Abstract
Most DNA-based microbial source tracking (MST) approaches target host-associated organisms within the order Bacteroidales, but the gut microbiota of humans and other animals contain organisms from an array of other taxonomic groups that might provide indicators of fecal pollution sources. To discern between human and nonhuman fecal sources, we compared the V6 regions of the 16S rRNA genes detected in fecal samples from six animal hosts to those found in sewage (as a proxy for humans). We focused on 10 abundant genera and used oligotyping, which can detect subtle differences between rRNA gene sequences from ecologically distinct organisms. Our analysis showed clear patterns of differential oligotype distributions between sewage and animal samples. Over 100 oligotypes of human origin occurred preferentially in sewage samples, and 99 human oligotypes were sewage specific. Sequences represented by the sewage-specific oligotypes can be used individually for development of PCR-based assays or together with the oligotypes preferentially associated with sewage to implement a signature-based approach. Analysis of sewage from Spain and Brazil showed that the sewage-specific oligotypes identified in U.S. sewage have the potential to be used as global alternative indicators of human fecal pollution. Environmental samples with evidence of prior human fecal contamination had consistent ratios of sewage signature oligotypes that corresponded to the trends observed for sewage. Our methodology represents a promising approach to identifying new bacterial taxa for MST applications and further highlights the potential of the family Lachnospiraceae to provide human-specific markers. In addition to source tracking applications, the patterns of the fine-scale population structure within fecal taxa suggest a fundamental relationship between bacteria and their hosts.
INTRODUCTION
Microbial source tracking (MST) is used to determine sources of fecal pollution in surface waters and recreational beaches with the goal of minimizing the risk to human health (1–3). Transmission of bacterial, viral, and zoonotic diseases occurs through feces-contaminated water (4), and identification of the type of host inputs (e.g., sewage, wildlife, agricultural) can provide a more accurate assessment of the risks to human health and better direct management actions to reduce likely sources of pollution (5). Identification of fecal sources is based on the assumption that some microorganisms exhibit host-specific distribution patterns (6). The majority of microbial population studies performed to identify members that specifically or preferentially associate with particular animal hosts have used the 16S rRNA gene as a marker (7–9).
Bacteria within the order Bacteroidales have been the major focus of molecular MST efforts (10–16), as they have many qualities that make them an effective indicator. These organisms are abundant in the gastrointestinal tract of many animals, some species exhibit an association with particular hosts, and many have persistence and survival characteristics similar to those of pathogens (17). Bacteroidales markers for humans, ruminants, pigs, and other animals have been used extensively with much success (9, 12, 15). However, additional markers are needed to support the results of these assays, to provide alternative indicators in regions where Bacteroidales are a minor component of human fecal pollution, and to resolve contributions from multiple hosts in environments with complex pollution sources (16, 18). Only a few studies have proposed alternatives to Bacteroidales markers for the identification of mammalian fecal sources (7, 19–21), and even fewer studies have compared the performance of marker genes from the Bacteroidales with that of marker genes from other major taxonomic groups (6, 19, 22). Recent microbiome studies have identified an array of taxonomic groups within the dominant fecal phyla Bacteroidetes and Firmicutes (23–25) that are abundant in humans and other mammals; genera within these phyla might also contain host-specific strains that could serve as novel alternative indicators. Employing multiple indicators could be more useful for obtaining an understanding of the ecology of fecal organisms in the environment and establishing more definitive relationships with pathogens.
Next-generation sequencing (NGS) platforms now yield on the order of 104 to 106 sequence reads per sample and provide nearly comprehensive descriptions of microbial communities (26). Such deep sequencing allows the in silico analysis of the potential overlap of similar or identical sequences from different hosts, even if the sequences are present at a relatively low abundance (19). NGS-based approaches have successfully been used in MST studies to allocate fecal contributions from specific hosts in contaminated water using comparisons with source fecal samples (27, 28). Despite the volume of information gained from an increased depth of sequencing, NGS can be limited by the sensitivity of the methods used to analyze these large and complex data sets due to computational challenges. The 16S rRNA gene is somewhat limited in its ability to discriminate between very closely related organisms, and most NGS technologies additionally produce relatively short sequences that contain reduced amounts of information that are insufficient for assigning a fine-level taxonomy or for resolving sequences in cluster analyses on the basis of a similarity threshold (29). Oligotyping, a supervised computational method of sequence analysis, uses Shannon entropy to exploit the information contained in short read sequences by identifying relevant nucleotide changes (30). Sequences are therefore grouped not by a fixed sequence similarity threshold but by the occurrence of certain nucleotides in key positions. This method produces greater taxonomic homogeneity within clusters, reduces error-based diversity, and can distinguish ecologically distinct organisms whose rRNA sequences differ from each other by as few as 1 nucleotide (30).
Newton et al. (31) recently used oligotyping to demonstrate that the composition of bacteria within six fecal families in sewage accurately reflects the human gut microbiome at the population level. Here we examined sewage samples as a proxy for human fecal microbiota and used oligotyping as a novel approach to identify targets for their potential use as alternative indicators. We examined the fine-level population structures within 10 fecal taxa to identify sequences that distinguish human and nonhuman fecal sources with a high degree of sensitivity and specificity.
MATERIALS AND METHODS
Sample collection.
Sewage serves as a good representative sample with which to identify human-specific indicators, as human fecal pollution concerns are typically related to sewage releases (combined/sanitary sewer overflows, failing sanitary sewer infrastructure) rather than individual inputs (32). Sewage has previously been shown to contain a consistent suite of human fecal bacteria (31, 33) that occur at a relatively high abundance in untreated influent, in addition to the larger portion of bacteria that are associated with the sewerage infrastructure (34). Primary influent sewage samples were collected over a 24-h period from nine municipal wastewater treatment facilities in eight cities across the United States in August 2012 and January 2013. These facilities serve populations ranging from ∼1,500 to >500,000 and include both combined and separated sewage systems to include a wide range of potential inputs. Samples were shipped on ice to the University of Wisconsin—Milwaukee lab and filtered (25 ml per sample; pore size, 0.22 μm; filter diameter, 47 mm; mixed cellulose ester filters; Millipore, Billerica, MA, USA) within 48 h of collection. The filters were stored in cryovials at −80°C until the DNA extraction procedure. We previously sequenced DNA extracted from 53 fecal samples from animals (cats, chickens, cows, deer, dogs, swine) and two primary influent sewage samples and reported on the fine-scale population structure within the genus Blautia (35). The environmental samples and additional sewage samples analyzed in this study were also sequenced previously, and methods for their collection and processing are reported elsewhere (18, 36).
DNA extraction, sequencing, and data quality screening.
We extracted DNA from crushed frozen filters with a FastDNA Spin kit for soil (MP Biomedicals, Santa Ana, CA) according to the manufacturer's instructions as previously described (31) and assessed the DNA concentration and purity using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). To minimize the impact of early-round PCR errors, we prepared three independent PCR libraries for the hypervariable V6 region of the bacterial 16S rRNA gene (∼60 bp) using a set of primers that target all major bacterial groups and 25 cycles of amplification (35). Subsequent to pooling of the reaction mixtures for each sample, we performed 5 cycles of PCR using custom fusion primers (an Illumina adaptor and 8 different inline bar codes for the forward primers plus 12 specific indices for reverse primers annealed to conserved regions flanking the V6 sequence) as previously described (35). Samples were sequenced on one lane of an Illumina HiSeq 1000 sequencing system as a paired-end run with 100 cycles. The quality-filtering method described elsewhere (37) minimized sequencing errors in the final data set. Global Alignment for Sequence Taxonomy (GAST) (38) assigned a taxonomy to our high-quality reads. Previous publications have described in detail the methods for DNA sequencing, quality control, and bioinformatic trimming related to Illumina amplicon sequencing (35, 37).
Oligotyping.
Singletons and doubletons were removed to further reduce the error-based diversity from the high-quality sequence and taxonomic count data (37). We analyzed only the fecal portion of the sewage samples, using the six dominant fecal families (Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, Rikenellaceae, Porphyromonadaceae, and Prevotellaceae), which comprise >95% of the total fecal community, as described by Newton et al. (31). We randomly subsampled animal data sets prior to oligotyping to normalize the sequence reads to 500,000 per sample (with the exception of eight samples from chickens and one sample from a dog that contained <500,000 reads; see Table S1 in the supplemental material). GAST taxonomic classification (38) at the genus level or the highest level of resolution available (e.g., unclassified Lachnospiraceae) identified sets of sequences for oligotyping. We implemented the oligotyping pipeline (30) to determine the oligotypes within 10 abundant human fecal taxa. The entropy analysis script in the oligotyping pipeline calculated the Shannon entropy at each base along the length of the sequences. Starting with the nucleotide positions with the highest entropy, we supervised the oligotyping process by selecting additional nucleotide positions until all high-entropy peaks in individual oligotypes were minimized, following the steps shown in the supplemental flowchart presented elsewhere (30). Each oligotype was required to be present in at least three samples (−s 3) and have a minimum substantive abundance (−M) of N/5,000, where N equals the total number of sequence reads in the data set for each individual oligotyping run. Table S2 in the supplemental material reports the total number of sequence reads, the number of reads retained after the application of noise filtering parameters –s and –M, and the total number of oligotypes generated for each of the 10 fecal taxa analyzed.
Oligotype data analysis.
We performed network analysis using Gephi software (39) to determine the distribution of all oligotypes from the 10 fecal taxa using a force-directed graph algorithm (ForceAtlas2 in Gephi software). Oligotypes present in ≥1 sample from each host group were counted as positive for that host. We used the LEfSe program (v1.0) (40) with default parameters (number of linear discriminant analysis [LDA] bootstrap iterations, 30; minimum effect size, 2.0) to identify the oligotypes most strongly associated with sewage. LEfSe uses nonparametric statistical tests and LDA to estimate the statistical significance of the presence of individual oligotypes in one host compared to its presence in others. The effect size of an oligotype reflects its significance in defining one community over another; thus, a highly significant oligotype would have a consistent presence and a high relative abundance in one host type but might be sporadically present at a low abundance in several other hosts as well.
To compare the sequences of sewage-specific oligotypes to previously published sequences obtained by human-specific quantitative PCR (qPCR) assays, we used the V6 sequence that was employed to design the reverse primer and probe of the Lachno2 assay (19). The reverse primer and probe dictate the specificity of this assay. For the HF183 assay (9), we used the type strain of Bacteroides dorei, which contains the HF183-specific primer sequence (8), to infer the V6 sequence of this organism and match it to our data set. We identified the V6 region of this sequence by primer matching to the primers used to create our amplicons and found the matching oligotype sequence from our data set within a sequence count table using the VLOOKUP function in Excel software.
We used BLAST analysis to compare the sequence of the V6 region of the 16S rRNA gene from sewage-specific oligotypes and oligotypes preferentially associated with sewage (sewage-preferred oligotypes) to full-length sequences in the National Center for Biotechnology Information (NCBI) nr database (accessed 17 April 2015). We limited the results to a maximum of 100 perfect matches (100% identity over the entire length of the query, which was 60 nucleotides). The accession numbers of all sequences with a perfect match were used to acquire full GenBank records through Batch Entrez, and matching sequences were binned as human or nonhuman on the basis of their reported isolation source.
Assessment of candidate indicators and a sewage-specific signature.
The 20 most abundant human-associated sewage-specific and -preferred oligotypes from sewage were selected as candidate targets. We combined these 20 oligotypes to create a sewage signature and compared the collective oligotype abundance and the ratios of the oligotypes within this signature to those in sewage from other countries (Spain and Brazil) and environmental samples. The percent abundance of an oligotype in sewage was expressed relative to the abundance only in the fecal portion of sewage rather than the total number of reads (31). Environmental samples with known human fecal or sewage contamination were obtained from a Brazilian river with direct human fecal inputs from a small village, stormwater with high copy numbers of human fecal indicators Lachno2 and HF183, and surface water samples taken from Lake Michigan after a combined sewer overflow (CSO) event (18, 36, 41). Stormwater samples with very low copy numbers or results below the detection limit from the Lachno2 and HF183 assays and Lake Michigan water collected during the baseflow were used as comparison samples with low levels of contamination.
Statistical analyses.
We conducted all statistical tests in R (42); the adonis function in the vegan package (43) was used to evaluate the sources of variance in sewage and animal groups and for sewage versus all animals treated as a single group. We analyzed the variance within groups using the betadisper function on the basis of the distance from the median and determined significance using analysis of variance. A paired t test was used to assess whether the variance of oligotypes of all animals grouped together was different from oligotype variance within each animal group. Spearman correlations provided statistical support for the significance of relationships between the sewage signature in U.S. sanitary sewage and that in other sewage or environmental samples.
Nucleotide sequence accession numbers.
All sequences from sewage, animal fecal samples, and Brazilian river water are available in the NCBI Sequence Read Archive under accession number SRP041262; stormwater and lake water sequences are associated with project SRP056973.
RESULTS
Identification of sewage-specific patterns within individual fecal taxa.
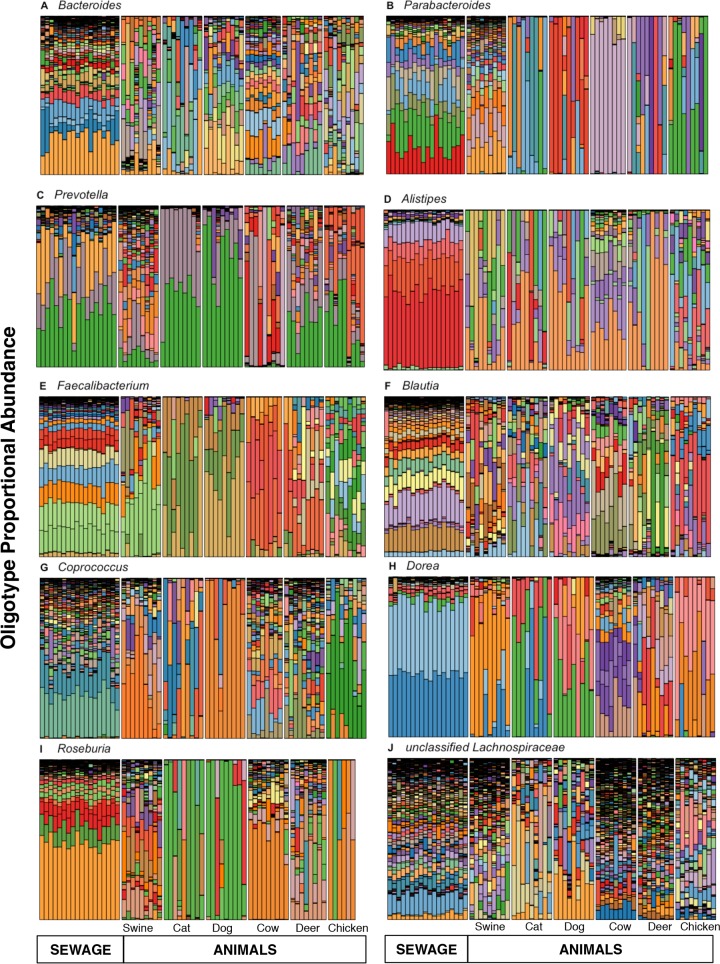
We compared the amplicon sequences from 18 untreated sewage samples and fecal samples from 53 individual animals (cats, chicken, cows, deer, dogs, and swine) to investigate the potential of various fecal taxa to provide new alternative indicator targets. The fecal portion of our sewage samples comprised an average of 19% ± 9.5% of the total community sequence reads per sample. Ten genus-level taxonomic groups (Alistipes, Bacteroides, Blautia, Coprococcus, Dorea, Faecalibacterium, unclassified Lachnospiraceae, Parabacteroides, Prevotella, and Roseburia; see Fig. S1 in the supplemental material) were particularly abundant, accounting for ∼87% of the feces-associated organisms in sewage. In contrast, these taxa comprised a smaller proportion in animal fecal samples and accounted for only 19% ± 7.8% of the sequences. Sewage and fecal samples from animals also displayed distinct trends in the relative abundance of these taxonomic groups individually. Other dominant members of the animal fecal communities included unclassified Ruminococcaceae and Rikenellaceae, Lactobacillus, and Peptostreptococcus (data not shown). Sewage samples contained higher proportions of Bacteroides, Parabacteroides, Prevotella, and Roseburia than most animal fecal samples, but none of the 10 taxa were found exclusively in sewage.
We used oligotyping to investigate the distribution of sequences within these 10 taxa to better resolve the differences between the fecal microbial communities of sewage versus those of animals. Decomposition of the Shannon entropy in selected nucleotide positions from the V6 hypervariable region defined oligotypes for each of the 10 taxa (see Table S2 in the supplemental material). Figures 1A to J show the relative oligotype abundances within each of the taxa for sewage and animals. Oligotypes displayed patterns of relative abundance in sewage that were distinct from those in all animal hosts, and overall, the oligotype distributions varied more among host types than within groups (adonis R2 > 0.50, P < 0.001; see Table S3 in the supplemental material). The oligotypes in sewage were significantly different from those in both individual animal groups and all animals pooled as a single group, but individual animal groups better explained the variation due to the higher variance of the oligotypes within the animal group (adonis R2 <0.22, P < 0.001; see Table S3 in the supplemental material). For most of the taxa analyzed, a few dominant oligotypes accounted for the majority of the sequence reads in an individual sample; oligotypes from the unclassified Lachnospiraceae, which were highly diverse in all hosts, provided a notable exception to this general trend. Overall, many oligotypes abundant in sewage were absent or present at a low relative abundance in animals.
FIG 1.
Stacked bar charts show the patterns of oligotype proportions in 10 abundant fecal taxa from sewage and animal fecal samples. (A) Bacteroides; (B) Parabacteroides; (C) Prevotella; (D) Alistipes; (E) Faecalibacterium; (F) Blautia; (G) Coprococcus; (H) Dorea; (I) Roseburia; (J) unclassified Lachnospiraceae. Oligotypes were generated with the parameters s equal to 3 and M equal to N/5,000 using the oligotyping pipeline program (version 0.96), available at http://github.com/meren/oligotyping.
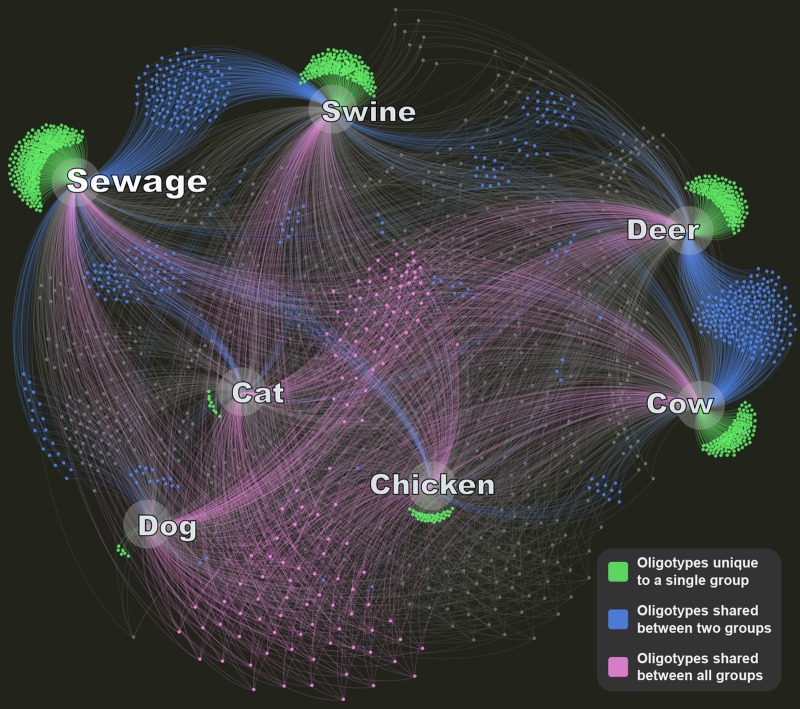
Distribution of oligotypes shared among and unique to host groups.
Although the patterns of oligotype distribution within each taxon were clearly different in sewage and animals, many oligotypes were shared as well. Network analysis allowed visualization of the specificity of the oligotypes, i.e., how they were distributed among sewage and animals, to show how many oligotypes were strictly associated with sewage. We identified oligotypes that were either unique to a single host group, shared by two host groups, or present in all host groups (Fig. 2). Host-specific oligotypes (found only in sewage or a single type of animal) accounted for the largest fraction (820/1,846), while oligotypes shared by all groups made up a small portion of the total (93/1,846). Sewage shared the most oligotypes with swine (n = 118), in addition to some overlap with only cats (n = 27) or only dogs (n = 20), but also contained many oligotypes that were not found in any animal fecal samples and were therefore considered specific to sewage.
FIG 2.
A network analysis of the oligotypes present in each host group was performed using Gephi (39). Every dot identifies an oligotype present in at least one sample from a given host group, and each edge on the network connects an oligotype to one or more host groups. Green, blue, and purple dots represent oligotypes present in only one, only two, or all host groups, respectively; gray dots represent all other oligotypes.
The use of less restrictive criteria for affiliation with only sewage (<100% specificity) can identify a wider range of indicators that may also have a greater sensitivity for detection of sewage due to a higher abundance. Genetic markers that are differentially distributed in humans compared to their distribution in animals could be used collectively as a sewage signature which considers both the presence of multiple organisms and their abundance patterns. Therefore, a collection of genetic markers for these organisms would be expected to covary in water contaminated with sewage. For oligotypes that were shared between or among sewage and one or more animal hosts, LEfSE analysis identified sewage oligotypes that were consistently present and highly abundant in sewage compared to their presence and abundance in animals. LDA scores provided statistical support for the patterns observed among oligotypes, with scores greater than 2.0 (equivalent to an effect size of 2 orders of magnitude for the differentiation of groups) signifying a significant association with a particular group. Over 450 oligotypes had a significant association with sewage, with representatives being present in all 10 taxa. Furthermore, 289 oligotypes showed a high sensitivity, as evidenced by their presence in all 18 sewage samples. Table 1 summarizes the distribution of specific and preferred oligotypes across the 10 taxa examined. Overall, 159 oligotypes found in all sewage samples were present only in sewage, while 130 were preferentially associated with sewage. Unclassified Lachnospiraceae, Bacteroides, and Blautia were rich in sewage-specific and -preferred oligotypes based on both incidence and abundance of sequence reads found in these oligotypes. Table S4 in the supplemental material highlights the sewage-preferred and -specific oligotypes and their taxonomic association, rank in sewage, average percent abundance in sewage (normalized to the fecal reads), and average abundance in each animal host.
TABLE 1.
Distribution and abundance of specific and preferred oligotypes in sewage
| Taxon | No. of oligotypes |
Sequence read abundance (%)a |
|||||
|---|---|---|---|---|---|---|---|
| Total | In sewage | Sewage specific | Sewage preferred | Sewage specific | Sewage preferred | Total | |
| Alistipes | 52 | 19 ± 2 | 3 | 4 | 0.0668 | 1.62 | 2.06 |
| Bacteroides | 187 | 91 ± 3 | 38 | 22 | 8.34 | 18.8 | 32.9 |
| Blautia | 152 | 85 ± 7 | 20 | 20 | 2.27 | 2.75 | 6.29 |
| Coprococcus | 242 | 65 ± 9 | 14 | 10 | 0.0960 | 0.385 | 0.742 |
| Dorea | 89 | 25 ± 5 | 1 | 5 | 0.0160 | 1.04 | 1.20 |
| Faecalibacterium | 101 | 56 ± 6 | 13 | 8 | 0.863 | 1.22 | 3.49 |
| Lachnospiraceae (unclassified) | 613 | 144 ± 11 | 28 | 40 | 1.50 | 3.81 | 8.26 |
| Parabacteroides | 103 | 49 ± 4 | 19 | 2 | 7.30 | 2.01 | 14.3 |
| Prevotella | 121 | 61 ± 5 | 8 | 7 | 1.23 | 2.86 | 12.6 |
| Roseburia | 186 | 69 ± 8 | 15 | 12 | 0.349 | 3.94 | 4.74 |
| Total | 1,846 | 159 | 130 | ||||
The percentages are relative to the fecal portion of sewage.
A BLAST search of the sequences within the NCBI nr database revealed that 215 of the 289 sewage-specific and -preferred oligotypes were primarily associated with human feces, with 99 human-associated oligotypes being sewage specific and 116 oligotypes being sewage preferred (see Table S4 in the supplemental material). The remaining oligotypes were solely or additionally from nonhuman or nonfecal sources, such as soil, animal feces, human skin, or industrial wastewater; three oligotypes had no match with greater than 95% identity in the database. We considered only the human-associated oligotypes for further analyses. Table 2 lists the 20 most abundant human-associated sewage oligotypes, their percent abundance in the sewage fecal community, and the animal hosts that showed low levels of that oligotype. In general, the preferred oligotypes tended to have a higher relative abundance in sewage, with 16 of the top 20 oligotypes being sewage preferred rather than sewage specific. The sewage-preferred oligotypes were found in trace abundance in one or more animals. The majority of the top 20 oligotypes came from the genera Blautia and Bacteroides; however, Roseburia, Alistipes, Dorea, and Faecalibacterium also had representatives. Two of the oligotypes that were ubiquitous in sewage samples had sequences identical to the sequence of the V6 region of organisms targeted by the human-specific markers currently used in qPCR assays, HF183 and Lachno2. Lachno2, from the genus Blautia, ranked 17th in relative abundance in sewage and was also detected in multiple fecal samples from animals, particularly cats and dogs. While we could not make a direct comparison to the HF183 assay since it targets the V2 region, the V6 region of the type strain of B. dorei, which contains the HF183 sequence, was used for comparison. The oligotype matching B. dorei ranked 7th and was found at low levels in all animals except cows, where it was absent.
TABLE 2.
Sewage signature oligotypes: the 20 most abundant human-associated preferred and specific oligotypes from sewage
| Oligotype namea | Rank in sewage | Sewage fecal % |
Presence of oligotypec in: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Avgb | SD | Cat | Chicken | Cow | Deer | Dog | Swine | ||
| Bacteroides_CGAGCAGAATTACGGCCGTAACCCATTG | 1 | 7.27 | 1.62 | + | + | + | + | + | + |
| Bacteroides_CGGGCAACATGATGCATGTCACCCAGTG | 4 | 2.74 | 0.861 | + | + | − | + | + | + |
| Roseburia_AGCTTCCCGTCCCACCTCTCAGT | 6 | 2.19 | 0.639 | − | − | − | − | − | + |
| Bacteroides_CGAGCACTATGATCCGGTTCACGAGCAG | 7 | 1.95 | 0.492 | + | + | − | + | + | + |
| Bacteroides_CGAGCAACATATATCAGTATACTCTGTG | 9 | 1.67 | 0.468 | + | + | + | + | − | − |
| Blautia_AATCTCACCGTCTTTACCTCTTTGCAGT | 13 | 1.10 | 0.442 | − | − | − | − | − | − |
| Alistipes_GCGATCTAGGTCTCGG | 16 | 0.953 | 0.314 | + | − | − | + | + | − |
| Blautia_AGTGCCACCATCCTCATCTTCTTACGCA | 17 | 0.884 | 0.352 | + | + | + | + | + | + |
| Lachnospiraceae_TGCTTCCCTACCGGCAATGTCTTCCTTGACAGT | 18 | 0.808 | 0.328 | + | + | + | + | + | + |
| Bacteroides_CGAGCAACATATGGCGCCATACGAGCGT | 20 | 0.715 | 0.211 | + | − | − | − | − | − |
| Bacteroides_CGAGCAAAATTATGCCCATACCCCATTG | 21 | 0.605 | 0.173 | − | − | − | − | − | − |
| Blautia_AGTCTCACCACTCTCGTCTTCTTACAGA | 22 | 0.586 | 0.184 | − | − | − | − | − | + |
| Dorea_GCAACGCCTAAGCTACT | 23 | 0.534 | 0.196 | + | − | − | − | − | + |
| Roseburia_AGCTTCCCGTCTCACCTCTCAGT | 24 | 0.516 | 0.155 | − | − | − | − | − | + |
| Faecalibacterium_TCTACGATGCGACATATTCG | 25 | 0.502 | 0.152 | − | − | − | − | − | + |
| Bacteroides_CGAGCAACATATGGCGCTATACGAGCGT | 26 | 0.500 | 0.138 | − | − | − | − | − | − |
| Blautia_AATCTCACCGGACTCCCCTTCTTACGGA | 27 | 0.481 | 0.176 | − | − | − | − | − | − |
| Bacteroides_CGAGCATTATATATCAGTATACTCAATG | 28 | 0.468 | 0.180 | + | + | + | + | + | + |
| Dorea_GCAACGCCTAAGTTACT | 29 | 0.459 | 0.160 | + | + | − | − | − | + |
| Alistipes_CTGATCGAGGTCTCAG | 31 | 0.410 | 0.171 | + | + | − | − | + | + |
Sewage-specific oligotypes are shown in bold.
Average for all sewage samples as a percentage of the fecal portion of sewage for that sample.
Oligotype presence in nonsewage samples. +, present; −, absent. For oligotypes present in animal sources, the percent abundance was significantly higher in sewage, as determined by LEfSE analysis.
Global applicability of sewage oligotypes and detection of the oligotypes in environmental samples.
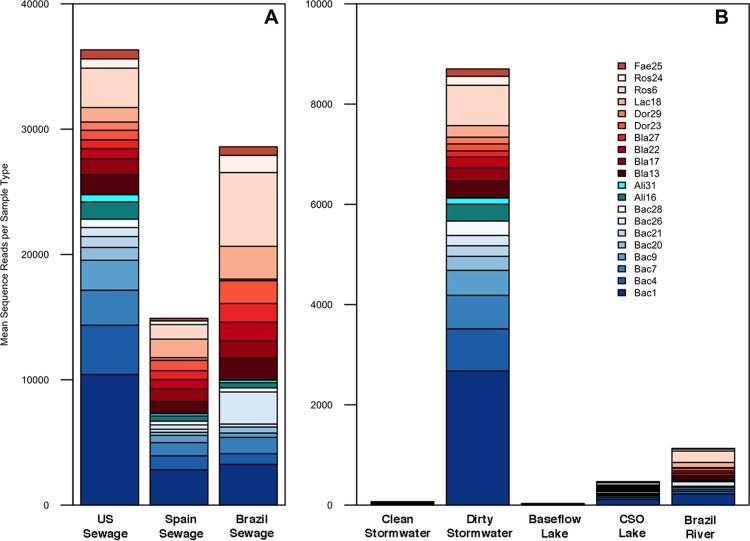
We chose the 20 most abundant human-origin oligotypes that either had 100% specificity or were host preferred within our sewage samples for assessment of the sensitivity for their detection in sewage samples from other countries (Brazil and Spain) and environmental samples (from the United States and Brazil; see Table S5 in the supplemental material). We considered both individual indicators and the ratios of the 20 oligotypes within a given sample using a signature approach. All oligotypes from the sewage signature were observed in samples known to contain human fecal pollution, and the oligotype proportions found in contaminated environmental samples were similar to those found in pure sewage.
Figure 3 shows the abundance of 20 candidate indicators in sewage from the United States, Spain, and Brazil as well as environmental samples. The indicator signatures in sanitary sewage from the United States and Spain were highly similar to each other, having high proportions of Bacteroides oligotypes, while sewage from Brazil contained higher proportions of oligotypes from the family Lachnospiraceae, including the genera Blautia, Dorea, and Roseburia. All candidate indicators from the signature were found in sewage from both Spain and Brazil; most were additionally found in all environmental samples. Figure S2 in the supplemental material shows the relationships, the correlation coefficients, and the significance of the correlations between the sewage signature of U.S. sanitary sewage and that of sewage from Brazil and Spain, stormwater with sewage contamination, stormwater with only background or mixed fecal contamination, lake water after a CSO, and lake water under baseflow conditions (no rain). Figure S2 in the supplemental material also compares the sewage signature of Brazilian river water samples to the ratio of the oligotypes in that signature in Brazilian sewage or U.S. sewage. The stronger correlation of the sewage signature of Brazilian river water to Brazilian sewage demonstrates that the signature oligotypes are present, but at ratios different from those in U.S. sewage. The percent abundance value for each candidate oligotype indicator in individual samples is given in Table S5 in the supplemental material. Combined sequence reads for the sewage signatures comprised >24% of the fecal portion of U.S. sewage, 3 to 40% of other sewage samples, and up to >1% of the entire bacterial community in contaminated environmental samples. The samples with background or low-level contamination contained the sewage signature oligotypes at a low relative abundance (0.001% to 0.01%).
FIG 3.
Sewage signature oligotypes in sanitary sewage and environmental samples. The relative abundance of the 20 most abundant specific and preferred oligotypes of human origin are shown for sewage from the United States, Spain, and Brazil (A) and clean and dirty stormwater nearshore Lake Michigan water under baseflow and CSO conditions, and water from a Brazilian river with fecal inputs from a nearby village (B). Signature oligotypes comprised >0.1% of the total community sequence reads, on average, for all sample types except clean stormwater and baseflow lake water samples. The oligotype identifiers given in the legend represent the abbreviated genus name (Fae, Faecalibacterium; Ros, Roseburia; Lac, unclassified Lachnospiraceae; Dor, Dorea; Bla, Blautia; Ali, Alistipes; Bac, Bacteroides) and the oligotype's rank in sewage (e.g., Bac1 is from the genus Bacteroides and was the most abundant of the sewage-preferred and sewage-specific oligotypes). Oligotypes are grouped by phylum and color coded by family, as follows: Bacteroidetes-Bacteroidaceae, blue; Rikenellaceae, cyan; Firmicutes-Lachnospiraceae, red; and Ruminococcaceae, brown.
DISCUSSION
Next-generation sequencing approaches to identifying alternative indicators.
Fecal indicators were formerly limited to organisms that could easily be grown and counted (2). The shift toward more molecular-based surveys has alleviated the need to utilize cultivable bacteria as indicators and allowed a focus on the difficult-to-grow but highly abundant anaerobes that dominate the microbiota of the vertebrate gut (9, 17, 44). The discovery of new genetic markers for MST is now limited only by the depth and breadth of the data used to create and test DNA-based tools (9, 28). This study sought to employ the basic steps typically used for the development of alternative indicators (44) by using in silico analysis of NGS data sets to identify candidate targets from multiple taxa capable of differentiating human from nonhuman fecal pollution. As microbiome studies grow more common, a wealth of data on fecal sources will become available to further facilitate the exploration and validation of new indicators. However, a framework is needed to evaluate sequence data for use in a signature approach or to create a single target endpoint or quantitative PCR assays.
We used oligotyping, a recently described method of sequence analysis (30), to move beyond characterizing taxonomic distributions and systematically assess sequences that mapped to the genera of human fecal bacteria abundantly present in sewage. Oligotyping is more refined than the simple identification of unique sequences because it groups sequences on the basis of high-entropy nucleotide positions alone, which reduces the error-driven inflation of diversity while retaining the resolution that is sensitive to even a single nucleotide difference between two populations of organisms at the marker gene level (30). As the phylogenetic heterogeneity in a resolved oligotype is minimal, its representative sequence would be more suitable for use in downstream analyses. Our recent analysis of fecal community profiles using this approach with sequences that mapped to the genus Blautia revealed a high degree of host preference and host specificity for both humans and other animals at the oligotype level (35). Another recent study by Menke et al. also demonstrated the efficacy of Blautia at distinguishing two free-ranging sympatric carnivore host species (45). Our current study expanded upon this effort and examined eight fecal taxa, in addition to Blautia (32, 35) and the widely studied Bacteroides (8, 13, 15, 16, 44), that are relatively unexplored in terms of source tracking to determine the potential of each of these taxa to provide novel targets for use as alternative indicators. Within each of the 10 taxa we found distribution patterns that can be described as host preferred and strictly host specific (35). Host-preferred organisms are those that are dominant in a host source but occur at low levels in other hosts, whereas strictly host-specific organisms are found in a single host. In general, we found that strictly host-specific organisms were slightly less abundant than host-preferred organisms. For the detection of suspected low-level contamination, indicator selection criteria could be adjusted to allow the detection of host-preferred oligotypes with a higher relative abundance for which the detection potential is greater but the specificity is lower.
The host-related structure within taxa differentiates fecal sources.
The bacterial genera analyzed in this study had a consistent presence and high relative abundance in the sewage samples evaluated in the present study as well as sewage samples characterized in previous studies (31, 33, 46). Although this study examined samples from a limited number of individual animals, the relative proportions of the major taxonomic groups determined from their community profiles were similar to those of cats and dogs (47), chickens (48), cows (49), and swine (50) described in previous microbiome studies. The compositions of animal fecal microbiomes were distinctly different from those of sewage, a proxy for human fecal pollution (31), as evidenced by both the proportions of the fecal genera present and the oligotypes within those genera. Oligotyping identified groups of sequences within each of the taxa that displayed ecological relevance, as demonstrated by reproducible associations with sewage versus a variety of animal host species.
Because sewage samples contain feces from many individuals, dominant oligotypes from this group appeared consistently throughout all samples, allowing us to clearly differentiate the signatures of human feces in sewage from those of common animal fecal sources. However, some of the oligotypes unique to sewage associated most strongly with organisms or cloned sequences from nonhuman origins, as was previously observed for select oligotypes abundant in sewage (31). The original source of these oligotypes in sewage is unclear, but they may represent organisms that selectively grow in sewage or nonfecal inputs from environmental contributions (terrestrial or aquatic bacteria). Despite their ubiquitous presence in sewage, we disregarded these oligotypes as potential indicators due to their lack of a traceable association with human feces and, therefore, the pathogen risk associated with sewage (6, 44).
Our recent analysis of the genus Blautia identified oligotypes preferentially or exclusively found in humans, cows, chickens, swine, and deer (35). In addition to Blautia, all nine of the other taxa analyzed in the current study contained sewage-specific oligotypes of human origin, and some were also relatively abundant within the community as a whole. In particular, Blautia, Bacteroides, and unclassified Lachnospiraceae contained sewage-specific marker genes. However, the observed similarities between the oligotypes from sewage and those from feces of swine, cats, and dogs suggest that any indicators developed should be tested thoroughly against feces from a wider selection of these hosts to ensure their specificity or high degree of preference for sewage. Although the differentiation of animal sources was not the focus of this study, similarities (shared oligotypes) were common among hosts with similar diets and/or physiologies, such as humans (sewage) and swine or deer and cows, as has been observed in other studies (10, 25, 45). This suggests that a more comprehensive sampling effort with respect to particular animals of interest would also yield host-specific oligotypes for tracking animal fecal pollution. We previously hypothesized that the adaptation of Blautia strains to different host gut environments may drive the fine-level differences in population structure and suggested that host specificity may reflect adaptation to subtle differences in host physiology or fulfillment of a keystone metabolic activity by certain organisms within a taxon (35). In turn, knowledge related to how these fundamental processes shape the fine-scale genetic structure within taxa may aid in determining better genetic markers more truly specific for different host groups. The emergence of host patterns in these data soundly supports the underlying hypothesis that drives microbial source tracking studies. Further sampling of feces from animals from a range of geographic locations and on various diets would allow additional fine-scale genetic analysis to identify mutualistic taxa that are common to animal hosts of interest.
Global applicability and validation of candidate indicators.
Differences in diet, geography, and many other factors can shape the fecal bacterial community in human populations (51–53). Of particular note for MST purposes is the relatively lower Bacteroidaceae:Prevotellaceae ratios in fecal samples from people with a non-Western diet (18, 53) and the higher abundances of Lachnospiraceae in those who eat a more plant-based diet (54). The currently used genetic markers may therefore be less effective in regions of the world where populations lack high proportions of Bacteroidaceae in their fecal communities (16, 18). We observed a shift from a Bacteroides-dominated signature to a Lachnospiraceae-dominated signature in U.S. versus Brazilian sewage and water samples, but the majority of our candidate indicators were present in Brazilian sewage, even though the ratios differed from those in U.S. sewage. Oligotype ratios within the fecal signature for Brazilian sewage did match well with those obtained by use of a composite data set for humans in Brazil (18), again highlighting the similarities between human and sewage fecal oligotypes and the geographic patterns of gut microbiomes on different continents (31, 32).
We observed a traceable signature of 20 abundant sewage-specific organisms in lake water following a CSO and in stormwater with evidence of sewage contamination (41), as previously determined by both the HF183 and Lachno2 qPCR assays. The cleaner stormwater samples showed evidence of low-level chronic fecal contamination common to urban stormwater (41), and the levels of fecal contamination were 2 orders of magnitude lower than those of the highly contaminated stormwater. These samples had a skewed ratio of the signature oligotypes, suggesting that some of the members in the signature may occur in urban wildlife, like squirrels, rabbits, and raccoons, or that select members of the suite of organisms used as the sewage signature persist in the environment. A similar pattern was observed in water from Lake Michigan, where water collected during a CSO contained the sewage signature near the limits of detection, but the ratios of signature oligotypes in the water correlated well with the ratios found in sanitary sewage. Tenfold lower levels of the sewage signature oligotypes were found in the lake during baseflow than during the CSO. Ratios of signature oligotypes from baseflow lake samples also had a lower correlation with sanitary sewage, again suggesting the persistence of some oligotypes that contribute to chronic sewage pollution or the presence of a nonhuman source of pollution (34).
Other sources of fecal pollution may contain some of the sewage signature organisms, but in this case the signature will no longer covary in the same way (i.e., the proportions will no longer be preserved) and the pollution signal will appear as mixed sources, as in the cleaner stormwater and lake samples. The occurrence of a mixture of sources rather than an effect of the aging of fecal pollution could be distinguished by the detection of fecal organisms not associated with the sewage signature in the sample. Additionally, the sewage signature might change as the pollution input ages with differential die-off (55). The distinctly different oligotype ratios in environments with known sewage contamination and in samples with low-level, background, or mixed fecal contamination highlight the need for the use of a multiple-indicator approach rather than an approach that relies on a single target.
The HF183 and Lachno2 genetic markers are present at high abundances in human fecal communities, sewage, and urban waterways; they are typically present at the same order of magnitude and are highly correlated (19). The organisms targeted by these genetic markers have V6 sequences identical to two highly abundant sewage-preferred oligotypes. The Lachno2 assay employs the V6 region, allowing a direct match to the corresponding V6 region of the oligotype. The V6 region corresponding to B. dorei was used as a proxy for the HF183-specific primer, which targets the V2 region (9). The HF183 assay is known to cross-react with fecal samples from chickens and dogs in particular (56), and the inferred V6 oligotype was found in our study in the feces of all animals except cows. However, as V6 is not the direct target of the HF183 assay, the V6 sequence from B. dorei may match those of other Bacteroides species as well. The results of in silico analysis from this study also showed that the oligotype corresponding to the Lachno2 assay was highly abundant in feces from dogs and cats and present at a low abundance in feces from other animals. In these cases, different organisms may be present in the feces of different hosts, which could be verified by obtaining full-length sequences from the populations present in the feces of these hosts. The crossover of the oligotypes that represent the organisms targeted by current assays with multiple animals highlights the pros and cons of using highly abundant but not strictly specific markers, as well as the trade-off of using short sequence tags to track populations, and confirms the need for additional alternative indicators to better resolve complex fecal pollution.
Multiple indicators allow the development of additional single-target assays or a signature approach based on the proportional abundances of multiple sequences from a suite of different taxa. Analysis of data sets using oligotyping provides the means to optimize the information contained within short sequences (30), and the depth of sequencing provided by NGS allows in silico evaluation of the sensitivity and specificity of potential targets (19). We highlight the idea of using a sewage signature, given the dropping costs of sequencing and advances in bioinformatics; algorithms could be developed to directly analyze sequence data for specific sequences in a range of predetermined proportions. As massive sequencing projects continue, establishment of a searchable database of bacterial marker genes could greatly facilitate development and testing of additional targets for MST for pollution from humans or from specific animals as well. Our methodology represents a promising approach to identifying candidate bacterial groups for MST applications; however, further analysis, such as targeted cloning of longer regions of the 16S rRNA gene, may be needed to translate host-specific oligotypes into usable assays. The findings presented here also independently confirm the utility of the genus Bacteroides in MST applications for several host groups, as determined by previous studies (11, 15), and further highlight the potential of the family Lachnospiraceae to provide human-specific markers (19, 32).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this project was provided by a National Institutes of Health grant (R01AI091829-01A1) to S.L.M.
We thank the Milwaukee Metropolitan Sewerage District and Veolia Water for providing sewage samples. We also thank Deb Dila and Danielle Cloutier for their comments on drafts of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01524-15.
REFERENCES
- 1.Yan T, Sadowsky MJ. 2007. Determining sources of fecal bacteria in waterways. Environ Monit Assess 129:97–106. doi: 10.1007/s10661-006-9426-z. [DOI] [PubMed] [Google Scholar]
- 2.Scott TM, Rose JB, Jenkins TM, Farrah SR, Lukasik J. 2002. Microbial source tracking: current methodology and future directions. Appl Environ Microbiol 68:5796–5803. doi: 10.1128/AEM.68.12.5796-5803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nnane DE, Ebdon JE, Taylor HD. 2011. Integrated analysis of water quality parameters for cost-effective faecal pollution management in river catchments. Water Res 45:2235–2246. doi: 10.1016/j.watres.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Leclerc H, Schwartzbrod L, Dei-Cas E. 2002. Microbial agents associated with waterborne diseases. Crit Rev Microbiol 28:371–409. doi: 10.1080/1040-840291046768. [DOI] [PubMed] [Google Scholar]
- 5.Santo Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res 41:3539–3552. [DOI] [PubMed] [Google Scholar]
- 6.Field KG, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 7.Zheng G, Yampara-Iquise H, Jones JE, Carson CA. 2009. Development of Faecalibacterium 16S rRNA gene marker for identification of human faeces. J Appl Microbiol 106:634–641. doi: 10.1111/j.1365-2672.2008.04037.x. [DOI] [PubMed] [Google Scholar]
- 8.Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst Appl Microbiol 33:348–357. doi: 10.1016/j.syapm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick LK, Bernhard AE, Brodeur TJ, Santo Domingo JW, Simpson JM, Walters SP, Field KG. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl Environ Microbiol 71:3184–3191. doi: 10.1128/AEM.71.6.3184-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res 41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 12.Krentz CA, Prystajecky N, Isaac-Renton J. 2013. Identification of fecal contamination sources in water using host-associated markers. Can J Microbiol 59:210–220. doi: 10.1139/cjm-2012-0618. [DOI] [PubMed] [Google Scholar]
- 13.Fogarty LR, Voytek MA. 2005. Comparison of Bacteroides-Prevotella 16S rRNA genetic markers for fecal samples from different animal species. Appl Environ Microbiol 71:5999–6007. doi: 10.1128/AEM.71.10.5999-6007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schriewer A, Miller WA, Byrne BA, Miller MA, Oates S, Conrad PA, Hardin D, Yang HH, Chouicha N, Melli A, Jessup D, Dominik C, Wuertz S. 2010. Presence of Bacteroidales as a predictor of pathogens in surface waters of the central California coast. Appl Environ Microbiol 76:5802–5814. doi: 10.1128/AEM.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fremaux B, Gritzfeld J, Boa T, Yost CK. 2009. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res 43:4838–4849. doi: 10.1016/j.watres.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 16.Reischer GH, Ebdon JE, Bauer JM, Schuster N, Ahmed W, Astrom J, Blanch AR, Bloeschl G, Byamukama D, Coakley T, Ferguson C, Goshu G, Ko G, de Roda Husman AM, Mushi D, Poma R, Pradhan B, Rajal V, Schade MA, Sommer R, Taylor H, Toth EM, Vrajmasu V, Wuertz S, Mach RL, Farnleitner AH. 2013. Performance characteristics of qPCR assays targeting human- and ruminant-associated Bacteroidetes for microbial source tracking across sixteen countries on six continents. Environ Sci Technol 47:8548–8556. doi: 10.1021/es304367t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreader CA. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol 61:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koskey AM, Fisher JC, Eren AM, Ponce-Terashima R, Reis MG, Blanton RE, McLellan SL. 2014. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep 6:696–704. doi: 10.1111/1758-2229.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton RJ, Vandewalle JL, Borchardt MA, Gorelick MH, McLellan SL. 2011. Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Appl Environ Microbiol 77:6972–6981. doi: 10.1128/AEM.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ufnar JA, Wang SY, Christiansen JM, Yampara-Iquise H, Carson CA, Ellender RD. 2006. Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J Appl Microbiol 101:44–52. doi: 10.1111/j.1365-2672.2006.02989.x. [DOI] [PubMed] [Google Scholar]
- 21.Lamendella R, Santo Domingo JW, Kelty C, Oerther DB. 2008. Bifidobacteria in feces and environmental waters. Appl Environ Microbiol 74:575–584. doi: 10.1128/AEM.01221-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston C, Byappanahalli MN, Gibson JM, Ufnar JA, Whitman RL, Stewart JR. 2013. Probabilistic analysis showing that a combination of Bacteroides and Methanobrevibacter source tracking markers is effective for identifying waters contaminated by human fecal pollution. Environ Sci Technol 47:13621–13628. doi: 10.1021/es403753k. [DOI] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, Corthier G, Furet JP. 2009. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methe BA, Huttenhower C. 2012. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol 10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unno T, Jang J, Han D, Kim JH, Sadowsky MJ, Kim OS, Chun J, Hur HG. 2010. Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Environ Sci Technol 44:7777–7782. doi: 10.1021/es101500z. [DOI] [PubMed] [Google Scholar]
- 28.McLellan SL, Eren AM. 2014. Discovering new indicators of fecal pollution. Trends Microbiol 22:697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claesson M, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:1–13. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. 2015. Sewage reflects the microbiomes of human populations. mBio 6(2):e02574–14. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLellan SL, Newton RJ, Vandewalle JL, Shanks OC, Huse SM, Eren AM, Sogin ML. 2013. Sewage reflects the distribution of human faecal Lachnospiraceae. Environ Microbiol 15:2213–2227. doi: 10.1111/1462-2920.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai L, Feng J, Zhang T. 2014. Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biotechnol 98:3317–3326. doi: 10.1007/s00253-013-5402-z. [DOI] [PubMed] [Google Scholar]
- 34.Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL. 2013. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb Ecol 65:1011–1023. doi: 10.1007/s00248-013-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eren A, Sogin M, Morrison H, Vineis J, Fisher J, Newton R, McLellan S. 2015. A single genus in the gut microbiome reflects host preference and specificity. ISME J 9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher J, Newton RJ, Dila DK, McLellan SL. 2015. Urban microbial ecology of a freshwater estuary of Lake Michigan. Elementa Sci Anthropocene 3:000064. doi: 10.12952/journal.elementa.000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eren AM, Vineis JH, Morrison HG, Sogin ML. 2013. A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks, p 361–362. In Proceedings of the Third International ICWSM Conference. AAAI Publications, Palo Alto, CA. [Google Scholar]
- 40.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLellan SL, Dila DK. 2014. Greater Milwaukee watersheds stormwater report. MMSD, Milwaukee, WI. http://home.freshwater.uwm.edu/mclellanlab/files/2014/05/2008-2012-SW-Report.pdf. [Google Scholar]
- 42.Development Core Team R. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 43.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H. 2013. vegan: community ecology package. R package version 2.0-8. R Foundation for Statistical Computing, Vienna, Austria: http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 44.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 45.Menke S, Wasimuddin Meier M, Melzheimer J, Mfune JKE, Heinrich S, Thalwitzer S, Wachter B, Sommer S. 2014. Oligotyping reveals differences between gut-microbiomes of free-ranging sympatric Namibian carnivores (Acinonyx jubatus, Canis mesomelas) on a bacterial species-like level. Front Microbiol 5:526. doi: 10.3389/fmicb.2014.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. 2013. Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 48.Yeoman C, Chia N, Jeraldo P, Sipos M, Goldenfeld ND, White B. 2012. The microbiome of the chicken gastrointestinal tract. Animal Health Res Rev 13:89–99. doi: 10.1017/S1466252312000138. [DOI] [PubMed] [Google Scholar]
- 49.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H, Borewicz K, White B, Singer R, Sreevatsan S, Tu Z, Isaacson R. 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol 153:124–133. doi: 10.1016/j.vetmic.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 51.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 52.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balleste E, Blanch AR. 2010. Persistence of Bacteroides species populations in a river as measured by molecular and culture techniques. Appl Environ Microbiol 76:7608–7616. doi: 10.1128/AEM.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, Shanks OC. 2014. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl Environ Microbiol 80:3086–3094. doi: 10.1128/AEM.04137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.