Abstract
In contrast to the honey bee gut, which is colonized by a few characteristic bacterial clades, the hive of the honey bee is home to a diverse array of microbes, including many lactic acid bacteria (LAB). In this study, we used culture, combined with sequencing, to sample the LAB communities found across hive environments. Specifically, we sought to use network analysis to identify microbial hubs sharing nearly identical operational taxonomic units, evidence which may indicate cooccurrence of bacteria between environments. In the process, we identified interactions between noncore bacterial members (Fructobacillus and Lactobacillaceae) and honey bee-specific “core” members. Both Fructobacillus and Lactobacillaceae colonize brood cells, bee bread, and nectar and may serve the role of pioneering species, establishing an environment conducive to the inoculation by honey bee core bacteria. Coculture assays showed that these noncore bacterial members promote the growth of honey bee-specific bacterial species. Specifically, Fructobacillus by-products in spent medium supported the growth of the Firm-5 honey bee-specific clade in vitro. Metabolic characterization of Fructobacillus using carbohydrate utilization assays revealed that this strain is capable of utilizing the simple sugars fructose and glucose, as well as the complex plant carbohydrate lignin. We tested Fructobacillus for antibiotic sensitivity and found that this bacterium, which may be important for establishment of the microbiome, is sensitive to the commonly used antibiotic tetracycline. Our results point to the possible significance of “noncore” and environmental microbial community members in the modulation of honey bee microbiome dynamics and suggest that tetracycline use by beekeepers should be limited.
INTRODUCTION
The gut of the European honey bee (Apis mellifera) is host to a characteristic microbial community composed predominantly of three major phyla (Firmicutes, Proteobacteria, and Actinobacteria) within which several honey bee-specific families and genera are taxonomically classified (1–3). The adult bee gut has been characterized as being colonized by a small number of bacterial clades, some with genus and species designations (2). These core clades (and named genera), found within the previously mentioned bacterial phyla, are as follows: Firm-4, Firm-5 (within the Firmicutes), Bifido (within the Actinobacteria), and Alpha-2.1, Alpha-2.2 (Parasaccharibacter sp.), Alpha-1, Beta (Snodgrassella), Gamma-1 (Gilliamella sp.), and Gamma-2 (Frischella sp.) (within the Proteobacteria) (2). The development of a honey bee microbiome inclusive of these core clades requires interaction between kin and/or with hive components (4, 5). The honey bee is a eusocial insect that lives in a dense population of individuals that make up the colony. The worker caste of bees performs different tasks in the hive, dependent on their age. Younger bees (“nurse bees”) are generally constrained to the hive, feed on protein and lipid rich processed pollen (“bee bread”) and participate in the rearing of brood. Older bees (“foragers”) fly out of the hive in search of nectar and pollen (6). When food is brought back to the hive, it is passed from bee to bee via trophallaxis, a mechanism for food exchange, and made into the food products honey and bee bread. Lactobacillus sp. commonly associated with pollen, have also been identified in the crop of adult honey bees (7, 8). Social transmission of the honey bee microbiome has been tested previously; bees allowed access to bee bread alone, but kept in isolation, were found to lack some members of the core microbial clades later in life (4). In addition, newly eclosed worker bees that were exposed to hive components acquired some of the characteristic bacterial phylotypes, suggesting that social transmission is not necessary for colonization of the honey bee by some clades but that interaction with comb might facilitate inoculation (4). Similarly, more recent results also suggest that full transmission of the characteristic gut microbiota requires the physical interaction of honey bees with hive environments, and perhaps with fecal material, and cannot be completed through trophallaxis alone (5). It seems that natural hive rearing, including interactions with other bees and hive components, is critical to the colonization by Gram-negative core bacteria (such as Gilliamella species) while exposure to comb or trophallaxis alone resulted in gut communities which contained other core microbial members (Firm-4, Firm-5, Gamma-2, Bifido, Snodgrassella, and Alpha-2.1) (5).
Directional change in species composition in an environment, over time, is referred to as ecological succession. From studies of bacterial succession in the digestive tract of mammals, we know that young are relatively uncolonized and that the first members to colonize the gut are often facultative anaerobes, such as Escherichia coli (9–12). These “pioneering species” pave the way for colonization of the gut by obligate anaerobes by consuming oxygen, producing carbon dioxide, and changing the pH (10). During early stages of succession, bacterial diversity is often low, and the community changes rapidly; this result has been observed in the mammalian digestive tract and also in the insect gut and in other, non-host-associated environments (9–17). However, after this period of early succession, the bacterial community reaches a steady state often referred to as the “climax community.” The climax community of a bacterial population would be reached when there is an equilibrium that can be maintained of a specific mix of bacteria (9, 10). The event often coincides with a later developmental stage of the organism harboring the bacterial community (9, 11, 12, 15). Bacterial diversity and the total number of bacteria are higher in a climax community than during early succession (9, 10, 17). The honey bee is a holometabolous insect and moves through a life cycle marked by the metamorphic transition from larval stage into a developed imago form. During the course of this event, a matured larva is enclosed in its brood cell by worker bees, pupates, and develops into a honey bee. It is understood that, during this metamorphic period, the larval gut is shed (18). Consequently, newly eclosed worker bees (NEWs) retain none of the characteristic microbiota associated with the larval gut and, over the span of a few days, are colonized with bacterial phylotypes characteristic of an adult honey bee (5). However, because larval bees mature and pupate in the same space—the brood cell—it is possible that they are reinoculated with the same microbes they were exposed to as larvae upon completion of their metamorphic transition. In addition, because NEWs interact with hive components such as comb and processed food, colonization of these hive components by bacterial community members may impact community succession.
Although there has been substantial investment into profiling the microbes most commonly associated with the honey bee gut, research directed toward determining the microbes commonly associated with honey bee-related environments has been more limited in scope. For example, 16S rRNA profiling of pollen and bee bread found sequences homologous to the Pasteurelaceae family, as well as Lactobacillus and Bifidobacteriaceae phylotypes (8). Another study, using quantitative PCR (qPCR), determined that bee bread was deficient in the characteristic phylotypes commonly found associated with the honey bee gut, although bacteria belonging to the phylotype Alpha-2.2 were identified (4). In addition, culture based studies have identified the presence of Bacillus species in bee bread samples (12). Relatively little research has been devoted to identifying bacteria associated with other hive components, such as propolis and the comb itself. For example, phospholipid based analyses have revealed a diversity of bacteria associated with hive components but did not resolve strain-specific signatures (19).
We attempted here to determine the environments from which honey bees may be inoculated and interactions between bacterial community members that may shape the microbiome. Although workers shed their gut lining during metamorphosis, bacteria present during the developmental process, in the food produced by the bees, or in the comb, may persist or may facilitate colonization by core microbiome members. We therefore looked to identify possible trends in microbial communities found in the larvae, nurse bees, and hive components. We cultured and sequenced a subset of the bacterial community, the lactic acid bacteria (LAB), an abundant and ubiquitous clade of microbes found associated with bees throughout development and in other external hive environments. We examined LAB community composition across the hive and in the honey bees themselves. Examination of pairwise comparisons between environments containing identical 99% operational taxonomic units (OTUs) suggests a homogenous distribution of microbes between the hive environments. Finally, we identify a strain of Fructobacillus found in microbial hubs, able to promote the growth of honey bee-specific LAB, and able to utilize the complex plant carbohydrate lignin, providing a potential candidate as a pioneering species in the development of the honey bee gut community.
MATERIALS AND METHODS
Bee sampling and microbiological protocols.
Samples were obtained from three healthy, established hives located in Bloomington, IN. All sampling was performed aseptically (sterilized collection equipment and cryogenic vials were used and gloves were worn throughout). Young worker bees, associated with brood cells and observed actively feeding a larva, were identified as nurse bees and collected, along with the associated larva. A sterile swab was used to sample the brood cell contents previously inhabited by the larva. In addition, a sample of pollen, found in the nearby comb, was taken from each hive. For two hives, we additionally sampled nectar by pipetting 100 μl of volume out of cells and into a sterile cryogenic vial. Nectar was identified for sampling as fresh honey, regurgitated by forager bees but not yet desiccated or capped for maturation. Each of these five samples (nurse, larvae, cell, pollen, and nectar) were collected from the same frame, within each individual hive. All samples were transported on ice and directly plated on media within one-half hour of collection.
Nurse bee guts were removed via aseptic hindgut dissection and homogenized using a plastic, sterile pestle in 1× phosphate-buffered saline (PBS; pH 8.0). Bee bread and whole larval samples were similarly homogenized. Each homogenate was plated on de Man, Rogosa, and Sharpe (MRS) agar at 1:100 dilutions. Swabs taken from cells were streaked directly, without dilution. Cultures were incubated anaerobically at 37°C for 24 h. The resulting cultures of bacteria on each plate were scraped from the plate in 1 ml of PBS and then pipetted into 1.5-ml microcentrifuge tubes. The use of pH neutral PBS did not bias our results, since we were are able to culture representative isolates of Fructobacillus, Firm-4, Firm-5, Bifido, and Lactobacillus on neutral to slightly basic media (Luria-Bertani [LB], brain heart infusion [BHI], and tryptic soy agar [TSA]) in addition to MRS.
DNA extraction, amplification, and sequencing.
DNA was extracted from the bacterial homogenates using the MoBio PowerSoil DNA extraction kit. DNA concentration from each sample was quantified spectrophotometrically, normalized, and amplified via PCR (using Earth Microbiome barcoded primers 515F and 806R and tags rcbc1 to rcbc20) (20). Earth Microbiome amplification protocols were followed, except for the polymerase used (NEB HF Phusion). Reactions were performed in triplicate, using 100 ng of template DNA for each 25-μl reaction. Each reaction was visualized on a 1% agarose gel to confirm amplification. Replicate amplicons were pooled and then cleaned with a Qiagen PCR cleanup kit. Picogreen protocol was used to quantify DNA concentration for each pooled sample. Samples were then normalized and pooled collectively for sequencing. Sequencing was performed on an Illumina Miseq, using 300 PE cycles.
Bioinformatics and OTU-based analyses.
All sequence processing was performed using the Mothur microbial ecology suite (21). Reads from each sample were combined into contiguous sequences and screened for quality (maxambig = 0, maxlength = 275). Sequences were then aligned with the Silva reference database (silva.bacteria.fasta), preclustered, and examined for chimeras via the uchime function. After removal of chimeric sequences, sequences were taxonomically classified using a honey bee-specific training set as a reference (1) and binned into operational taxonomic units (OTUs) based upon 99% sequence identity (see Files S1 and S2 in the supplemental material). The data set was also subsampled to the smallest sample size of 4186 sequences, in order to normalize results across all environments. The 10 OTUs with the highest sequence abundance in this subsampled data set were identified (see Table S1 in the supplemental material). The data from these 10 OTUs, from the three sampled hives, were averaged for each environment, and the standard errors were calculated. In addition, relative sequence abundance was explored for each hive independently. Diversity metrics (such as Simpson indices and Bray-Curtis dissimilarities) were also calculated within Mothur.
Network analysis.
The presence of each of the top 10 OTUs was calculated for each sampling environment (nodes) and used to generate an interaction network in Cytoscape. For visualization purposes only, the connections between nodes (edges) were weighted based on relative abundance of the shared OTUs making up that edge. The network was constructed using OTU data from two of the three sampled hives, for which we had data for all six sampled environments. To identify important hubs in the network, centrality was assessed. “Betweenness centrality” measures how often a path passes through a specific node while moving from one node to another (22).
Bacterial culture, antibiotic sensitivity, and coculture assays.
The Newton Laboratory honey bee bacterial strain bank was utilized as a source of bacterial isolates for this portion of the work. Briefly, bacteria from the honey bee gut and bee bread were cultured on either MRS, LB, BHI, or TSA agar (at 37°C for 48 h under anaerobic conditions), and individual colonies were massively isolated using a robotic colony picker (QPExpression; Genetix). The classification of each isolate was based on 16S rRNA gene sequencing and classification using the Naive Bayesian Classifier and the honey bee-specific training set (1). For the present study, we chose LAB isolates identified in each of the sampled environments (Bifidobacteria, Firm-4, Firm-5, and Fructobacillus). Each isolate was cultured for 48 h in MRS broth at 37°C under anaerobic conditions. After 48 h, measurements of the culture optical density at 600 nm (OD600) were taken, and each was normalized to the lowest OD. The bacteria were cultured alone or in coculture in triplicate, parallel experimental replicates under all pairwise combinations. In addition, bacterial supernatants were used in lieu of cultures to determine whether metabolic by-products of a specific organism (Fructobacillus) stimulated the growth of isolates. To analyze the coculture data, the expected optical density of cocultures was first calculated based on the growth of each isolate alone. If the isolates grew better in coculture, the expected OD would be significantly greater than (i.e., outside of the standard deviation) that of the calculated expected OD. Similarly, if one of the isolates inhibited the growth of the other, the OD would be below the expected value.
OD measured above or below the standard deviation of the expected value was considered significant. Experiments using culture supernatants were similarly analyzed (using Microsoft Excel). To examine a specific interaction between Fructobacillus spent medium and the honey bee-specific Firm-5 isolate, Firm-5 was grown to an OD of 1.4 and subcultured to an OD of 0.01 in either MRS or Fructobacillus spent medium (MRS medium in which Fructobacillus had been cultured at 30C, aerobically, to an OD600 of 1.5 and then spun at 15,000 rpm for 10 min to remove cells). After 48 h of growth at 37°C under anaerobic conditions, OD600 measurements were taken, and the cultures were diluted and plated. All results were normalized to starting OD600 values. Evolutionary analyses were conducted in MEGA6 using the 16S rRNA gene sequences and using a maximum-likelihood method based on the general time reversible model with a gamma distribution, invariable sites, and 100 bootstrap replicates (23). For antibiotic sensitivity tests, overnight cultures of Fructobacillus sp. were mixed with soft MRS agar and overlaid onto MRS plates, onto which an antibiotic impregnated disc had been placed. Diameters of zones of inhibition measured around each antibiotic disc using a ruler. The antibiotics used were as follows: BBL Sensi-Disc tetracycline, 30 μg; ampicillin with sulbactam, 20 μg; rifampin, 5 μg; ciprofloxacin, 5 μg; and vancomycin, 30 μg.
Carbon source utilization assay.
The Fructobacillus isolate used for the coculture assays was grown for 72 h in MRS broth at 30°C under aerobic conditions. Cells were pelleted via centrifugation, and the supernatant was removed. The pellet was resuspended in a buffer of 0.1 M Tris-HCl (pH 6.5). This centrifugation and resuspension process was performed twice to ensure removal of all residual MRS. Then, 15 μl of cell suspension was added to each well of an MT2 plate (Biolog, Inc.). To this, 150 μl of 2% carbohydrate solutions was added to the wells, and the experiment for each carbohydrate was repeated in triplicate. In addition, water control wells were established in triplicate and inoculated with cell suspension and filtered, autoclaved water. The carbon sources examined in here were fructose, glucose, sucrose, maltose, galactose, sorbitol, xylan, pectin, and lignin (all from Sigma-Aldrich). Plates were read via spectrophotometry at an A590 immediately after inoculation in order to establish a baseline. The plates were incubated aerobically at 30°C, and A590 readings were taken every 24 h. The assay was deemed complete when a maximum A590 was observed for the plates. To assess whether a carbon source was utilized, absorbance readings from time points with peak absorbance were compared to the absorbance of the initial time point. The absorbance in water control wells was subtracted from the absorbance in carbohydrate-containing wells. These differences were averaged. Using standard unpaired t tests, differences in growth compared to the water control were compared between the initial and final time points. A carbon source was determined to have been utilized by Fructobacillus if the difference was statistically significant with a P value of ≤0.001.
GenBank accession numbers.
For the LAB isolates identified in each of the sampled environments (Bifidobacteria, Firm-4, Firm-5, and Fructobacillus), the GenBank accession numbers are KT598287 to KT598296.
RESULTS
LAB community profiles across environments.
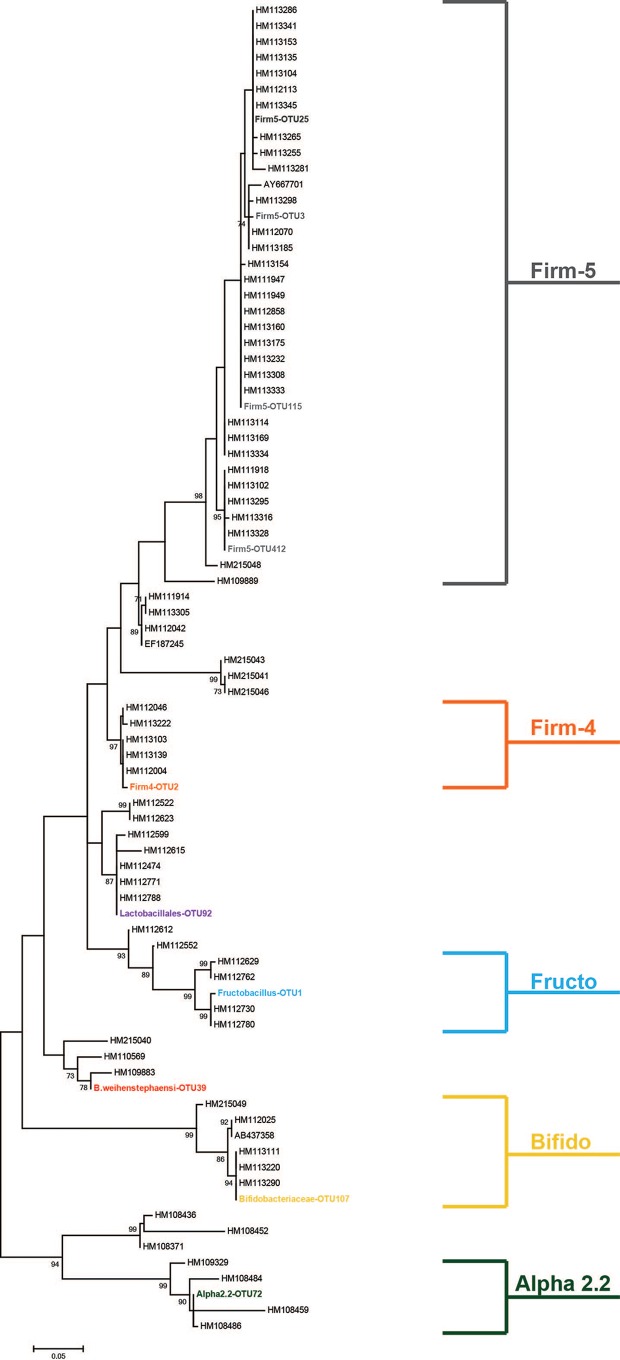
We chose to query the lactic acid bacteria (LAB) associated with the honey bee as a representative community, through which we could begin to examine potential trends in microbial transmission between honey bee associated environments. Processing 2,040,169 total sequences resulted in 1,519,195 unique sequences, grouped into 4,005 individual OTUs when binned at a 99% sequence identity. The rationale behind using a 99% identity threshold was to reach strain level resolution when examining each environment. This facilitated the ability to determine whether specific microbes were being transferred between environments or whether the appearance of the same taxa in two locations was merely an artifact of homology. An abundance threshold of 1% of total sequence abundance was applied to the data set, yielding 10 OTUs that met the criteria. These 10 OTUs dominated the data set, containing 89.7% of total sequence abundance. The other 10.3% of OTUs in the sequence data were similarly classified as the 10 largest OTUs (as Firm-5, Fructobacillus, Bacillus weinhenstephaensi, Bifidobacteriaceae, Firm-4, Lactobacillales, or Alpha-2.2) but did not meet our abundance threshold. To confirm that these top 10 OTUs were members of a group of bacteria previously identified as associated with the honey bee gut, a phylogenetic analysis was performed. Utilizing representative sequences taken from each OTU (the most abundant sequence in that OTU), as well as sequences from a honey bee-specific training set (1), a maximum-likelihood tree was constructed. The phylogeny confirmed the classification of each of the top 10 OTUs, with each member forming a clade with previously identified sequences (Fig. 1). Therefore, our combined culture and amplicon sequencing method identified previously known, honey bee-associated bacteria.
FIG 1.
Phylogenetic analysis of top 10 OTUs, based on 16S rRNA sequences. A maximum-likelihood tree was constructed using a Jukes-Cantor correction model, with 1,000 bootstrap replicates. Sequence names beginning with “AB” or “HM” are published sequences taken from a honey bee-specific training set (1).
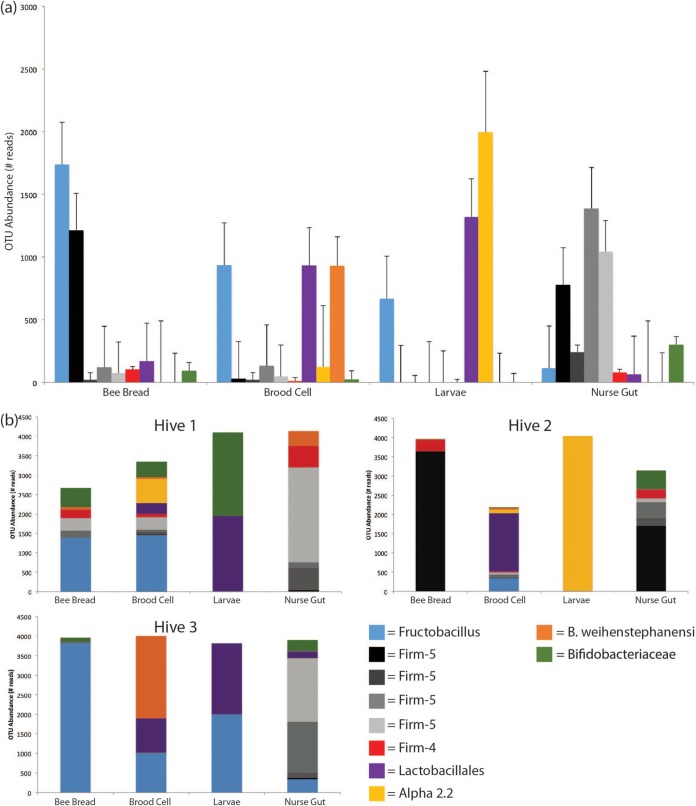
Community richness and diversity were assessed for each sampled environment, using the Chao1 richness index and the inverse Simpson index, respectively. Upon averaging across each of the three sampled hives, we found that no sampled library contained a significantly richer or more diverse culturable LAB community (Table 1). To identify trends in culturable LAB composition across sampling environments, abundances of the top 10 OTUs were averaged and analyzed for all hives together as well as independently (Fig. 2). The culturable LAB community profile in larvae was different compared to nurse bees: Lactobacillales, Alpha-2.2, and Fructobacillus, which were largely representative of larval samples, in contrast to a predominantly Firm-5 and Bifidobacteriaceae LAB community culturable from the nurse gut (Fig. 2a). The same Lactobacillales and Fructobacillus OTUs found in larvae were also identified in the bee bread and the brood cell (Fig. 2), as well as in nectar samples (number of sequences/total; Fructobacillus = 2,257/4,186; Lactobacillales = 1,320/4,186). Interestingly, whereas a significant amount of variation was observed in larval LAB communities from different hives, nurse guts demonstrated largely consistent compositions (Bray-Curtis dissimilarity [mean ± the standard deviation] = 0.192 ± 0.0.029) (Table 1).
TABLE 1.
Microbial community richness and diversity measures for each sampled environment in the honey bee hivea
| Sample | Microbial community richness and diversity measurement |
|||||
|---|---|---|---|---|---|---|
| Chao1 |
Inverse Simpson |
Bray-Curtis |
||||
| Mean | SD | Mean | SD | Mean | SD | |
| Bee bread | 37.667 | 23.544 | 2.2372 | 1.7716 | 0.8008 | 0.2033 |
| Brood cell | 90.503 | 66.414 | 4.1760 | 1.6069 | 0.5985 | 0.0996 |
| Larvae | 12.083 | 13.220 | 1.7666 | 0.5928 | 0.6842 | 0.2725 |
| Nurse gut | 10.000 | 8.8882 | 1.4677 | 0.3536 | 0.1921 | 0.0286 |
Each metric was calculated for individual replicates and averaged within each environment. Chao1 estimates show the expected taxon richness within each environment. Inverse Simpson results are a measure of diversity, with a higher number indicating greater diversity. The Bray-Curtis results represent the dissimilarity found in pairwise comparisons between samples from the same environment and highlight the consistency of the nurse gut replicates.
FIG 2.
Sequence abundance of the top 10 LAB OTUs cultured from each of four subsampled honey bee environment, in three hives, binned at 99% identity based on 16S rRNA sequencing. (a) Average OTU abundances for each sampled environment for each of the top 10 OTUs. The data were compiled and averaged from each of three sampled hives. Error bars are the results of three independent biological replicates. (b) Sequence abundance for the top 10 OTUs found in each of three sampled hives across four environments.
Environmental habitats may facilitate transfer of bacteria across the colony.
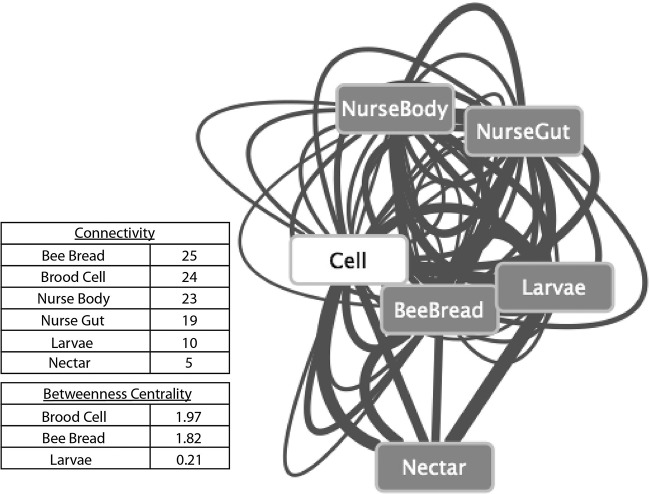
To examine OTU-based relationships between sampling environments, an interaction network was generated. The presence of the same OTU in two different habitats might suggest microbial exchange between the two habitats. By using a classification threshold of 99%, we increased the likelihood that we were observing the same strain in the two different environments; it is not possible to assign directionality to observed interactions through these methods alone. However, if two habitats undergo frequent and extensive microbial exchange, we would expect to observe a large number of shared OTUs. Through pairwise comparisons of environments containing identical OTUs, connections (edges), weighted based on proportion of total sequence abundance observed, were made between environments (nodes). Visual inspection and connectivity analysis suggest the presence of homogeneity in interactions; microbes sampled were found across virtually all hive environments, suggesting that these environments are highly connected (see Fig. 4). As expected, environments that are behaviorally connected, such as the nurse gut and the brood cell, are also connected in this interaction network. In addition, betweenness centrality metrics point to both the brood cell and bee bread as central hubs of the network, through which the OTUs are connected between environments (betweenness centrality: brood cell = 1.97; bee bread = 1.822) (Fig. 3). These environments may act as microbial hubs through which honey bees obtain, deposit, and propagate lactic acid bacteria within the hive and to the next generation.
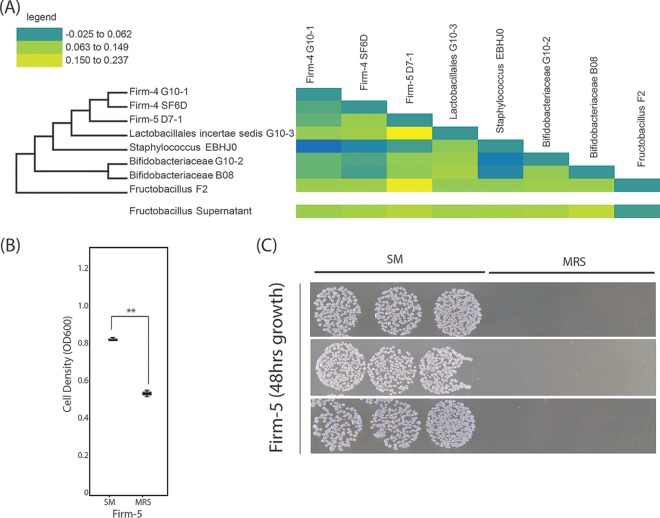
FIG 4.
Coculture of Fructobacillus sp. with honey bee-associated microbes. When honey bee-associated microbes are grown with Fructobacillus or with spent medium from Fructobacillus, they exhibit a growth advantage compared to growth alone. (A) Phylogeny and heat map of isolates used in a coculture interaction assay. The change in optical density from expected values was plotted as a heat map (yellow = more growth than expected; blue = less growth than expected). Lactobacillus and Fructobacillus isolates significantly increased the growth of a Firm-5 isolate. (B) Optical density measurements suggest that Firm-5 grows more robustly in Fructobacillus spent medium (SM) than in MRS medium (t = 11.196, df = 4, P < 0.001). (C) Difference in optical density corresponding to a dramatic difference in colony numbers when Firm-5 is grown on MRS plates postincubation for 48 h in MRS or SM (106 dilutions of cultures plated in triplicate).
FIG 3.
The cell acts as a centralized hub, connecting OTUs between bee-associated environments. The network generated through pairwise examination of environments sharing 99% identical OTUs. Edges weighted based on the proportion of total sequence abundance were observed in that interaction within the subsampled data set. Metrics were not weighted. Connectivity measurements (the number of OTUs shared between environments) show that the honey bee-associated environments sampled are all equally well connected. Centrality measurements (the number of shortest paths from one node to another that passes through this particular node) suggest that the cell serves as a hub through which bacteria may be transferred across environments.
The network analysis was also performed using a 97% classification threshold in order to reinforce results yielded from the 99% classified network. Although the network was less well resolved, and connectivity and centrality measurements were quantitatively different, trends in the data remained unchanged (brood cell and bee bread maintained the highest betweeness centrality, whereas the connectivity was equally distributed across environments). Therefore, our results were not biased based on the OTU divergence threshold used.
Fructobacillus is present early in bee development, found in bee-associated environments, and promotes the growth of honey bee-specific microbes.
Larvae are in contact with both the brood cell and the bee bread during development, and these hive components are known to efficiently transmit Firmicutes (4, 5). Since Fructobacillus and Lactobacillus sp. were found in the brood cell and bee bread, both microbial hubs based on our analyses, we sought to determine whether honey bee core members interacted in vitro with these two taxa. We used coculture assays and found that the “noncore,” yet predominant taxa found associated with the microbial hubs (the brood cell and the bee bread) promoted the growth of bee-specific “core” members (Fig. 4). Specifically, Fructobacillus F2 in coculture with five isolates (Firm-5 D7-1, Firm-4 G10-1, Bifido G10-2, Firm-4 SF6D, and Bifido B08) significantly promoted growth above the expected OD. In addition, Lactobacilliales incertae sedis G10-3 was also associated with positive growth of four isolates (Firm-5 D7-1, Firm-4 G10-1, Firm-4 SF6D, and Bifido B08). In contrast, a honey bee isolate from the Newton lab strain bank not found associated with the hub (Staphylococcus EBHJ0) was associated with the negative growth of two isolates when grown in coculture (Firm-4 G10-1 and Bifido G10-2). To determine whether Fructobacillus F2 mediated interactions with core phyla via by-products of metabolism, we cultured these same core strains with the cell-free supernatant of Fructobacillus F2 spent cultures. The results with Fructobacillus F2 supernatant recapitulated a subset of the results from Fructobacillus F2 cocultures; Fructobacillus F2 supernatant had a similar, positive effect on growth of Bifido G10-2, Firm-4 SF6D, Firm-5 D7-1, and Bifido B08 compared to growth of these isolates alone. Because Firm-5 species are known to associate with second-instar honey bee larvae (24) and therefore are present early in development, we further characterized the potential interaction between a Firm-5 strain (Firm-5 D7-1) and Fructobacillus F2. Spent medium from Fructobacillus F2 significantly increased the differential optical density reached by Firm-5 compared to growth in MRS alone based on both the optical density and the CFU (Fig. 4).
Carbon source utilization of Fructobacillus.
Because Fructobacillus and its spent media promoted the growth other honey bee microbiome members, specifically Firm-5, we reasoned that Fructobacillus could play a syntrophic role, interacting with other bacterial members via metabolic by-products. We therefore characterized the isolate's ability to utilize an array of single carbohydrate sources. Using Biolog MT2 plates inoculated in triplicate with our Fructobacillus isolate and a panel of single simple or complex carbohydrates typically found in the honey bee's diet (see Materials and Methods), we were able to determine that Fructobacillus is capable of utilizing the simple sugars fructose and glucose, in addition to lignin, a plant-derived complex polysaccharide (compared to water-only controls, unpaired t test; P ≤ 0.001 for OD590 measurements postincubation).
Fructobacillus is sensitive to antibiotics.
Honey bees are commonly treated with oxytetracycline, prophylactically, for the prevention of foulbrood diseases caused by the bacteria Melissococcus plutonius and Paenibacillus larvae. Our data suggested that Fructobacillus may produce by-products that promote the growth of honey bee gut core microbiome members. We reasoned it would be important to identify whether this bacterial strain was resistant to antibiotics since treatment might alter the abundance of Fructobacillus, if sensitive. Using soft agar overlays, we exposed Fructobacillus cultures to five different antibiotics (tetracycline, ampicillin [with sulbactam], rifampin, ciprofloxacin, and vancomycin) and measured the resulting zones of inhibition (Table 2). In three of five cases, we found Fructobacillus to be susceptible to these antibiotics. It was most sensitive (the largest zone of inhibition was produced) when exposed to tetracycline or ampicillin. Since these two antibiotics are commonly used in agriculture, they are particularly relevant.
TABLE 2.
Antibiotic sensitivity of isolated Fructobacillus spp.a
| Antibiotic | Concn (μg/ml) | Zone of inhibition (mm) | Interpretation |
|---|---|---|---|
| Tetracycline | 30 | 32 | Susceptible |
| Ampicillin with sulbactam | 20 | 36 | Susceptible |
| Rifampin | 5 | 25 | Susceptible |
| Ciprofloxacin | 5 | 9 | Resistant |
| Vancomycin | 30 | 0 | Resistant |
The zones of inhibition and the concentrations of antibiotics used on Fructobacillus soft agar overlays are shown where the results demonstrated marked sensitivity to most antibiotics.
DISCUSSION
A large number of diverse lactic acid bacteria are found associated with insects, including bees, and have been suggested to provide health benefits in various contexts (8, 25–27). Bacteria that comprise the lactic acid bacteria are not a phylogenetically cohesive designation, based on 16S rRNA evolution, but instead are grouped based on common metabolic characteristics, including the production of lactic acid through fermentative metabolism. Within the honey bee, several different LAB clades have been found associated with the hymenopteran host through culture, clone library sequencing, or amplicon-based sequencing (3, 7, 24). Some of these LAB clades are described as part of the core honey bee microbiome and include Firm-4, Firm-5, and Bifido. In our study, we found OTUs corresponding to each of these honey bee-specific LAB, as well as other noncore OTUs in environmental samples taken from hives, including the bees themselves, as well as bee bread and brood cells (Fig. 1). The genus Fructobacillus has been previously identified as a contributing member to the larval gut community (28) but is not considered a significant contributor to the adult honey bee microbiome; this bacterium is commonly associated with flowers, although it is sometimes identified in association with bees (29–31). However, contigs with homology to Fructobacillus can be found in previously published, culture-independent metatranscriptomic and metagenomic analyses of the honey bee gut (32, 33). It is reasonable to hypothesize that Fructobacillus is a transient member of the bee-associated gut microbiome, primarily associated with foraged foods. However, this does not a priori mean that Fructobacillus, or any transient member of the community, has no effect (direct or otherwise) on the honey bee and its microbiome. In coculture, Fructobacillus has a beneficial effect on the growth of “core” honey bee bacteria. Specifically, Fructobacillus had a significant positive effect on the growth of one Firm-5, two Firm-4, and two Bifido isolates. Fructobacillus was found to utilize the simple sugars fructose and glucose (previously determined to be utilized within the bee gut by the microbial community [34]), as well as the plant carbohydrate lignin. The utilization of lignin by Fructobacillus is particularly interesting; Fructobacillus is associated with pollen and flowers, and this complex plant carbohydrate is found in pollen collected by the honey bee (35–38). Fructobacillus seems therefore well positioned to begin the breakdown of this important plant-derived food.
Fructobacillus supernatants significantly promoted the growth of a subset of honey bee microbiome isolates, and although the exact mechanism has not yet been elucidated, we can hypothesize both direct and indirect interactions. For example, Fructobacillus, a member of the Leuconostocaceae, may produce fermentative products, such as lactic acid, CO2, ethanol, or acetate (39); these products may selectively promote the growth of the honey bee-specific microbial community. Indeed, selective culture of many of the “core” microbiome members requires elevated atmospheric CO2 and acidic media such as MRS (40). Importantly, these effects do not require the presence of Fructobacillus in the bee gut but may be mediated by metabolic products ingested by the bee. Interactions between Fructobacillus and other honey bee-specific microbes may include quorum sensing (via AI-2) or the production of antimicrobials active against noncore members. These potential interactions between honey bee bacteria deserve future study since they could impact community succession.
Early successors of the microbiota in the honey bee gut may have positive effects on bee health due to the promotion of the “core” characteristic bacterial community. Through continued characterization of bee-associated microbes, a number of beneficial interactions with their insect host have been proposed. One such interaction hypothesizes that gut microbes bestow increased metabolic functionality to the honey bee by means of degrading complex polysaccharides that are otherwise inaccessible to the host organism (33). Honey bees subsist on a plant-based diet, consisting of carbohydrates built on strings of β(1–4) glycosidic bonds, in addition to simple sugars such as glucose and fructose (6). Indeed, while genomic studies of the honey bee uncover the presence of several genes coding for β-glucosidases, transcriptomic analysis reveals a pronounced lack in gene expression for these particular enzymes (34). However, upon query of the bacterial community, β-glucosidase genes specific to the breakdown of cellulose were found to be present within and actively transcribed by community members belonging to Actinobacteria, Betaproteobacteria, and Gammaproteobacteria (33, 34). This suggests that members of the gut community may be assisting in the breakdown of plant-based carbohydrates into monomeric subunits, which can be subsequently absorbed by the host. Gut bacteria may also provide immune benefits for their honey bee host. Previous work has suggested that the presence of Bifidobacterium and Lactobacillus in larvae may act to facilitate a more rapid and vigorous immune response against invading pathogens (41). In addition, well-established community members may provide exclusionary effects against potential pathogens. For example, stratified biofilms composed of core community members provide a competitive advantage for these organisms, allowing them to outcompete harmful microbes for important nutrients (4). Exclusionary effects have been previously documented, correlating the presence of Bifidobacterium and other LAB strains with the absence of the pathogens Melissococcus plutonis and Paenibacillus larvae, respectively (42, 43).
We found that honey bee larvae are colonized with Fructobacillus, Alpha-2.2, and Lactobacillales. Other studies of honey bee larvae also identified Alpha-2.2 and Lactobacilliales as early colonizers (24, 44, 45), suggesting that these clades may be commonly found with young, larval bees. It is important to note that these culture-based studies are dependent on the medium used to select for microbial members and that the medium used here (and in previous work) selects for acetic and lactic acid bacteria. Therefore, it is still possible that other community members (such as facultative enteric anaerobes) may be present among early colonizers but cannot be detected through these methods. Regardless, the fact that microbes can consistently be isolated from surface sterilized honey bee larvae suggests bacterial presence, even if methods such as qPCR fail to detect them (2, 5). In addition, the fact that newly eclosed workers, even when exposed to hive components alone, develop a mature gut microbiome containing many of the core members (Firm-5, Firm-4, Frischella, Bifido, Snodgrasella, and Alpha-2.1) (5) supports our observation that comb, including the brood cell, may serve as a source of inoculum for microbial transmission in the hive.
Our results have demonstrated that Fructobacillus and its by-products promote the growth of honey bee gut community members in vitro. Fructobacillus is present in the microbial hubs identified here (the brood cell and bee bread) and able to utilize lignin. This environmental microbe may therefore affect early microbiome development. In addition, we have shown there exists a marked level of sensitivity in Fructobacillus isolates to tetracycline, the most commonly used antibiotic for prophylactic treatment of hives (46) (Table 1). As such, we hypothesize that antibiotic treatment of hives may act to decrease populations of environmental microbes such as Fructobacillus, potentially disrupting the succession of important community members. Our work supports further investigation of “noncore” honey bee community members and suggests that hive environments, including the food prepared by the bees, might impact development of the gut microbiome.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fredrick Lee and Kathy Sheehan for helpful discussions during the design of this experiment and for feedback on the manuscript. Z.P.R. was funded by a grant from the Indiana Academy of Sciences, as well as the IU Biology L. S. McClung and Microbiology Undergraduate Fellowships. M.A.H. was funded by the IU Floyd Microbiology Summer Fellowship and the George Hudock Fellowship. I.L.G.N. was funded through generous startup funds from Indiana University.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01259-15.
REFERENCES
- 1.Newton ILG, Roeselers G. 2012. The effect of training set on the classification of honey bee gut microbiota using the naive Bayesian classifier. BMC Microbiol 12:221. doi: 10.1186/1471-2180-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 3.Moran NA, Hansen AK, Powell E, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol 78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell JE, Martinson VG, Urban-Mead K, Moran NA. 2014. Routes of acquisition of the gut microbiota of Apis mellifera. Appl Environ Microbiol 80:7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydak MH. 1970. Honey bee nutrition. Annu Rev Entomol 15:143. doi: 10.1146/annurev.en.15.010170.001043. [DOI] [Google Scholar]
- 7.Vasquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. 2012. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7:e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasquez A, Olofsson TC. 2009. The lactic acid bacteria involved in the production of bee pollen and bee bread. J Apic Res 48:189–195. doi: 10.3896/IBRA.1.48.3.07. [DOI] [Google Scholar]
- 9.Danzeisen JL, Calvert AJ, Noll SL, McComb B, Sherwood JS, Logue CM, Johnson TJ. 2013. Succession of the turkey gastrointestinal bacterial microbiome related to weight gain. PeerJ 1:e237. doi: 10.7717/peerj.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillilland MG, Erb-Downward JR, Bassis CM, Shen MC, Toews GB, Young VB, Huffnagle GB. 2012. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl Environ Microbiol 78:2359–2366. doi: 10.1128/AEM.05239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favier CF, de Vos WM, Akkermans ADL. 2003. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe 9:219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Jangid K, Whitman WB, Condron LM, Turner BL, Williams MA. 2013. Soil bacterial community succession during long-term ecosystem development. Mol Ecol 22:3415–3424. doi: 10.1111/mec.12325. [DOI] [PubMed] [Google Scholar]
- 14.Schutte UME, Abdo Z, Bent SJ, Williams CJ, Schneider GM, Solheim B, Forney LJ. 2009. Bacterial succession in a glacier foreland of the High Arctic. ISME J 3:1258–1268. doi: 10.1038/ismej.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brucker RM, Bordenstein SR. 2012. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 66:349–362. doi: 10.1111/j.1558-5646.2011.01454.x. [DOI] [PubMed] [Google Scholar]
- 17.Carrasco P, Pérez-Cobas AE, van de Pol C, Baixeras J, Moya A, Latorre A. 2014. Succession of the gut microbiota in the cockroach Blattella germanica. Int Microbiol 17:79–109. [DOI] [PubMed] [Google Scholar]
- 18.Hakim RS, Baldwin K, Smagghe G. 2010. Regulation of midgut growth, development and metamorphosis. Annu Rev Entomol 55:593–608. doi: 10.1146/annurev-ento-112408-085450. [DOI] [PubMed] [Google Scholar]
- 19.Grubbs KJ, Scott JJ, Budsberg KJ, Read H, Balser TC, Currie CR. 2015. Unique honey bee (Apis mellifera) hive component-based communities as detected by a hybrid of phospholipid fatty-acid and fatty-acid methyl ester analyses. PLoS One 10:e0121697. doi: 10.1371/journal.pone.0121697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szalay-Beko M, Palotai R, Szappanos B, Kovacs IA, Papp B, Csermely P. 2012. ModuLand plug-in for Cytoscape: determination of hierarchical layers of overlapping network modules and community centrality. Bioinformatics 28:2202–2204. doi: 10.1093/bioinformatics/bts352. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vojvodic S, Rehan SM, Anderson KE. 2013. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS One 8:e72106. doi: 10.1371/journal.pone.0072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsgren E, Olofsson TC, Vasquez A, Fries I. 2010. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41:99–108. doi: 10.1051/apido/2009065. [DOI] [Google Scholar]
- 26.McFrederick QS, Wcislo WT, Hout MC, Mueller UG. 2014. Host species and developmental stage, but not host social structure, affects bacterial community structure in socially polymorphic bees. FEMS Microbiol Ecol 88:398–406. doi: 10.1111/1574-6941.12302. [DOI] [PubMed] [Google Scholar]
- 27.McFrederick QS, Cannone JJ, Gutell RR, Kellner K, Plowes RM, Mueller UG. 2013. Specificity between lactobacilli and hymenopteran hosts is the exception rather than the rule. Appl Environ Microbiol 79:1803–1812. doi: 10.1128/AEM.03681-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojvodic S, Rehan SM, Anderson KE. 2013. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS One 8:e72106. doi: 10.1371/journal.pone.0072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endo A, Irisawa T, Futagawa-Endo Y, Sonomoto K, Itoh K, Takano K, Okada S, Dicks LM. 2011. Fructobacillus tropaeoli sp. nov., a fructophilic lactic acid bacterium isolated from a flower. Int J Syst Evol Microbiol 61:898–902. doi: 10.1099/ijs.0.023838-0. [DOI] [PubMed] [Google Scholar]
- 30.Anderson KE, Sheehan TH, Mott BM, Maes P, Snyder L, Schwan MR, Walton A, Jones BM, Corby-Harris V. 2013. Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS One 8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch H, Schmid-Hempel P. 2011. Bacterial communities in central European bumblebees: low diversity and high specificity. Microb Ecol 62:121–133. doi: 10.1007/s00248-011-9854-3. [DOI] [PubMed] [Google Scholar]
- 32.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton IL. 2015. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 33.Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee FJR, DB; Stewart FJ; Mattila HR; Newton ILG. 2015. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 35.Brooks J, Shaw G. 1968. Chemical structure of exine of pollen walls and a new function for carotenoids in nature. Nature 219:532–533. [DOI] [PubMed] [Google Scholar]
- 36.Graven P, DeKoster CG, Boon JJ, Bouman F. 1996. Structure and macromolecular composition of the seed coat of the Musaceae. Ann Bot London 77:105–122. doi: 10.1006/anbo.1996.0013. [DOI] [Google Scholar]
- 37.Liu ZH, Ger MJ. 1997. Changes of enzyme activity during pollen germination in maize, and possible evidence of lignin synthesis. Aust J Plant Physiol 24:329–335. doi: 10.1071/PP96015. [DOI] [Google Scholar]
- 38.Weng JK, Mo HP, Chapple C. 2010. Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J 64:898–911. doi: 10.1111/j.1365-313X.2010.04391.x. [DOI] [PubMed] [Google Scholar]
- 39.Nieminen TT, Sade E, Endo A, Johansson P, Björkroth J. 2014. The family Leuconostocaceae, p 215–240. In Rosenberg E, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: firmicutes and tenericutes. Springer, Berlin, Germany. [Google Scholar]
- 40.Engel P, James RR, Koga R, Kwong WK, McFrederick QS, Moran NA. 2013. Standard methods for research on Apis mellifera gut symbionts. J Apic Res 52(4). doi: 10.3896/IBRA.1.52.4.07. [DOI] [Google Scholar]
- 41.Evans JD, Lopez DL. 2004. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J Econ Entomol 97:752–756. doi: 10.1093/jee/97.3.752. [DOI] [PubMed] [Google Scholar]
- 42.Olofsson TC, Vasquez A. 2008. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol 57:356–363. doi: 10.1007/s00284-008-9202-0. [DOI] [PubMed] [Google Scholar]
- 43.Mattila HR, Rios D, Walker-Sperling V, Roeselers G, Newton I. 2012. Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS One 7:e32962. doi: 10.1371/journal.pone.0032962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corby-Harris V, Snyder LA, Schwan MR, Maes P, McFrederick QS, Anderson KE. 2014. Origin and effect of Acetobacteraceae Alpha 2.2 in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl Environ Microbiol 80:7460–7472. doi: 10.1128/AEM.02043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarpy DR, Mattila HR, Newton ILG. 2015. Development of the honey bee gut microbiome throughout the queen-rearing process. Appl Environ Microbiol 81:3182–3191. doi: 10.1128/AEM.00307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans JD. 2003. Diverse origins of tetracycline resistance in the honey bee bacterial pathogen Paenibacillus larvae. J Invertebr Pathol 83:46–50. doi: 10.1016/S0022-2011(03)00039-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.