpredictable. However, NHEJ is active throughout the cell cycle and is the predominant DSB repair mechanism, whereas HDR occurs less frequently and is significantly down-regulated outside of S and G2 cell cycle phases. Therefore recent efforts have focused on promoting HDR by inhibition of NHEJ events to afford more precise genome editing (Chu et al. 2015; Maruyama et al. 2015).
Because NHEJ and HDR can be used to incorporate specific sequence changes into genomes, the capability to induce DSBs at target loci holds great potential for genome engineering (Carroll 2014; Kim and Kim 2014). Although ZFs and TALEs do not intrinsically cleave target DNA, they can be directly fused to the catalytic domain of the type II restriction endonuclease FokI (Li et al. 1992) to create ZF and TALE nucleases (ZFNs and TALENs). Engineering new DNA-binding specificities into the ZFs and TALEs allows programmed DSB induction (Kim et al. 1996; Christian et al. 2010). FokI domains must dimerize to cleave target DNA (Bitinaite et al. 1998; Vanamee et al. 2001), necessitating the engineering of two ZFN or TALEN monomers for each DSB. Modifications to the FokI domain have also been created to increase nuclease activity and specificity (Miller et al. 2007; Szczepek et al. 2007; Guo et al. 2010; Doyon et al. 2011b). Moreover, fusion of the FokI domain to the nuclease-inactivated Cas9 protein (dCas9) has also been used to increase the specificity of the CRISPR gene editing system (Guilinger et al. 2014b; Tsai et al. 2014). Optimizing the length of the ZF or TALE array can also increase nuclease activity and specificity (Bhakta et al. 2013; Guilinger et al. 2014a).
As an alternative to introducing DSBs and inducing DNA repair pathways, catalytic domains of site-specific recombinases can also be fused to synthetic ZFs (Akopian et al. 2003) and TALEs (Mercer et al. 2012) to excise genomic DNA segments or integrate exogenous DNA at targeted genomic sites (Gordley et al. 2009; Gersbach et al. 2011). CRISPR/Cas-based recombinase fusions have not yet been reported but may also prove useful. Additionally, transposases catalyze the rearrangement of endogenous elements and have been used for artificial genome manipulation (Ivics et al. 2009). Targeting of transposase activity with ZF, TALE, or CRISPR/Cas scaffolds is also an active area of research (Yant et al. 2007; Voigt et al. 2012; Galvan et al. 2014). Importantly, targeted recombination or transposition may reduce cellular toxicity relative to the introduction of DSBs and their subsequent resolution through NHEJ or HDR. These alternate mechanisms to genome engineering may also increase efficiencies by decoupling editing from endogenous DNA repair mechanisms.
Synthetic regulation of transcription
In addition to the editing of DNA sequences, these genome engineering technologies can be used to manipulate endogenous gene expression. Early work with an engineered ZF protein directly fused to the herpes simplex viral VP16 transactivator demonstrated proof-of-principle of targeted transcriptional activation by inducing the expression of an extrachromosomal transgene in human cells (Liu et al. 1997). Subsequent studies showed that tandem repeats of VP16 were even more robust transcriptional activators than VP16 alone when linked to ZFs (Beerli et al. 1998), TALEs (Zhang et al. 2011), and nuclease-null deactivated Cas9 (dCas9) (Fig. 1E; Gilbert et al. 2013; Konermann et al. 2013; Maeder et al. 2013b; Mali et al. 2013a; Perez-Pinera et al. 2013a). Tetrameric VP16 domains (termed “VP64”) have exhibited the most widespread application as transcriptional activation domains, although larger multimers of VP16 have also been reported (Cheng et al. 2013). VP16 domains recruit cellular cofactors, such as components of the basal transcriptional machinery and chromatin remodelers (Hirai et al. 2010). Other transcriptional activation domains function similarly and have also been used in engineered transcription factors (Kim et al. 1997; Liu et al. 2001; Bikard et al. 2013; Anthony et al. 2014; Chavez et al. 2015; Konermann et al. 2015). Synergistic effects among multiple activators have been frequently observed with this class of activation domains, both when these effectors are localized in high density at adjacent sequences (Maeder et al. 2013b,c; Mali et al. 2013a; Perez-Pinera et al. 2013a,b) and when combined in cis as multimolecular complexes (Cheng et al. 2013; Chakraborty et al. 2014; Gao et al. 2014; Tanenbaum et al. 2014; Konermann et al. 2015). This synergy is likely related to enhanced subunit recruitment and/or effective decreases in cofactor dissociation rates at targeted loci.
Synthetic DBPs can also function as programmable transcriptional repressors. Localization of dCas9 near transcription start sites (TSSs) can repress active gene expression (Bikard et al. 2013; Qi et al. 2013). However, when fused to repressive domains, such as the KRAB domain (Margolin et al. 1994), the inhibitory effect of ZFs (Beerli et al. 1998), TALEs (Cong et al. 2012), or dCas9 (Gilbert et al. 2013) on transcription is markedly enhanced. Transcriptional repression by these methods is often accompanied by changes in chromatin structure (Groner et al. 2010; Kearns et al. 2015), which is likely a reflection of secondary KRAB-mediated recruitment of chromatin remodelers (Ying et al. 2015). Other domains have also been used for programmed transcriptional repression (Beerli et al. 1998; Snowden et al. 2002; Cong et al. 2012; Mahfouz et al. 2012; Gilbert et al. 2013; Konermann et al. 2013). In contrast to artificial gene activation with effectors and recruited cofactors, repression using synthetic DBPs has not been observed to function synergistically. In addition, the degree of repression by different tools varies dramatically, even within the same DNA-targeting platform and when targeting sequences in close proximity. Additional work is needed to characterize the factors that determine the potency of gene repression, which could be related to variable targeting affinities, interactions with endogenous factors, and/or local chromatin architecture.
Next generation genome engineering: epigenome editing
The ability to readily toggle epigenetic states holds tremendous value for basic research and potentially for human therapies. Efforts aimed at editing the epigenome using synthetic DBPs are rapidly evolving (Jurkowski et al. 2015). These methods can be used to provide evidence of the causality of epigenetic marks such as DNA methylation and histone subunit modifications. Furthermore chromatin-remodeling domains fused to DBPs have also expanded our ability to modulate genomic regulatory regions. For instance, transcriptional manipulation mediated by synthetic DBPs has been most well-characterized when targeted to within 300 base pairs of TSSs. However, directed modulation of distal regulatory elements, such as enhancers, has recently been shown to be possible, albeit with varying efficacy (Gao et al. 2013, 2014; Mendenhall et al. 2013; Ji et al. 2014; Frank et al. 2015; Hilton et al. 2015; Kearns et al. 2015). Enhancers represent dynamic genomic regulatory modules with differential functionality during cell lineage specification, and perturbation of enhancer function has been strongly associated with disease (Heinz et al. 2015; Roadmap Epigenomics Consortium et al. 2015). Similar to other regions of eukaryotic genomes, enhancers are controlled by dynamic epigenetic states, including methylated DNA and post-translational modification of histone subunits (Shlyueva et al. 2014; Heinz et al. 2015). Thus, epigenome editing tools for manipulating these epigenetic modifications are critical to facilitating our understanding of the links between gene regulation, development, and disease.
High levels of 5-methylcytosine (5mC) at enhancers and promoter regions are frequently correlated with transcriptional repression (Jones 2012; Schübeler 2015). Initial work with designer ZFs fused to prokaryotic DNA methyltransferases demonstrated targeted methylation of DNA in vitro (Xu and Bestor 1997; McNamara et al. 2002; Nomura and Barbas 2007; Smith and Ford 2007), on extrachromosomal (Nomura and Barbas 2007) or integrated plasmid DNA (Smith and Ford 2007), and at endogenous eukaryotic targets (Carvin et al. 2003). Furthermore, targeting of mammalian DNA methyltransferases with synthetic DBPs has established that site-specific DNA methylation at promoters can repress endogenous gene expression (Li et al. 2007; Rivenbark et al. 2012; Siddique et al. 2013). Currently, direct and targeted DNA methyltransferase activity to endogenous genes has only been applied using engineered ZF and TALE (Bernstein et al. 2015) protein scaffolds, However, it is probable that similar strategies could be adapted to CRISPR/Cas platforms.
The targeted demethylation of genomic DNA has also been used to activate gene expression using artificial DBPs. The TET family of proteins catalyzes oxidation of 5mC in eukaryotic genomes, leading to reversion to unmethylated cytosine following DNA replication (Lu et al. 2015). Direct fusion of the TET1 catalytic domain to TALEs targeting regions near endogenous human genes decreased DNA methylation, leading to increased mRNA expression (Maeder et al. 2013a). In addition, the catalytic domain of murine TET2, and to a lesser extent TET1, decreased DNA methylation when targeted to human promoter regions by engineered ZF proteins (Chen et al. 2014). Artificial localization of murine thymidine DNA glycosylase, an enzyme involved in cytosine demethylation (Cortellino et al. 2011) has also been shown to decrease DNA methylation and augment gene expression from an endogenous target promoter (Gregory et al. 2013). Collectively, these results demonstrate that the targeted manipulation of DNA methylation is possible, and cytosine methylation is functionally linked to controlling gene expression.
Certain modifications on the histone subunit tails of nucleosomes are highly correlated with genomic regulatory activity (Zhou et al. 2011; Shlyueva et al. 2014; Heinz et al. 2015). In order to take advantage of this mode of transcriptional regulation and also to develop tools to better understand its roles in gene regulation, there has been a recent emphasis on targeted perturbation of histone modifications. Acetylation at lysine residues 27 and 9 of histone subunit H3 (H3K27ac and H3K9ac, respectively) are generally enriched at loci associated with high transcriptional activity such as active promoters and enhancers. A fusion of the acetyltransferase core domain of the human EP300 protein robustly activated endogenous human genes when targeted to promoter or enhancer loci using ZFs, TALEs, and dCas9 variants (Hilton et al. 2015). This activation was accompanied by enrichment for H3K27ac at a targeted promoter and at a targeted enhancer. Notably, targeted H3K27 acetylation at the well-characterized human beta-globin HS2 enhancer using dCas9 fused to the catalytic core of EP300 also led to H3K27ac enrichment and transcriptional induction from HS2-responsive promoters. Together, these results support a model in which acetylation plays a casual role in gene activation. Furthermore, this suggests that H3K27ac enrichment at human enhancers may precede and coordinate distal H3K27ac deposition. Whether this deposition occurs through physical genomic contacts and/or other endogenous factors is the subject of ongoing study. Moreover, the direct manipulation of chromatin signatures using a chromatin acetyltransferase domain appeared to be mechanistically distinct from effectors requiring other cofactors for activity, such as VP64 (Hilton et al. 2015). Thus, improvements in these programmable epigenomic modifiers may enhance the synthetic engineering of gene activation.
The acetylation of histone subunit tails can be reversed by histone deacetylases (HDACs). Recent studies have used full proteins or truncated protein domains with HDAC activity fused to ZFs (Keung et al. 2014) or TALEs (Konermann et al. 2013) to silence gene expression. Histone subunits are also dynamically regulated through methylation and demethylation of lysine residues. The catalytic regions of histone H3K9 methyltransferases EHMT2 (also known as G9A) and SUV39H1 have been found to repress transcription and alter chromatin status at targeted promoters when fused to ZFs (Snowden et al. 2002; Falahi et al. 2013; Heller et al. 2014) and TALEs (Konermann et al. 2013). Targeted H3K4 demethylation has also been applied using TALEs or dCas9 fused to the KDM1A protein (also known as LSD1) (Mendenhall et al. 2013; Kearns et al. 2015), enabling the characterization of known and putative enhancers.
In addition to modulation of DNA methylation and histone residues, synthetic DBPs have also been used to manipulate chromosomal architecture. ZFs designed to artificially coordinate genomic looping between the HS2 enhancer of the globin locus control region and the beta-globin promoter activate gene expression in mouse cells (Deng et al. 2012) and similar designs can direct differential gene expression patterns between HS2 and globin genes in human and mouse cell lines (Deng et al. 2014). These results suggest that the physical interactions between enhancers and promoters can have a causal effect on gene expression. Although artificially generated chromosomal contacts have not been reported yet using TALEs or dCas9 platforms, similar approaches are likely feasible and would provide useful expansions to the genome engineering toolbox for rapid characterization of the role of chromatin conformation.
Specificity of ZFs, TALEs, and CRISPR/Cas9 systems
Understanding the target specificity of synthetic DBPs is central to their efficacy as biotechnological tools and therapeutics. Some studies using artificial ZFs fused to transcriptional effector domains indicate relatively high specificity for target gene modulation (Snowden et al. 2003). However, other results suggest widespread genomic interactions with these proteins that can lead to off-target transcriptional dysregulation (Falahi et al. 2013; Grimmer et al. 2014). Surveys of ZF nuclease specificities demonstrate that off-target effects can be prevalent (Cornu et al. 2008; Pattanayak et al. 2011), although this off-target activity may be mitigated by optimized design tools that minimize confounding factors such as context-dependent ZF domain effects (Isalan et al. 2001; Maeder et al. 2008; Sander et al. 2011; Gupta et al. 2012; Persikov et al. 2015).
As the RVDs of each synthetic TALE repeat dictate base-pair recognition, TALEs can theoretically target any genomic sequence of interest. However, engineered TALE RVDs exhibit a quantifiable variance in nucleotide recognition as well as positional effects that can lead to localization at unintended sequences (Cermak et al. 2011; Miller et al. 2011, 2015; Mali et al. 2013a; Meckler et al. 2013; Juillerat et al. 2014). Although the frequency of generating highly active TALE nucleases is typically higher than that of ZF nucleases (Kim and Kim 2014), certain limitations exist, such as apparent difficulty targeting methylated DNA (Valton et al. 2012) and requirements for thymine bases at 5′ targeting sites (Mak et al. 2012; Lamb et al. 2013). In addition, the larger size and proclivity for recombination of repetitive sequences in TALE proteins may present difficulties in certain applications, such as viral delivery (Holkers et al. 2013), although this issue has been addressed by optimizing the codon usage of repetitive RVDs (Yang et al. 2013b). Studies of genome-wide DNA-binding, gene regulation, and chromatin remodeling suggest a high level of specificity of TALE-based transcriptional activators, although binding to off-target sites is measurable (Polstein et al. 2015).
The interaction between Cas9 and a gRNA leads to conformational changes that activate surveillance for specific target sites by Cas9 (Jinek et al. 2014; Nishimasu et al. 2014; Jiang and Doudna 2015). However, off-target interactions between dCas9 and genomic DNA have been observed in human cells, even in the absence of gRNAs (Kuscu et al. 2014; Wu et al. 2014a; O'Geen et al. 2015; Polstein et al. 2015). Although off-target binding events likely occur with dCas9-based transcriptional/epigenetic modifiers, assessments of global gene expression suggest that changes are largely restricted to the intended target sites (Gilbert et al. 2013; Perez-Pinera et al. 2013a; Hilton et al. 2015; Polstein et al. 2015). Cas9 nucleases have also been found to cause DSBs at unintended sites (Fu et al. 2013; Hsu et al. 2013; Mali et al. 2013a; Pattanayak et al. 2013), and this activity is currently thought to be related to factors including gRNA composition, chromatin accessibility, and gRNA seed/PAM sequence abundance. Several algorithms exist that allow researchers to predict potential off-target Streptococcus pyogenes gRNA binding sites and aid in optimal gRNA design (Hsu et al. 2013; Bae et al. 2014; Cradick et al. 2014; Heigwer et al. 2014; Singh et al. 2015), However, this is clearly an area where significant future research is needed.
Applications of modern genome engineering technologies
Gene knockouts
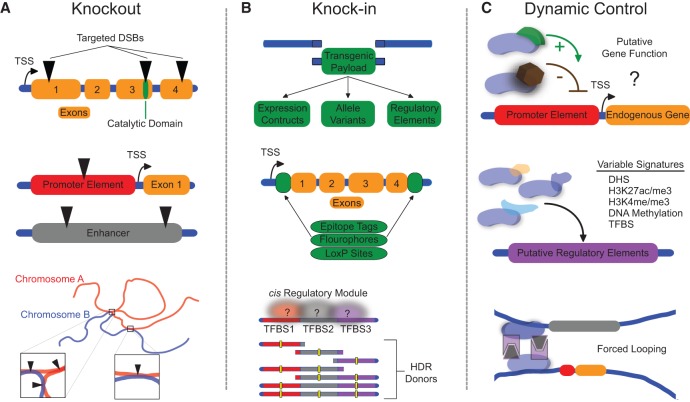
The most established application of modern genome engineering technologies is the disruption of loci through targeted nuclease activity (Fig. 2A; Urnov et al. 2010; Joung and Sander 2013; Hsu et al. 2014; Sander and Joung 2014). Genetic knockouts in eukaryotes through NHEJ-mediated disruption both in cell culture (Porteus and Baltimore 2003; Perez et al. 2008; Miller et al. 2011; Cho et al. 2013; Cong et al. 2013; Jinek et al. 2013; Mali et al. 2013b; Liao et al. 2015b) and in animals (Bibikova et al. 2002, 2003; Doyon et al. 2008; Meng et al. 2008; Cui et al. 2011; Wood et al. 2011; Hwang et al. 2013) can lead to complete loss of gene function. Moreover, genetic knockouts applied to agriculturally relevant plants and animals using genome engineering methodologies are poised to revolutionize the nutrition content and the availability of food crops and livestock (Hsu et al. 2014; Ni et al. 2014; Cyranoski 2015). In addition, these techniques to disrupt genetic information or interrogate gene function offer advantages over others, such as RNAi, which may have substantial off-target effects and incomplete abrogation of mRNA (Shalem et al. 2015). Deletions of genomic regions are also useful for removing entire genes or portions of the genome using the concurrent action of two targeted nucleases flanking the region to be excised (Lee et al. 2010; Kim et al. 2013; Essletzbichler et al. 2014; Ousterout et al. 2015b).
Figure 2.
Manipulation of endogenous loci using genome engineering tools. (A) Genetic knockout of coding regions, protein catalytic domains, promoters, enhancers, or genomic contact points is possible using targeted nuclease platforms. (B) Genetic knock-in using appropriate donor repair templates can be applied to deliver various transgenic payloads, including epitope tags, regulatory components, or disrupted motifs such as mutant transcription factor binding sites (TFBSs) within cis regulatory modules, to decipher endogenous regulatory element activity. (C) Dynamic regulation of endogenous loci using nuclease-null genome engineering tools fused to effector domains can be used to interrogate gene function or the activity of putative regulatory elements enriched with varying epigenomic signatures. Additionally, these tools can be used to artificially direct physical interactions between distal endogenous loci. (DSB) double-strand break; (TSS) transcription start site; (HDR) homology-directed repair; (DHS) DNase hypersensitivity site.
The complete and precise deletion of a gene or genetic segment may obviate potential confounding factors of gene knockout with a single nuclease that might still lead to functionally active truncated or frame-shifted proteins (Shi et al. 2015). Disruption or deletion of promoter or enhancer regions could also be used to knockout or diminish gene function, and enhancer elements have been characterized and validated through such methods (Bauer et al. 2013; Li et al. 2014; Mansour et al. 2014; Zhou et al. 2014a). Deletion of boundaries between topologically associating domains can also reveal important mechanistic properties of genome structure (Nora et al. 2012; Dowen et al. 2014; Crane et al. 2015b; Lupiáñez et al. 2015). However, deletion of genomic sequences may have disadvantageous pleiotropic effects, such as unintended alterations in the native architecture of bystander regulatory elements.
Gene knock-ins
When appropriate donor DNA repair templates are provided, HDR can lead to defined integration events such as the inclusion of epitope tags, reporter genes, and regulatory units, such as LoxP sites, at endogenous loci (Fig. 2B; Hockemeyer et al. 2009, 2011; Doyon et al. 2011a; Yang et al. 2013a). These methodologies allow for epitope-based detection of proteins and interacting partners for which high quality or specific antibodies are not available, as well as tracking of cellular proteins in real time on a single-cell basis. In addition, conditional alleles are useful in instances in which genetic knockout results in embryonic lethality or when assessments of gene function at different developmental stages and lineages are desired. HDR-mediated introduction of natural genetic variation into an isogenic background can also be used to elucidate the in vivo contributions of specific regulatory elements and DNA-interacting proteins. For instance, the targeted alteration of specific transcription factor binding site motifs in otherwise intact loci could reveal the functional contribution of transcription factor binding to regulatory element activity. This approach could also be extended to dissect cis regulatory modules in which several transcription factor binding sites are putatively involved in regulatory specificity (Hardison and Taylor 2012; Hnisz et al. 2015).
Dynamic regulation of genomic activity and conformation
In addition to nuclease-mediated disruption or deletion, genes and associated regulatory regions can also be dynamically manipulated using targeted ZF, TALE, or dCas9-based transcription factors or epigenome editing tools (Fig. 2C). This approach is particularly useful for avoiding the stochastic and cell-type– and cell-cycle–dependent DNA repair pathways involved in nuclease-mediated genome editing. Such methods can elucidate potential endogenous gene function without exogenous overexpression or permanent sequence disruption. These tools are also especially applicable in targeting the multitude of putative regulatory regions that contain epigenetic hallmarks correlated with activity in certain settings, such as differentially active enhancers (Shlyueva et al. 2014; Farh et al. 2015; Heinz et al. 2015; Roadmap Epigenomics Consortium et al. 2015; Leung et al. 2015). For example, recent efforts have identified signatures, such as DNase hypersensitivity, H3K27ac, and H3K4 methylation that are associated with active enhancers and promoters. However, validation of the potential functionality of these elements is a major scientific bottleneck to our understanding of the epigenetics of gene regulation. Selective and targeted writing or erasure of appropriate epigenetic modifications can establish the causality of respective marks in determining gene expression and may also define their relevance in organismal development and in cellular responses to distinct stimuli. Furthermore, targeted activation or suppression of regulatory loci involved in lineage specification or reprogramming could have enormous biotechnological utility (Gao et al. 2013; Chakraborty et al. 2014; Ji et al. 2014; Chavez et al. 2015). Additionally, programmed looping to connect distal genomic regions can serve to define the function of specific genomic contacts (Deng et al. 2014). Ultimately, the most comprehensive and valuable definitions of regulatory element functionality will use several independent approaches, including sequence disruption and manipulation of activity across varied biological contexts.
Genome engineering to model disease and develop therapeutics
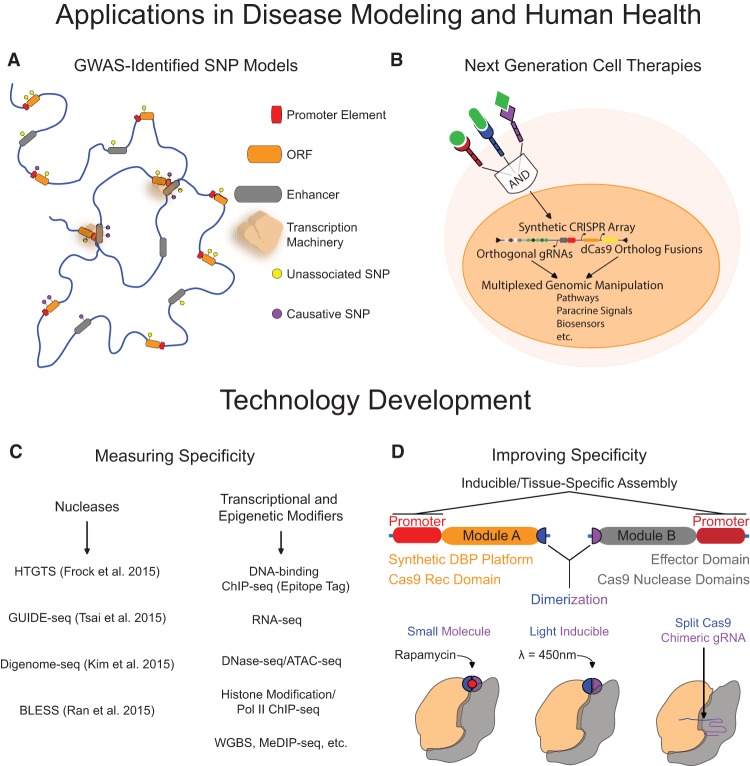
ZFs, TALEs, and the CRISPR/Cas9 system are also important tools for understanding and modeling disease. In Mendelian disorders, in which single gene products are implicated in disease or development, nuclease-aided disruption or deletion can be used to determine causal relationships between genes and phenotypes (Soldner et al. 2011; Toscano et al. 2013). In addition, specific SNPs or corrections can be introduced into coding regions to identify and validate variants associated with disease (Kiskinis et al. 2014; Wienert et al. 2015). SNPs in noncoding elements, such as enhancers, are also associated with many diseases; hence, similar modeling and validation methods outside of coding regions will also be extremely useful. Notably, genome engineering is unique in its ability to perturb regulatory elements in their endogenous genomic context in contrast to other tools, such as RNAi and small molecules that modulate mRNA and protein activity. Large-scale genomic models of complex diseases, based on genome-wide associations, could also be created with these strategies in order to discriminate germane versus immaterial genetic variance (Fig. 3A; Bauer et al. 2013). Some disease phenotypes are also the result of fusion proteins generated during aberrant genomic translocations (Bunting and Nussenzweig 2013). Programmable nucleases could also be used to insert deleterious fusion proteins at endogenous loci to quantify causal effects and develop drug targets or to recapitulate chromosomal translocations to mimic disease (Maddalo et al. 2014).
Figure 3.
Outlook for genome engineering technologies. (A) Genome editing technologies can be used to incorporate genome-wide association study (GWAS)-identified single nucleotide polymorphisms (SNPs) into models of complex diseases and to ascertain casual disease variants. (B) Human cells can be programmed using genome engineering to produce the next generation of advanced cell therapies. (C) Techniques designed to assess potential off-target activity of programmable nucleases and transcriptional/epigenetic modifiers will facilitate technology improvements. (D) Increased control of genome engineering tools can be achieved with chemical and optogenetic regulation to develop highly articulated systems with increased specificity and dynamic properties.
Programmable DBPs can also be applied for genome-wide phenotypic screening. This is especially relevant for CRISPR/Cas9-based screens, owing to the relative ease of multiplexing by simply using Cas9 in tandem with libraries of gRNAs that can be synthesized at high throughput (Shalem et al. 2015). Cas9 nuclease-based knockout screens have been recently used in combination with both positive and negative selection strategies in mammalian cell lines. These techniques have revealed genes that are essential for certain cell states and sensitivity to toxins or drugs (Koike-Yusa et al. 2014; Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014b; Chen et al. 2015). In addition, nuclease-inactivated dCas9-based repressors and activators can be used in loss-of-function and gain-of-function screens, respectively (Gilbert et al. 2014; Konermann et al. 2015). Importantly, these screening strategies have not only validated previously implicated genes, but have also identified novel drivers of selected cellular phenotypes. The relative ease of construction of these screening platforms should make these technologies broadly useful to numerous research laboratories investigating multiple different pathologies and phenotypes.
The advent of modern genome engineering tools has also stimulated persistent and warranted optimism in the field of gene therapy. Knockout of pathological genes with Mendelian phenotypes is possible using ZFs, TALEs, and Cas9 nucleases. Similar approaches could also be used to correct deleterious copy number variation. The knockout of portions of genes or reading frame correction is also a useful strategy for pathologies in which truncated genetic variants provide amelioration, such as Duchenne muscular dystrophy (Li et al. 2015a; Ousterout et al. 2015a). Furthermore, as methods for HDR-mediated gene correction and integration continue to improve, the replacement of causative SNPs at coding regions and regulatory elements may become routine (Genovese et al. 2014; Crane et al. 2015a; Hoban et al. 2015). Similarly, targeted addition of exogenous genetic payloads may also become a valuable tool to correct loss-of-function, provide dosage compensation, or create novel cellular phenotypes (Li et al. 2011; Genovese et al. 2014). Engineered DBPs may also have utility as vaccination and immunotherapeutic agents. For instance, targeted disruption of viral entry molecules, such as CCR5 for HIV, have been shown to be efficacious as a prophylactic measure (Holt et al. 2010; Tebas et al. 2014). Viral loads have also been decreased using nucleases targeted to other viral genomes (Kennedy et al. 2014, 2015; Wang and Quake 2014; Liao et al. 2015a). In addition, genome engineering efforts to enhance human T cell immunotherapy have demonstrated substantial promise (Provasi et al. 2012; Torikai et al. 2013; Beane et al. 2015) and may pave the way toward customized patient-specific prophylactics and therapies.
The therapeutic use of ZFs, TALEs, and Cas9 as artificial transcriptional/epigenetic modifiers also has tremendous potential. The precise genetic repression of detrimental genes may be a useful strategy to mitigate pathology, such as repressing the huntingtin gene for Huntington's disease (Garriga-Canut et al. 2012). Similarly, targeted activation of aberrantly silenced endogenous loci may provide therapeutic benefits (Lara et al. 2012). Additionally, the manipulation of irregular epigenetic modifications or genomic contacts could be useful to prevent or correct disease states. Notably, in contrast to permanent nuclease-based sequence modification, synthetic transcriptional or epigenetic modifications are dynamic, tunable, and reversible. In certain circumstances, limited durations or transmission of artificial manipulation may be preferable. Furthermore, the versatility and multiplex capacity of these genome engineering tools combined with the recent advances in synthetic eukaryotic genetic circuit designs (Khalil et al. 2012; Slusarczyk et al. 2012; Esvelt et al. 2013; Farzadfard et al. 2013; Daringer et al. 2014; Kabadi et al. 2014; Kiani et al. 2014; Nielsen and Voigt 2014; Nissim et al. 2014; O'Connell et al. 2014; Li et al. 2015b; Zalatan et al. 2015) may lead to the next generation of cell therapies (Fig. 3B).
Future outlook
Genome engineering tools have been widely implemented for editing and understanding eukaryotic genomes. However, technological improvements are still needed to fulfill the potential of these technologies. Ideal genome engineering tools would have completely predictable effects, lack toxicity, be easily designed and constructed, and be easily deliverable with high efficiency in vitro and in vivo. Although ZFs, TALEs, and CRISPR/Cas9 platforms have propelled the field of genome engineering, they still suffer potential limitations in all of these areas. The comprehensive characterization and optimization of targeting specificities and in vivo delivery parameters of modern genome engineering tools are arguably the most pressing concerns (Kim and Kim 2014; Cox et al. 2015).
The successful application of genome engineering in diverse organisms demonstrates that the technologies are effective with limited negative side effects in vivo, However, the translation of these tools to clinical settings requires a greater resolution of any off-target effects than has previously been possible. Some examples of the possible off-target activity of the ZF, TALE, and Cas9-based platforms have been published (Perez et al. 2008; Fu et al. 2013; Hsu et al. 2013; Mali et al. 2013a; Pattanayak et al. 2013; Guilinger et al. 2014a; Kim and Kim 2014). An important recent advance has been the development of unbiased approaches for detecting off-target effects with a much greater level of sensitivity than previous methods (Fig. 3C; Gabriel et al. 2011; Frock et al. 2015; Kim et al. 2015; Ran et al. 2015; Tsai et al. 2015). The use of large sets of negative control gRNAs in high-throughput CRISPR/Cas9-based screening applications is also enabling reproducible and highly quantitative analyses of off-target gRNA activity (Gilbert et al. 2014; Sanjana et al. 2014). However, despite the importance of these advances, their detection limits are still restrained by the accuracy of current DNA sequencing technologies, and this will be a critical area for improvement moving forward.
The protein engineering of nuclease domains to improve activity and specificity will also continue to be a focus area, particularly as the methods for computational design and experimental selection of proteins with novel properties improve. For example, engineered “nickases” have been developed to stimulate HDR but minimize off-target indels generated by the creation of double-strand breaks by nucleases, but the nickase-mediated HDR frequencies are generally lower (Doyon et al. 2011b; Miller et al. 2011; Kim et al. 2012; Wang et al. 2012; Mali et al. 2013a; Ran et al. 2013; Guilinger et al. 2014b; Tsai et al. 2014; Wu et al. 2014b). Protein engineering may also be used to alter the specificity of sequence recognition requirements of DNA-binding proteins, such as the directed evolution of Cas9 variants to recognize alternate PAMs (Kleinstiver et al. 2015) or of TALEs to change the requirement of a 5′ thymine (Lamb et al. 2013). Continued efforts to generate effective guidelines for optimal gRNA design will also be extremely important for future Cas9-based technologies (Briner et al. 2014; Cho et al. 2014; Doench et al. 2014; Fu et al. 2014; Chari et al. 2015; Farboud and Meyer 2015; Singh et al. 2015; Xu et al. 2015). For example, the development of algorithms that incorporate biological phenomena into optimal gRNA design are an important area for future work (Singh et al. 2015). Finally, the continued development of targeted recombinases and transposases could eventually supplant nuclease-based genome editing if the challenges of efficiently and specifically targeting highly active versions of these proteins to new sites can be overcome.
The specificity of the genome engineering technologies is also pertinent for transcriptional and epigenetic modification using these tools. Although genome-wide measurements of gene expression and chromatin structure suggest low frequencies of off-target effects (Hilton et al. 2015; Polstein et al. 2015), a thorough and controlled assessment of unintended epigenetic modifications is still necessary for many of the newly described effector domains. The duration and heritability of these synthetically deposited modifications should also be the subject of future investigation (Kungulovski et al. 2015). Optimal concentrations and spatiotemporal control by chemical or optogenetic regulation will also aid in mitigating any inadvertent effects (Fig. 3D; Polstein and Gersbach 2012, 2015; Konermann et al. 2013; Davis et al. 2015; Nihongaki et al. 2015; Wright et al. 2015; Zetsche et al. 2015). In addition, the direct delivery of purified ZFs, TALEs, or preassembled Cas9/gRNA complexes can minimize off-target activity (Gaj et al. 2012; Chen et al. 2013b; Ru et al. 2013; Kim et al. 2014; Ramakrishna et al. 2014; Liu et al. 2015; Zuris et al. 2015), which may further safeguard against aberrant effects.
Despite these areas of potential optimization and improvement, genome engineering technologies are clearly ready to have a significant impact on genomics and medicine. Functional characterization of the epigenetic modifications associated with gene regulation is now possible using these tools. This will allow high resolution functional annotation and indexed categorization of genomic regulatory elements. The assignment of functional data to these regulatory regions will also benefit from analyses across lineages and cell states, thereby providing regulatory atlases across genomic space and organismal development. Ongoing improvements in delivery systems for primary cells and tissues will continue to facilitate this work (Kabadi et al. 2014; Ran et al. 2015; Zuris et al. 2015). Similarly, the manipulation of epigenetic states at specific loci will also allow determinations of causal effects of these marks and the role of epigenomic signatures at specific regulatory elements in disease and lineage specification.
The precise perturbation of genomic contact points may also be useful to determine the functionality of physical connections and to understand the reported stochasticity associated with these interactions (Kind et al. 2013; Nagano et al. 2013). Imaging of genomic loci using engineered DBPs is also possible (Chen et al. 2013a; Miyanari et al. 2013), and this could provide an independent approach to demonstrate colocalization of genomic regions in real time. In addition, directed localization of specified genomic regulatory elements to nuclear regions associated with repression, such as the nuclear lamina (Amendola and van Steensel 2014; Pombo and Dillon 2015), could aid in validating the functional relevance of nuclear subcompartmentalization. Therefore the challenge of mapping the complex and dynamic four-dimensional genome is positioned to be a major future research area enabled by these new genome engineering technologies.
The functional characterization of the expanding sets of putative regulatory regions is nontrivial but will be facilitated by modern genome engineering. Likewise, determination of the causal variants in complex non-Mendelian diseases could be accomplished using precise recapitulation of the variants in otherwise isogenic cell cultures and animal models. Developing custom genomic models will continue to enable drug development and models of potential drug resistance (Kasap et al. 2014; Smurnyy et al. 2014). This will facilitate rapid identification of drugs with therapeutic efficacy and realize the potential of personalized and precision medicine by connecting genomics, therapeutic targets, and disease phenotypes. Modern genome engineering platforms are now established as indispensable research tools for diverse areas of biotechnology, and promising areas for their direct and indirect application to improving human health are rapidly expanding. This new era of genomic understanding is likely to continue to create new possibilities for genome research for the foreseeable future.
Competing interest statement
I.B.H. and C.A.G. are inventors on patent applications related to genome engineering. C.A.G. is a scientific advisor to Editas Medicine, a company engaged in therapeutic development of genome engineering technologies.
Acknowledgments
We thank Gregory Crawford and Timothy Reddy for helpful discussions on the topics of this Perspective and Matthew Gemberling and Sandi Wong for critical reading of the manuscript and helpful comments. This work was supported by US National Institutes of Health (NIH) grants R01DA036865, U01HG007900, R21AR065956, P30AR066527, UH3TR000505, and an NIH Director's New Innovator Award (DP2OD008586), and a National Science Foundation (NSF) Faculty Early Career Development (CAREER) Award (CBET-1151035).
Footnotes
Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.190124.115.
Freely available online through the Genome Research Open Access option.
References
- Akopian A, He J, Boocock MR, Stark WM. 2003. Chimeric recombinases with designed DNA sequence recognition. Proc Natl Acad Sci 100: 8688–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. 2014. Mechanisms and dynamics of nuclear lamina–genome interactions. Curr Opin Cell Biol 28: 61–68. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony K, More A, Zhang X. 2014. Activation of silenced cytokine gene promoters by the synergistic effect of TBP-TALE and VP64-TALE activators. PLoS One 9: e95790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, et al. 2013. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane JD, Lee G, Zheng Z, Mendel M, Abate-Daga D, Bharathan M, Black M, Gandhi N, Yu Z, Chandran S, et al. 2015. Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol Ther 23: 1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF III. 1998. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci 95: 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DL, Le Lay JE, Ruano EG, Kaestner KH. 2015. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J Clin Invest 125: 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakta MS, Henry IM, Ousterout DG, Das KT, Lockwood SH, Meckler JF, Wallen MC, Zykovich A, Yu Y, Leo H, et al. 2013. Highly active zinc-finger nucleases by extended modular assembly. Genome Res 23: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. 2013. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 41: 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. 1998. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci 95: 10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Boissel S, Jarjour J, Astrakhan A, Adey A, Gouble A, Duchateau P, Shendure J, Stoddard BL, Certo MT, Baker D, et al. 2014. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res 42: 2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. 2014. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell 56: 333–339. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Nussenzweig A. 2013. End-joining, translocations and cancer. Nat Rev Cancer 13: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. 2012. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109: 17382–17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. 2014. Genome engineering with targetable nucleases. Annu Rev Biochem 83: 409–439. [DOI] [PubMed] [Google Scholar]
- Carvin CD, Parr RD, Kladde MP. 2003. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res 31: 6493–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. 2014. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep 3: 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chari R, Mali P, Moosburner M, Church GM. 2015. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. 2015. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013a. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jaafar L, Agyekum DG, Xiao H, Wade MF, Kumaran RI, Spector DL, Bao G, Porteus MH, Dynan WS, et al. 2013b. Receptor-mediated delivery of engineered nucleases for genome modification. Nucleic Acids Res 41: e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. 2014. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 42: 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160: 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. [DOI] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. 2012. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun 3: 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. 2008. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther 16: 352–358. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. 2015. Therapeutic genome editing: prospects and challenges. Nat Med 21: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. 2014. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids 3: e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane AM, Kramer P, Bui JH, Chung WJ, Li XS, Gonzalez-Garay ML, Hawkins F, Liao W, Mora D, Choi S, et al. 2015a. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Rep 4: 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015b. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. 2011. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol 29: 64–67. [DOI] [PubMed] [Google Scholar]
- Cyranoski D. 2015. Super-muscly pigs created by small genetic tweak. Nature 523: 13–14. [DOI] [PubMed] [Google Scholar]
- Daringer NM, Dudek RM, Schwarz KA, Leonard JN. 2014. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth Biol 3: 892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. 2015. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11: 316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. 2013. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 14: 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. 2014. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159: 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. 2008. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon JB, Zeitler B, Cheng J, Cheng AT, Cherone JM, Santiago Y, Lee AH, Vo TD, Doyon Y, Miller JC, et al. 2011a. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol 13: 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. 2011b. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SM, Bürckstümmer T. 2014. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res 24: 2059–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. 2013. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahi F, Huisman C, Kazemier HG, van der Vlies P, Kok K, Hospers GA, Rots MG. 2013. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol Cancer Res 11: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Farboud B, Meyer BJ. 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. 2013. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, Vockley CM, Safi A, Gersbach CA, Crawford GE, et al. 2015. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci 18: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. 2015. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. 2011. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823. [DOI] [PubMed] [Google Scholar]
- Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF III. 2012. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 9: 805–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF III. 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan DL, Kettlun CS, Wilson MH. 2014. Targeting piggyBac transposon integrations in the human genome. Methods Mol Biol 1114: 143–161. [DOI] [PubMed] [Google Scholar]
- Gao X, Yang J, Tsang JC, Ooi J, Wu D, Liu P. 2013. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Rep 1: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tsang JC, Gaba F, Wu D, Lu L, Liu P. 2014. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res 42: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga-Canut M, Agustín-Pavón C, Herrmann F, Sánchez A, Dierssen M, Fillat C, Isalan M. 2012. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc Natl Acad Sci 109: E3136–E3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, Holmes MC, et al. 2014. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF III. 2011. Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res 39: 7868–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersbach CA, Gaj T, Barbas CF III. 2014. Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies. Acc Chem Res 47: 2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordley RM, Gersbach CA, Barbas CF III. 2009. Synthesis of programmable integrases. Proc Natl Acad Sci 106: 5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory DJ, Zhang Y, Kobzik L, Fedulov AV. 2013. Specific transcriptional enhancement of inducible nitric oxide synthase by targeted promoter demethylation. Epigenetics 8: 1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer MR, Stolzenburg S, Ford E, Lister R, Blancafort P, Farnham PJ. 2014. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res 42: 10856–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, Bucher P, Trono D. 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6: e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Pattanayak V, Reyon D, Tsai SQ, Sander JD, Joung JK, Liu DR. 2014a. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods 11: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. 2014b. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF III. 2010. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol 400: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. 2012. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods 9: 588–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Taylor J. 2012. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet 13: 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M. 2014. E-CRISP: fast CRISPR target site identification. Nat Methods 11: 122–123. [DOI] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, Golden SA, Herman JP, Walsh JJ, Mazei-Robison M, et al. 2014. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci 17: 1720–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Tani T, Kikyo N. 2010. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int J Dev Biol 54: 1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. 2015. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 58: 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban MD, Cost GJ, Mendel MC, Romero Z, Kaufman ML, Joglekar AV, Ho M, Lumaquin D, Gray D, Lill GR, et al. 2015. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125: 2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T, Gonçalves MA. 2013. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res 41: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, et al. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M, Klug A, Choo Y. 2001. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol 19: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mátés L, Boeke JD, Nagy A, Bradley A, Izsvák Z. 2009. Transposon-mediated genome manipulation in vertebrates. Nat Methods 6: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Fischer AL, Brown CR, Eastlund ER, Dvash T, Zhong B, Gerber MA, Lyons I, Knight SW, Kreader CA. 2014. Engineered zinc-finger transcription factors activate OCT4 (POU5F1), SOX2, KLF4, c-MYC (MYC) and miR302/367. Nucleic Acids Res 42: 6158–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. 2015. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol 30C: 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. 2013. RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343: 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD. 2013. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat A, Dubois G, Valton J, Thomas S, Stella S, Maréchal A, Langevin S, Benomari N, Bertonati C, Silva GH, et al. 2014. Comprehensive analysis of the specificity of transcription activator-like effector nucleases. Nucleic Acids Res 42: 5390–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski TP, Ravichandran M, Stepper P. 2015. Synthetic epigenetics—towards intelligent control of epigenetic states and cell identity. Clin Epigenetics 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. 2014. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res 42: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasap C, Elemento O, Kapoor TM. 2014. DrugTargetSeqR: a genomics- and CRISPR-Cas9–based method to analyze drug targets. Nat Chem Biol 10: 626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U. 2007. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651. [DOI] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. 2015. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12: 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. 2014. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol 88: 11965–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. 2015. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology 476: 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keung AJ, Bashor CJ, Kiriakov S, Collins JJ, Khalil AS. 2014. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell 158: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. 2012. A synthetic biology framework for programming eukaryotic transcription functions. Cell 150: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li Y, Weiss R. 2014. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat Methods 11: 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. 2014. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15: 321–334. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. 1996. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Kim J, Cepek KL, Sharp PA, Pabo CO. 1997. Design of TATA box-binding protein/zinc finger fusions for targeted regulation of gene expression. Proc Natl Acad Sci 94: 3616–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. 2012. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22: 1327–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. 2013. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31: 251–258. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. 2015. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12: 237–243, 1 p following 243. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, et al. 2014. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 14: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32: 267–273. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. 2013. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kungulovski G, Nunna S, Thomas M, Zanger UM, Reinhardt R, Jeltsch A. 2015. Targeted epigenome editing of an endogenous locus with chromatin modifiers is not stably maintained. Epigenetics Chromatin 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. 2014. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32: 677–683. [DOI] [PubMed] [Google Scholar]
- Lamb BM, Mercer AC, Barbas CF III. 2013. Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res 41: 9779–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara H, Wang Y, Beltran AS, Juárez-Moreno K, Yuan X, Kato S, Leisewitz AV, Cuello Fredes M, Licea AF, Connolly DC, et al. 2012. Targeting serous epithelial ovarian cancer with designer zinc finger transcription factors. J Biol Chem 287: 29873–29886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. 2010. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS. 2012. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res 22: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, Yen CA, Lin S, Lin Y, Qiu Y, et al. 2015. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 518: 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wu LP, Chandrasegaran S. 1992. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci 89: 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. 2007. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res 35: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. 2011. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rivera CM, Ishii H, Jin F, Selvaraj S, Lee AY, Dixon JR, Ren B. 2014. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One 9: e114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H, et al. 2015a. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep 4: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jiang Y, Chen H, Liao W, Li Z, Weiss R, Xie Z. 2015b. Modular construction of mammalian gene circuits using TALE transcriptional repressors. Nat Chem Biol 11: 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, et al. 2015a. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6: 6413. [DOI] [PubMed] [Google Scholar]
- Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. 2015b. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 47: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Segal DJ, Ghiara JB, Barbas CF III. 1997. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci 94: 5525–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PQ, Rebar EJ, Zhang L, Liu Q, Jamieson AC, Liang Y, Qi H, Li PX, Chen B, Mendel MC, et al. 2001. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem 276: 11323–11334. [DOI] [PubMed] [Google Scholar]
- Liu J, Gaj T, Wallen MC, Barbas CF III. 2015. Improved cell-penetrating zinc-finger nuclease proteins for precision genome engineering. Mol Ther Nucleic Acids 4: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhao BS, He C. 2015. TET family proteins: oxidation activity, interacting molecules, and functions in diseases. Chem Rev 115: 2225–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. 2014. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516: 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. 2008. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. 2013a. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol 31: 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. 2013b. CRISPR RNA–guided activation of endogenous human genes. Nat Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Reyon D, Angstman JF, Fu Y, Sander JD, Joung JK. 2013c. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods 10: 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy DK, Zhu JK. 2012. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol 78: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. 2012. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. 2013a. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013b. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. 2014. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346: 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ III. 1994. Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci 91: 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara AR, Hurd PJ, Smith AE, Ford KG. 2002. Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships. Nucleic Acids Res 30: 3818–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsäesser J, et al. 2013. Quantitative analysis of TALE–DNA interactions suggests polarity effects. Nucleic Acids Res 41: 4118–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. 2013. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 31: 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. 2008. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC, Gaj T, Fuller RP, Barbas CF III. 2012. Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res 40: 11163–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Miller JC, Zhang L, Xia DF, Campo JJ, Ankoudinova IV, Guschin DY, Babiarz JE, Meng X, Hinkley SJ, Lam SC, et al. 2015. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods 12: 465–471. [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. 2013. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol 20: 1321–1324. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155(Pt 3): 733–740. [DOI] [PubMed] [Google Scholar]
- Morbitzer R, Römer P, Boch J, Lahaye T. 2010. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci 107: 21617–21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M, Polejaeva IA, Chen C. 2014. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS One 9: e106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AA, Voigt CA. 2014. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol 10: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. 2015. CRISPR-Cas9-based photoactivatable transcription system. Chem Biol 22: 169–174. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. 2014. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 54: 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura W, Barbas CF III. 2007. In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc 129: 8676–8677. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. 2014. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. 2015. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res 43: 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. 2015a. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 6: 6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousterout DG, Kabadi AM, Thakore PI, Perez-Pinera P, Brown MT, Majoros WH, Reddy TE, Gersbach CA. 2015b. Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol Ther 23: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. 2011. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods 8: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. 1991. Zinc finger–DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252: 809–817. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. 2013a. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. 2013b. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods 10: 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Wetzel JL, Rowland EF, Oakes BL, Xu DJ, Singh M, Noyes MB. 2015. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res 43: 1965–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. 2012. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc 134: 16480–16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. 2015. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11: 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein L, Perez-Pinera P, Kocak D, Vockley C, Bledsoe P, Song L, Safi A, Crawford G, Reddy T, Gersbach C. 2015. Genome-wide specificity of DNA-binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res 25: 1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Dillon N. 2015. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16: 245–257. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. 2003. Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed] [Google Scholar]
- Provasi E, Genovese P, Lombardo A, Magnani Z, Liu PQ, Reik A, Chu V, Paschon DE, Zhang L, Kuball J, et al. 2012. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med 18: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. 2014. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. 2012. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7: 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. 2007. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318: 645–648. [DOI] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14: 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru R, Yao Y, Yu S, Yin B, Xu W, Zhao S, Qin L, Chen X. 2013. Targeted genome engineering in human induced pluripotent stem cells by penetrating TALENs. Cell Regen (Lond) 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. 2011. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature 517: 321–326. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15: 272–286. [DOI] [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. 2013. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 425: 479–491. [DOI] [PubMed] [Google Scholar]
- Singh R, Kuscu C, Quinlan A, Qi Y, Adli M. 2015. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarczyk AL, Lin A, Weiss R. 2012. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet 13: 406–420. [DOI] [PubMed] [Google Scholar]
- Smith AE, Ford KG. 2007. Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Res 35: 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. 1985. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 317: 230–234. [DOI] [PubMed] [Google Scholar]
- Smurnyy Y, Cai M, Wu H, McWhinnie E, Tallarico JA, Yang Y, Feng Y. 2014. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat Chem Biol 10: 623–625. [DOI] [PubMed] [Google Scholar]
- Snowden AW, Gregory PD, Case CC, Pabo CO. 2002. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol 12: 2159–2166. [DOI] [PubMed] [Google Scholar]
- Snowden AW, Zhang L, Urnov F, Dent C, Jouvenot Y, Zhong X, Rebar EJ, Jamieson AC, Zhang HS, Tan S, et al. 2003. Repression of vascular endothelial growth factor A in glioblastoma cells using engineered zinc finger transcription factors. Cancer Res 63: 8968–8976. [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. 2011. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146: 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. 2014. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob DNA 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. 2007. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786–793. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, et al. 2014. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Folger KR, Capecchi MR. 1986. High frequency targeting of genes to specific sites in the mammalian genome. Cell 44: 419–428. [DOI] [PubMed] [Google Scholar]
- Torikai H, Reik A, Soldner F, Warren EH, Yuen C, Zhou Y, Crossland DL, Huls H, Littman N, Zhang Z, et al. 2013. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood 122: 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano MG, Anderson P, Muñoz P, Lucena G, Cobo M, Benabdellah K, Gregory PD, Holmes MC, Martin F. 2013. Use of zinc-finger nucleases to knock out the WAS gene in K562 cells: a human cellular model for Wiskott-Aldrich syndrome. Dis Model Mech 6: 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. 2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. 2015. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Valton J, Dupuy A, Daboussi F, Thomas S, Maréchal A, Macmaster R, Melliand K, Juillerat A, Duchateau P. 2012. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem 287: 38427–38432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee ES, Santagata S, Aggarwal AK. 2001. FokI requires two specific DNA sites for cleavage. J Mol Biol 309: 69–78. [DOI] [PubMed] [Google Scholar]
- Voigt K, Gogol-Döring A, Miskey C, Chen W, Cathomen T, Izsvák Z, Ivics Z. 2012. Retargeting sleeping beauty transposon insertions by engineered zinc finger DNA-binding domains. Mol Ther 20: 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Quake SR. 2014. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci 111: 13157–13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, et al. 2012. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res 22: 1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Funnell AP, Norton LJ, Pearson RC, Wilkinson-White LE, Lester K, Vadolas J, Porteus MH, Matthews JM, Quinlan KG, et al. 2015. Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat Commun 6: 7085. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29: 183–212. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. 2015. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci 112: 2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. 2014a. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Gao T, Wang X, Hu Y, Hu X, Hu Z, Pang J, Li Z, Xue J, Feng M, et al. 2014b. TALE nickase mediates high efficient targeted transgene integration at the human multi-copy ribosomal DNA locus. Biochem Biophys Res Commun 446: 261–266. [DOI] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, et al. 2013. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Bestor TH. 1997. Cytosine methylation targetted to pre-determined sequences. Nat Genet 17: 376–378. [DOI] [PubMed] [Google Scholar]
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, et al. 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. 2013a. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, et al. 2013b. Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41: 9049–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Huang Y, Akache B, Kay MA. 2007. Site-directed transposon integration in human cells. Nucleic Acids Res 35: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Yang X, Zhao K, Mao J, Kuang Y, Wang Z, Sun R, Fei J. 2015. The Krüppel-associated box repressor domain induces reversible and irreversible regulation of endogenous mouse genes by mediating different chromatin states. Nucleic Acids Res 43: 1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. 2015. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33: 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. 2011. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29: 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. [DOI] [PubMed] [Google Scholar]
- Zhou HY, Katsman Y, Dhaliwal NK, Davidson S, Macpherson NN, Sakthidevi M, Collura F, Mitchell JA. 2014a. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev 28: 2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. 2014b. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509: 487–491. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. 2015. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]