Ku et al. show that loss of one Gata3 allele leads to diminished expansion of immature T cells as well as aberrant induction of myeloid transcription factor PU.1. Gata3 is monoallelically expressed in hematopoietic stem cells and early T-cell progenitors. Half of the developing cells switch to biallelic Gata3 transcription abruptly at midthymopoiesis.
Keywords: GATA3, T-cell development, monoallelic transcription, biallelic transcription
Abstract
Protein abundance must be precisely regulated throughout life, and nowhere is the stringency of this requirement more evident than during T-cell development: A twofold increase in the abundance of transcription factor GATA3 results in thymic lymphoma, while reduced GATA3 leads to diminished T-cell production. GATA3 haploinsufficiency also causes human HDR (hypoparathyroidism, deafness, and renal dysplasia) syndrome, often accompanied by immunodeficiency. Here we show that loss of one Gata3 allele leads to diminished expansion (and compromised development) of immature T cells as well as aberrant induction of myeloid transcription factor PU.1. This effect is at least in part mediated transcriptionally: We discovered that Gata3 is monoallelically expressed in a parent of origin-independent manner in hematopoietic stem cells and early T-cell progenitors. Curiously, half of the developing cells switch to biallelic Gata3 transcription abruptly at midthymopoiesis. We show that the monoallelic-to-biallelic transcriptional switch is stably maintained and therefore is not a stochastic phenomenon. This unique mechanism, if adopted by other regulatory genes, may provide new biological insights into the rather prevalent phenomenon of monoallelic expression of autosomal genes as well as into the variably penetrant pathophysiological spectrum of phenotypes observed in many human syndromes that are due to haploinsufficiency of the affected gene.
Precise control of protein abundance—achieved through balanced transcriptional activation and repression, mRNA generation and stability, translation and protein stability, and localization—is vital for normal development. In metazoan organisms, most genes are encoded by paternal and maternal alleles, and cells are thought to most often express autosomal genes from both alleles. Trisomy can result in miscarriage or physiological abnormalities (e.g., Down syndrome, Edwards syndrome, Patau syndrome, Warkany syndrome 2, and more) (Hassold and Hunt 2001), while haploinsufficiency has been shown to result in numerous autosomal dominant disorders (e.g., Williams syndrome, autosomal dominant Zinsser-Cole-Engman syndrome, Ehlers-Danlos syndrome, Marfan syndrome, and others) (Inoue and Lupski 2002). Thus, the number of active alleles can be a principal determinant used to modulate the level of protein expression within a cell.
Transcription factor GATA3 (originally named NF-E1c) (Yamamoto et al. 1990; Ko et al. 1991) is required throughout T-cell development (Hosoya et al. 2010). T lymphocytes develop in the thymus from multipotential hematopoietic progenitors that reside in the bone marrow. Bone marrow-derived progenitors then home to the thymus, where they undergo cell expansion to become early T-cell progenitors (ETPs) that are specified to the T-cell lineage (Allman et al. 2003; Porritt et al. 2004; Sambandam et al. 2005; Tan et al. 2005; Bell and Bhandoola 2008). ETPs sequentially develop through double-negative (DN) stages (DN2–DN4)—during which cells do not express either the CD4 or CD8 coreceptor (CD4− CD8−)—into a double-positive stage (DP; CD4+ CD8+) and finally resolve into either CD4 single-positive (SP) helper T cells or CD8 SP cytotoxic T cells. These mature SP cells then exit the thymus via the circulatory system to seed secondary lymphoid organs, where they acquire immune competence (Fowlkes and Mathieson 1985; Penit and Vasseur 1988).
Patients with GATA3 haploinsufficiency suffer from HDR (hypoparathyroidism, deafness, and renal dysplasia) syndrome and present variable clinical symptoms, including hypoparathyroidism, heart defects, deafness, renal malformation, and reduced T-cell numbers (Daw et al. 1996; van Esch et al. 2000). These developmental anomalies are also reproduced when Gata3 is disrupted in mice (Lim et al. 2000; Grigorieva et al. 2010), while a twofold increase in the abundance of GATA3 protein results in thymic lymphoma (Nawijn et al. 2001). Thus, GATA3 abundance must be precisely controlled during T-lymphocyte development as well as in multiple organs. While it has been reported that such stringent control over GATA3 abundance exists (e.g., see Scripture-Adams et al. 2014), how such precision is achieved is currently speculative.
Results
T cells with a single Gata3 allele exhibit diminished GATA3 mRNA, impaired cell expansion, and elevated PU.1 expression
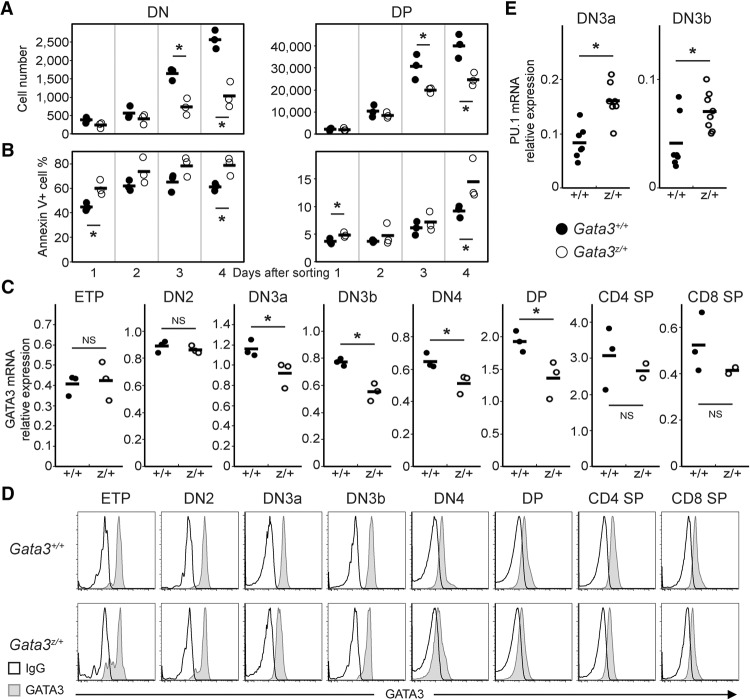
To examine the consequences of Gata3 haploinsufficiency on T-cell development, we analyzed heterozygous mice bearing one wild-type (Gata3+) and one null mutant (Gata3z) allele (van Doorninck et al. 1999). The absolute number of CD4 SP thymocytes in Gata3z/+ mice was 77% of Gata3+/+ (wild-type) mice, which was statistically significant, while the reduction in the number of DP (85%) and CD8 SP (92%) thymocytes was modest and statistically insignificant (Supplemental Fig. S1A). A similar reduction in the number of DP (79%), CD4 SP (56%), and CD8 SP (72%) thymocytes was also observed in Gata3flox/+:TgCd2Cremice (Supplemental Fig. S1B) in which one Gata3 allele was removed in only B and T lymphocytes, and thus the effect is lymphoid cell-autonomous. Since TgCd2Crealone has no effect on T-cell development (Shi and Petrie 2012), compromised T-cell development in the thymi of Gata3flox/+:TgCd2Cre mice could be directly attributed to the lymphocyte-specific inactivation of one Gata3 allele. Both Gata3z/+ (Supplemental Fig. S2A) and Gata3g/+ (Supplemental Fig. S2B) heterozygotes had a reduced number (∼50%) of ETP in comparison with wild-type mice, while the number of ETP in Gata3flox/+:TgCd2Cre heterozygous mice was not statistically different from the number in controls (Supplemental Fig. S2C). The difference in ETP number between these different mutant alleles suggests that the 50% reduced number of ETP (in Gata3z/+ [Supplemental Fig. S2A] or Gata3g/+ [Supplemental Fig. S2B] heterozygous mutant mice) is because there is only one active Gata3 allele in either prethymic progenitors or other nonlymphoid cells. However, this reduced number of ETP was compensated, possibly by excess proliferation in these immature cells, such that no significant reduction in thymocyte number was observed in heterozygous mutant mice through the DN2 to DN4 stages (Supplemental Fig. S2).
To confirm the intrinsic developmental potential of Gata3 heterozygous mutant cells, we isolated DN4 stage T cells from Gata3z/+ thymi and cocultured them on OP9-DL1 feeder cells. OP9-DL1 feeder cells promote Notch signaling, thereby supporting further T-cell development (Schmitt and Zuniga-Pflucker 2002). This method has some advantages as an assay for cell yield and viability, as phagocytes may conceal differences by clearing dead cells in the thymus in vivo. Equal numbers of flow-sorted Gata3z/+ or Gata3+/+ DN4 stage cells were cocultured on OP9-DL1 in the presence of Flt3 ligand and IL7. T cells were recovered, stained, and analyzed by flow cytometry for four consecutive days following the initiation of the cocultures. The significant reduction in DN and DP T cells in cocultures seeded with Gata3z/+ DN4 cells was most evident after 3 or 4 d of coculture (Fig. 1A), when compared with cocultures seeded with wild-type DN4 stage thymocytes. The frequency of apoptotic and dead (Annexin V+) cells on each day in Gata3z/+ DN and DP cell populations was higher in comparison with Gata3+/+ cells (Fig. 1B). Hence, a reduction in Gata3 gene dosage attenuates DN4 cell proliferation, viability, and differentiation.
Figure 1.
Reduced activity of Gata3 alleles results in reduced expansion of immature T cells and elevated expression of myeloid transcription factor PU.1. (A,B) DN4 stage thymocytes were isolated from Gata3z/+ (○) or Gata3+/+ (•) mice and then cocultured on OP9-DL1 feeder cells. Cells were harvested and stained for CD4 and CD8 coreceptors followed by flow cytometric analysis at the indicated time points (after 1–4 d of coculture initiation) for cell number (A) and apoptosis (by Annexin V staining) (B). The horizontal bar in each genotype panel represents the average value for each analyzed genotype group. Representative data are shown from one experiment examining three mice of each genotype; similar results were collected from at least six mice of each genotype in two independent experiments. (C,E) Quantitative RT–PCR (qRT–PCR) in staged T cells of wild-type (Gata3+/+; •) or heterozygous mutant (Gata3z/+; ○) mice. GATA3 (C) and PU.1 (E) mRNA abundance was quantified using the total reverse-transcribed product from 100 live-cell equivalents of RNA normalized to HPRT mRNA. Each circle represents an individual mouse, and the black bars represent the averages. (*) P < 0.05; (NS) not significant. The data summarize duplicate measurements of three to eight mice of each genotype from at least two independent experiments. (D) Intracellular GATA3 (filled curve) protein abundance in staged T cells from wild-type (Gata3+/+) (top panel) and heterozygous mutant (Gata3z/+) (bottom panel) thymi was monitored by flow cytometry. The open curve represents background (IgG) staining in each sample. Representative histograms are shown as characterized in at least three mice of each genotype.
To evaluate whether the different outcomes of Gata3z/+ and Gata3+/+ DN4 OP9-DL1 cocultures could be attributed to altered kinetics in GATA3 mRNA accumulation during T-lineage progression, cells at different T-cell developmental stages (ETP to SP) were isolated from Gata3z/+ and Gata3+/+ thymi and analyzed by quantitative RT–PCR (qRT–PCR). We found that the levels of GATA3 mRNA in Gata3+/+and Gata3z/+ were virtually identical at the ETP and DN2 stages, while its abundance in DN3a and later stage thymocytes was reduced by between 15% and 30% (Fig. 1C). We next tested whether the differences in mRNA levels coincide with protein accumulation at different stages. Thymocytes were isolated from Gata3z/+ and Gata3+/+ thymi, and GATA3 protein abundance was quantified by flow cytometry as described (Scripture-Adams et al. 2014). GATA3 protein abundance in Gata3z/+ was virtually identical to wild type at the ETP and DN2 stages, while its abundance in DN3a and later stage thymocytes was reduced (Fig. 1D), in keeping with the reduction in GATA3 mRNA (Fig. 1C). These data indicate that, during late thymopoiesis, only cells bearing two wild-type Gata3 alleles can elevate GATA3 to wild-type levels and that the failure to attain this temporally critical GATA3 surge (in Gata3z/+ DN4 cells) perturbs their development in vitro and in vivo. The data suggested that GATA3 protein abundance is at least in part regulated at the transcriptional level and that GATA3 abundance critically controls T-cell expansion and the development of thymocytes. This conclusion is entirely consistent with the modest deficiency in T-cell number observed in human GATA3 haploinsufficient patients (Daw et al. 1996; Lichtner et al. 2000).
During the transition from the DN2 to the DN3 stage, when a quantitative difference in GATA3 mRNAs and protein between heterozygous and wild-type mice (Fig. 1C,D) was first detected, GATA3 was shown to block apoptosis and repress transcription factor PU.1 (Scripture-Adams et al. 2014), a key protein that promotes myeloid gene expression (Nerlov and Graf 1998). Mice in which the Gata3 gene was conditionally ablated at the DN3 stage exhibited increased apoptosis and reduced TCRβ protein, but not mRNA, expression (Pai et al. 2003). To address the possibility that the differential Gata3 abundance observed between Gata3z/+ and Gata3+/+ animals might account for these observations, Gata3z/+ heterozygotes were further analyzed.
We first assessed the intracellular abundance of TCRβ by flow cytometry and found that, contrary to the study by Pai et al. (2003), there was only very modest variation in intracellular TCRβ abundance in Gata3+/+ compared with Gata3z/+ DN3 stage thymocytes (Supplemental Fig. S3). In contrast, PU.1 mRNA abundance increased by approximately twofold in Gata3z/+ T cells at both the DN3a and DN3b stages when compared with the equivalent wild-type Gata3 stages (Fig. 1E). Hence, these data show that only T cells with two intact Gata3 alleles can promote normal expansion and development as well as repress PU.1 in DN3a and DN3b thymocytes but also that one active Gata3 allele is sufficient for normal levels of TCRβ protein accumulation.
Gata3 is monoallelically expressed in early DN thymocytes but is biallelically expressed in a subpopulation of late stage thymocytes
The previous data demonstrate that the molecular basis for regulating GATA3 abundance at different developmental stages is at least in part dictated by transcription. To shed light on possible mechanisms by which this reduction in abundance of GATA3 between the DN2 and DN3a stages (Fig. 1C,D) might be elicited, we initially hypothesized that this could be most directly explained by either enhanced transcription from both chromosomes or, although unprecedented in developmental mechanisms, switching from monoallelic to biallelic Gata3 transcription.
Three mechanisms have been shown to lead to the epigenetic activation of genes on only one of two equivalent chromosomes. X-chromosome inactivation, first described more than half a century ago, generates one randomly inactivated X chromosome in females so that both mammalian sexes express equivalent amounts of X-encoded gene product (Ohno et al. 1959; Lyon 1961). In contrast to the inactivation of a whole chromosome, mammalian genomic imprinting selectively inactivates specific autosomal loci in a male or female parent of origin-dependent manner (Bartolomei et al. 1991; DeChiara et al. 1991). Finally, the discovery that multiple autosomal genes, including Il2 (Hollander et al. 1998; Rhoades et al. 2000) and Tlr4 (Pereira et al. 2003), are monoallelically expressed provided yet a third class of unequally expressed genes. Even more recently, this category was extended to include widely dispersed genes comprising 8%–24% of all autosomal transcripts that are monoallelically regulated (Gimelbrant et al. 2007; Deng et al. 2014). From these studies, there appeared to emerge two distinct types of autosomal monoallelic expression: (1) a subset that is expressed in an unstable, stochastic pattern (Deng et al. 2014) and (2) a subset in which the pattern is stably maintained (Gimelbrant et al. 2007; Zwemer et al. 2012). The latter observations imply that the number of active alleles is precisely regulated in order to achieve alternative levels of gene product encoded by some unknown number of key genes, although this commonly held maxim has not been directly challenged.
To address whether Gata3 is monoallelically or biallelically expressed in ETP and DN2 stage thymocytes (Fig. 1C,D), we exploited a silent single nucleotide polymorphism (SNP) in the third Gata3 exon that differs between inbred mouse strains C57Bl/6J (B6) and FVB/NJ (FVB) F1 progeny using single-cell RT–PCR followed by Sanger sequencing. To distinguish between the paternal and maternal alleles, F1 (B6 × FVB) hybrid mice were generated by crossing B6 males to FVB females in order to determine whether one or both alleles in individual thymocytes were expressed and whether that expression was dependent on parental gender (genomic imprinting). Hematopoietic stem cells (HSCs) in the bone marrow and ETP through SP F1 thymocytes were individually isolated by flow sorting and lysed, total RNA was reverse-transcribed, and the cDNA was amplified by PCR and sequenced (Supplemental Fig. S4A). To test the sensitivity of the method, cDNAs from FVB and B6 strains were mixed in different ratios and analyzed (Supplemental Fig. S4B). The detection limit of the method was ∼5%, and therefore, when >95% dominance of either transcript was detected, that was defined as monoallelic expression, while a mixture of both alleles (between 5% and 95%) was defined as biallelic.
As summarized in Table 1, individual cells recovered from adult mice at all early stages exhibited a profound bias for monoallelic Gata3 expression: HSC (100%), ETP (97%), DN2 (67%), and DN3a (74%). In contrast, from the DN3b stage onward, approximately half of the cells expressed Gata3 from only one allele, while the other half expressed both mRNAs (Table 1). The observed gender-independent activation of either the paternal (B6) or maternal (FVB) allele (modestly biased in favor of generation of the paternal allele) indicates that Gata3 monoallelic expression is not sex-dependent or parent of origin-dependent, and thus the monoallelic to biallelic GATA3 expression bias that is exhibited during midthymopoiesis is likely to be regulated by a mechanism distinct from the ones used in X inactivation or genomic imprinting.
Table 1.
GATA3 mRNA expression in single, staged adult bone marrow cells and thymocytes (from GATA3 RT–PCR SNP sequence)

To confirm the conclusion from the single-cell mRNA analysis, we also examined transcript localization in developmentally staged T-cell nuclei by nascent transcript fluorescence in situ hybridization (RNA-FISH). Cells recovered from the bone marrow or thymi of wild-type adult B6 mice were flow-sorted according to developmental stage and then hybridized to FISH probes that detect CD45 (expressed on all hematopoietic progenitor cells) as well as GATA3 (expressed at all stages of T-cell development) (David-Fung et al. 2006; Ku et al. 2012). Cells were then counterstained with 4′,6-diamidion-2-phenylindole (DAPI) to define the nucleus and finally visualized by fluorescence microscopy. Only cells with visually complete, intact nuclei were imaged.
The pan-hematopoietic marker CD45 was expressed from two chromosomal alleles in all cells imaged (Fig. 2, pseudo-red spots in each panel). In all HSCs (13 of 13) and ETPs (13 of 13) as well as nearly two-thirds of the DN2 (10 of 15) and DN3a (13 of 20) stage T cells, GATA3 primary transcripts could be detected only at a single chromosomal location (Fig. 2, green spots; quantified in Table 2). However, in DN3b and later stage thymocytes, monoallelic and biallelic expression among the cells was essentially equally distributed (Fig. 2; Table 2).
Figure 2.

Gata3 is transcribed from only one allele in HSCs and ETPs and then switches to biallelic transcription in half of DN3b and later stage T cells. Adult bone marrow cells and thymocytes were isolated using conventional cell surface markers (Hosoya et al. 2009; Ku et al. 2012), hybridized in situ to fluorescently labeled probes to detect either GATA3 (green spots) or CD45 (red spots), and analyzed by RNA-FISH (Materials and Methods). Bar, 3 μm. Representative images are shown from at least four mice of each developmental stage from five independent experiments and are summarized in Table 2. The images shown were taken as a single focal plane, but Z-stack images were recorded to confirm that no overlapping dots were misrepresented as a single dot on the single-focal-plane images.
Table 2.
GATA3 mRNA expression in single, staged adult bone marrow cells and thymocytes (from GATA3 RNA-FISH)

Genes expressed at very low levels have been theorized to exhibit transcription of only one allele due to completely random events (Hume 2000; Kuznetsov et al. 2002), and experimentally it has been shown that Il4 and Il13 are transcribed from either one or two alleles in Th2 cells by probabilistic stochastic regulation (Guo et al. 2005). If the availability of transcriptional machinery is the limiting cause of monoallelic expression at the Gata3 locus, then one should observe a higher monoallelic cell frequency in CD8 SP thymocytes, which express the lowest levels of GATA3 at all thymocyte stages (Hernández-Hoyos et al. 2003; Ku et al. 2012). The data shown here suggest the opposite of this mode of monoallelic expression; GATA3 is expressed lower in CD8 SP cells than in ETPs or HSCs, but, nonetheless, CD8 SP cells exhibit a higher frequency of Gata3 biallelic cells (Tables 1, 2). If the Gata3 locus were regulated by probabilistic stochastic regulation like Il4 or Il13, a higher frequency of biallelic expression should be observed at stages of development where GATA3 is most abundant. On the contrary, stage-specific and GATA3 transcript abundance-independent monoallelic expression was observed only at the HSC and ETP stages in the Gata3 locus. These data demonstrate that Gata3 monoallelic expression and the switch to biallelic expression differ from a monoallelic expression pattern that would be generated in a stochastic manner (Eckersley-Maslin and Spector 2014). Thus, we conclude that Gata3 is monoallelically expressed in early stages of T-cell development and that half of the cells then switch to biallelic expression at intermediate stages of T-cell thymic development.
The commitment to Gata3 monoallelic or biallelic transcriptional status is regulated by a stable mechanism
GATA3 activity is required for initial T-cell generation as well as several intermediate maturational stages (Hosoya et al. 2010; Yui and Rothenberg 2014). In considering experiments that might shed further light on these phenomena, we reasoned that a hypomorphic Gata3-eGFP fusion allele (Gata3g) (Hosoya et al. 2009) might not generate adequate GATA3 functional activity to allow T-cell intermediates to survive and thereby confer a selective survival or developmental advantage in adult T cells that initially express the wild-type allele at the ETP and DN2 stages in Gata3g/+ mice (Fig. 3A). Again, this hypothesis is consistent with the recent demonstration that progression through early T-cell development is exquisitely sensitive to GATA3 abundance (Scripture-Adams et al. 2014). We discovered support for this hypothesis by determining that eGFP fluorescence was absent in Gata3g/+ adult ETP and DN2 stage thymocytes (Fig. 3B), but, as cells mature and switch to biallelic transcription at the DN3 stage, eGFP-positive (eGFP+) thymocytes began to appear (Fig. 3B). This observation was confirmed by examining a mouse bearing a different Gata3(-null) mutant allele in which the bacterial lacZ gene was inserted at the Gata3 start codon (Gata3z/+) (van Doorninck et al. 1999); thymocytes only began to transcribe the lacZ allele after the DN2 stage (Supplemental Fig. S5).
Figure 3.
The commitment to Gata3 monoallelic or biallelic transcriptional status is mechanistically stable. (A) A hypothetical model. The GATA3-eGFP fusion (Gata3g; red) allele-expressing adult Gata3g/+ ETP cells are not viable because the reduced hypomorphic GATA3 monoallelic activity is not sufficient to allow ETPs to survive and they are eliminated, since GATA3 is vital for early T-cell generation, while wild-type (Gata3+; blue) monoallelic-expressing cells survive and continue to develop. Later during T cell development, after the DN3b stage, the second allele is activated in about half of the surviving cells and expresses both hypomorphic (Gata3g) and wild-type (Gata3+) alleles. (B) Expression of a GATA3-eGFP fusion protein. eGFP expression from Gata3+/+ (open curve) and Gata3g/+ (filled curve) heterozygotes was monitored by flow cytometry in immature thymocytes to track transcription from the Gata3g allele (Hosoya et al. 2009). Numbers above the bars indicate the percentage of eGFP-positive cells compared with wild-type controls from the same ancestral population. These histograms are representative of data recovered for at least five mice of each genotype from three independent experiments. (C) Hypothetical gradual (left) versus bimodal (right) switching models. (D) Gata3 monoallelic cells expressing the Gata3g allele survive and develop in the absence of the Gata3+ allele. Gata3g/g homozygous hypomorphic mutant thymocytes express eGFP in a cell-autonomous manner. Gata3g/g or Gata3+/+ HSCs were recovered from fetal livers and adoptively transferred into sublethally irradiated adult mice. Sixteen weeks after HSC transplantation, thymocytes were analyzed for eGFP expression. The diagram presents representative histograms in which seven to eight recipients were examined for each genotype in two independent experiments. (E) DN4 stage thymocytes were sorted into eGFP-negative (eGFP−; open curve) or eGFP+ (filled curve) populations from Gata3g/+ mice and then cocultured on OP9-DL1 feeder cells. The eGFP expression profiles depicted immediately after sorting (left panels) or after 4 d of coculture and development into DP stage cells (right panels) are shown. The diagrams are representative of a total of eight mice examined in two independent experiments.
To determine whether Gata3 is transcribed from the wild-type allele in eGFP-negative (eGFP−) Gata3g/+ T cells, eGFP− and eGFP+ DN4 stage cells were isolated from Gata3g/+ thymi by flow cytometry (Supplemental Fig. S6A), fixed and separated into equal portions to determine intracellular GATA3 and IgG (background) staining, and finally analyzed by flow cytometry (Supplemental Fig. S6B). Since eGFP fluorescence is diminished after fixation, we used a two-step method. We initially observed that eGFP− DN4 cells express GATA3 but also that eGFP+ cells express much more GATA3 (resulting from the sum of the wild-type and GATA3-eGFP fusion proteins). A similar result was observed in CD4 SP stage cells (Supplemental Fig. S6C,D). We also confirmed that GATA3 mRNA is expressed at all stages in eGFP− immature T cells (Supplemental Fig. S7). Based on these data, we conclude that eGFP− thymocytes in Gata3g/+ animals express monoallelic Gata3, while eGFP+ cells that express GATA3 more abundantly transcribe both Gata3 alleles.
We next asked whether the monoallelic-to-biallelic expression switch at the Gata3 locus represents a gradual change (Fig. 3C, model A) or a bimodal event (Fig. 3C, model B). One model might posit that, as the demand for Gata3 mRNA increases and progenitor cells mature, the total transcriptional activity from the Gata3 gene increases in 100% of immature cells, at which point roughly half of the cells stochastically activate both alleles (Fig. 3C, model A). For example, some component of the machinery required for Gata3 transcription may be limiting, and thus T cells could have an equal probability of activating either one or both Gata3 alleles as cells traverse DN2 to DN3 maturation to allow increased GATA3 abundance. This would result in a shift of a single peak of eGFP intensity to greater overall fluorescence in the thymocytes of Gata3g/+ mice. However, the bimodal eGFP expression profile detected at the DN4 stage in Gata3g/+ thymocytes (Fig. 3B) best supports a model in which Gata3 is transcribed from only one allele at the beginning of thymopoiesis, and, at some time during the DN2/DN3 transition, half of the cells begin to transcribe the second Gata3 allele (Fig. 3C, model B).
To remove any possible bias between expression of the wild-type and mutant alleles, we also analyzed thymocytes recovered from homozygous Gata3g/g fetal liver HSC-reconstituted animals; these T cells again exhibited bimodal GATA3 expression at the DN2 stage and beyond (Fig. 3D) and never exhibited a single shifted peak from lower to greater fluorescence intensity as the thymocytes matured and as would be predicted from the first model. The fact that a more robust eGFP peak in Gata3g/g mice is first detected at the DN2 stage (Fig. 3D) is entirely consistent with the observation that the Gata3 monoallelic-to-biallelic switch initiates around the DN2 stage in approximately one-third of all thymocytes (Fig. 2; Tables 1, 2). Thus, in Gata3g/+ thymi, we could identify cells that express biallelic Gata3 (eGFP+) and monoallelic Gata3 (eGFP−) at the DN4 and later stages. These data validate the conclusions that, first, Gata3 is monoallelically transcribed in ETPs, and, second, the expression pattern switches to biallelic transcription in about half of the T cells during late thymocyte development.
To test the hypothesis that the commitment to Gata3 monoallelic or biallelic transcriptional status is regulated by a stable mechanism, we next asked whether, after the switch in transcription occurs at DN2/DN3, late stage monoallelic cells continue to express from only one chromosome or can switch to biallelic transcription and, conversely, whether cells that express both alleles can revert to single-allele expression. To address this question, we isolated an equal number of Gata3 biallelic (eGFP+) and Gata3 monoallelic (eGFP−) DN4 stage thymocytes from Gata3g/+ animals (Fig. 3E, left panel) and cocultured them separately on OP9-DL1 feeder cells to induce T-cell development to the DP stage. After 4 d of coculture, a majority of DN4 stage cells develop to the DP stage. DP cells that had developed from biallelic (eGFP+) DN4 cells did not exhibit a bimodal expression pattern (i.e., none of the cells reverted to monoallelic transcription), although eGFP intensity was significantly reduced (Fig. 3E, bottom right panel), as was also observed in vivo (Hosoya et al. 2009). This reduction in intensity of fluorescence is probably attributable to dilution of the fluorochrome that results from the extensive proliferation during culture for 4 d (see Fig. 1A for the remarkable increase in cell number after 4 d of coculture; Supplemental Fig. S8). Similarly, DP cells that were differentiated from monoallelic (eGFP−) DN4 stage cells did not develop a bimodal expression pattern (i.e., they failed to switch to biallelic transcription) (Fig. 3E, top right panel). Both patterns were consistent over all 4 d of coculture (data not shown). These data demonstrate that once cells are set to express either one or both Gata3 alleles at the DN2/DN3 stage, they stably maintain their monoallelic or biallelic Gata3 transcriptional identity during T-cell development at least to the DP stage.
Maintenance of the Gata3 monoallelic-to-biallelic switch
To explore one aspect of the mechanism that might regulate the early thymocyte Gata3 monoallelic-to-biallelic switch, we performed chromatin immunoprecipitation (ChIP)-qPCR in the Gata3 locus to determine the methylation status of two key epigenetic marks: H3K27me3 and H3K4me3. H3K27me3 is a histone mark for gene repression (Plath et al. 2003; Umlauf et al. 2004) and is often found near monoallelicly expressed genes (Nag et al. 2013), while H3K4me3 modifications are commonly coincident with actively transcribed genes (Bernstein et al. 2005; Kim et al. 2005). Therefore, we hypothesized that H3K27me3 could be observed in or near the Gata3 locus in monoallelic (eGFP−) Gata3g/+ DN4 stage cells but not biallelic eGFP+ cells (since both alleles would be active). Similarly, we speculated that the H3K4me3-activating mark would be higher in the neighborhood of the Gata3 locus in biallelic eGFP+ cells (from Gata3g/+ DN4 stage) when compared with monoallelic eGFP− cells. Since H3K27me3 ChIP-seq (ChIP combined with deep sequencing) data for DN3 and DP stage cells have been reported (Zhang et al. 2012), PCR primer sets for ChIP-qPCR were designed that would detect the H3K27me3 mark that was previously observed 5′ to the Gata3 gene in DN3 and DP cells (Supplemental Fig. S9). Unexpectedly, we found that there was no enrichment of the H3K27me3 mark near the Gata3 locus in monoallelic eGFP− cells when compared with (biallelic) eGFP+ cells (Supplemental Fig. S9). We also found no difference in the activating H3K4me3 mark in these two cell populations (Supplemental Fig. S9). These data suggest that these two specific histone modifications do not play an active role in maintaining the silencing of the inactive locus in lymphocytes that transcribe only one Gata3 allele. However, the data do not address the possibility that other epigenetic modifications may be involved in the switch.
Monoallelic or biallelic expression in Th2 or nonhematopoietic cells
Circulating Th2 cells express the highest levels of GATA3 in T lymphocytes, much more abundantly than in any thymocyte. We wondered whether those cells all express both alleles of Gata3, or perhaps the increased GATA3 levels were generated simply by increased transcription of a single allele. The bimodal eGFP profile in Gata3g/+ Th2 cells (Hosoya-Ohmura et al. 2011) suggests that both Gata3 monoallelic expression and biallelic expression are observed in Th2 stage lymphocytes and thus that any increase in GATA3 expression at the Th2 stage is achieved by augmented transcription from either one or both Gata3 loci but additionally that the monoallelic-to-biallelic switch remains stable and intact.
Gata3 is vital for not only T-cell development but also the generation or maintenance of multiple other tissues; for example, the sympathetic nervous system (Lim et al. 2000; Moriguchi et al. 2006), kidney development (Hasegawa et al. 2007), breast epithelial development (Kouros-Mehr et al. 2006), and a growing list of other organs. This provoked the question of whether Gata3 might also be monoallelically transcribed in tissues other than T cells. To examine this hypothesis, we interrogated Gata3 expression in embryos in tissues where prominent Gata3 expression has been reported previously (Hasegawa et al. 2007).
When we originally generated the Gata3g allele, a full-length mouse GATA3 cDNA was fused in-frame to the eGFP cDNA (Hosoya et al. 2009). In Gata3g/+ tissues, we could easily visualize individual cells expressing the chimeric GATA3-gfp fusion protein using an anti-GFP antibody. However, this would not allow distinction of cells that solely expressed the wild-type Gata3 allele, since an anti-GATA3 antibody would recognize both the wild-type and chimeric GATA3 proteins. Given this technical constraint, we therefore attempted to address whether monoallelic or biallelic GATA3 expression occurred in tissues other than T cells by breeding together two mutant alleles: one a lacZ knock-in null allele (directed by normal Gata3 regulatory elements) (van Doorninck et al. 1999) and the second the hypomorphic Gata3g allele. Gata3g/z embryos did not survive to late gestation (K-C Lim and C-J Ku, unpubl.).
Cells lining the nephric duct (Supplemental Fig. S10, top panels, dashed circle) of a wild-type embryonic day 10.5 (E10.5) Gata3 embryo were devoid of fluorescence (except for autofluorescencing erythrocytes) (Supplemental Fig. S10, bottom right), demonstrating the specificity of both the anti-GFP and anti-β-galactosidase (anti-β-gal) antibodies used in these experiments. In an E10.5 Gata3g/z littermate (Supplemental Fig. S10, middle panels), all nephric duct cells displayed immunoreactivity with both the anti-GFP and anti-β-gal antibodies, indicating exclusive biallelelic GATA3 expression in the E10.5 nephric duct. However, we also observed that, while most cells coexpressed both the Gata3g and Gata3z alleles (for example, in E10.5 branchial arches [BAs] [data not shown] and the V2 interneurons of the E13.5 developing spinal cord [SC] [Supplemental Fig. S10, bottom panels]), a few cells appeared to be very faintly stained for either GFP or β-gal; however, upon close inspection, every cell that labeled with the GFP antibody also stained with the anti-β-gal antibody. This was also true when we immunostained Gata3z/+ adult bladders and kidneys (data not shown) with anti-GATA3 and anti-β-gal antibodies. We tentatively concluded that, if Gata3 is monoallelically expressed in the embryonic urothelium, sympathetic ganglia, or BAs or adult bladders or kidneys, such monoallelic Gata3-expressing cells are, at most, very rare.
Discussion
We showed previously that cells can be quite sensitive to modest changes in transcription factor abundance (Motohashi et al. 2000; Shimizu et al. 2004; Hasegawa et al. 2007). Here, we showed that an increase in GATA3 protein and transcript abundance accompanies the monoallelic-to-biallelic genetic switch, which appears to be biologically required for both the repression of PU.1 and apoptosis during thymic T-cell development. Since progression through early T-cell development is exquisitely sensitive to GATA3 abundance (Scripture-Adams et al. 2014) and since forced expression of GATA3 in ETP to DN2 stage thymocytes leads to thymic lymphoma (Nawijn et al. 2001), diploid organisms appear to have evolved a mechanism by which they can exquisitely control gene expression by regulating monoallelic versus biallelic transcription. Although B-cell transcription factor Pax5 was originally reported by RNA-FISH to be monoallelically expressed (Nutt et al. 1999), this was later disproven using a more sensitive single-cell RT–PCR assay (Rhoades et al. 2000). Here, we used both methods to ensure the veracity of the results. We propose that both Gata3 T-cell-specific enhancer-mediated regulation (Hosoya-Ohmura et al. 2011) and the monoallelic-to-biallelic switch must collaborate to achieve precise control of transcription factor GATA3 abundance during T-cell development.
Why is the switch to biallelic expression observed in only half of the cells while the other half remain stably monoallelic? The most intriguing hypothesis concerns the correlation with allelic exclusion at the Tcrβ locus, which happens during the DN3 stage within the same differentiation window in which cells must surmount β selection. Another viable hypothesis is that DN3 stage thymocytes already “know” whether they are committed to a CD4 SP versus CD8 SP cell fate, since GATA3 is more abundantly expressed in CD4 SP cells than in CD8 SP cells and is required for CD4 SP cell development. We are in the process of testing these and several alternative hypotheses at the present time.
With the knowledge that the monoallelic-to-biallelic switch of a tissue-restricted transcription factor, GATA3, is intimately involved in several different aspects of thymic T-cell development, this raises the question of whether Gata3 monoallelic or biallelic transcriptional status mediates processes in other tissues. While Gata3 monoallelic or biallelic transcription in the many other cell types and organ systems where it is normally expressed has not yet been analyzed (e.g., in the CNS, peripheral nervous system [PNS], and kidneys) (Yamamoto et al. 1990; George et al. 1994; Lim et al. 2000; Moriguchi et al. 2006), Gata3 haploinsufficiency is known to lead to variably penetrant HDR syndrome in humans as well as other partially penetrant pathophysiological effects (Van Esch et al. 2000; Usary et al. 2004; Grigorieva et al. 2010). We therefore speculate that Gata3 monoallelic expression and/or a monoallelic-to-biallelic switch in tissues other than T cells could play critical roles in their normal development or homeostatic maintenance. It will be extremely interesting to determine, for example, whether the quite variable kidney dysplasia observed in human HDR patients might arise as a consequence of early monoallelic expression of GATA3 followed by apoptotic failure of a variable number of cells in this developing organ, since this aspect of the disease can be readily modeled in the kidneys of mice (Hasegawa et al. 2007).
It remains to be determined how frequently similar genetic switch mechanisms are employed among the growing number of monoallelic autosomal genes that have been documented in diploid organisms. Although Nanog was originally reported to be monoallelic in early preimplantation embryos, with the proportion of biallelism increasing progressively from 2% at the eight-cell stage to 70% of the inner cells in the late blastocyst (Miyanari and Torres-Padilla 2012), that conclusion has been challenged recently (Faddah et al. 2013; Filipczyk et al. 2013). Furthermore, the active allelic selection appears to be reversible during culture of embryonic stem cells for 6 d (Chambers et al. 2007). In contrast, Gata3 monoallelic (eGFP−) cells stably maintain expression from the active allele during 4 d of in vitro culture as the cells developed from the DN4 stage to the DP stage. The imprinted gene Kcnq1 (also known as Kvlqt1) is expressed from only the maternal allele early during embryogenesis, while the paternal allele becomes active as this gene acquires tissue-specific expression (Lee et al. 1997; Gould and Pfeifer 1998); in contrast, Gata3 monoallelic expression is not parent of origin-dependent. These differences suggest that the mechanism that regulates the Gata3 monoallelic-to-biallelic expression switch is fundamentally different from the mechanism exploited by these genes. If this phenomenon is prevalent, the number of genes controlled by monoallelic regulation could be even greater than previously estimated (8%–24%) simply because all of the genes that are expressed in a monoallelic fashion might not have been detected under the specific experimental conditions used (Gimelbrant et al. 2007; Deng et al. 2014). Given the present findings coupled with the fact that there are a large number of autosomal genes that are monoallelically expressed, it does not seem unreasonable to suggest that defects in monoallelic-to-biallelic conversion in some subset of these could contribute to many variably penetrant phenotypes that result from human haploinsufficiency.
Materials and methods
Mice
The Gata3z allele (van Doorninck et al. 1999) and Gata3g allele (Hosoya et al. 2009) have been described previously and were maintained on a B6 genetic background. Wild-type B6 mice and FVB mice were purchased from Jackson Laboratory. All mice were analyzed at 6–8 wk of age after backcrossing onto the B6 background for at least seven generations. All mice used in this study were housed in the Unit for Laboratory Animal Medicine at the University of Michigan, and all experiments were approved by the University Committee on Use and Care of Animals.
Cell preparation and flow cytometry
Cells were analyzed and sorted by flow cytometry (LSRFortessa and FACSAria III, BD Biosciences) as previously described (Hosoya et al. 2009; Ku et al. 2012). Sorted cells were reanalyzed to confirm that they were of >95% purity before subsequent study. Thymocytes were resuspended in propidium iodide (Calbiochem) or DAPI (Roche) solution to discriminate between live and dead cells. Depending on the specific analysis, the following antibodies were used: B220 (RA3-6B2), CD2 (RM2-5), CD3 (17A2), CD4 (RM4-5), CD5 (53-7.3), CD8 (53-6.7), CD11c (N418), CD19 (6D5), CD25 (PC61.5), CD27 (LG.7F9), CD41 (MWReg30), CD48 (HM48.1), CD150 (TC15-12F12.2), c-Kit (2B8), GATA3 (16E10A23), Gr1 (RB6-8C5), Mac-1 (M1/70), NK1.1 (PK136), Sca-1 (D7), TCRβ (H57-597), TCRγδ (GL3), and TER119 (TER-119). All antibodies were purchased from BD Biosciences, BioLegend, or eBioscience. After cell surface staining, apoptosis was analyzed using Annexin V and DAPI staining in Annexin V-binding buffer, per the manufacturer's instructions (BD Pharmingen). Data were analyzed using FlowJo software (Tree Star) or FACSDiva software (BD Biosciences).
OP9-DL1 coculture
Confluent OP9-DL1 stromal cells were prepared prior to coculture initiation. DN4 stage thymocytes (2 × 105) were cultured on a feeder layer of OP9-DL1 cells (Schmitt and Zuniga-Pflucker 2002) in the presence of 5 ng/mL IL7 and 5 ng/mL Flt3 ligand (R&D Systems). Suspension and semiadherent cells were harvested by pipetting and stained for flow cytometry for four consecutive days after coculture initiation.
qRT–PCR
Total RNA was isolated from sorted cells by RNeasy micro kit (Qiagen) with DNase treatment and used to synthesize cDNA with SuperScript III first strand synthesis kit (Invitrogen). qRT–PCR was performed with SYBR Green dye using the StepOne real-time PCR system (Applied Biosystems). The primers used were previously described as follows: GATA3 (Yu et al. 2009), HPRT (Maillard et al. 2008), PU.1 (Sarrazin et al. 2009), and β-actin (Esteghamat et al. 2013). All results of qRT–PCR were performed using the total RT product from 100 cell equivalents and quantified relative to the mRNA expression of endogenous reference genes Hprt and β-actin. Data normalized to HPRT as the control are shown in the figures; data normalized to β-actin (data not shown) exhibited a similar tendency.
Allele-specific single-cell RT–PCR
Allele-specific single-cell RT–PCR is presented diagrammatically in Supplemental Figure S3. cDNA synthesis was carried out by reverse transcription using the iScript cDNA synthesis kit (Bio-Rad). A SNP was identified in the Gata3 third exon (rs27150232), which allows discrimination between alleles of the Mus musculus inbred strains B6 and FVB. First, flow-sorted cell populations from F1 hybrid (B6 × FVB) mice were resorted as single cells into 1.8 μL of 0.14% Triton X-100 (Sigma-Aldrich) followed by the addition of 0.5 μL of 5× iScript reaction mix and 0.2 μL of iScript reverse transcriptase, resulting in a 2.5-μL RT reaction volume. This was then incubated for 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C per the manufacturer's instructions. cDNA was amplified by polymerase chain reaction in a 25-μL reaction mixture consisting of 2.5 μL of cDNA, 1× PCR buffer (Qiagen), 200 μM dNTPs, 200 nmol of each primer, and 0.75 U of Taq DNA polymerase (Qiagen). The PCR condition was 5 min at 95°C followed by 42 cycles of 20 sec at 95°C, 30 sec at 55°C, and 45 sec at 72°C and then a final extension for 7 min at 72°C. Primers used were 5′-GAGGTGGACGTACTTTTTAACATCG-3′ and 5′-CGGGAAGGTGAAGAGATGAGG-3′. β-Actin (Esteghamat et al. 2013) was used as a control to confirm for the presence of a cell in each well and the efficiency of single-cell sorting, which was 65% ± 15% overall. Ten percent to 20% of the PCR products were visualized on a 2% agarose gel, and positives were then purified using QIAquick gel kit (Qiagen) or GeneJET get extraction kit (Fisher Scientific) and sequenced on an Applied Biosystems 3730XL DNA sequencer at the University of Michigan Sequencing Core facility. The primer used for sequencing was 5′-ATGGCGGCAAAGCCCTGAGC-3′.
RNA-FISH
RNA-FISH was performed as described previously (Maclary et al. 2014). In detail, RNA probes were prepared by randomly priming DNA templates using the BioPrime DNA labeling system (Invitrogen). Probes were labeled with FITC-dUTP (Agilent) or Cy3-dCTP (GE Healthcare). Labeled probes were column-purified (GE Healthcare) and then precipitated in 0.3 M sodium acetate with 300 μg of yeast transfer RNA (Invitrogen) and 150 μg of sheared, boiled salmon sperm DNA (Invitrogen). The probe solution was spun down at 4°C and then washed in 70% ethanol followed by 100% ethanol. The pellet was dried and resuspended in deionized formamide (Amresco). The resuspended probe was denatured for 10 min at 95°C followed immediately by a 5-min incubation on ice. A 2× hybridization solution (4× SSC, 20% Dextran sulfate [Millipore]) was added to the denatured probe solution and stored at −20°C until use. The GATA3 (WIBR1-2318H06) and receptor-type tyrosine protein phosphatase C (WIBR1-2452A19; CD45) clones used to prepare probes were purchased from Children's Hospital Oakland Research Institute BACPAC Resource Center. Cells deposited on coverslips were fixed in freshly prepared cold 4% paraformaldehyde (Sigma-Aldrich) for 10 min, washed in PBS followed by permeabilization on ice in PBS with 0.5% Triton X-100 and 10 mM Ribonucleoside Vanadyl Complex (New England Biolabs) for 8 min, and then stored in 70% ethanol for up to 2 wk at 4°C before use. Samples were dehydrated through 2-min incubations in 70%, 85%, 95%, and 100% ethanol, subsequently air-dried, and then hybridized to the probe overnight at 37°C in a humid chamber (humidity provided by 2× SSC/50% formamide). After hybridization, samples were washed three times with 2× SSC/50% formamide for 7 min each at 39°C while shaking, twice with 2× SSC, and twice with 1× SSC. A 1× DAPI solution was added to the first 1× SSC wash. Cells were then mounted in ProLong Gold anti-fade (Invitrogen) and imaged using a 60× (air) or 100× (oil) objective on Nikon Eclipse TiE inverted microscope with a Photometrics CCD camera. Neuroblastoma cell line C1300 (George et al. 1994) was used as a positive control for GATA3 expression and as a negative control for CD45 expression. All images were deconvolved and processed using NIS-Elements software.
Immunohistochemistry
Embryos and adult tissues were fixed in 4% paraformaldehyde for 1–2 h at 4°C and then equilibrated in 30% sucrose prior to cryosectioning. Sagittal sections (10 μm) of Gata3+/+ and Gata3g/z embryos (E10.5 and E13.5) were first stained with rabbit anti-β-gal antibody (1:2000) (Moriguchi et al. 2006) for 2 h at room temperature followed by goat anti-rabbit Cy3-conjugated secondary antibody (1:400; Invitrogen) for 30 min at room temperature. These sections were then incubated overnight at 4°C with rabbit anti-GFP Alexa fluor 488 antibody (1:1500; Invitrogen). Sagittal sections (10 μm) of adult Gata3z/+ bladders and kidneys were similarly treated sequentially with rabbit anti-β-gal antibody followed by goat anti-rabbit Alexa fluor 488-conjugated secondary antibody (1:400; Invitrogen) and then incubated overnight at 4°C with mouse anti-GATA3 Alexa fluor 647 antibody (1:3; BD Bioscience). All tissue sections were counterstained with DAPI and then imaged using a Nikon A1 confocal microscope in the University of Michigan Microscopy and Image Analysis Laboratory.
Statistical analysis
Statistical significance was determined by Student t-test. Data were considered statistically significant when P < 0.05.
Supplementary Material
Acknowledgments
We are grateful to Stephen J. Weiss for numerous thoughtful clarifying comments, Thomas Graf for positing a significant question addressed here, and Maria Figueroa and the University of Michigan Epigenetics Core for assistance with the histone ChIP assays. This work was supported by National Institute of Health grants AI094642 (to T.H. and J.D.E.) and AI091627 (to I.M.), the Leukemia and Lymphoma Society (I.M.), and the Lymphoma Research Foundation (C.-J.K.). The research was also supported in part by the National Cancer Institute through the University of Michigan Comprehensive Cancer Center Support Grant (P30 CA046592) and the use of the Flow Cytometry, Morphology, Microscopy and Image Analysis Laboratory, Experimental Irradiation, and Sequencing cores at University of Michigan. C.-J.K. designed the study, performed experiments, analyzed the data, and wrote the manuscript. K.-C.L. designed the study, performed experiments, and edited the manuscript. S.K. designed the FISH study and edited the manuscript. I.M. designed the study and edited the manuscript. J.D.E. designed the study, analyzed the data, and wrote the paper. T.H. designed the study, performed experiments, analyzed the data, and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.265025.115.
References
- Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol 4: 168–174. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM. 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155. [DOI] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. 2008. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452: 764–767. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234. [DOI] [PubMed] [Google Scholar]
- David-Fung E-S, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, Rothenberg EV. 2006. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol Rev 209: 212–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw SC, Taylor C, Kraman M, Call K, Mao J, Schuffenhauer S, Meitinger T, Lipson T, Goodship J, Scambler P. 1996. A common region of 10p deleted in DiGeorge and velocardiofacial syndromes. Nat Genet 13: 458–460. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859. [DOI] [PubMed] [Google Scholar]
- Deng Q, Ramskold D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Spector DL. 2014. Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet 30: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteghamat F, Gillemans N, Bilic I, van den Akker E, Cantu I, van Gent T, Klingmuller U, van Lom K, von Lindern M, Grosveld F, et al. 2013. Erythropoiesis and globin switching in compound Klf1::Bcl11a mutant mice. Blood 121: 2553–2562. [DOI] [PubMed] [Google Scholar]
- Faddah DA, Wang H, Cheng AW, Katz Y, Buganim Y, Jaenisch R. 2013. Single-cell analysis reveals that expression of nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell Stem Cell 13: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A, Gkatzis K, Fu J, Hoppe PS, Lickert H, Anastassiadis K, Schroeder T. 2013. Biallelic expression of nanog protein in mouse embryonic stem cells. Cell Stem Cell 13: 12–13. [DOI] [PubMed] [Google Scholar]
- Fowlkes BJ, Mathieson BJ. 1985. Intrathymic differentiation: thymocyte heterogeneity and the characterization of early T-cell precursors. Surv Immunol Res 4: 96–109. [DOI] [PubMed] [Google Scholar]
- George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. 1994. Embryonic expression and cloning of the murine GATA-3 gene. Development 120: 2673–2686. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. 2007. Widespread monoallelic expression on human autosomes. Science 318: 1136–1140. [DOI] [PubMed] [Google Scholar]
- Gould TD, Pfeifer K. 1998. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet 7: 483–487. [DOI] [PubMed] [Google Scholar]
- Grigorieva IV, Mirczuk S, Gaynor KU, Nesbit MA, Grigorieva EF, Wei Q, Ali A, Fairclough RJ, Stacey JM, Stechman MJ, et al. 2010. Gata3-deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor Gcm2. J Clin Invest 120: 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Hu-Li J, Paul WE. 2005. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity 23: 89–99. [DOI] [PubMed] [Google Scholar]
- Hasegawa SL, Moriguchi T, Rao A, Kuroha T, Engel JD, Lim K-C. 2007. Dosage-dependent rescue of definitive nephrogenesis by a distant Gata3 enhancer. Dev Biol 301: 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Hernández-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19: 83–94. [DOI] [PubMed] [Google Scholar]
- Hollander GA, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Simpson S, Terhorst C, Wishart W, Golan DE, Bhan AK, et al. 1998. Monoallelic expression of the interleukin-2 locus. Science 279: 2118–2121. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. 2009. GATA-3 is required for early T lineage progenitor development. J Exp Med 206: 2987–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Maillard I, Engel JD. 2010. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev 238: 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, Lim KC, Hosoya T, Engel JD. 2011. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Mol Cell Biol 31: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA. 2000. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood 96: 2323–2328. [PubMed] [Google Scholar]
- Inoue K, Lupski JR. 2002. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3: 199–242. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Yamamoto M, Leonard MW, George KM, Ting P, Engel JD. 1991. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor δ gene enhancer. Mol Cell Biol 11: 2778–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CJ, Hosoya T, Maillard I, Engel JD. 2012. GATA-3 regulates hematopoietic stem cell maintenance and cell-cycle entry. Blood 119: 2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov VA, Knott GD, Bonner RF. 2002. General statistics of stochastic process of gene expression in eukaryotic cells. Genetics 161: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, Hu RJ, Johnson LA, Feinberg AP. 1997. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet 15: 181–185. [DOI] [PubMed] [Google Scholar]
- Lichtner P, Konig R, Hasegawa T, Van Esch H, Meitinger T, Schuffenhauer S. 2000. An HDR (hypoparathyroidism, deafness, renal dysplasia) syndrome locus maps distal to the DiGeorge syndrome region on 10p13/14. J Med Genet 37: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. 2000. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet 25: 209–212. [DOI] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Maclary E, Buttigieg E, Hinten M, Gayen S, Harris C, Sarkar MK, Purushothaman S, Kalantry S. 2014. Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation. Nat Commun 5: 4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I, Koch U, Dumortier A, Shestova O, Xu L, Sai H, Pross SE, Aster JC, Bhandoola A, Radtke F, et al. 2008. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell 2: 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Torres-Padilla ME. 2012. Control of ground-state pluripotency by allelic regulation of Nanog. Nature 483: 470–473. [DOI] [PubMed] [Google Scholar]
- Moriguchi T, Takako N, Hamada M, Maeda A, Fujioka Y, Kuroha T, Huber RE, Hasegawa SL, Rao A, Yamamoto M, et al. 2006. Gata3 participates in a complex transcriptional feedback network to regulate sympathoadrenal differentiation. Development 133: 3871–3881. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. 2000. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103: 865–875. [DOI] [PubMed] [Google Scholar]
- Nag A, Savova V, Fung HL, Miron A, Yuan GC, Zhang K, Gimelbrant AA. 2013. Chromatin signature of widespread monoallelic expression. eLife 2: e01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, Grosveld F, Hendriks RW. 2001. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol 167: 715–723. [DOI] [PubMed] [Google Scholar]
- Nerlov C, Graf T. 1998. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 12: 2403–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Vambrie S, Steinlein P, Kozmik Z, Rolink A, Weith A, Busslinger M. 1999. Independent regulation of the two Pax5 alleles during B-cell development. Nat Genet 21: 390–395. [DOI] [PubMed] [Google Scholar]
- Ohno S, Kaplan WD, Kinosita R. 1959. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp Cell Res 18: 415–418. [DOI] [PubMed] [Google Scholar]
- Pai S-Y, Truitt ML, Ting C-N, Leiden JM, Glimcher LH, Ho I-C. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19: 863–875. [DOI] [PubMed] [Google Scholar]
- Penit C, Vasseur F. 1988. Sequential events in thymocyte differentiation and thymus regeneration revealed by a combination of bromodeoxyuridine DNA labeling and antimitotic drug treatment. J Immunol 140: 3315–3323. [PubMed] [Google Scholar]
- Pereira JP, Girard R, Chaby R, Cumano A, Vieira P. 2003. Monoallelic expression of the murine gene encoding Toll-like receptor 4. Nat Immunol 4: 464–470. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. [DOI] [PubMed] [Google Scholar]
- Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20: 735–745. [DOI] [PubMed] [Google Scholar]
- Rhoades KL, Singh N, Simon I, Glidden B, Cedar H, Chess A. 2000. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT–PCR analysis. Curr Biol 10: 789–792. [DOI] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. 2005. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol 6: 663–670. [DOI] [PubMed] [Google Scholar]
- Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L, Kelly Modis L, Kastner P, Chan S, Duprez E, et al. 2009. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell 138: 300–313. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17: 749–756. [DOI] [PubMed] [Google Scholar]
- Scripture-Adams DD, Damle SS, Li L, Elihu KJ, Qin S, Arias AM, Butler RR III, Champhekar A, Zhang JA, Rothenberg EV. 2014. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol 193: 3470–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Petrie HT. 2012. Activation kinetics and off-target effects of thymus-initiated cre transgenes. PLoS One 7: e46590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R, Ohneda K, Engel JD, Trainor CD, Yamamoto M. 2004. Transgenic rescue of GATA-1-deficient mice with GATA-1 lacking a FOG-1 association site phenocopies patients with X-linked thrombocytopenia. Blood 103: 2560–2567. [DOI] [PubMed] [Google Scholar]
- Tan JB, Visan I, Yuan JS, Guidos CJ. 2005. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol 6: 671–679. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, et al. 2004. Mutation of GATA3 in human breast tumors. Oncogene 23: 7669–7678. [DOI] [PubMed] [Google Scholar]
- van Doorninck JH, van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI. 1999. GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J Neurosci 19: RC12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, Harding B, Beetz R, Bilous RW, Holdaway I, et al. 2000. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 406: 419–422. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD. 1990. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev 4: 1650–1662. [DOI] [PubMed] [Google Scholar]
- Yu Q, Sharma A, Oh SY, Moon H-G, Hossain MZ, Salay TM, Leeds KE, Du H, Wu B, Waterman ML, et al. 2009. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-γ. Nat Immunol 10: 992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Rothenberg EV. 2014. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14: 529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwemer LM, Zak A, Thompson BR, Kirby A, Daly MJ, Chess A, Gimelbrant AA. 2012. Autosomal monoallelic expression in the mouse. Genome Biol 13: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.