Abstract
Corals harbor diverse bacterial associations that contribute to the health of the host. Using 16S rRNA pyrosequencing, we compared the bacterial communities of red and orange morphs of the Hawaiian coral Montipora capitata. Although both color morphs shared dominant bacterial genera, weighted and unweighted UniFrac analyses showed distinct bacterial communities. A single operational taxonomic unit (OTU), classified as Vibrio, represented the largest driver of differences between the color morphs. This OTU comprised 35.4% (±5.5%) of the orange morph bacterial community yet comprised 1.1% (±0.6%) of the red morph bacterial community. Cultivable bacteria from the two color morphs were also compared and tested for antibacterial activity. Cultured isolates represented 14 genera (7% of the total genera identified from sequencing data), and all but two cultured isolates had a matching OTU from the sequencing data. Half of the isolates tested (8 out of 16) displayed antibacterial activity against other cultured isolates but not against two known bacterial pathogens of M. capitata. The results from this study demonstrate that the specificity of coral-bacterial associations extends beyond the level of coral species. In addition, culture-dependent methods captured bacterial diversity that was representative of both rare and abundant members of the associated bacterial community, as characterized by culture-independent methods.
INTRODUCTION
Scleractinian (reef-building) corals thrive in the oligotrophic waters of the tropics due to the symbiotic relationship of the coral animal with photosynthetic, single-celled algae (genus Symbiodinium), which reside within the coral's tissues. Symbiodinium contributes significantly to coral nutrition by providing up to 90% of the coral's daily energy needs (1). In addition, corals harbor a diversity of microorganisms, including bacteria, archaea, and fungi. Microbes inhabit various coral niches, including the surface mucus layer (2), tissue layers (3), and the coral skeleton (4). Coral-associated bacteria are distinct from those in the surrounding seawater and marine sediment (5–7) and have been shown to be species specific within similar environments (8, 9).
The importance of microbes in host health is recognized in a multitude of organisms from both terrestrial and marine ecosystems, including corals. Coral-associated bacteria are thought to contribute to coral nutrition through the biogeochemical cycling of nitrogen, carbon, and sulfur compounds (10–15) and contribute to coral health by acting as the primary defense against pathogen invasion of host tissues (16). Bacteria on corals are hypothesized to act as an ecological and/or a physical barrier against pathogen entry (8) and to produce antibacterial compounds that prevent the dominance of a single, potentially pathogenic bacterium (17). Disease has emerged as a worldwide threat to coral reef ecosystems and is predicted to increase over time (18), so an understanding of factors that contribute to coral defenses, such as associated bacterial communities, is of interest.
Montipora capitata is a common Hawaiian reef coral that exists in two color morphs, red and orange (Fig. 1), which harbor different dominant clades of Symbiodinium (19). Both color morphs occur in the same environment on fringing reefs in Kaneohe Bay, Oahu, HI, and so we were able to compare the bacterial communities of these two color morphs without the influence of environment. Our objective was to characterize and compare the compositions of bacterial communities associated with the two color morphs of M. capitata using culture-independent and culture-dependent methods. In addition, we examined the antibacterial activity of cultured isolates to determine if antagonistic interactions are present in M. capitata bacterial communities and to determine if any were capable of inhibiting known bacterial pathogens of M. capitata. Using culture-independent methods, we found that although red and orange morphs shared dominant bacterial genera, UniFrac analysis showed that each color morph had distinct bacterial communities. Bacterial communities of orange morphs were more variable than those of red morphs (low evenness) and were dominated by Vibrio species. Cultured bacterial isolates from both color morphs displayed selective antibacterial activity and were representative of both rare and abundant genera found in the overall bacterial community analysis.
FIG 1.
Color morphologies of Montipora capitata. Two neighboring colonies of M. capitata display red (right colony) and orange (left colony) coloration.
MATERIALS AND METHODS
Sample collection and processing.
Fragments of M. capitata (n = 5 for the red morph; n = 5 for the orange morph) were collected at depths of 1 to 3 m on a single fringing reef from Moku O Loe, Kaneohe Bay, Oahu, HI. Coral fragments were placed into individual bags at depth and brought to the laboratory for immediate processing. Coral fragments were washed with 0.2-μm-filtered seawater (FSW) to remove loosely attached microbes and then crushed in 10 ml FSW with a mortar and pestle (“coral crushate”). The homogenized coral crushate was immediately used for DNA extraction and culturing of bacteria. Coral fragments were collected under special activity permit SAP2011-68 from the Hawaii Department of Aquatic Resources.
DNA sequencing and processing of data.
To compare the bacterial communities of red and orange fragments of M. capitata, total DNA from the coral crushate was extracted by using the PowerSoil DNA extraction kit (MoBio Laboratories, Inc.) according to the manufacturer's instructions, with one exception: the lysis step was modified by increasing the heat lysis incubation period to 15 min with 1 min of vortexing every 5 min. The V3 hypervariable region of the 16S rRNA gene was targeted by using universal bacterial primers 341F and 534R (20) and was amplified by using a bar-coded primer set constructed for pyrosequencing (see Table S1 in the supplemental material). To verify that amplification was not due to DNA present in the FSW used to create the coral crushate, FSW was also treated with the PowerSoil DNA extraction kit and subjected to 16S rRNA gene amplification using one of the bar-coded primer sets. Filtered seawater failed to give PCR products and thus did not contribute to bacterial communities in this study. PCR products from the coral crushate were visualized on 2% agarose—TAE (Tris-acetate-EDTA) gels and purified by using the QIAquick gel extraction kit (Qiagen, Inc.). Purified PCR products were submitted to the Advanced Studies of Genomics, Proteomics, and Bioinformatics sequencing facility at the University of Hawaii at Manoa for high-throughput sequencing on a Roche 454 next-generation GSFLX system.
Low-quality sequences and sequences <150 nucleotides (nt) long were removed, and all sequences were checked for chimera formation by using the RDPII pyrosequencing pipeline (http://pyro.cme.msu.edu/). Operational taxonomic units (OTUs) were clustered at 97% identity by using the UCLUST OTU picking method in QIIME (Quantitative Insights into Microbial Ecology) (21). OTUs within each sample were used to generate rank abundance curves and to calculate alpha diversity by using Chao1 indices. OTU taxonomy was assigned by using the RDPII classifier (22). Representative sequences from each OTU were merged into one file and used as the input for the PyNAST (Python Nearest Alignment Spaced Termination) alignment tool to create a nearest-neighbor phylogenetic tree for weighted and unweighted UniFrac beta diversity analysis using QIIME. The UniFrac distance matrix was used to construct a principal coordinate analysis (PCoA) plot to assess differences in bacterial community structure in a taxonomy-independent manner. Weighted UniFrac analysis takes into account the abundance of OTUs present within each sample, whereas unweighted UniFrac analysis takes into account only presence/absence data for OTUs.
Identification of cultured bacteria and antibacterial assays.
To obtain bacterial isolates for antibacterial assays, the coral crushate was plated in triplicate onto glycerol artificial seawater (GASW) solid medium (23). Bacterial colonies were selected after 48 h of incubation at 28°C. Bacterial colonies that varied in size, pigmentation, and growth morphology were selected to maximize diversity. A total of 360 bacterial isolates, designated isolates H1 through H360, were streaked to purity and identified by amplification of the full 16S rRNA gene with universal primers 8F and 1513R (see Table S1 in the supplemental material) (24). A small sample of the 1,505-bp PCR product was verified on a 1% agarose—TAE gel. The remaining PCR product was purified by using the High Pure Ultra Filtration Cleanup kit (Roche Life Sciences). The purified PCR product was sequenced with the same primers as those used for PCR amplification. Forward and reverse sequences were aligned by using BioEdit (25) and analyzed by using the Basic Local Alignment Search Tool (BLAST).
After the identification of the 360 cultured isolates, a subset was chosen for further analysis. One representative isolate from each bacterial genus was chosen for screening of antibacterial activity. Bacillus and Psychrobacter had two representative isolates for antibacterial assays because both genera were dominated by two species each. Conditioned medium (1 ml of the cell-free supernatant from a culture of the tester bacterium grown overnight) was combined 1:1 with fresh medium before inoculation with 5 μl from a culture of the target bacterium grown overnight. Antibacterial assays were conducted in triplicate. As a negative control, the target bacterium replaced the tester bacterium during the preparation of conditioned medium. Growth inhibition was defined as an average optical density at 600 nm (OD600) of <0.05 after incubation overnight at 25°C.
Contribution of cultured bacteria to bacterial communities.
The contribution of cultured isolates to the overall bacterial community was estimated. OTUs that were classified into the same genera as cultured isolates were manually aligned in BioEdit to the 16S rRNA gene sequences of representative isolates. OTUs with sequences with 100% identity to the V3 region of their corresponding isolate were used to calculate the average proportion of that OTU in the bacterial communities of red and orange fragments. In the case of the two Psychrobacter isolates (H2 and H217), the 16S rRNA sequences contained 10 mismatches within the V3 region, which were used to differentiate the two isolates when aligning Psychrobacter OTUs. For the two Bacillus isolates (H276 and H282), the 16S rRNA sequences had no mismatches in the V3 region. Therefore, these two isolates were grouped together for subsequent calculations.
Statistics.
Unless otherwise stated, data are presented as means ± standard errors of the means (SEM). Analysis of similarities (ANOSIM) was performed by using weighted and unweighted UniFrac matrices to test for significant differences in bacterial communities between red and orange morphs. PRIMER-E v6 (Primer-E Ltd.) was used to conduct similarity percentage (SIMPER) analysis, which identified OTUs that contributed to differences between red and orange bacterial communities (see Table S2 in the supplemental material). Shannon's H′, Pielou's J′, rank abundance, and Chao1 rarefaction measures were calculated to assess alpha diversity, evenness, and richness of bacterial communities. Shannon's H′ index (loge), which measures diversity, ranges from 0 (no diversity) to 4.6 (maximum diversity). Pielou's J′ index, which measures evenness, ranges from 0 (no evenness) to 1 (maximum evenness). The Mann-Whitney nonparametric test was used to evaluate differences in diversity metrics, OTU abundances, and bacterial genus abundances between red and orange morphs.
Accession numbers.
Sequences from 454 pyrosequencing were submitted to the NCBI Sequence Read Archive under accession no. SRP047463. The 16S rRNA gene sequences from cultured isolates used in antibacterial assays were submitted to the GenBank database under accession no. KP640578 to KP640594.
RESULTS
Red and orange morphologies have distinct bacterial communities.
Bacterial communities from red and orange morphs assessed by using high-throughput sequencing are here referred to as “red communities” and “orange communities,” respectively. After removal of low-quality, chimeric, and short (<150-bp) reads, 173,027 high-quality sequences with an average length of 192 bp (minimum of 159 bp and maximum of 204 bp) were obtained from all 10 coral fragments. Totals of 77,981 and 95,046 sequences were analyzed for red and orange fragments, respectively, with an average of 17,302 sequences per coral fragment. The total pool of sequences was clustered into 2,320 operational taxonomic units (OTUs) at the level of 97% identity. Red communities had 1,172 OTUs present, and orange communities had 1,697 OTUs present (Table 1). Overall, there was 23.6% overlap between the two color morphs: 549 out of 2,320 OTUs were present in at least one fragment from both color morphs (Table 1). Eight OTUs were present in all 10 red and orange fragments, which were identified as three species of Vibrio and one species each of Listonella, Enhydrobacter, Bacillus, Ralstonia, and Microbulbifer.
TABLE 1.
Summary description of samples and sequence analysis
| Sample | Color morphology | Shannon H′a | Pielou J′b | No. of OTUsc | No. of unique OTUs | No. of sequences analyzed |
|---|---|---|---|---|---|---|
| O1 | Orange | 2.989 | 0.4628 | 639 | 33 | 20,272 |
| O2 | Orange | 3.284 | 0.5060 | 658 | 47 | 16,441 |
| O3 | Orange | 2.905 | 0.4660 | 510 | 157 | 19,928 |
| O4 | Orange | 2.615 | 0.4308 | 433 | 128 | 18,242 |
| O5 | Orange | 3.545 | 0.5418 | 694 | 209 | 20,163 |
| R1 | Red | 3.300 | 0.5475 | 415 | 36 | 14,443 |
| R2 | Red | 3.409 | 0.5471 | 532 | 56 | 17,938 |
| R3 | Red | 3.753 | 0.5753 | 681 | 99 | 12,893 |
| R4 | Red | 3.357 | 0.5367 | 521 | 58 | 18,314 |
| R5 | Red | 3.441 | 0.5502 | 520 | 51 | 14,393 |
| Total | ||||||
| Orange | 1,697 | 574 | 95,046 | |||
| Red | 1,172 | 300 | 77,981 | |||
| Combined | 2,320d | 874 | 173,027 |
Shannon's H′ values (loge) range from 0 (no diversity) to 4.6 (maximum diversity).
Pielou's J′ values range from 0 (no evenness) to 1 (maximum evenness).
OTUs were defined as having 97% similarity.
A total of 549 OTUs are shared between red and orange morphs.
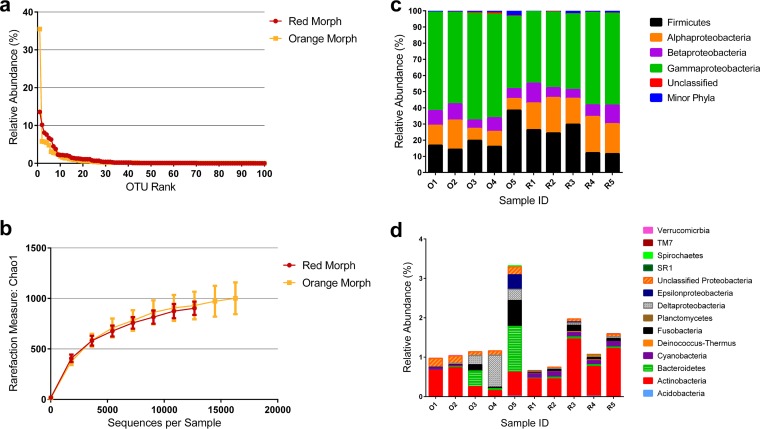
The overall diversity of bacterial communities was high and was similar between both color morphs (W = 36; P = 0.09 [determined by a Mann-Whitney test]), with red communities having an average H′ value of 3.4520 (±0.079) and orange communities having an H′ value of 3.068 (±0.160) (Table 1). However, red communities displayed higher evenness (J′) than orange communities, 0.5506 (±0.0065) versus 0.4815 (±0.0192) (W = 39.0; P = 0.02 [determined by a Mann-Whitney test]) (Table 1). Rank abundance curves (Fig. 2a), which plot relative OTU abundance, also showed lower evenness in orange communities, which were dominated by a single OTU (Vibrio) that comprised on average 35.4% (±5.5%) of orange communities. In contrast, the highest-ranked OTU (Listonella) in red communities comprised on average 13.1% (±2.5%) of red communities. Richness, as estimated by Chao1 rarefaction curves, showed no difference between color morphs in estimated OTU richness (Fig. 2b).
FIG 2.
Diversity of bacterial communities in red and orange morphs. (a and b) Alpha diversity in red and orange communities illustrated by using rank abundance (a) and Chao1 estimator (b) analyses and colored according to the corresponding morphology. (c and d) Relative abundances of phyla and Proteobacteria classes that represent >1% (c) and <1% (d) of total the bacterial community in each Montipora capitata fragment. R1 to R5 represent red morph fragments. O1 to O5 represent orange morph fragments.
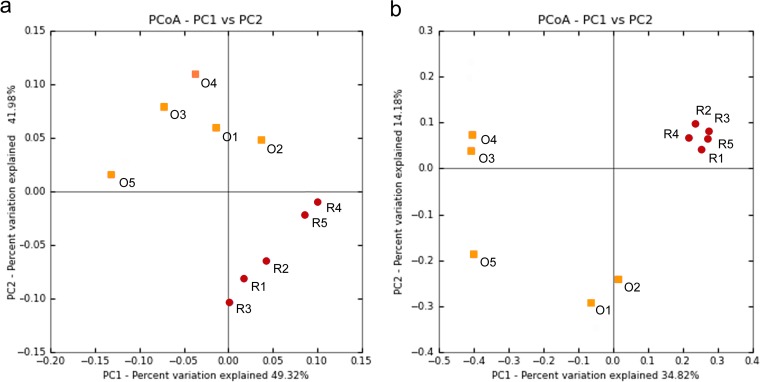
There were significant differences in overall bacterial community structure between red and orange morphs of M. capitata, as indicated by a weighted UniFrac distance matrix (global R = 0.544; P < 0.01 [determined by ANOSIM]) (Fig. 3a). An unweighted UniFrac distance matrix, which takes into account only presence/absence data for OTUs, also displayed significant differences between red and orange communities (global R = 0.740; P < 0.01 [determined by ANOSIM]) (Fig. 3b). Both weighted and unweighted UniFrac measures indicated that orange communities are more variable than red communities. The OTU that contributed the most (28.8% of total dissimilarity) (see Table S2 in the supplemental material) to differences between red and orange communities was classified as Vibrio and comprised 35.4% (±5.5%) of the total orange community yet only 1.1% (±0.6%) of the total red community.
FIG 3.
Partitioning of red and orange M. capitata bacterial communities. Shown are data from PCoA of the weighted (a) and unweighted (b) UniFrac distance matrices. Each dot corresponds to a coral fragment. Red circles, red morph fragments; orange squares, orange morph fragments.
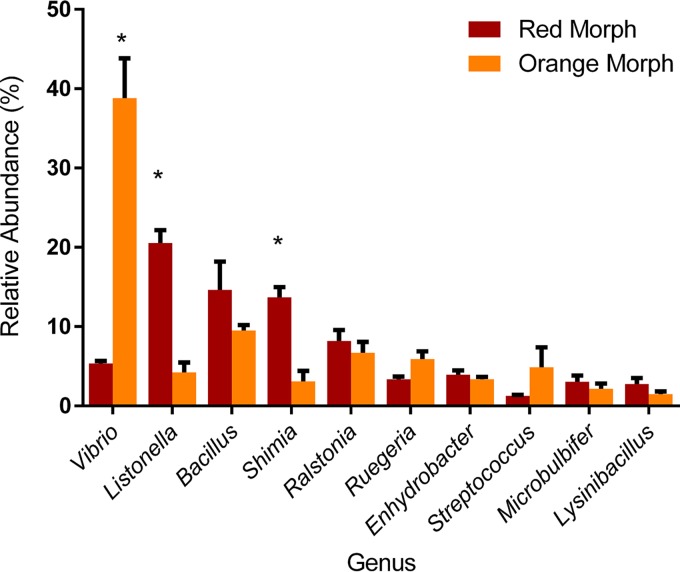
The bacterial community of M. capitata was represented by 14 phyla and 198 genera (Fig. 2c and d). Two phyla, Deinococcus-Thermus and SR1, had no representation in orange communities. Bacterial communities of both color morphs were dominated by Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, and Firmicutes (Fig. 2c). Minor phyla were defined as individually representing <1% of sequences per fragment (Fig. 2d). Actinobacteria, Bacteroidetes, Fusobacteria, and Deltaproteobacteria were present in all fragments of both color morphs. Cyanobacteria were present in all red fragments but in only two out of five of orange fragments. Taxonomic differences that were not obvious at the level of bacterial phylum were very pronounced at the level of bacterial genus. There were 10 dominant bacterial genera that amounted to >70% of the sequences in red and orange communities. Three of the 10 dominant genera were significantly different between color morphologies. Vibrio was more abundant in orange communities, whereas Listonella and Shimia were more abundant in red communities (P < 0.05 [determined by a Mann-Whitney test]) (Fig. 4).
FIG 4.
Dominant members of bacterial communities are present in both red and orange M. capitata corals. Shown are relative abundances of the top 10 most abundant genera in red and orange communities. The RDPII classifier was used to assign taxonomic groups at the genus level. Data represent means ± SEM and are colored according to morphology. *, P < 0.05, as determined by a Mann-Whitney test.
Cultured isolates display selective antibacterial activity and represent rare and abundant genera of M. capitata bacterial communities.
To characterize the community of culturable bacteria of M. capitata and to test for antibacterial activity, 360 bacterial isolates from red and orange fragments (36 isolates each from 10 fragments) were collected and identified. Many isolates had identical 16S rRNA gene sequences (100% identity over a range of 1,386 to 1,405 bp) and were dereplicated into 23 distinct phylotypes that were classified into 14 genera. Cultured bacteria represent 7.1% (14 out of 198) of the bacterial genera found in the bacterial communities characterized by high-throughput sequencing.
Sixteen representative isolates were screened for antibacterial activity in a pairwise fashion as both tester and target strains against all other representative isolates. Isolates were also tested against the M. capitata pathogens Vibrio owensii strain OCN002 (23) and Vibrio coralliilyticus strain OCN008 (26). Representative isolates included one isolate from each bacterial genus cultured from M. capitata. Two phylotypes were commonly observed for Bacillus and Psychrobacter, so two representative isolates from these genera were used in antibacterial assays. Eight out of 16 isolates (50%) displayed antibacterial activity against one or more of the other strains (Table 2). None of the representative isolates inhibited the growth of the coral pathogen OCN002 or OCN008 (Table 2).
TABLE 2.
Antibacterial activity of M. capitata-associated bacteria
| Isolate | Closest match in GenBank |
Matching OTUb (no. of aligned bp) | Isolate inhibiteda |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organism | Accession no. | H2 | H16 | H17 | H18 | H38 | H39 | H43 | H206 | H217 | H223 | H236 | H254 | H269 | H276 | H282 | H291 | OCN002 | OCN008 | ||
| H2 | Psychrobacter celer | FJ613610.1 | 439 (195) | + | |||||||||||||||||
| H16 | Erythrobacter sp. | HG008905.1 | 1836 (169) | ||||||||||||||||||
| H17 | Kocuria rosea | EU660350.1 | 2265 (159) | ||||||||||||||||||
| H18 | Micrococcus luteus | AB697159.1 | 1998 (174) | + | + | ||||||||||||||||
| H38 | Microbulbifer variabilis | JN128259.1 | 143 (195) | + | + | + | + | ||||||||||||||
| H39 | Pseudovibrio sp. | KC429855.1 | 1188 (169) | + | |||||||||||||||||
| H43 | Ruegeria sp. | HQ908677.1 | 1115 (169) | ||||||||||||||||||
| H206 | Ferrimonas balearica | JQ799133.1 | NA | + | + | + | + | ||||||||||||||
| H217 | Psychrobacter piscidermidis | EU127295.1 | 65 (195) | + | + | + | + | + | |||||||||||||
| H223 | Lysinibacillus fusiformis | JN416567.1 | 1572 (195) | + | |||||||||||||||||
| H236 | Chromohalobacter sp. | HQ683730.1 | 1348 (178) | ||||||||||||||||||
| H254 | Stenotrophomonas sp. | AM400231.1 | NA | ||||||||||||||||||
| H269 | Pseudomonas stutzeri | HF571099.1 | 696 (194) | ||||||||||||||||||
| H276 | Bacillus hwajinpoensis | JN208078.1 | 414 (195) | + | + | ||||||||||||||||
| H282 | Bacillus baekryungensis | JF721989.1 | 414 (195) | ||||||||||||||||||
| H291 | Pseudoalteromonas sp. | FJ981833.1 | 2063 (194) | ||||||||||||||||||
+ indicates growth inhibition at an OD600 of <0.05 after 16 h of incubation at 25°C.
Representative isolates that matched an OTU from high-throughput sequencing data with 100% identity. Numbers in parentheses represent the number of base pairs aligned between the 16S rRNA sequence of the isolate and the OTU sequence. NA, not applicable.
The contribution of the representative isolates to the overall bacterial community was estimated by aligning the 16S rRNA sequences of each representative isolate to OTU sequences. Each representative isolate, except H206 and H254, had a single OTU with 100% identity over a range of 159 bp to 195 bp (Table 2). Nine representative isolates (H2, H16, H17, H18, H39, H217, H236, H269, and H291) had matching OTUs that were rare members of the M. capitata community, individually comprising <1% of the sequences per fragment. Five representative isolates had matching OTUs that were dominant members (>1%) of red and orange communities (H38, H43, H223, H276, and H282).
DISCUSSION
We found that red and orange morphs of M. capitata displayed significantly different bacterial communities. Sampling of the two color morphs of Montipora capitata from the same reef controlled for major environmental factors. This allowed us to investigate whether intraspecific differences in the coral host or Symbiodinium clade affected the associated bacterial communities. Although color morphs shared the same most abundant genera, the relative proportions of these shared OTUs differed. The different bacterial communities found in the two color morphs of M. capitata show that the specificity of coral-bacterial associations can go beyond the level of coral species. Differences in bacterial communities were detected even though a relatively small sample size was used for analysis (n = 5 colonies sampled per color morph). The tight clustering of red communities in the unweighted PCoA plot (Fig. 3b) suggests that there is a conserved community associated with this color morph. In contrast, the high variability seen within the orange morph may suggest that there is a less consistent bacterial community associated with this color morph. Further studies involving deeper sampling of M. capitata color morphs across different sites and times will be needed to assess whether differences in bacterial communities are widespread and enduring.
The red morph of M. capitata, harboring clade C Symbiodinium, had more consistent bacterial communities (lower variability and higher evenness) than the bacterial communities of the orange morph with clade D Symbiodinium. We also found that the major difference was in the relatively high proportion of Vibrio in the orange morphs. Similarly, Littman et al. (27) showed that juvenile corals infected with clade C Symbiodinium had more diverse and even bacterial communities than did juveniles infected with clade D Symbiodinium. In addition, corals infected with clade D also had higher proportions of Vibrio. Symbiodinium is thought to indirectly influence bacterial communities by contributing to the amount and composition of the external mucus layer covering corals (28–30). Variation within the coral host (independent of the Symbiodinium clade) may also be important in structuring coral-bacterial associations. A cold-water coral that lacks Symbiodinium (Lophelia pertusa) has been shown to have two color morphs that harbor distinct bacterial communities (31). While we cannot separate the influence of color morph versus Symbiodinium in this study, one or both are likely to contribute to differences in bacterial communities of red and orange morphs of M. capitata.
The bacterial communities of red and orange morphs of M. capitata were comprised mostly of heterotrophic bacteria with a few dominant taxa and many rare taxa, which is consistent with data from other studies of coral-associated bacterial communities (9, 32). Vibrio was a dominant genus in both red and orange communities. Vibrio is often described as being detrimental to coral health because some species are coral pathogens (23, 26, 33, 34), and increases in Vibrio abundance have been correlated with stressors, including temperature (3, 35), eutrophication (36), and disease (23). However, Vibrio is also a dominant member of bacterial communities from healthy corals (9, 37; this study). Olson et al. (38) found Vibrio-derived nifH gene sequences in M. capitata samples and concluded that Vibrio may have the potential for nitrogen fixation in this coral species.
Eight isolates from M. capitata of both color morphs inhibited the growth of other cultured isolates. These isolates suggest that antagonistic interactions between members of the bacterial community of M. capitata are present and could be important in influencing overall community composition. These findings are consistent with data from other studies that investigated the inhibitory activity of bacterial communities in other coral species (17, 39). Representative isolates chosen for antibacterial assays were selected based on 16S rRNA sequence diversity. However, core genes such as 16S rRNA cannot reflect whole-genome diversity. Thus, the antibacterial activity of representative isolates is not likely to reflect the full antibacterial capacity of all the cultured isolates from M. capitata. Interestingly, none of the representative isolates were able to inhibit two known pathogens of M. capitata, Vibrio coralliilyticus strain OCN008 (26) and Vibrio owensii strain OCN002 (23). Many other studies have found bacterial isolates from coral that inhibit relevant bacterial pathogens (17, 35, 39, 40). This suggests that the bacterial community of M. capitata may not be able to effectively control these two particular pathogens.
We found a large overlap of data from culture-dependent and culture-independent methods. The 14 genera found in cultured isolates were also found in high-throughput sequencing data, and all except two representative isolates had a matching OTU with 100% identity. The two isolates without 100% identity to an OTU (Ferrimonas and Stenotrophomonas) show that this comparative approach has limitations. Additionally, these isolates may be contaminants derived from seawater because Ferrimonas isolates have been detected in seawater that has been passed through a 0.2-μm-pore-size filter (41, 42). Previous studies that investigated the functional role of cultured bacteria in the coral holobiont lacked the ability to estimate the contribution of these isolates to the overall bacterial community (16, 35). This study was able to address this issue by comparing the 16S rRNA gene sequences of isolates to high-throughput sequencing data. We found that the representative isolates had matching OTUs that were dominant members as well as rare members of the overall bacterial community of M. capitata. Generalizations about the role of any of these representative isolates (especially in terms of antibacterial activity) in the context of the whole bacterial community are tenuous since the alignment of sequences of cultured isolates to sequences of OTUs spans only a small region of the 16 rRNA gene and may not extend to other isolates. For example, H276 and H282 displayed different patterns of antibacterial activity, but both isolates had 100% identity to OTU 414. However, comparison of 16S rRNA gene sequences of isolates to high-throughput sequencing data may help prioritize which cultured isolates are to be further characterized based on their estimated abundance in the overall bacterial community.
Reduced diversity and higher proportions of Vibrio in bacterial communities have been linked to higher mortality rates of corals in response to elevated temperatures (37). The differences in bacterial communities found among color morphs in M. capitata may also help explain the pattern of disease occurrence commonly found in the field with healthy and diseased colonies adjacent to each other (43). The red and orange colonies that were sampled in this study were not monitored over time to see if they developed signs of disease, but work is under way to determine whether red and orange morphs of M. capitata also differ in their susceptibility to disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation (NSF) (grant no. OCE0961814).
We thank Tyler Culpepper for bioinformatics support. We also thank two anonymous reviewers for their valuable comments and suggestions to improve the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01935-15.
REFERENCES
- 1.Muscatine L, McCloskey L, Marian R. 1981. Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol Oceanogr 26:601–611. doi: 10.4319/lo.1981.26.4.0601. [DOI] [Google Scholar]
- 2.Ritchie K, Smith G. 2004. Microbial communities of coral surface mucopolysaccharide layers, p 259–264. In Rosenberg E, Loya Y (ed), Coral health and disease. Springer, Berlin, Germany. [Google Scholar]
- 3.Koren O, Rosenberg E. 2006. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol 72:5254–5259. doi: 10.1128/AEM.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweet M, Croquer A, Bythell J. 2011. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30:39–52. doi: 10.1007/s00338-010-0695-1. [DOI] [Google Scholar]
- 5.Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N. 2001. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20:85–91. doi: 10.1007/s003380100138. [DOI] [Google Scholar]
- 6.Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microbiol 68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne DG, Munn CB. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol 7:1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- 8.Rohwer F, Azam F, Knowlton N, Seguritan V. 2002. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 9.Sunagawa S, Woodley CM, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. 2004. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- 11.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 12.Siboni N, Ben-Dov E, Sivan A, Kushmaro A. 2008. Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ Microbiol 10:2979–2990. doi: 10.1111/j.1462-2920.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- 13.Raina J-B, Tapiolas D, Willis BL, Bourne DG. 2009. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75:3492–3501. doi: 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiore CL, Jarett JK, Olson ND, Lesser MP. 2010. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol 18:455–463. doi: 10.1016/j.tim.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Kimes NE, Van Nostrand JD, Weil E, Zhou J, Morris PJ. 2010. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ Microbiol 12:541–556. doi: 10.1111/j.1462-2920.2009.02113.x. [DOI] [PubMed] [Google Scholar]
- 16.Shnit-Orland M, Kushmaro A. 2009. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol 67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 17.Rypien KL, Ward JR, Azam F. 2010. Antagonistic interactions among coral-associated bacteria. Environ Microbiol 12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 18.Maynard J, van Hooidonk R, Eakin CM, Puotinen C, Garren M, Williams G, Heron S, Lamb J, Weil E, Willis BL, Harvell CD. 2015. Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat Clim Chang 5:688–694. doi: 10.1038/nclimate2625. [DOI] [Google Scholar]
- 19.LaJeunesse T, Thornhill D, Cox E, Stanton F, Fitt W, Schmidt G. 2004. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23:596–603. [Google Scholar]
- 20.Muyzer G, de Waal E, Uitterlinden A. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso J, Kuczynski J, Stombaugh J. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ushijima B, Smith A, Aeby GS, Callahan SM. 2012. Vibrio owensii induces the tissue loss disease Montipora white syndrome in the Hawaiian reef coral Montipora capitata. PLoS One 7:e46717. doi: 10.1371/journal.pone.0046717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aebischer T, Fischer A, Walduck A, Schlötelburg C, Lindig M, Schreiber S, Meyer TF, Bereswill S, Göbel UB. 2006. Vaccination prevents Helicobacter pylori-induced alterations of the gastric flora in mice. FEMS Immunol Med Microbiol 46:221–229. doi: 10.1111/rp10.1016-j.femsim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95–98. [Google Scholar]
- 26.Ushijima B, Videau P, Burger AH, Shore-Maggio A, Runyon CM, Sudek M, Aeby GS, Callahan SM. 2014. Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl Environ Microbiol 80:2102–2109. doi: 10.1128/AEM.03463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Littman R, Willis B, Bourne D. 2009. Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades. Mar Ecol Prog Ser 389:45–59. doi: 10.3354/meps08180. [DOI] [Google Scholar]
- 28.Stat M, Morris E, Gates RD. 2008. Functional diversity in coral-dinoflagellate symbiosis. Proc Natl Acad Sci U S A 105:9256–9261. doi: 10.1073/pnas.0801328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raina J, Dinsdale E, Willis B, Bourne D. 2010. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol 18:101–108. doi: 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Deschaseaux E, Beltran V. 2014. Comparative response of DMS and DMSP concentrations in Symbiodinium clades C1 and D1 under thermal stress. J Exp Mar Biol Ecol 459:181–189. doi: 10.1016/j.jembe.2014.05.018. [DOI] [Google Scholar]
- 31.Neulinger SC, Järnegren J, Ludvigsen M, Lochte K, Dullo W-C. 2008. Phenotype-specific bacterial communities in the cold-water coral Lophelia pertusa (Scleractinia) and their implications for the coral's nutrition, health, and distribution. Appl Environ Microbiol 74:7272–7285. doi: 10.1128/AEM.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barott KL, Rodriguez-Brito B, Janouškovec J, Marhaver KL, Smith JE, Keeling P, Rohwer FL. 2011. Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis. Environ Microbiol 13:1192–1204. doi: 10.1111/j.1462-2920.2010.02419.x. [DOI] [PubMed] [Google Scholar]
- 33.Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. 2001. Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol 51:1383–1388. [DOI] [PubMed] [Google Scholar]
- 34.Ben-Haim Y, Thompson C, Cnockaert M, Thompson F, Swings J, Hoste B, Rosenberg E. 2003. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53:309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie K. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 36.Garren M, Raymundo L, Guest J, Harvell CD, Azam F. 2009. Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS One 4:e7319. doi: 10.1371/journal.pone.0007319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Littman R, Bourne DG, Willis BL. 2010. Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol Ecol 19:1978–1990. doi: 10.1111/j.1365-294X.2010.04620.x. [DOI] [PubMed] [Google Scholar]
- 38.Olson ND, Ainsworth TD, Gates RD, Takabayashi M. 2009. Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J Exp Mar Biol Ecol 371:140–146. doi: 10.1016/j.jembe.2009.01.012. [DOI] [Google Scholar]
- 39.Nissimov J, Rosenberg E, Munn CB. 2009. Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol Lett 292:210–215. doi: 10.1111/j.1574-6968.2009.01490.x. [DOI] [PubMed] [Google Scholar]
- 40.Sabdono A, Radjasa O. 2006. Anti-bacterial property of a coral-associated bacterium Bacillus sp. against coral pathogenic BBD (black band disease). J Coast Dev 9:175–182. [Google Scholar]
- 41.Haller C, Rölleke S, Vybiral D, Witte A, Velimirov B. 2000. Investigation of 0.2 μm filterable bacteria from the Western Mediterranean Sea using a molecular approach: dominance of potential starvation forms. FEMS Microbiol Ecol 31:153–161. doi: 10.1111/j.1574-6941.2000.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 42.Velimirov B. 2001. Nanobacteria, ultramicrobacteria and starvation forms: a search for the smallest metabolizing bacterium. Microbes Environ 16:67–77. doi: 10.1264/jsme2.2001.67. [DOI] [Google Scholar]
- 43.Gochfeld D, Aeby G. 2008. Antibacterial chemical defenses in Hawaiian corals provide possible protection from disease. Mar Ecol Prog Ser 362:119–128. doi: 10.3354/meps07418. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.