Abstract
Enterohemorrhagic Escherichia coli (EHEC) is the causative agent of bloody diarrhea and extraintestinal sequelae in humans, most importantly hemolytic-uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (TTP). Besides the bacteriophage-encoded Shiga toxin gene (stx), EHEC harbors the locus of enterocyte effacement (LEE), which confers the ability to cause attaching and effacing lesions. Currently, the vast majority of EHEC infections are caused by strains belonging to five O serogroups (the “big five”), which, in addition to O157, the most important, comprise O26, O103, O111, and O145. We hypothesize that these four non-O157 EHEC serotypes differ in their phylogenies. To test this hypothesis, we used multilocus sequence typing (MLST) to analyze a large collection of 250 isolates of these four O serogroups, which were isolated from diseased as well as healthy humans and cattle between 1952 and 2009. The majority of the EHEC isolates of O serogroups O26 and O111 clustered into one sequence type complex, STC29. Isolates of O103 clustered mainly in STC20, and most isolates of O145 were found within STC32. In addition to these EHEC strains, STC29 also included stx-negative E. coli strains, termed atypical enteropathogenic E. coli (aEPEC), yet another intestinal pathogenic E. coli group. The finding that aEPEC and EHEC isolates of non-O157 O serogroups share the same phylogeny suggests an ongoing microevolutionary scenario in which the phage-encoded Shiga toxin gene stx is transferred between aEPEC and EHEC. As a consequence, aEPEC strains of STC29 can be regarded as post- or pre-EHEC isolates. Therefore, STC29 incorporates phylogenetic information useful for unraveling the evolution of EHEC.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) has emerged as a serious public health concern, since it is responsible for a large number of food-borne outbreaks of diarrheal diseases in humans (1, 2). Bloody diarrhea, hemorrhagic colitis (HC), and the potentially fatal hemolytic-uremic syndrome (HUS) belong to a wide spectrum of disease presented by EHEC infections (2, 3). The common virulence trait defining EHEC strains is the production of Shiga toxin (Stx), which makes them a highly pathogenic subgroup of Stx-producing E. coli (STEC). The Stx-encoding gene (stx), uniquely defining the STEC pathotype, is encoded on a lambdoid bacteriophage that is integrated into the bacterial chromosome and functions as a mobile genetic element. Another important virulence factor in EHEC is a pathogenicity island designated the locus of enterocyte effacement (LEE), which encodes proteins causing the formation of attaching and effacing (A/E) lesions on intestinal epithelial cells through actin rearrangement (2, 3). While humans can develop serious illness when infected with EHEC, its main reservoir, cattle, remains symptomless when the gut is colonized with these bacteria (4). Other small ruminants, such as sheep or goats, and even wildlife ruminants, are also known to carry and excrete EHEC (5, 6). EHEC is transmitted to humans via the fecal-oral infection route, from animal feces to the environment, water, and food of animal and organic origin. The transmission from animals to humans, in addition to human-to-human transmission, marks EHEC as a zoonotic agent (3, 7).
EHEC of serotype O157:H7 has caused several distinct outbreaks in South America, Europe, Africa, North America, Japan, and China, and sporadic cases have been surveyed worldwide (8–12). Shortly after the first observation of an O157:H7 EHEC infection, other serotypes were also connected with diarrheal diseases in humans. Among the >150 non-O157 EHEC O serogroups, O26, O103, O111, and O145 occur most frequently in severe human diseases (3, 13). It is a challenge for clinicians to identify high-risk non-O157 serotypes, since the genetic basis that leads to the diverse clinical outcomes of diseases caused by this EHEC group remains unsolved (14). Comparison of genome sequences of O157, O26, O103, and O111 revealed high similarities within their whole gene repertoire and other factors specific for the EHEC pathotype. Nevertheless, independent acquisition of mobile genetic elements, such as pathogenicity islands (PAIs), bacteriophages, and plasmids, via horizontal gene transfer has been identified as the primary driving force for a parallel evolution that leads to different EHEC phylogenies (15). Phylogenetic analyses from several studies suggest shared ancestors or common lineages from which the different serotypes have evolved. For instance, a group consisting of O103:H11 isolates was found to have the same ancestor as EHEC O26:H11 when variants of two virulence genes (fliC and eae) were analyzed (16). Also, since O26:H11 and O111:H11 harbor highly similar PAI virulence profiles, it was suggested that H11 isolates are closely related (17). Furthermore, complete genome analyses of two O145 EHEC isolates demonstrated that they had a common EPEC ancestor with O157:H7 before they evolved further into another sublineage (18).
Another fact hinting at a more complex phylogeny than the arrangement in serogroups and evolutionary background of EHEC strains is the frequent isolation of so-called atypical enteropathogenic E. coli (aEPEC) strains, yet another subgroup of intestinal pathogenic E. coli, which also belong to the same non-O157 O serogroups as EHEC strains (19, 20). aEPEC strains do not harbor the stx-converting bacteriophage but are similar to EHEC strains capable of the A/E lesion formation caused by the LEE. These strains are classified as “atypical” EPEC because they lack the EAF plasmid, an adherence factor plasmid present in EPEC strains (2). aEPEC is a common cause of nonbloody diarrhea in infants but is also frequently isolated from patients with bloody diarrhea and HUS, as well as from ruminants (21). aEPEC strains are thought to have developed during human infection via loss of the stx gene and were therefore designated EHEC-LST (EHEC that lost Stx) (22); they may convert back to EHEC upon the acquisition of stx genes (23, 24).
In this study, the gold standard method multilocus sequence typing (MLST) was used on randomly chosen O26, O103, O111, and O145 E. coli strains in order to gain insight into their phylogeny. We tested the hypothesis that these four non-O157 O-serogroups are phylogenetically related.
Interestingly, our results showed that more than 40% of the isolates examined were assigned to sequence types (STs) that form the ST complex (STC) STC29. In addition, several aEPEC strains from our strain collection also clustered within STC29, proving their phylogenetic similarity.
MATERIALS AND METHODS
Bacterial isolates.
A total of 250 Escherichia coli isolates were investigated. Most of the bovine E. coli strains of O serogroups O26, O103, O111, and O145 were chosen from strain collections of the Institute of Microbiology and Epizootics (IMT), Freie Universität, Berlin, Berlin, Germany, as well as the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (25). The human isolates of the relevant non-O157 O serogroups were sampled within an exploratory clinical study of the “FBI-Zoo” research consortium (www.fbi-zoo.de). In that study, stool samples of patients with diarrhea were screened for STEC/EHEC and for Yersinia, Salmonella, and Campylobacter spp. at the University Hospital Münster, the Hannover Medical School, and the Max von Pettenkofer-Institute for Hygiene and Medical Microbiology (Ludwig-Maximilians-Universität München).
In addition, 14 isolates of the HUS-associated EHEC (HUSEC) collection (13) assigned to the non-O157 O serogroups O26, O103, O111, and O145 were included. Therefore, the greater part of the E. coli isolates under investigation were sampled in Germany. Isolates obtained from other E. coli studies, such as studies conducted in Australia, Canada, the United States, and European countries such as Sweden, the United Kingdom, and Belgium, were also included in the study. The strains under investigation had been sampled between the years 1952 and 2009. The strains were isolated mainly from cattle and humans, but three isolates from sheep and one from a food source were also included in the analysis. After serotyping, 76 isolates were assigned to O serogroup O26 (NM, H11, H19, K60), 96 isolates to O103 (NM, H2, H3, H11, H18, H21, H25, H31, H43), 36 isolates to O111 (NM, H2, H8, H10), and 42 isolates to O145 (NM, H18, H25, H28, H34). Furthermore, 41 aEPEC isolates, of human (n = 10), bovine (n = 28), and ovine (n = 2) origin, as well as one isolate from a food source (n = 1), were included in our analysis. The isolates belonged to the non-O157 O serogroups and clustered within STs of STC29 (see Table S1 in the supplemental material).
Serotyping.
Serotyping was performed using a microtiter method with antisera against E. coli O antigens 1 to 182 and against H antigens 1 to 56 according to the method of Prager et al. (26).
Pathotyping.
Apart from the clinical data, the pathotypes of individual strains were determined on the basis of the presence or absence of the virulence genes stx1, stx2, escV, and bfpB, which were identified by PCR using published primer pairs and multiplex PCR protocols (27). LEE-positive strains (aEPEC, EHEC) were identified by the detection of escV, a LEE-located translocator gene. In addition, we also screened for the intimin-encoding gene eae to confirm the presence of the LEE (28). aEPEC strains do not harbor the EAF plasmid and therefore are bfpB negative.
The stx genes of strains positive for any stx gene were subtyped according to the work of Scheutz et al. (2012) by using the published primers and PCR protocols (29) (see also Fig. S2 in the supplemental material).
MLST.
Multilocus sequence typing (MLST) was performed by analyzing internal fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) (25). The alleles and sequence types (STs) were assigned in accordance with the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) using Ridom SeqSphere software (Ridom GmbH, Münster, Germany) in order to generate a database. The genetic relationships between different STs were determined on the basis of the allele profiles obtained using Bionumerics, version 7.1 (Applied Maths, Sint-Martens-Latem, Belgium). Bionumerics was also used to construct minimum spanning trees (MSTs).
Statistical analysis.
Host, O type, H type, and serotype information for isolates was summarized by sequence type complex and was statistically analyzed for STC10, STC20, STC29, and STC32 with multinomial logistic regression using IBM SPSS Statistics software (version 20). The results are shown in Table S2 in the supplemental material, where significant P values are highlighted.
RESULTS
Two-hundred fifty STEC strains belonging to O serogroups O26, O103, O111, and O145, of different origins, including human, animal, and food sources, were screened for typical virulence-associated genes using published primer pairs.
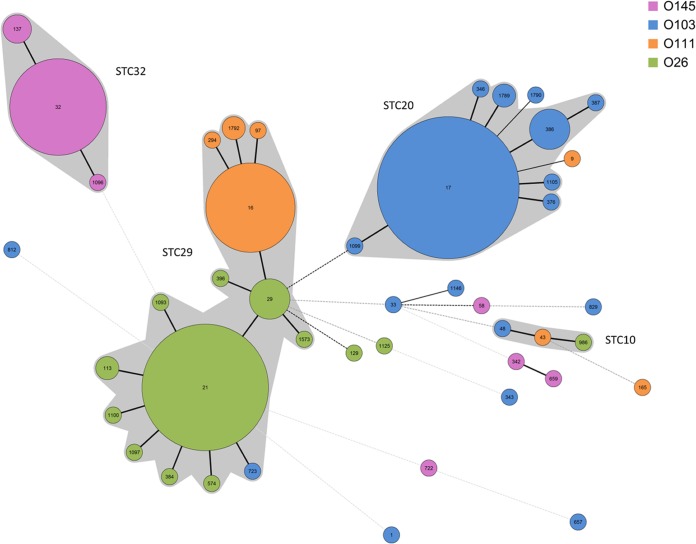
Next, we analyzed the non-O157 isolates using MLST. The resulting phylogenetic relationships of the isolate population are displayed in a minimum spanning tree (MST) (Fig. 1). The vast majority of isolates (n = 234 [93.6%]) were assigned to four different ST complexes (STCs): STC10, STC20, STC29, and STC32. STC29 comprised 42.8% (n = 107) of the isolates and 15 STs. Within STC29, ST16, ST21, and ST29 were the major STs, incorporating the largest number of isolates: a total of 93 (86.9%). ST21, ST29, and the majority of their single-locus variants (slv's) belonged to O serogroup O26 (Fig. 1), whereas ST16 and its slv's (ST97, ST1792, ST294) belonged to O serogroup O111 (NM, H2, H8) and comprised a total of 33 isolates. Only three O111 isolates belonged to STs unrelated to STC29; these were ST9 (O111:H2; isolated from cattle), ST165 (O111:K58; isolated from a wild rabbit with enteritis), and ST43 (STC10; O111:H10; isolated from human with HUS [strain HUSEC001]) (Fig. 1). Three O26 isolates showed a more distant relation to STC29. An isolate typed as ST129 (O26:H not typed; isolated from cattle) differed in three alleles from ST29; one typed as ST1125 (O26:H19; isolated from cattle) differed in four alleles from ST29; and the O26 isolate typed as ST986 (O26:H11; isolated from cattle) clustered within STC10. Only a single isolate of serotype O103:H11 was located within STC29; it was typed as ST723. Besides the clustering of O103 (NM, H2, H3) isolates within STC20, single isolates of this O serogroup had a variety of H antigens (such as H2, H18, H21, H25, H31, and H43) and MLST types. The same is true for O145 (NM, H28), where the majority of isolates clustered within STC32, but a minority of isolates clustered in distant, unrelated STs. The flagellar H antigens of these isolates belonging to distant STs were H18, H25, and H34, and one was nonmotile (see Fig. S1 in the supplemental material).
FIG 1.
Minimum spanning tree (MST) calculated with Bionumerics, version 7.1 (Applied Maths), displaying the population structure of 250 Shiga toxin-harboring Escherichia coli isolates based on allele sequence combinations of the adk, fumC, gyrB, icd, mdh, purA, and recA genes. The O serotypes of the isolates are color coded. Bold lines between the STs indicate a distinction in one allele; dotted lines indicate differences in more than one allele. The size of each circle representing an ST is scaled to the number of isolates included. Four main sequence type complexes (STCs) can be recognized, namely, STC29 (107 isolates), STC20 (86 isolates) STC32 (38 isolates), and STC10 (3 isolates), leaving just 16 isolates (6.4%) unassigned to these STCs.
An in silico analysis of the relationship between O serogroups O26 and O111 did not show a close relatedness of the respective O antigens (Geneious alignment; Geneious, version 7.0.2). The O-antigen gene clusters of O111 (GenBank accession no. AF078736) and O26 (GenBank accession no. AF529080) revealed an identity of only 45% (data not shown). Furthermore, in the Escherichia coli O-antigen Database (EcoDAB; http://nevyn.organ.su.se/ECODAB/), only a high similarity with a significant E value of 2e−66 between glycosyltransferases WdbH of O111 and WbtG of O103 is indicated, whereas for glycosyltransferases of O111 and O26, no similarity at all is mentioned.
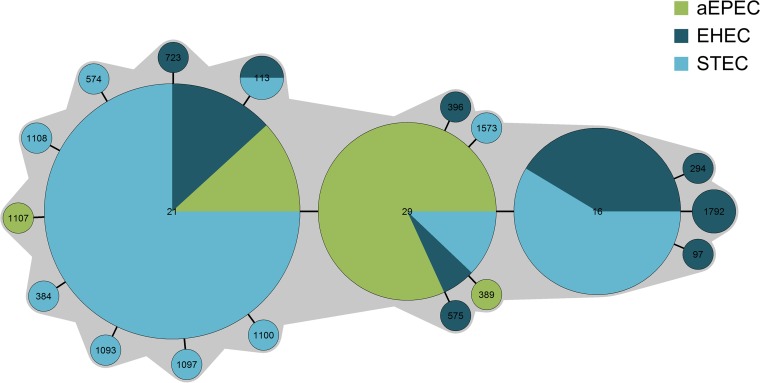
The unexpected clustering of mainly EHEC isolates from O serogroups O26 and O111 in STC29 prompted us to analyze atypical EPEC strains from these serotypes as well. Of a total of 41 aEPEC strains included in our analysis, we identified human (n = 10), bovine (n = 28), ovine (n = 2), and food (n = 1) isolates that belonged to the non-O157 O serogroups investigated and clustered into STC29. The majority (37 isolates [90.2%]) shared the STs with the EHEC/STEC isolates; only four additional STs occurred as slv's of ST21 or ST29: ST389, ST575, ST1107, and ST1108 (Fig. 2).
FIG 2.
MST displaying the population structure of 148 E. coli isolates belonging to pathotypes STEC, EHEC, and aEPEC and assigned to sequence type complex STC29. The distribution of the pathotypes of isolates is illustrated by different colors.
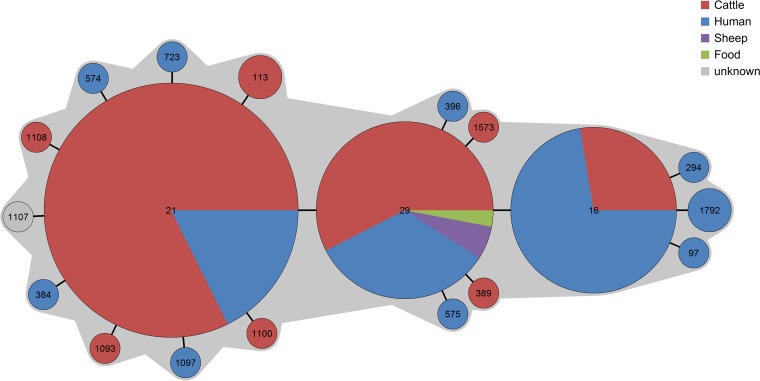
Analysis of the origins of the 148 isolates of STC29 demonstrated no clear association between origin and ST, since the three most common STs (ST16, ST21, ST29) were found in humans as well as in cattle. Because isolates from food or sheep were quite rare, no valid conclusion could be drawn here (Fig. 3).
FIG 3.
MST showing the population structure of 148 E. coli isolates assigned to sequence type complex STC29. This graph illustrates the origin of each isolate from different hosts as well as from food.
DISCUSSION
Several non-O157 O serogroups are important causes of diarrheal diseases producing clinical symptoms of similar severity to those for serotype O157:H7. The most frequent non-O157 causative agents belong to O serogroups O26, O103, O111, and O145 (30), which are capable of causing significant diarrheal outbreaks (31–34). Since they were first associated with human disease, the detection rate of non-O157 strains in diarrheal diseases has increased. This could be due to better detection techniques and increased awareness of the non-O157 serogroup-associated risk of infection, as well as to a higher frequency of these pathogens occurring in human and animal infections (11, 35, 36). In this study, we analyzed a collection of 250 non-O157 STEC and EHEC isolates by use of MLST. The MLST results revealed a close relationship of the two most prevalent non-O157 EHEC O serogroups, O26 and O111, since they clustered together within a single STC, STC29. This was also published for three EHEC isolates of O26:H11 that clustered together with three EPEC isolates of serotype O111:H8 in distinct STs of STC29 (25).
The common classification of E. coli isolates into different O serogroups is thought to indicate a distant relationship of these isolates. Thus, an O serogroup would also suggest a detached phylogeny of this lineage with accompanying differences in its biological characteristics. The divergence in the O-antigen genes of O111 and O26 could be due to lateral gene transfer of the rfb-like region, which caused the evolution of O157:H7 isolates from a common O55:H7-like ancestor, where this region is not present (37). Comparable recombination events of the O-antigen coding regions could be responsible for the evolution of EHEC and aEPEC strains of O serogroups O26 and O111 from a common ancestor, explaining the differences in the somatic antigen. The usage of O serogroups as descriptive indicators for pathogenicity results in distinct phylogenetic lineages, although the isolates investigated might be polyphyletic.
In contrast, phylogenetic analyses of the flagellar H antigens described them as monophyletic and revealed a close relatedness of O111:H11 and O26:H11 strains in a parsimony phylogenetic tree based on genome-wide single nucleotide polymorphisms (SNPs) (38). This led to the hypothesis that O26:H11 evolved from an ancestral O111:H11 strain by an antigenic shift from O111 to O26 and generally that STEC strains with the same H antigens might share an ancestor (16). In our study, the genes encoding flagellar H antigens showed a similar result in the MLST as the O antigens. Those isolates that were distributed outside the major STCs were assigned to various H types (H10, H19, H11, H18, H21, H25, H31, H34, and H43), which fits their designation as distinct STs (see Fig. S1 in the supplemental material). This corroborates the finding mentioned above that typing of H antigens is more informative than O serotyping (16). Therefore, analysis based on the O:H serotype reflects the phylogeny more properly.
The phylogenetic relatedness represented by STC29 gives new insights into the evolutionary scenario of non-O157 STEC and EHEC strains above and beyond those obtained by analysis of the O and H surface antigens. Furthermore, MLST population analysis of isolates of human and animal origins have revealed independent evolution of pathogenic E. coli isolates due to vast amounts of homologous recombination and allelic variations in the lineages examined (25, 39). Therefore, the HUSEC collection compiled by Karch and coworkers (13) includes strains of different non-O157 serogroups and different STs. Among the 42 isolates in the HUSEC collection, 10 were assigned to STs in STC29, indicating the importance of STC29 and the isolates it harbors.
By exploring the characteristics of the STC29 isolates, we found that isolates originating from human and animal hosts cluster together within single STs (Fig. 3). Some EHEC serotypes are thought to be more host adapted, because they are isolated more frequently from certain species than from others. For instance, serotypes O5:NM, O6:H10, O91:NM, O128:H2, and O146:H8/H21 occur often among isolates from sheep, whereas serotypes O22:H8, O26:H11, O91:H21, O113:H4/H21, O118:H16, and OX3:H2/H21 are most frequently found in cattle, even if these animals have been kept together in pens (40, 41).
In contrast to these studies, we could demonstrate that there is no clear-cut grouping of certain STs with certain hosts but rather that isolates recovered from humans are also recovered from cattle, sheep, or food. We conclude that these isolates pose a particular zoonotic risk.
Another result supporting this suggestion is the pathotype distribution within STC29 (Fig. 2). The isolates capable of producing Shiga toxin (EHEC and STEC) cluster together with stx-negative isolates that harbor the LEE PAI (aEPEC) and therefore illustrate the close relationship of these pathotypes. aEPEC strains share virulence factors, including the LEE-encoded type III secretion system, with EHEC strains. Neither of these pathotypes harbors the EAF plasmid, which is responsible for the development of a bundle-forming pilus that is characteristic of typical EPECs. Nevertheless, only STEC and EHEC are capable of Stx production, since they are lysogenized with lambdoid stx-converting bacteriophages. Bacteriophages, including the lambdoid stx-converting bacteriophage, are mobile genetic elements that belong to the flexible gene pool of microorganisms such as E. coli and contribute to their adaptation under special circumstances. Deletion and insertion of horizontally transferred genes contribute to the development of new pathotypes or modify existing characteristics (42). For O26 isolates, the concept of interconversion between O26 aEPEC and O26 EHEC has been suggested by the loss, as well as the gain, of stx prophages through lysogenic conversion (23). stx-negative isolates of EHEC O serogroups have been found in follow-up checks of patients previously diagnosed with EHEC infections. Since these isolates shared a range of non-stx virulence genes and the phylogenetic background with EHEC isolates, these stx-negative isolates were designated EHEC strains that had lost their Shiga toxin (EHEC-LST) (22). Anjum et al. demonstrated in 2003, by comparative genomic indexing, that 87% to 94% of the coding sequence was conserved between stx-negative and stx-positive O26 E. coli strains, indicating greater genetic homogeneity within this O serogroup than previously proposed (43). In another study, 12 ruminant as well as 27 human O26:H11/NM isolates were analyzed by MLST and were identified either as ST21 (16 human isolates, 9 ruminant isolates) or as ST29 (11 human isolates, 3 ruminant isolates). Interestingly, nine bovine isolates of ST21 were identified as aEPEC, and one stx2a-positive bovine isolate of ST29 showed virulence characteristics of the emerging human-pathogenic clone, hinting at interconversion between stx-negative O26 and STEC strains and the emergence of new STEC clones with human-pathogenic potential in cattle (44).
Whole-genome sequencing of aEPEC and EHEC isolates of STC29 will help verify the homogeneity of these pathotypes and further determine the bacteriophage integration sites. Furthermore, whole-genome data of a considerable number of non-O157 isolates sampled over a definite time period will allow long-term studies that might unravel whether aEPEC strains have lost the stx phage (EHEC-LST) or have been transformed to EHEC strains via lysogenic conversion.
Applying the concept of bidirectional conversion (the possible loss and gain of mobile genetic elements) (24) to the isolates within STC29, we can hypothesize that aEPEC strains might function as pre-EHEC strains that again integrate the stx prophage into their genomes. On the other hand, aEPEC strains may also be EHEC-LST strains, which have lost the stx bacteriophage and, simultaneously, the ability to secrete Stx. This has to be proven in further lysogenic conversion experiments, lysogenizing aEPEC strains of STC29 with stx-converting bacteriophages.
In conclusion, STC29 incorporates highly virulent non-O157 EHEC and aEPEC O serogroups. Thus, STC29 is a highly interesting group with which to analyze the microevolution of EHEC pathogens. Future genomic as well as functional studies will provide greater insight into the evolutionary mechanisms shaping isolates of this phylogenetic lineage into important human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the BMBF network “FBI-Zoo” (Food-Borne Zoonotic Infections of Humans, grant 01KI1012A) and by grant GRK1673/1 from the German Research Foundation (DFG).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01921-15.
REFERENCES
- 1.Levine MM. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis 155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karch H, Mellmann A, Bielaszewska M. 2009. Epidemiology and pathogenesis of enterohaemorrhagic Escherichia coli. Berl Munch Tierarztl Wochenschr 122:417–424. [PubMed] [Google Scholar]
- 4.Lowe RM, Baines D, Selinger LB, Thomas JE, McAllister TA, Sharma R. 2009. Escherichia coli O157:H7 strain origin, lineage, and Shiga toxin 2 expression affect colonization of cattle. Appl Environ Microbiol 75:5074–5081. doi: 10.1128/AEM.00391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofer E, Cernela N, Stephan R. 2012. Shiga toxin subtypes associated with Shiga toxin-producing Escherichia coli strains isolated from red deer, roe deer, chamois, and ibex. Foodborne Pathog Dis 9:792–795. doi: 10.1089/fpd.2012.1156. [DOI] [PubMed] [Google Scholar]
- 6.Zweifel C, Blanco JE, Blanco M, Blanco J, Stephan R. 2004. Serotypes and virulence genes of ovine non-O157 Shiga toxin-producing Escherichia coli in Switzerland. Int J Food Microbiol 95:19–27. doi: 10.1016/j.ijfoodmicro.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Beutin L. 2006. Emerging enterohaemorrhagic Escherichia coli, causes and effects of the rise of a human pathogen. J Vet Med B Infect Dis Vet Public Health 53:299–305. doi: 10.1111/j.1439-0450.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- 8.Akashi S, Joh K, Mori T, Tsuji A, Ito H, Hoshi H, Hayakawa T, Ihara J, Abe T, Hatori M, Nakamura T, Akashi S. 1994. A severe outbreak of haemorrhagic colitis and haemolytic uraemic syndrome associated with Escherichia coli O157:H7 in Japan. Eur J Pediatr 153:650–655. doi: 10.1007/BF02190685. [DOI] [PubMed] [Google Scholar]
- 9.Cole D, Griffin PM, Fullerton KE, Ayers T, Smith K, Ingram LA, Kissler B, Hoekstra RM. 2014. Attributing sporadic and outbreak-associated infections to sources: blending epidemiological data. Epidemiol Infect 142:295–302. doi: 10.1017/S0950268813000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Effler E, Isaacson M, Arntzen L, Heenan R, Canter P, Barrett T, Lee L, Mambo C, Levine W, Zaidi A, Griffin PM. 2001. Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerg Infect Dis 7:812–819. doi: 10.3201/eid0705.017507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gormley FJ, Little CL, Rawal N, Gillespie IA, Lebaigue S, Adak GK. 2011. A 17-year review of foodborne outbreaks: describing the continuing decline in England and Wales (1992–2008). Epidemiol Infect 139:688–699. doi: 10.1017/S0950268810001858. [DOI] [PubMed] [Google Scholar]
- 12.Xiong Y, Wang P, Lan R, Ye C, Wang H, Ren J, Jing H, Wang Y, Zhou Z, Bai X, Cui Z, Luo X, Zhao A, Zhang S, Sun H, Wang L, Xu J. 2012. A novel Escherichia coli O157:H7 clone causing a major hemolytic uremic syndrome outbreak in China. PLoS One 7:e36144. doi: 10.1371/journal.pone.0036144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellmann A, Bielaszewska M, Kock R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg Infect Dis 14:1287–1290. doi: 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombes BK, Gilmour MW, Goodman CD. 2011. The evolution of virulence in non-O157 Shiga toxin-producing Escherichia coli. Front Microbiol 2:90. doi: 10.3389/fmicb.2011.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iguchi A, Iyoda S, Ohnishi M. 2012. Molecular characterization reveals three distinct clonal groups among clinical Shiga toxin-producing Escherichia coli strains of serogroup O103. J Clin Microbiol 50:2894–2900. doi: 10.1128/JCM.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ju W, Rump L, Toro M, Shen J, Cao G, Zhao S, Meng J. 2014. Pathogenicity islands in Shiga toxin-producing Escherichia coli O26, O103, and O111 isolates from humans and animals. Foodborne Pathog Dis 11:342–345. doi: 10.1089/fpd.2013.1696. [DOI] [PubMed] [Google Scholar]
- 18.Cooper KK, Mandrell RE, Louie JW, Korlach J, Clark TA, Parker CT, Huynh S, Chain PS, Ahmed S, Carter MQ. 2014. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genomics 15:17. doi: 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokete TN, Whittam TS, Wilson RA, Clausen CR, O'Callahan CM, Moseley SL, Fritsche TR, Tarr PI. 1997. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis 175:1382–1389. doi: 10.1086/516470. [DOI] [PubMed] [Google Scholar]
- 20.Eklund M, Bielaszewska M, Nakari UM, Karch H, Siitonen A. 2006. Molecular and phenotypic profiling of sorbitol-fermenting Escherichia coli O157:H− human isolates from Finland. Clin Microbiol Infect 12:634–641. doi: 10.1111/j.1469-0691.2006.01478.x. [DOI] [PubMed] [Google Scholar]
- 21.Wieler LH, Sobjinski G, Schlapp T, Failing K, Weiss R, Menge C, Baljer G. 2007. Longitudinal prevalence study of diarrheagenic Escherichia coli in dairy calves. Berl Munch Tierarztl Wochenschr 120:296–306. [PubMed] [Google Scholar]
- 22.Bielaszewska M, Kock R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. doi: 10.1371/journal.pone.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bielaszewska M, Prager R, Kock R, Mellmann A, Zhang W, Tschape H, Tarr PI, Karch H. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol 73:3144–3150. doi: 10.1128/AEM.02937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellmann A, Lu S, Karch H, Xu JG, Harmsen D, Schmidt MA, Bielaszewska M. 2008. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol 74:67–72. doi: 10.1128/AEM.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prager R, Strutz U, Fruth A, Tschape H. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int J Med Microbiol 292:477–486. doi: 10.1078/1438-4221-00226. [DOI] [PubMed] [Google Scholar]
- 27.Müller D, Greune L, Heusipp G, Karch H, Fruth A, Tschape H, Schmidt MA. 2007. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 73:3380–3390. doi: 10.1128/AEM.02855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedrich AW, Borell J, Bielaszewska M, Fruth A, Tschape H, Karch H. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J Clin Microbiol 41:2448–2453. doi: 10.1128/JCM.41.6.2448-2453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielaszewska M, Karch H. 2000. Non-O157:H7 Shiga toxin (verocytotoxin)-producing Escherichia coli strains: epidemiological significance and microbiological diagnosis. World J Microbiol Biotechnol 16:711–718. doi: 10.1023/A:1008972605514. [DOI] [Google Scholar]
- 31.Bielaszewska M, Zhang W, Mellmann A, Karch H. 2007. Enterohaemorrhagic Escherichia coli O26:H11/H−: a human pathogen in emergence. Berl Munch Tierarztl Wochenschr 120:279–287. [PubMed] [Google Scholar]
- 32.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 33.Mariani-Kurkdjian P, Denamur E, Milon A, Picard B, Cave H, Lambert-Zechovsky N, Loirat C, Goullet P, Sansonetti PJ, Elion J. 1993. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J Clin Microbiol 31:296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor EV, Nguyen TA, Machesky KD, Koch E, Sotir MJ, Bohm SR, Folster JP, Bokanyi R, Kupper A, Bidol SA, Emanuel A, Arends KD, Johnson SA, Dunn J, Stroika S, Patel MK, Williams I. 2013. Multistate outbreak of Escherichia coli O145 infections associated with romaine lettuce consumption, 2010. J Food Prot 76:939–944. doi: 10.4315/0362-028X.JFP-12-503. [DOI] [PubMed] [Google Scholar]
- 35.Bettelheim KA. 2003. Non-O157 verotoxin-producing Escherichia coli: a problem, paradox, and paradigm. Exp Biol Med (Maywood) 228:333–344. [DOI] [PubMed] [Google Scholar]
- 36.Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH, White PL, Wymore K, Gierke RE, Mahon BE, Griffin PM. 2013. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000–2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis 10:453–460. doi: 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 37.Feng P, Lampel KA, Karch H, Whittam TS. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis 177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 38.Ju W, Cao G, Rump L, Strain E, Luo Y, Timme R, Allard M, Zhao S, Brown E, Meng J. 2012. Phylogenetic analysis of non-O157 Shiga toxin-producing Escherichia coli strains by whole-genome sequencing. J Clin Microbiol 50:4123–4127. doi: 10.1128/JCM.02262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 40.Urdahl AM, Beutin L, Skjerve E, Zimmermann S, Wasteson Y. 2003. Animal host associated differences in Shiga toxin-producing Escherichia coli isolated from sheep and cattle on the same farm. J Appl Microbiol 95:92–101. doi: 10.1046/j.1365-2672.2003.01964.x. [DOI] [PubMed] [Google Scholar]
- 41.Wieler LH, Busse B, Steinruck H, Beutin L, Weber A, Karch H, Baljer G. 2000. Enterohemorrhagic Escherichia coli (EHEC) strains of serogroup O118 display three distinctive clonal groups of EHEC pathogens. J Clin Microbiol 38:2162–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellmann A, Bielaszewska M, Karch H. 2009. Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology 136:1925–1938. doi: 10.1053/j.gastro.2008.12.072. [DOI] [PubMed] [Google Scholar]
- 43.Anjum MF, Lucchini S, Thompson A, Hinton JC, Woodward MJ. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect Immun 71:4674–4683. doi: 10.1128/IAI.71.8.4674-4683.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zweifel C, Cernela N, Stephan R. 2013. Detection of the emerging Shiga toxin-producing Escherichia coli O26:H11/H− sequence type 29 (ST29) clone in human patients and healthy cattle in Switzerland. Appl Environ Microbiol 79:5411–5413. doi: 10.1128/AEM.01728-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.