Abstract
Microbiological, genomic and transcriptomic analyses were used to examine three species from the bacterial genus Caldicellulosiruptor with respect to their capacity to convert the carbohydrate content of lignocellulosic biomass at 70°C to simple sugars, acetate, lactate, CO2, and H2. Caldicellulosiruptor bescii, C. kronotskyensis, and C. saccharolyticus solubilized 38%, 36%, and 29% (by weight) of unpretreated switchgrass (Panicum virgatum) (5 g/liter), respectively, which was about half of the amount of crystalline cellulose (Avicel; 5 g/liter) that was solubilized under the same conditions. The lower yields with C. saccharolyticus, not appreciably greater than the thermal control for switchgrass, were unexpected, given that its genome encodes the same glycoside hydrolase 9 (GH9)-GH48 multidomain cellulase (CelA) found in the other two species. However, the genome of C. saccharolyticus lacks two other cellulases with GH48 domains, which could be responsible for its lower levels of solubilization. Transcriptomes for growth of each species comparing cellulose to switchgrass showed that many carbohydrate ABC transporters and multidomain extracellular glycoside hydrolases were differentially regulated, reflecting the heterogeneity of lignocellulose. However, significant differences in transcription levels for conserved genes among the three species were noted, indicating unexpectedly diverse regulatory strategies for deconstruction for these closely related bacteria. Genes encoding the Che-type chemotaxis system and flagellum biosynthesis were upregulated in C. kronotskyensis and C. bescii during growth on cellulose, implicating motility in substrate utilization. The results here show that capacity for plant biomass deconstruction varies across Caldicellulosiruptor species and depends in a complex way on GH genome inventory, substrate composition, and gene regulation.
INTRODUCTION
The genus Caldicellulosiruptor is comprised of Gram-positive, anaerobic bacteria that ferment a variety of simple and complex carbohydrates to primarily H2, CO2, acetate, and lactate at temperatures at or above 70°C (1). To date, Caldicellulosiruptor species have been isolated globally from terrestrial hot springs and thermal features in locations including the United States (C. owensensis [2, 3] and C. obsidiansis [4, 5]), Russia (C. bescii [6, 7]), C. kronotskyensis [2, 8], and C. hydrothermalis [2, 8]), New Zealand (C. saccharolyticus [9–11]), and Iceland (C. kristjanssonii [2, 12] and C. lactoaceticus [2, 13]). Characteristic of Caldicellulosiruptor species are multidomain extracellular and S-layer-associated glycoside hydrolases (GHs) that mediate the microbial conversion of complex carbohydrates (14–16). Sequenced genomes for Caldicellulosiruptor species (2, 4, 6, 10) indicate that some, but not all, encode GH48-containing enzymes (17), and these appear to be essential for crystalline cellulose degradation (18). The cellulolytic Caldicellulosiruptor species also utilize novel binding proteins (tāpirins) to adhere to plant biomass (19). Several Caldicellulosiruptor species can extensively degrade crystalline cellulose (Avicel), and at least one species, Caldicellulosiruptor bescii, has been reported to utilize unpretreated switchgrass (20, 21) and, furthermore, to be metabolically engineered to produce ethanol from plant biomass (22).
Given the promise of cellulolytic Caldicellulosiruptor species as consolidated bioprocessing microorganisms for the production of liquid transportation fuels from lignocellulose (23), it is important to determine those species that are most effective for plant biomass deconstruction. These species then become targets for metabolic engineering efforts focusing on direct fermentation products to biofuels. In addition, to improve upon the capacity of the wild-type strains for lignocellulose utilization, an understanding of the factors that are most important in degrading and processing complex carbohydrates is essential. Genome sequences can be used to determine differences among these species with respect to inventories of key elements for carbohydrate utilization, such as ATP binding cassette (ABC) sugar transporters and glycoside hydrolases. However, this information is more insightful if viewed through complementary perspectives, developed through microbiological, metabolite, and transcriptional analysis. With this in mind, three cellulolytic Caldicellulosiruptor species (C. bescii, C. kronotskyensis, and C. saccharolyticus) were examined to determine differentiating features related to their capacity for lignocellulose deconstruction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Caldicellulosiruptor species used in this study were obtained as freeze-dried cultures from the German Collection of Microorganisms and Cell Cultures (DSMZ [http://www.dsmz.de]) were revived in modified DSMZ medium 640 containing (per liter) 0.9 g of NH4Cl, 0.9 g of NaCl, 0.4 g of MgCl2·6H2O, 0.75 g of KH2PO4, 1.5 g of K2HPO4, 1.0 g of yeast extract, 1.0 g of cysteine HCl·H2O, 0.5 mg of resazurin, and 1.0 ml of trace element solution SL-10 with 1 g of cellobiose as the carbon source. Medium pH was adjusted to 7.2 with 10 M NaOH and prepared anaerobically under 100% N2 headspace. Prior to all experiments species were transferred onto a defined modified version of DSMZ medium 671 (671d) containing cellulose or unpretreated switchgrass as the sole carbon source. Medium 671d contained (per liter) 1.0 g of NH4Cl, 0.1 g of NaCl, 0.1 g of MgCl2·6H2O, 0.05 g of CaCl2·2H2O, 0.3 g of K2HPO4, 2.6 g of NaHCO3, 1 g of cysteine HCl·H2O, 0.5 mg of resazurin, 1.0 ml of trace element solution SL-10, and 1 ml of vitamin solution. Medium 671d pH was adjusted to 7.2 with 10 M NaOH and filter sterilized through 0.2-μm filters prior to addition to cellulose (5 g/liter) or unpretreated switchgrass (5 g/liter) and prepared anaerobically under N2-CO2 (80%-20% [vol/vol]) headspace. Cellulose and unpreatreated switchgrass were prepared as described below. The trace element SL-10 solution contained (per liter) 10 ml of HCl (7.7 M), 1.5 g of FeCl2·4H2O, 70 mg of ZnCl2, 100 mg of MnCl2·4H2O, 6 mg of H3BO3, 190 mg of COCl2·6H2O, 2 mg of CuCl2·2H2O, 24 mg of NiCl2·6H2O, and 36 mg of Na2MoO4·2H2O. The vitamin solution contained (per liter) 20 mg of biotin, 20 mg of folic acid, 100 mg of pyridoxine hydrochloride, 50 mg of pantothenic acid, 100 mg of thiamine hydrochloride, 50 mg of nicotinamide, 50 mg of cobalamin, 50 mg of p-aminobenzoic acid, 50 mg of lipoic acid, and 50 mg of riboflavin. Unless otherwise specified, cultures were grown as 50-ml batch cultures in 125-ml serum bottles at 70°C and 150 rpm. Prior to solubilization, carbon balance, and transcriptional response experiments, each species were passaged on medium 671d with either cellulose or unpretreated switchgrass 3 or 4 times by cell growth for 1 to 2 days and passaging immediately.
Preparation of substrates.
Unpretreated switchgrass (Panicum virgatum) cultivar Cave-in-Rock field grown in Monroe County, IA, and obtained from the National Renewable Energy Laboratory (NREL) was mechanically ground and sieved to 20/80 mesh. Sieved switchgrass was washed with water at 25°C through coarse filtering crucibles (40 to 60 μm; Corning) until the permeate ran clear to remove easily solubilized switchgrass components. Switchgrass was then oven dried overnight at 70°C and used as a growth substrate in all experiments. Cellulose (Avicel; FMC Bio-Polymer) was used as received. Cellulose and unpretreated switchgrass were not autoclaved, to avoid any type of pretreatment.
Solubilization assays.
Small batch cultures (50 ml) were prepared in triplicate on 671d medium with either cellulose or unpretreated switchgrass (5 g/liter), inoculated with 1 × 106 cells/ml, agitated at 150 rpm, incubated at 70°C for 7 days, and harvested immediately. Cellulose-grown cultures were harvested by centrifugation at 6,000 × g for 15 min, while switchgrass cultures were harvested by vacuum filtering through coarse filtering crucibles (40 to 60 μm; Corning). The residual substrate was washed with 2 volumes (100 ml) of 70°C sterile water and oven dried at 70°C until it reached a constant mass. The extent of solubilization was determined from the mass difference between cellulose or switchgrass used to prepare cultures and the insoluble substrate remaining after harvest.
Preparation of cultures for carbon balances.
Small batch cultures (250 ml), containing 671d medium and cellulose or unpretreated switchgrass, were inoculated with 1 × 106 cells/ml and incubated 70°C and 150 rpm for 7 days. Cultures were harvested for spent substrate (see above), and the supernatant was stored at −20°C for subsequent metabolite analysis.
Determination of switchgrass composition.
The carbohydrate content of switchgrass, before and after supporting growth of Caldicellulosiruptor species, was analyzed using a modified version of the National Renewable Energy Laboratory procedure (http://www.nrel.gov/biomass/analytical_procedures.html) described in Determination of Structural Carbohydrates and Lignin in Biomass (24). Sulfuric acid (600 μl of 72% [wt/wt]) was added to 60 mg of switchgrass and mixed using a glass stir rod. Samples were shaken in a 30°C (constant temperature) water bath at 150 rpm for 1 h and were mixed with a glass rod every 10 min. Sulfuric acid was diluted to 4% (wt/wt) with 16.8 ml of deionized (DI) water. Tubes were sealed and autoclaved for 1 h on the liquid cycle. Sugar concentrations were determined using high-performance liquid chromatography (HPLC) (Alliance e2695 separations module; Waters), equipped with photodiode array (model 2998; Waters) and refractive index (model 2414; Waters) detectors. Acetate, cellobiose, glucose, xylose, and arabinose were quantified using an Aminex-87H column (300 mm by 7.8 mm; Bio-Rad) operated with a mobile phase of 5 mM H2SO4 at 0.6 ml/min and 60°C. The inert components (lignin and ash) were taken to be the difference between the mass of switchgrass hydrolyzed and the organic acids and carbohydrates measured. Unpretreated switchgrass was found to contain 40.1% ± 1.4% glucan, 26.9% ± 0.7% xylan, 2.7% ± 0.2% arabinan, and 27.8% ± 3.1% inert components.
Analysis of fermentation products.
Total sugars in cell-free supernatants were determined by the phenol-sulfuric acid method (25), using glucose as the standard. Ethanol was determined by gas chromatography (Shimadzu GC-2014 equipped with a ZB-WAXplus [30-m length, 0.53-mm inner diameter, and 1.0-μm film thickness]; Phenomenex) with an injector temperature of 200°C, a 10:1 split ratio, a 30.3-cm/s linear velocity, and oven ramp at 20°C/min from 50 to 220°C. Trace amounts of lactate in switchgrass cultures were detected by derivitization with dibromoacetophenone (DBAP) to form a phenacyl ester and were assayed using reverse-phased HPLC. For each sample, 500 μl was acidified with 50% H2SO4 to pH <2 and extracted with 750 μl of diethyl ether to remove salts prior to derivitization. The ether fraction was separated by centrifugation at 6,000 × g for 10 min and neutralized with 50 μl of 20 mM sodium bicarbonate. Ether was removed by evaporation, with the remaining aqueous portion mixed with 50 μl of acetonitrile and a small amount of pH indicator (0.5 μl of 0.5% phenolphthalein). The solution was then alkalized with 1 M KOH until the sample turned pink (pH ∼9 to 10), after which 100 μl of acetonitrile, 50 μl of 1 μM 15-crown-5-ether, and 200 μl of 20 mM 2,4-dibromoacetophenone were added. The reaction mixture was heated to 80°C for 30 min and cooled. Samples were analyzed by HPLC using an Atlantis dC18 column (3 μm, 4.6 by 150 mm; Waters) operated at 30°C. The mobile phase, 1.5 ml/min, had an initial composition of 65% buffer A (0.1% formic acid) and 35% buffer B (acetonitrile). Products were eluted over a 15-min linear gradient to 30% buffer A and 70% buffer B and detected by absorption at 254 nm. Acetate and lactate were analyzed by HPLC using an Aminex HPX-87H column (300 mm by 7.8 mm; Bio-Rad), operated with a mobile phase of 5 mM H2SO4 at 0.6 ml/min and 60°C. Samples were acidified to 0.05% (wt/wt) H2SO4 and detected by absorption at 210 nm.
Determination of cell carbon.
Cultures were prepared in 671d medium with cellobiose (1 g/liter) as the carbon source, inoculated with 1 × 106 cells/ml, and grown well into stationary phase. Each stationary-phase culture (45 ml) was centrifuged at 6,000 × g for 10 min. Cell pellets were resuspended into aluminum weigh boats with deionized water and dried for 5 h at 90°C. Dry cell masses of 8.2 × 10−10 ± 2.9 × 10−10, 1.9 × 10−10 ± 0.5 × 10−10, and 5.8 × 10−10 ± 1.1 × 10−10 mg per cell were obtained for C. bescii, C. kronotskyensis, and C. saccharolyticus, respectively. The elemental composition for all Caldicellulosiruptor species was assumed to be CH1.62O0.46N0.23S0.0052P0.0071, corresponding to a molecular mass of 24.6 g per mole of cells as previously determined for C. saccharolyticus (26).
Isolation of RNA.
Total RNA was isolated from cells harvested at mid-logarithmic phase (2 × 107 to 5 × 107 cells/ml) on either switchgrass or cellulose (5 g/liter) at 70°C and 150 rpm. Cells were rapidly cooled to 4°C and then harvested by centrifugation at 6,000 × g prior to storage at −80°C until further processing. Total RNA was isolated using a modified TRIzol (Life Technologies) protocol in combination with an RNeasy kit (Qiagen). As an additional step, cells were sonicated after resuspension in TRIzol reagent. cDNA was produced from the collected RNA using Superscript III reverse transcriptase (Life Technologies), random primers (Life Technologies), and the incorporation of 5-(3-amino-allyl)-2′-deoxyurinidine-5′-triphosphate (Life Technologies), as described in reference 65.
Microarray hybridization.
A spotted whole-genome microarray was developed for each C. bescii, C. kronotskyensis, and C. saccharolyticus using 30- to 62-mer oligonucleotides, based on open reading frames (ORFs) of the respective species. cDNA was labeled with either Cy3 or Cy5 dye (GE Healthcare) and hybridized to GAPS II coated slides (Corning). Microarray slides were scanned using a 4000B scanner (Molecular Devices). GenePix Pro 7 (Molecular Devices) was used to quantitate signal intensities, prior to analysis with JMP Genomics 5 (SAS) using a mixed-effects analysis-of-variance model. ORFs that were differentially transcribed 2-fold or more and met the Bonferroni statistical criterion were considered to be up- or downregulated.
Microarray data accession number.
Transcriptional response data are deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE68810.
RESULTS AND DISCUSSION
Biosolubilization of cellulose and switchgrass.
Three strongly cellulolytic Caldicellulosiruptor species (18), C. bescii, C. kronotskyensis, and C. saccharolyticus, were assessed for the ability to degrade cellulose and unpretreated switchgrass. All three species were able to use either substrate as the sole carbon source for growth in defined medium containing only salts and vitamins (Fig. 1A). Planktonic growth on cellulose included an exponential phase (doubling time [td]<3 h), followed by an extended stationary phase. In contrast, growth on switchgrass was biphasic exponential. Initially, cell density increased rapidly to ∼2 × 107 cells per ml, with doubling times under 3 h, likely supported by readily solubilized sugars released by abiotic thermal factors. Switchgrass typically contains approximately 15% water-soluble material, 25 to 50% of which is carbohydrate (27, 28). After approximately 24 h, as the readily solubilized sugars were utilized by the bacteria, planktonic cells entered a second, slower growth phase, which was tracked over the next 200 h. During the slower growth phase, the Caldicellulosiruptor species can recruit carbohydrates by deploying extracellular, multidomain glycoside hydrolases (GHs), both free (29, 30) and attached to the cell surface via the S-layer (15). These GHs are thought to degrade the more recalcitrant complex carbohydrate fractions, thereby releasing the soluble sugars necessary for growth. The slower planktonic growth phase (Fig. 1A) corresponded to doubling times of 16 to 18 h for C. bescii and C. kronotskyensis and 34 h for C. saccharolyticus, although C. saccharolyticus ultimately reached planktonic cell densities comparable to those of the other two species. Since all three species grew at similar rates on cellulose, C. bescii and C. kronotskyensis apparently overcame the recalcitrance of the unpretreated switchgrass more effectively than C. saccharolyticus as indicated by this species' slower growth on unpretreated switchgrass.
FIG 1.

Growth of Caldicellulosiruptor species on cellulose and unpretreated switchgrass. (A) Growth of species on 5 g/liter of cellulose (Avi) or 5 g/liter of unpretreated switchgrass (SWG). The arrow indicates the 7-day harvest point for solubilization assay and carbon balance determination. (B) Extent of solubilization of cellulose and switchgrass. (C) Switchgrass composition before and after degradation by Caldicellulosiruptor species. Solubilization and switchgrass composition values are means ± standard deviations (n = 3). Cbes, C. bescii; Ckro, C. kronotskyensis; Csac, C. saccharolyticus.
The extent of total mass solubilization of cellulose and switchgrass was determined for each species following a 7-day incubation on these substrates (Fig. 1B; Table 1). Cellulose solubilizations by C. bescii and C. kronotskyensis were comparable, at 77.1% ± 2.0% and 71.7% ± 2.6%, respectively, while solubilization by C. saccharolyticus was significantly lower, at 58.0% ± 1.2%. Since no cellulose solubilization was noted in the abiotic thermal control, enzymatic and microbial processes were responsible for all apparent degradation. Overall, switchgrass solubilization was approximately half of that observed on cellulose for each species, if the thermal abiotic contribution is included. Solubilizations of switchgrass after 7 days by C. bescii and C. kronotskyensis were again comparable and also very reproducible, at 40.3% ± 1.0% and 39.6% ± 1.1%, respectively. For C. saccharolyticus, impaired growth indicated a decreased capacity for switchgrass solubilization. Solubilization by C. saccharolyticus was only slightly higher (23.5% ± 2.3%) than levels achieved by abiotic thermal factors (19.6% ± 2.0%), indicating that microbial action by C. saccharolyticus did not liberate significantly more sugars than extended incubation at high temperatures (70°C). Fermentation products and residual substrate composition were determined for all three species following a 7-day incubation with cellulose and switchgrass. Unpretreated switchgrass before incubation with Caldicellulosiruptor species was found to contain 40.1% ± 1.4% glucan, 26.9% ± 0.7% xylan, 2.7% ± 0.2% arabinan, and 27.8% ± 3.1% inert components (lignin and ash) by weight. After degradation by Caldicellulosiruptor species, the composition of switchgrass did not change significantly; the relative fractions of carbohydrate and inert components remained largely unchanged following biosolubilization (Fig. 1C). This was consistent with previous studies with C. bescii that found that lignin and carbohydrate solubilizations were proportional during degradation (31).
TABLE 1.
Extent of solubilization and fermentation products during growth on cellulose (Avicel) and switchgrass by Caldicellulosiruptor speciesa
| Species | Sampleb | Solubilization (%) | Acetate (mM) | Lactate (mM) | Sugar (mM) | Ethanol (mM) |
|---|---|---|---|---|---|---|
| C. bescii | Avi | 77.1 ± 2.0 | 7.3 ± 1.6 | 17.4 ± 4.9 | 5.8 ± 0.7 | 0.07 ± 0.01 |
| SWG | 40.3 ± 1.0 | 7.4 ± 0.2 | 0.2 ± 0.04 | 0.8 ± 0.1 | 0.17 ± 0.08 | |
| C. kronotskyensis | Avi | 71.7 ± 2.6 | 6.4 ± 1.1 | 11.8 ± 0.7 | 6.1 ± 1.8 | 0.05 ± 0.04 |
| SWG | 39.6 ± 1.1 | 6.8 ± 0.8 | 0.2 ± 0.02 | 0.8 ± 0.04 | 0.22 ± 0.05 | |
| C. saccharolyticus | Avi | 58.0 ± 1.2 | 11.3 ± 1.7 | 12.1 ± 0.5 | 1.0 ± 0.6 | 0.84 ± 0.07 |
| SWG | 23.5 ± 2.3 | 2.7 ± 0.1 | 0.05 ± 0.02 | 0.7 ± 0.07 | 0.13 ± 0.01 |
Data are means ± standard deviations (n = 3).
Avi, Avicel; SWG, switchgrass.
Caldicellulosiruptor species have not been found to utilize lignin as a carbon source; accordingly, the carbohydrate fraction solubilized from cellulose and switchgrass could be accounted for by primary fermentation products (acetate, lactate, ethanol, and carbon dioxide), cellulose and switchgrass degradation products (acetate and soluble sugars), and Caldicellulosiruptor biomass to greater than 92% carbon balance closure, assuming that CO2 generation was equal to acetate generation on a molar basis (20) (Fig. 2). The acidic fermentation products for growth on cellulose and switchgrass were the same for all three species, but lactate and acetate concentrations varied (Table 1). For ∼75% solubilization of cellulose, C. bescii and C. kronotskyensis acetate levels reached approximately 6 to 7 mM, with trace amounts of ethanol (<0.1 mM), and similar amounts of unfermented soluble sugar, 6 mM. The formation of lactate is related to the impact of accumulating levels of molecular hydrogen on Caldicellulosiruptor metabolism, causing a shift toward this organic acid (32). Levels of lactate generation by C. bescii and C. kronotskyensis were also similar (∼12 mM), as expected for the same amount of solubilization. Although C. saccharolyticus solubilized <60% of cellulose, it also generated high levels of lactate (12 mM) and significantly more acetate (11.3 mM) and ethanol (0.8 mM) than the other two species. The residual unfermented soluble sugar for C. saccharolyticus was 1 mM, suggesting that C. saccharolyticus metabolized the C6 sugars released from cellulose solubilization to a greater extent than the other two species. On unpretreated switchgrass, the same major fermentation products were generated by each of the three species. Acetate levels for C. bescii and C. kronotskyensis were comparable (∼7 mM), but only trace amounts of lactate were produced. C. saccharolyticus, which solubilized switchgrass only slightly more than the abiotic control, grew slowly and produced correspondingly low levels of acetate (∼3 mM) and lactate (0.05 mM). For each of the three species on switchgrass, unlike the case for cellulose, only small amounts of unfermented sugar (<1 mM) remained at the time of harvest, reflecting the lower extents of carbohydrate solubilization.
FIG 2.
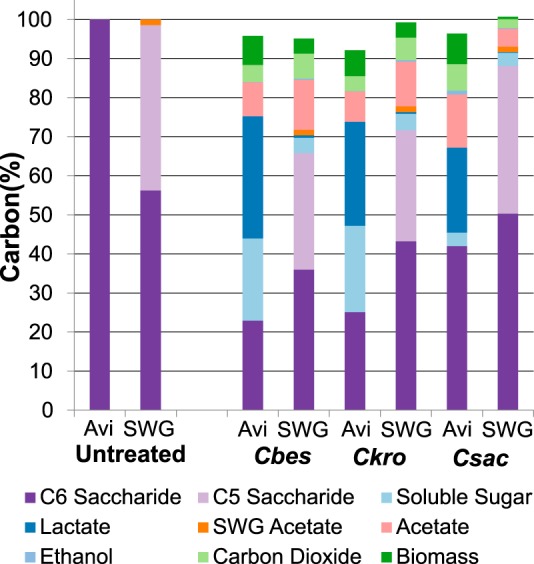
Carbon balance of carbohydrate substrates and products from Caldicellulosiruptor growth on cellulose and unpretreated switchgrass. Carbon balances were as follows: for cellulose, 95.8% ± 13.2% for C. bescii, 92.2% ± 3.9% for C. kronotskyensis, and 96.4% ± 1.5% for C. saccharolyticus; for switchgrass, 95.1% ± 3.4% for C. bescii, 99.3% ± 2.1% for C. kronotskyensis, and 100.7% ± 0.5% for C. saccharolyticus. Values are means ± standard deviations (n = 3).
The results here indicate that there are significant differences among the three species in the capacity to utilize and convert biomass substrates. The levels of organic acids produced on cellulose and switchgrass and the residual amounts of soluble sugars reflect the extent to which the breakdown of biomass through enzymatic processes is coordinated with conversion (carbohydrate transport and fermentative metabolism) by the three species. For example, it is interesting that although C. saccharolyticus solubilized less plant biomass, this species nonetheless produced high levels of organic acids with minimal amounts of unfermented sugars, suggesting a highly productive fermentative metabolism. These data illustrate potentially important physiological differences within the genus Caldicellulosiruptor in choosing candidates for consolidated bioprocessing through metabolic engineering. These differences likely reflect distinct roles of these bacteria in their communities of origin in terrestrial hot springs. The following genomic and transcriptomic analysis describes differences in cellulolytic capacity and metabolism between the three species.
Genomic comparison of C. bescii, C. kronotskyensis, and C. saccharolyticus.
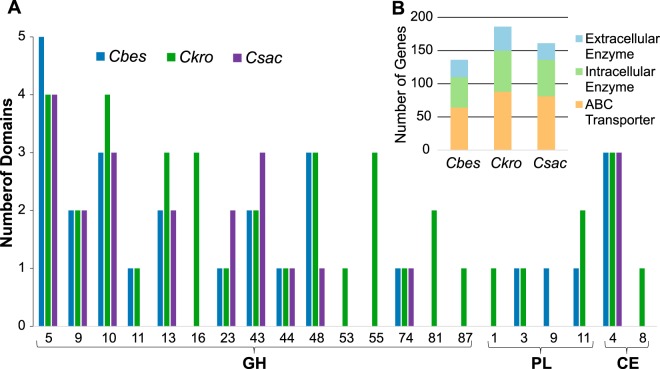
A comparative analysis of the sequenced genomes for C. bescii (2,776 ORFs), C. kronotskyensis (2,583 ORFs), and C. saccharolyticus (2,760 ORFs) was done to identify features that could be directly connected to observed differences in biomass solubilization for the three species (Table 2). Genome-wide homology (95.6% identity at the nucleotide level) is highest for the two Russian isolates (C. bescii and C. kronotskyensis). Homology of C. saccharolyticus to the other two species is lower but still relatively high, with 80.3% and 79.6% nucleotide identity to C. bescii and C. kronotskyensis, respectively (17). Homologous protein sequences were identified by sequence clustering using UCLUST software (drive5) (based on 50% identity and 50% coverage) (33), genes from all three species were organized into 3,223 families with 885 singletons (family with only a single gene). The relatedness among all three species (1,799 gene families in the core genome) was much higher than for either unique or common families for two species (Fig. 3). The unique gene families for each of three species, C. bescii (338 gene families), C. kronotskyensis (234 gene families), and C. saccharolyticus (346 gene families) relate at some level to their differential abilities to degrade and convert complex carbohydrates. For biomass deconstruction, the extracellular carbohydrate-active enzymes (CAZymes) are particularly important; there are 14 in the core genome and a total of 20, 31, and 19 for C. bescii, C. kronotskyensis, and C. saccharolyticus, respectively (Table 2). Note that unlike C. bescii and C. kronotskyensis, which have three GH48 domains, the C. saccharolyticus genome encodes only a single GH48 domain, represented in CelA (14); GH48 domains were found to be a determinant for cellulose hydrolysis capacity in Caldicellulosiruptor species (17, 18). As discussed below, the lack of GH48 domains in C. saccharolyticus's genome may relate to a decreased cellulolytic capacity compared to those of the other two species. In addition, although its genome encodes 50% more extracellular CAZymes than the other two species (Table 2), C. kronotskyensis did not solubilize the biomasses tested here any better than C. bescii (Table 1; Fig. 1B). There are 16 ABC carbohydrate transporters in the core genome (Table 2), with totals of 20, 28, and 24 in C. bescii, C. kronotskyensis, and C. saccharolyticus, respectively. Coordination between CAZymes and ABC transporters would seem to be important to optimize utilization of the carbohydrate content of plant biomass for Caldicellulosiruptor species. It is also worth noting that C. saccharolyticus has 16 gene families linked to amino acid transport and metabolism, which is 10 or more than for either C. bescii or C. kronotskyensis. It is likely that there are subtle differences among the three species that are not easily correlated with genome sequence. In particular, the regulation of carbon flux through central metabolism is key to high levels of carbohydrate conversion.
TABLE 2.
Transcriptional response of Caldicellulosiruptor species during growth on cellulose (Avicel) and switchgrass
| Species | Nucleotides (Mb) | Total no. of genes | Avia | SWGa | No. of extracellular CAZymes | Avia | SWGa | No. of ABC transporters | Avia | SWGa |
|---|---|---|---|---|---|---|---|---|---|---|
| C. bescii | 2.93 | 2,776 | 129 (98) | 243 (167) | 20 | 0 | 9 (8) | 20 | 2 (2) | 11 (8) |
| C. kronotskyensis | 2.84 | 2,583 | 64 (48) | 106 (81) | 31 | 0 | 8 (4) | 28 | 0 | 13 (9) |
| C. saccharolyticus | 2.97 | 2,760 | 211 (165) | 257 (160) | 19 | 1 (1) | 7 (6) | 24 | 1 (1) | 16 (11) |
| Core | NA | 1,922 | 9 | 18 | 14 | 0 | 4 | 16 | 0 | 6 |
Number of genes upregulated on cellulose or switchgrass. Numbers in parentheses indicate genes in core genome. Avi, Avicel; SWG, switchgrass; NA, not applicable.
FIG 3.
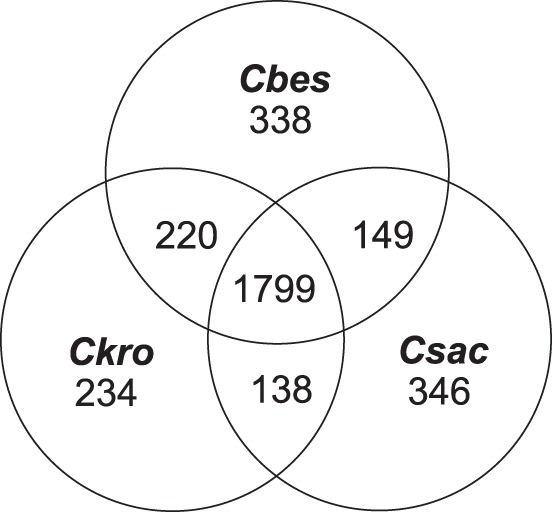
Homologous gene families for C. bescii, C. kronotskyensis, and C. saccharolyticus. Numbers of gene families, as determined by UCLUST, with 50% identity and 50% coverage are shown.
Core genome transcriptome for growth on biomass substrates.
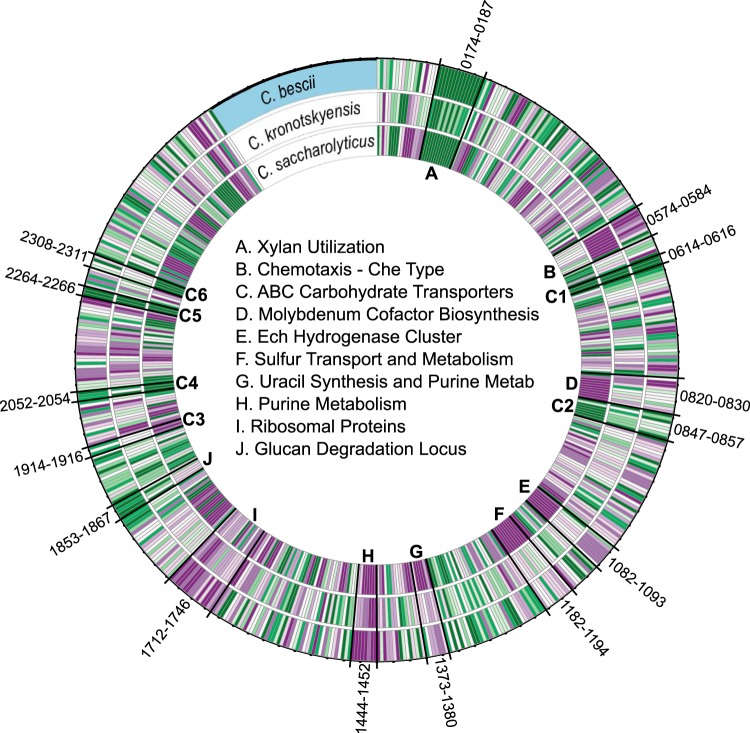
While differences in lignocellulose degradation capacity among the three Caldicellulosiruptor species can be viewed at the level of genome sequence, regulation of genes that relate to this capacity can provide additional insights into specific microbial strategies. In principle, the same carbohydrate deconstruction and utilization processes as utilized by the three Caldicellulosiruptor species for growth on cellulose should also be employed during growth on switchgrass, given that cellulose is a major component of lignocellulose. However, this was not the case in this study. Nor was there strong consensus among the three species for genes differentially transcribed during growth on the two substrates, even within the core genome. Differential transcription (2-fold or more) of genes that are conserved among the three species (core genome) is represented in Fig. 4 (referenced to the ORF numbers in the C. bescii genome), for growth on cellulose (purple) compared to switchgrass (green), with intensity of color indicative of level of gene transcription. C. saccharolyticus has the largest numbers of differentially transcribed genes on cellulose and switchgrass (211 and 257, respectively), followed by C. bescii (129 and 243, respectively) and then C. kronotskyensis (64 and 106, respectively). Extracellular GHs (CAZymes) were exclusively responsive to switchgrass (except for one core GH in C. saccharolyticus) (Table 2), reflecting the heterogeneity of this substrate. This differential response to switchgrass was also the case for carbohydrate ABC transporters, many of which are part of the core genome; 6 out of 16 of these responded to switchgrass in all three species. Three transporters were upregulated on cellulose in the core genome, but none were differentially regulated in all three species. Two transporters, upregulated only in C. bescii (Athe_0595-0598 [locus tags in a range of Athe_0595 to Athe_0598] and Athe_2552-2554), belong to the CUT1 family, a family considered to transport both di- and oligosaccharides (34) (see Table S1 in the supplemental material). Previous work proposed the substrates for these transporters to be xyloglucan and unknown, respectively (35). The other, the only di- or oligopeptide (DPP/OPP) transporter in the core genome (Athe_1913-1917, Calkro_0798-0802, Csac_1028-1032) (Fig. 4, C3), was upregulated on switchgrass in C. bescii and on cellulose in C. saccharolyticus and constitutively transcribed in C. kronotskyensis. DPP/OPP transporters are implicated in the transport of peptides, nickel, heme, and sugars (34). The broad specificity of this transporter and differences in regulation between the species imply that it could perform a range of transport functions in Caldicellulosiruptor species.
FIG 4.
Core genome transcriptome for C. bescii, C. kronotskyensis, and C. saccharolyticus. All genes in the core genome differentially regulated 2-fold or more in one or more species are shown. Green indicates upregulation on switchgrass, and purple indicates upregulation on cellulose. Gene clusters for specific functions are indicated by letters defined in the center. Locus tags in C. bescii (outer ring) are used for reference.
Certain genomic loci responded similarly in all three species during growth on cellulose compared to switchgrass. For example, the xylan utilization locus (Fig. 4, A) responded at high levels to switchgrass in all three species, as did many sets of ABC carbohydrate transporters (Fig. 4, C1 to C6), homologs of which were previously annotated in C. saccharolyticus (35). Growth on cellulose triggered expression of purine metabolism (Fig. 4, H) and ribosomal protein biosynthesis (Fig. 4, I) genes in all three species, likely the result of the higher growth rates on this less recalcitrant substrate. Nonetheless, although a total of 517 different genes in the core genome were differentially transcribed for the cellulose versus switchgrass contrast (Fig. 4), only 27 of these responded in all three species (Table 2). This suggests that at the level of gene regulation, significant differences exist among the species, which relate to their deconstruction and conversion of lignocellulose. Illustrative examples of these differences are discussed below.
Fermentative metabolism.
As reported in Table 1, fermentative product concentrations for cellulose and switchgrass conversion varied across the three species; some insights into these differences could be obtained from their transcriptomes. For example, C. saccharolyticus generated large amounts of organic acids and correspondingly low levels of unfermented soluble sugars for growth on cellulose (Table 1), indicative of extensive carbohydrate utilization. This metabolic activity was reflected in the transcriptome, where 8 out of 10 genes involved in pyruvate metabolism are transcribed at higher levels on cellulose in C. saccharolyticus than in C. bescii and C. kronotskyensis, in the respective transcriptomes (see Table S2 in the supplemental material). This is also indicated in Fig. 4, where the inner ring shows relatively high transcription levels of many genes on cellulose (purple). For C. saccharolyticus, much more so than for C. bescii and C. kronotskyensis, cellulose triggered genes related to molybdenum cofactor biosynthesis (Fig. 4, D), hydrogen metabolism (E), sulfur transport and metabolism (F), and uracil synthesis and purine metabolism (G). Hydrogen production in Caldicellulosiruptor is linked to acetate production (36) and plays an important role in metabolism by recycling reducing equivalents, NADH and reduced ferrodoxin (Fdred) generated during glycolysis. Genes encoding the reduced ferredoxin-dependent hydrogenase cluster (EchA to EchF; Csac_1534-1539) and genes required for its maturation (HypA to HypF; Csac_1540-1545) were upregulated on cellulose, especially in C. saccharolyticus (Fig. 4, E). The sulfate (cysteine) assimilation gene cluster (Csac_1631-1643) was differentially upregulated in C. saccharolyticus during growth on cellulose (Fig. 4, F), consistent with the fact that hydrogenases and ferredoxin contain Fe-S clusters (37). Note that the molybdenum cofactor biosynthesis proteins upregulated in C. saccharolyticus (Fig. 4, D) also contain Fe-S clusters (38). In contrast, homologs within the sulfate assimilation locus (Fig. 4, F) in C. bescii (Athe_1182-1194) and C. kronotskyensis (Calkro_1504-1516) were transcribed at low levels, possibly relating to lower levels of fermentative metabolism. The strong response of genes related to reducing power generation needed to drive metabolic processes in C. saccharolyticus is consistent with the higher levels of carbohydrate conversion observed (Table 1).
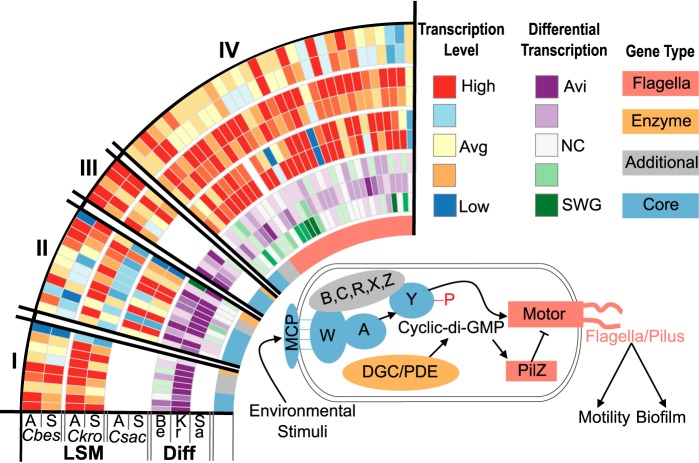
Chemotaxis and motility.
Processes related to the detection and pursuit of cellulose and hemicellulose as primary growth substrates have not been considered to any extent for lignocellulose-degrading microorganisms. Initially discovered in Escherichia coli, the Che-type chemotaxis system is known to sense extracellular chemicals and modulate cellular motility via flagellar rotation (39). The core set of proteins in this system consists of methyl-accepting chemotaxis proteins (MCPs), a scaffolding protein (CheW), a histidine kinase (CheA), and the response regulator (CheY). The methyl-accepting chemotaxis proteins (MCPs) sense environmental signals (Fig. 5), which are then transmitted through a scaffolding protein (CheW) to a histidine kinase (CheA) that phosphorylates the response regulator (CheY). Phosphorylated Che-Y then interacts with the flagellar motor to control motility. Most chemotactic systems contain additional components that modulate MCP methylation (CheR and CheB) and/or phosphatases (CheC, CheX, and/or CheZ) that aid in dephosphorylating CheY (40–43). The genomes of C. bescii and C. kronotskyensis each contain four loci containing the Che-type signal transduction pathway core components, while C. saccharolyticus contains only two (Fig. 5, II and IV). Loci I and II are associated with diguanylate cyclases/phosphodiesterases (DGC/PDE), and locus IV is associated with flagellum/pilus biosynthesis and structural genes (Fig. 5). This implies that the Che-type systems play an integral part in regulation of flagellum-based motility and cyclic di-3′,5′-guanylate (cyclic di-GMP) turnover in Caldicellulosiruptor species. This relationship is supported by the transcriptome, with genes in each locus highly transcribed and differentially upregulated on cellulose (Fig. 5; see also Tables S3 and S4 in the supplemental material), implicating these chemotaxis systems in how Caldicellulosiruptor species identify and utilize cellulosic substrates.
FIG 5.
Transcriptional response of Che-type chemotaxis systems and flagellar genes in Caldicellulosiruptor species. Gene type is indicated on the inner ring: core Che component (light blue), additional Che component (gray), enzyme (orange) or flagella (pink). Tracks from outer to inner: for transcription level (LSM), C. bescii, cellulose (A); C. bescii, switchgrass (S); C. kronotskyensis, cellulose; C. kronotskyensis, switchgrass; C. saccharolyticus, cellulose; C. saccharolyticus, switchgrass; for differential transcription (Diff), C. bescii (Be), C. kronotskyensis (Kr), and C. saccharolyticus (Sa). Locus tags and transcription values can be seen in Table S3 and S4 in the supplemental material.
Two of the Che-type chemotaxis systems are associated with diguanylate cyclases/phosphodiesterases (Fig. 5, I and II); these enzymes are responsible for the production and conversion of cyclic di-GMP and are known to be involved in regulating the switch from planktonic growth to biofilm formation (44). In fact, C. saccharolyticus generates elevated intracellular levels of cyclic di-GMP during biofilm formation (45), and another Caldicellulosiruptor species, C. obsidiansis, forms cellulose-degrading biofilms (46, 47). The precise relationship between the DGC/PDE and the Che-type system has not been studied for Caldicellulosiruptor. In other Firmicutes these domains are contained in a single protein. Likewise, the Proteobacteria Caulobacter crescentus and Pseudomonas aeruginosa are described to contain the Che-Y and DGC/PDE domains in a single protein, with Che-Y phosphorylation modulating DGC/PDE activity (48–50). The two DGC/PDE-associated Che loci (Fig. 5, I and II) are conserved and highly transcribed in both C. bescii and C. kronotskyensis. Furthermore, 13 of the 20 genes were differentially upregulated on cellulose in C. kronotskyensis, with only three differentially regulated in C. bescii and none in C. saccharolyticus. As such, chemotaxis systems may be integral to cellulose utilization by C. kronotskyensis and less so for C. bescii and C. saccharolyticus. The two DGC/PDE associated with Che-type loci are unique to the genus Caldicellulosiruptor and have differing architectures. The DGC/PDE associated with locus II contains a PAS sensor domain coupled with the GGDEF and EAL domains typically associated with DGC/PDE enzymes (51, 52). Transcription of this enzyme was very high on switchgrass in C. bescii and very low in C. saccharolyticus (Fig. 5, II; see also Table S3 in the supplemental material). Interestingly, in C. kronotskyensis, this gene is truncated and only contains the PAS domain, suggesting a complete loss in function as a DGC/PDE. Although there are no other DGC/PDE enzymes in C. kronotskyensis with identical PAS-GGDEF-EAL architecture, functionality may be supplemented by several other DGC/PDE enzymes in the C. kronotskyensis genome. The other DDC/PDE-associated locus, locus I, is missing in C. saccharolyticus yet is highly transcribed in the other two species (Fig. 5, I; see also Table S3), in which it contains only the GGDEF and EAL domains. It is unlikely that the presence of the second DGC/PDE-containing locus in C. bescii and C. kronotskyensis is redundant, as many of the genes are transcribed to transcription levels greater than 8-fold higher than average across the genome. Instead, the functional output of this locus may give C. bescii and C. kronotskyensis an advantage for cellulose utilization over the less cellulolytic C. saccharolyticus.
Caldicellulosiruptor species are known to contain flagella (3, 8, 12, 30). Locus IV (Fig. 5, IV) is associated with flagellar structural and biosynthesis genes. The type IV pilus assembly gene, pilZ, is directly adjacent to cheA, -W, -C, and -D. In addition to cell motility, flagella are known to play an integral role in biofilm formation (53). Both flagellar genes and Che system genes are highly transcribed on cellulose and switchgrass, with 36 out of 38 associated genes highly transcribed in one or more species (Fig. 5, IV; see also Tables S3 and S4 in the supplemental material). This transcriptional response implies that colonization of both cellulose and switchgrass is important for the deconstruction and utilization of these substrates by all three species. It is likely that flagellar regulation is more complex than the Che type system, in which CheY is responsible for modulating flagellar rotation (40). Other studies have shown that cyclic di-GMP plays a role in modulating flagellar rotation through interaction with the type IV pilus assembly, PilZ (54, 55). Furthermore, work with other Firmicutes found that cyclic di-GMP levels directly regulate transcription of type IV pilus genes (56, 57). In Caldicellulosiruptor, cellulose identification and biofilm formation likely involve a complex regulatory network including cyclic di-GMP, flagellar genes, and Che-type systems (Fig. 5). The occurrence and differential regulation of these systems in Caldicellulosiruptor species suggests a role for these systems in how these bacteria sense, colonize, and deconstruct cellulosic substrates.
In addition to cyclic di-GMP, genes related to biosynthesis of purines were triggered by growth on cellulose. These include a Che-type locus conserved in C. bescii and C. kronotskyensis associated with an inosine/guanosine/xanthosine phosphorylase family protein, a type of purine-nucleoside phosphorylase, the exact function of which is unknown. Similar to the diguanylate cyclase, the purine-nucleoside phosphorylase may play a role in intracellular signaling. Several other loci (Athe_1443-1453, Calkro_1257-1267, Csac_1990-2000) involved in purine metabolism were also upregulated on cellulose (Fig. 4, G and H) and could be involved in producing signaling molecules (cyclic di-GMP and cyclic AMP) or other important purine-containing molecules involved in energy transfer (NADH and ATP) and nucleotide structure (DNA and RNA).
Relationship of CAZyme genome inventory to biomass deconstruction.
Within the genus Caldicellulosiruptor, plant biomass deconstruction is driven to a large extent by CAZymes (1, 16). As mentioned above, the C. kronotskyensis genome encodes the most extracellular CAZymes, with 31, followed by C. bescii and C. saccharolyticus, with 20 and 19, respectively (Table 2). Fourteen of these CAZymes are present in all three species, implying that differences in solubilization are related to the other 23 CAZymes, some unique and some common to two species. Collectively, the three genomes contain 21 carbohydrate-active catalytic domains, including GHs, carbohydrate esterases (CEs), and polysaccharide lyases (PLs) (Fig. 6). GH domains are the majority, with GH family 5 (GH5), endo-1,4-β-glucanases and β-mannanases (58), being the most highly represented. The second most highly represented family is GH family 10 (GH10), endo-1,4-β-xylanases, indicative of the importance of both cellulose and hemicellulose deconstruction in Caldicellulosiruptor species.
FIG 6.
Carbohydrate-active extracellular enzyme domains encoded in the Caldicellulosiruptor genome. (A) Number of extracellular catalytic domains corresponding to CAZyme families (http://www.cazy.org) with signal peptides (http://www.cbs.dtu.dk/services/SignalP/). GH, glycoside hydrolase; PL, polysaccharide lyase; CE, carbohydrate esterase. (B) Total number of carbohydrate utilization genes in C. bescii, C. kronotskyensis, and C. saccharolyticus.
Carbohydrate esterases impact mechanical strength of the plant cell wall by removing a variety of polysaccharide side chain substitutions, including acetyl, feruloyl, and methoxy groups (59, 60). The three species each contain three CE family 4 (CE4) proteins, while the other CE family protein, CE8, is present only in C. kronotskyensis. None of these carbohydrate esterase domain-containing enzymes are highly transcribed in either species, suggesting a minor role in switchgrass deconstruction. PLs are responsible for cleaving uronic acid-containing polysaccharides, such as pectin (61). Differences in the PL inventory could play a role in switchgrass degradation, as several extracellular PLs are contained in the genomes of C. bescii and C. kronotskyensis but not C. saccharolyticus. Pectin forms a major component of primary cell wall structure in dicots (poplar) and, to a lesser extent, in monocots (switchgrass) (62). Many of these extracellular PLs are highly transcribed and differentially upregulated on switchgrass, implying an important role in lignocellulose deconstruction (Fig. 7 and 8; see also Table S5 in the supplemental material). Deletion of the extracellular PLs in C. bescii (Athe_1853-1854) impaired growth on switchgrass and poplar (63). These PLs were upregulated on switchgrass and may aid the removal of pectin. The absence of PLs in C. saccharolyticus's genome may leave certain cellulose and hemicellulose moieties concealed, impairing switchgrass deconstruction.
FIG 7.
Transcription of CAZymes and ABC carbohydrate transporters in C. bescii, C. kronotskyensis, and C. saccharolyticus core and noncore genomes. Gene type is indicated on the inner track: carbohydrate transporter (light purple), intracellular CAZyme (light orange), or extracellular CAZyme (light green). Xylan degradation locus (X) and glucan degradation locus (G) genes are indicated. Tracks from outer to inner: for transcription level (LSM), C. bescii, cellulose (A); C. bescii, switchgrass (S); C. kronotskyensis, cellulose; C. kronotskyensis, switchgrass; C. saccharolyticus, cellulose; C. saccharolyticus, switchgrass; for differential transcription (Diff), C. bescii (Be), C. kronotskyensis (Kr), and C. saccharolyticus (Sa). Locus tags and transcription values can be seen in Table S5 in the supplemental material.
FIG 8.
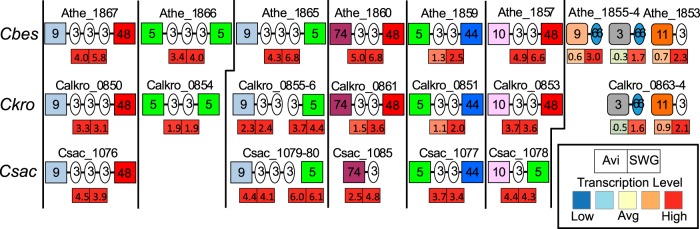
Glucan degradation locus in C. bescii, C. kronotskyensis, and C. saccharolyticus. Numbers indicate CAZy family; squares, glycoside hydrolases; ovals, carbohydrate binding modules; rounded squares, polysaccharide lyases. Genes are aligned by homologous families. Locus tags are written above, and boxes underneath represent transcription level on cellulose (left) and switchgrass (right). Red and blue represent high and low levels of transcription, respectively.
The xylan degradation locus (XDL) and glucan degradation locus (GDL) in Caldicellulosiruptor species are involved in processing C5 and C6 sugars, respectively (17, 35). The XDL contains two multidomain xylanases with GH10 and GH43 catalytic activities and carbohydrate binding module (CBM) family 6 and 22 binding domains (CBM22-CBM22-GH10 and GH43-CBM22-GH43-CBM6), two ABC carbohydrate transporters annotated for xylooligosaccharide transport, and several intracellular CAZymes (16, 35); as mentioned above, the genes in the XDL were highly responsive in all three species growing on switchgrass (Fig. 7). The GDL consists solely of extracellular multidomain CAZymes and was highly transcribed on both switchgrass and cellulose (Fig. 7 and 8), indicating the importance of GDL enzymes for the degradation of both homogenous and heterogenous cellulosic substrates. Inspection of the GDL more closely gives insight into differences between the three species that relate to cellulose and switchgrass solubilization (Fig. 8). C. bescii and C. kronotskyensis have three GH48 domain-containing enzymes, while C. saccharolyticus contains only one, CelA. Deletion of CelA in C. bescii inhibited growth on cellulose and switchgrass (64). The fact that all three species contain CelA but solubilize cellulose and switchgrass to different extents implies that this cellulase may be necessary but not solely responsible for high levels of lignocellulose deconstruction. The other two GH48-containing enzymes in C. bescii (Athe_1860 and Athe_ 1857) and C. kronotskyensis (Calkro_0861 and Calkro_0853) are paired with GH74 and GH10 domains, typically associated with xylanase and xyloglucanase activities, respectively (58). Homologous versions of these enzymes (Csac_1085 and Csac_1078) are found in C. saccharolyticus, although these do not contain GH48 domains. These enzymes were transcribed at very high levels in all three species, especially on switchgrass, where they are represented among the highest genes in the transcriptome (Fig. 8). The missing GH48 domains in the C. saccharolyticus enzymes raise the possibility that the lower C. saccharolyticus biomass solubilization capacity (Table 1) relates to the absence of these domains. Biochemical characterization of these enzymes from all three species is under way.
The fact that C. bescii and C. kronotskyensis achieved equivalent levels of cellulose and switchgrass solubilization implies that the additional GHs encoded in the C. kronotskyensis genome do not contribute significantly to this process. This is supported by the transcriptome data, as these additional GH genes are mostly transcribed at low levels on cellulose and switchgrass (Fig. 7, Ckro Only). The only exceptions are an extracellular GH13 (Calkro_2177) and a multidomain S-layer homology domain-containing protein (Calkro_0402), with a GH10. Several putative xylanases unique to C. saccharolyticus (Fig. 7, Csac Only) were highly transcribed on one or both substrates, although these did not seem to provide any advantage for switchgrass solubilization (Table 1).
Conclusion.
The difference in biomass solubilization and conversion capacity for C. saccharolyticus compared to the other two Caldicellulosiruptor species examined in this study is related not only to gene content but also to gene regulation. CAZyme inventory, more specifically the presence of additional GH48 domain-containing enzymes and polysaccharide lyases, appears to be advantageous for C. bescii and C. kronotskyensis in deconstruction of lignocellulosic substrates. But factors other than carbohydrate active enzymes likely play an important role since C. kronotskyensis, which has the most CAZymes, was not more effective in biomass deconstruction than C. bescii. A potential role for substrate sensing, signal transduction and chemotaxis in how Caldicellulosiruptor species utilize biomass is supported by genomic and transcriptomic evidence. Although C. saccharolyticus has an apparent deficiency in the ability to deconstruct lignocellulosic substrates, this is balanced with an increased fermentative intensity. It is important to keep in mind that in natural biotopes inhabited by Caldicellulosiruptor, plant biomass utilization is likely a community-based phenomenon, with individual species contributing in both general and specialized ways. Incorporating the most important features into a single organism through metabolic engineering needs to be weighed against using multiple species functioning synergistically. The approach employed in this study makes use of microbiological, genomic, transcriptomic, and chemical analyses and provides an insightful basis for comparing Caldicellulosiruptor species with respect to plant biomass deconstruction. The information obtained using this approach establishes a basis for choosing platform strains for metabolic engineering efforts.
Supplementary Material
ACKNOWLEDGMENTS
The BioEnergy Science Center (BESC) is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. L. L. Lee acknowledges support from an NIH Biotechnology Traineeship (NIH T32 GM008776-11), and J. M. Conway acknowledges support from a U.S. DoEd GAANN Fellowship (P200A100004-12).
Helpful discussions with Piyum Khatibi on cell motility are also acknowledged.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01622-15.
REFERENCES
- 1.Blumer-Schuette SE, Brown SD, Sander KB, Bayer EA, Kataeva I, Zurawski JV, Conway JM, Adams MW, Kelly RM. 2014. Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev 38:393–448. doi: 10.1111/1574-6976.12044. [DOI] [PubMed] [Google Scholar]
- 2.Blumer-Schuette SE, Ozdemir I, Mistry D, Lucas S, Lapidus A, Cheng JF, Goodwin LA, Pitluck S, Land ML, Hauser LJ, Woyke T, Mikhailova N, Pati A, Kyrpides NC, Ivanova N, Detter JC, Walston-Davenport K, Han S, Adams MW, Kelly RM. 2011. Complete genome sequences for the anaerobic, extremely thermophilic plant biomass-degrading bacteria Caldicellulosiruptor hydrothermalis, Caldicellulosiruptor kristjanssonii, Caldicellulosiruptor kronotskyensis, Caldicellulosiruptor owensensis, and Caldicellulosiruptor lactoaceticus. J Bacteriol 193:1483–1484. doi: 10.1128/JB.01515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang CY, Patel BK, Mah RA, Baresi L. 1998. Caldicellulosiruptor owensensis sp. nov., an anaerobic, extremely thermophilic, xylanolytic bacterium. Int J Syst Bacteriol 48:91–97. doi: 10.1099/00207713-48-1-91. [DOI] [PubMed] [Google Scholar]
- 4.Elkins JG, Lochner A, Hamilton-Brehm SD, Davenport KW, Podar M, Brown SD, Land ML, Hauser LJ, Klingeman DM, Raman B, Goodwin LA, Tapia R, Meincke LJ, Detter JC, Bruce DC, Han CS, Palumbo AV, Cottingham RW, Keller M, Graham DE. 2010. Complete genome sequence of the cellulolytic thermophile Caldicellulosiruptor obsidiansis OB47T. J Bacteriol 192:6099–6100. doi: 10.1128/JB.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamilton-Brehm SD, Mosher JJ, Vishnivetskaya T, Podar M, Carroll S, Allman S, Phelps TJ, Keller M, Elkins JG. 2010. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl Environ Microbiol 76:1014–1020. doi: 10.1128/AEM.01903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kataeva IA, Yang SJ, Dam P, Poole FL II, Yin Y, Zhou F, Chou WC, Xu Y, Goodwin L, Sims DR, Detter JC, Hauser LJ, Westpheling J, Adams MW. 2009. Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J Bacteriol 191:3760–3761. doi: 10.1128/JB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svetlichnyi VA, Svetlichnaya TP, Chernykh NA, Zavarzin GA. 1990. Anaerocellum thermophilum gen. nov. sp. nov.: an extremely thermophilic cellulolytic eubacterium isolated from hot-springs in the Valley of Geysers. Microbiology 59:598–604. [Google Scholar]
- 8.Miroshnichenko ML, Kublanov IV, Kostrikina NA, Tourova TP, Kolganova TV, Birkeland NK, Bonch-Osmolovskaya EA. 2008. Caldicellulosiruptor kronotskyensis sp. nov. and Caldicellulosiruptor hydrothermalis sp. nov., two extremely thermophilic, cellulolytic, anaerobic bacteria from Kamchatka thermal springs. Int J Syst Evol Microbiol 58:1492–1496. doi: 10.1099/ijs.0.65236-0. [DOI] [PubMed] [Google Scholar]
- 9.Rainey FA, Janssen PH, Morgan HW, Stackebrandt E. 1993. A biphasic approach to the determination of the phenotypic and genotypic diversity of some anaerobic, cellulolytic, thermophilic, rod-shaped bacteria. Antonie Van Leeuwenhoek 64:341–355. [DOI] [PubMed] [Google Scholar]
- 10.van de Werken HJ, Verhaart MR, VanFossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, van Niel EW, Stams AJ, Ward DE, de Vos WM, van der Oost J, Kelly RM, Kengen SW. 2008. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol 74:6720–6729. doi: 10.1128/AEM.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rainey FA, Donnison AM, Janssen PH, Saul D, Rodrigo A, Bergquist PL, Daniel RM, Stackebrandt E, Morgan HW. 1994. Description of Caldicellulosiruptor saccharolyticus gen. sp. nov.: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol Lett 120:263–266. doi: 10.1111/j.1574-6968.1994.tb07043.x. [DOI] [PubMed] [Google Scholar]
- 12.Bredholt S, Sonne-Hansen J, Nielsen P, Mathrani IM, Ahring BK. 1999. Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int J Syst Bacteriol 49(Part 3):991–996. [DOI] [PubMed] [Google Scholar]
- 13.Mladenovska Z, Mathrani IM, Ahring BK. 1995. Isolation and characterization of Caldicellulosiruptor lactoaceticus sp. nov, an extremely thermophilic, cellulolytic, anaerobic bacterium. Arch Microbiol 163:223–230. doi: 10.1007/BF00305357. [DOI] [Google Scholar]
- 14.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang SJ, Resch MG, Adams MWW, Lunin VV, Himmel ME, Bomble YJ. 2013. Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342:1513–1516. doi: 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- 15.Ozdemir I, Blumer-Schuette SE, Kelly RM. 2012. S-layer homology domain proteins C. saccharolyticus_0678 and C. saccharolyticus_2722 are implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol 78:768–777. doi: 10.1128/AEM.07031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. 2011. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng 108:1559–1569. doi: 10.1002/bit.23093. [DOI] [PubMed] [Google Scholar]
- 17.Blumer-Schuette SE, Giannone RJ, Zurawski JV, Ozdemir I, Ma Q, Yin Y, Xu Y, Kataeva I, Poole FL II, Adams MW, Hamilton-Brehm SD, Elkins JG, Larimer FW, Land ML, Hauser LJ, Cottingham RW, Hettich RL, Kelly RM. 2012. Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J Bacteriol 194:4015–4028. doi: 10.1128/JB.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blumer-Schuette SE, Lewis DL, Kelly RM. 2010. Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl Environ Microbiol 76:8084–8092. doi: 10.1128/AEM.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blumer-Schuette SE, Alahuhta M, Conway JM, Lee LL, Zurawski JV, Giannone RJ, Hettich RL, Lunin VV, Himmel ME, Kelly RM. 2015. Discrete and structurally unique proteins (tāpirins) mediate attachment of extremely thermophilic Caldicellulosiruptor species to cellulose. J Biol Chem 290:10645–10656. doi: 10.1074/jbc.M115.641480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basen M, Rhaesa AM, Kataeva I, Prybol CJ, Scott IM, Poole FL, Adams MW. 2014. Degradation of high loads of crystalline cellulose and of unpretreated plant biomass by the thermophilic bacterium Caldicellulosiruptor bescii. Bioresour Technol 152:384–392. doi: 10.1016/j.biortech.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 21.Yang SJ, Kataeva I, Wiegel J, Yin Y, Dam P, Xu Y, Westpheling J, Adams MW. 2010. Classification of ‘Anaerocellum thermophilum’ strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int J Syst Evol Microbiol 60:2011–2015. doi: 10.1099/ijs.0.017731-0. [DOI] [PubMed] [Google Scholar]
- 22.Chung D, Cha M, Guss AM, Westpheling J. 2014. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc Natl Acad Sci U S A 111:8931–8936. doi: 10.1073/pnas.1402210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zurawski JV, Blumer-Schuette SE, Conway JM, Kelly RM. 2014. The extremely thermophilic genus Caldicellulosiruptor: physiological and genomic characteristics for complex carbohydrate conversion to molecular hydrogen, p 177–195. In Zannoni D, De Philippis R (ed), Advances in photosynthesis and respiration, vol 38 Microbial bioenergy: hydrogen production; Springer Science+Business Media, Dordrecht, Netherlands. [Google Scholar]
- 24.Sluiter AHB, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. 2012. Determination of structural carbohydrates and lignin in biomass. Technical report NREL/TP-510-42618. National Renewable Energy Laboratory, Golden, CO: http://www.nrel.gov/biomass/pdfs/42618.pdf. [Google Scholar]
- 25.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 26.de Vrije T, Mars AE, Budde MA, Lai MH, Dijkema C, de Waard P, Claassen PA. 2007. Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biotechnol 74:1358–1367. doi: 10.1007/s00253-006-0783-x. [DOI] [PubMed] [Google Scholar]
- 27.Chen SF, Mowery RA, Sevcik RS, Scarlata CJ, Chambliss CK. 2010. Compositional analysis of water-soluble materials in switchgrass. J Agric Food Chem 58:3251–3258. doi: 10.1021/jf9033877. [DOI] [PubMed] [Google Scholar]
- 28.Thammasouk K, Tandjo D, Penner MH. 1997. Influence of extractives on the analysis of herbaceous biomass. J Agric Food Chem 45:437–443. doi: 10.1021/jf960401r. [DOI] [Google Scholar]
- 29.Lochner A, Giannone RJ, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Label-free quantitative proteomics for the extremely thermophilic bacterium Caldicellulosiruptor obsidiansis reveal distinct abundance patterns upon growth on cellobiose, crystalline cellulose, and switchgrass. J Proteome Res 10:5302–5314. doi: 10.1021/pr200536j. [DOI] [PubMed] [Google Scholar]
- 30.Lochner A, Giannone RJ, Rodriguez M Jr, Shah MB, Mielenz JR, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl Environ Microbiol 77:4042–4054. doi: 10.1128/AEM.02811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataeva I, Foston MB, Yang S-J, Pattathil S, Biswal AK, Poole Ii FL, Basen M, Rhaesa AM, Thomas TP, Azadi P, Olman V, Saffold TD, Mohler KE, Lewis DL, Doeppke C, Zeng Y, Tschaplinski TJ, York WS, Davis M, Mohnen D, Xu Y, Ragauskas AJ, Ding S-Y, Kelly RM, Hahn MG, Adams MWW. 2013. Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energ Environ Sci 6:2186. doi: 10.1039/c3ee40932e. [DOI] [Google Scholar]
- 32.van Niel EWJ, Claassen PAM, Stams AJM. 2003. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng 81:255–262. doi: 10.1002/bit.10463. [DOI] [PubMed] [Google Scholar]
- 33.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Koning SM, Albers SV, Konings WN, Driessen AJ. 2002. Sugar transport in (hyper)thermophilic archaea. Res Microbiol 153:61–67. doi: 10.1016/S0923-2508(01)01289-X. [DOI] [PubMed] [Google Scholar]
- 35.Vanfossen AL, Verhaart MR, Kengen SM, Kelly RM. 2009. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl Environ Microbiol 75:7718–7724. doi: 10.1128/AEM.01959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhaart MR, Bielen AA, van der Oost J, Stams AJ, Kengen SW. 2010. Hydrogen production by hyperthermophilic and extremely thermophilic bacteria and archaea: mechanisms for reductant disposal. Environ Technol 31:993–1003. doi: 10.1080/09593331003710244. [DOI] [PubMed] [Google Scholar]
- 37.Pawar SS, van Niel EWJ. 2014. Evaluation of assimilatory sulphur metabolism in Caldicellulosiruptor saccharolyticus. Bioresource Technol 169:677–685. doi: 10.1016/j.biortech.2014.07.059. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama K, Leimkuhler S. 2015. The role of FeS clusters for molybdenum cofactor biosynthesis and molybdoenzymes in bacteria. Biochim Biophys Acta 1853:1335–1349. doi: 10.1016/j.bbamcr.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segall JE, Ishihara A, Berg HC. 1985. Chemotactic signaling in filamentous cells of Escherichia coli. J Bacteriol 161:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ninfa EG, Stock A, Mowbray S, Stock J. 1991. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem 266:9764–9770. [PubMed] [Google Scholar]
- 41.Stock JB, Lukat GS, Stock AM. 1991. Bacterial chemotaxis and the molecular logic of intracellular signal transduction networks. Annu Rev Biophys Biophys Chem 20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- 42.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol 422:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 45.Pawar SS VT, Grey C, van Niel EW. 2015. Biofilm formation by designed co-cultures of Caldicellulosiruptor species as a means to improve hydrogen productivity. Biotechnol Biofuels 8:19. doi: 10.1186/s13068-015-0201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang ZW, Hamilton-Brehm SD, Lochner A, Elkins JG, Morrell-Falvey JL. 2011. Mathematical modeling of hydrolysate diffusion and utilization in cellulolytic biofilms of the extreme thermophile Caldicellulosiruptor obsidiansis. Bioresource Technol 102:3155–3162. doi: 10.1016/j.biortech.2010.10.104. [DOI] [PubMed] [Google Scholar]
- 47.Wang ZW, Lee SH, Elkins JG, Morrell-Falvey JL. 2011. Spatial and temporal dynamics of cellulose degradation and biofilm formation by Caldicellulosiruptor obsidiansis and Clostridium thermocellum. AMB Express 1:30. doi: 10.1186/2191-0855-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem 282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 51.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:1092–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11–21. [DOI] [PubMed] [Google Scholar]
- 53.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 56.Bordeleau E, Purcell EB, Lafontaine DA, Fortier LC, Tamayo R, Burrus V. 2015. Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol 197:819–832. doi: 10.1128/JB.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Chai Y, Guo JH, Losick R. 2012. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol 194:5080–5090. doi: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biely P. 2012. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol Adv 30:1575–1588. doi: 10.1016/j.biotechadv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 60.Williamson G, Kroon PA, Faulds CB. 1998. Hairy plant polysaccharides: a close shave with microbial esterases. Microbiology 144(Part 8):2011–2023. [DOI] [PubMed] [Google Scholar]
- 61.Garron ML, Cygler M. 2014. Uronic polysaccharide degrading enzymes. Curr Opin Struct Biol 28:87–95. doi: 10.1016/j.sbi.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 62.Mohnen D. 2008. Pectin structure and biosynthesis. Curr Opin Plant Biol 11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 63.Chung D, Pattathil S, Biswal AK, Hahn MG, Mohnen D, Westpheling J. 2014. Deletion of a gene cluster encoding pectin degrading enzymes in Caldicellulosiruptor bescii reveals an important role for pectin in plant biomass recalcitrance. Biotechnol Biofuels 7:147. doi: 10.1186/s13068-014-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young J, Chung D, Bomble YJ, Himmel ME, Westpheling J. 2014. Deletion of Caldicellulosiruptor bescii CelA reveals its crucial role in the deconstruction of lignocellulosic biomass. Biotechnol Biofuels 7:142. doi: 10.1186/s13068-014-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shockley KR, Ward DE, Chhabra SR, Conners SB, Montero CI, Kelly RM. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol 69:2365–2371. doi: 10.1128/AEM.69.4.2365-2371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.