Abstract
In this study, host-associated molecular markers and bacterial 16S rRNA gene community analysis using high-throughput sequencing were used to identify the sources of fecal pollution in environmental waters in Brisbane, Australia. A total of 92 fecal and composite wastewater samples were collected from different host groups (cat, cattle, dog, horse, human, and kangaroo), and 18 water samples were collected from six sites (BR1 to BR6) along the Brisbane River in Queensland, Australia. Bacterial communities in the fecal, wastewater, and river water samples were sequenced. Water samples were also tested for the presence of bird-associated (GFD), cattle-associated (CowM3), horse-associated, and human-associated (HF183) molecular markers, to provide multiple lines of evidence regarding the possible presence of fecal pollution associated with specific hosts. Among the 18 water samples tested, 83%, 33%, 17%, and 17% were real-time PCR positive for the GFD, HF183, CowM3, and horse markers, respectively. Among the potential sources of fecal pollution in water samples from the river, DNA sequencing tended to show relatively small contributions from wastewater treatment plants (up to 13% of sequence reads). Contributions from other animal sources were rarely detected and were very small (<3% of sequence reads). Source contributions determined via sequence analysis versus detection of molecular markers showed variable agreement. A lack of relationships among fecal indicator bacteria, host-associated molecular markers, and 16S rRNA gene community analysis data was also observed. Nonetheless, we show that bacterial community and host-associated molecular marker analyses can be combined to identify potential sources of fecal pollution in an urban river. This study is a proof of concept, and based on the results, we recommend using bacterial community analysis (where possible) along with PCR detection or quantification of host-associated molecular markers to provide information on the sources of fecal pollution in waterways.
INTRODUCTION
Fecal indicator bacteria (FIB), such as Escherichia coli and Enterococcus spp., have long been used as indirect measures of public health risks associated with environmental waters (1, 2). However, the use of FIB to identify the health risks associated with enteric viruses and protozoa has been questioned because of their poor cooccurrence or correlation (3–5). Some strains of FIB have been reported to have the ability to adapt in the environment and to persist in sediment and vegetation (6, 7). The major limitation of FIB is that they cannot be assigned to a specific original source due to their cosmopolitan nature (being frequently found in different warm-blooded and some cold-blooded animals) (8, 9). When environmental waters are polluted with FIB from multiple sources, it becomes extremely difficult to implement a robust management plan without identifying the potential sources of this pollution.
Over the past 2 decades, library-dependent and library-independent microbial source tracking (MST) methods have been developed to differentiate between sources of fecal pollution in environmental waters. The MST methods developed earlier were library dependent and required the collection and fingerprinting of FIB from host groups (fecal sources) and environmental waters to identify the dominant sources of fecal pollution (10, 11). However, limitations of these library-dependent methods in correctly assigning FIB to their host groups have been criticized in the literature (8, 12–14). In contrast, library-independent methods mainly involved identifying a specific DNA sequence or a target gene of a bacterial species found in human or animal feces. Over the past 15 years, numerous molecular markers have been developed to identify, and in some cases to quantify, the magnitude of fecal pollution in environmental waters from specific hosts (9). The validation of these markers is based on several performance characteristics, such as host specificity, host sensitivity, evenness in the feces, persistence in environments, and relevance to health risks (9, 11). None of the markers possesses all of the desirable performance characteristics, however, and it has been recommended that a “toolbox” approach should be used for accurate identification of polluting sources (15–17).
In recent years, bacterial community analyses using next-generation sequencing have emerged as promising library-dependent MST tools. These methods have allowed better characterization of bacterial communities from environmental waters (18–20). Unno and colleagues reported the development of a new library-dependent method using pyrosequencing-derived shared operational taxonomic units (OTUs) to identify the sources of fecal pollution in waterways in South Korea (21). Their results indicated that the majority of bacteria in the feces of humans and domesticated animals belonged to the phyla Bacteroidetes and Firmicutes, whereas the predominant bacteria in the feces of geese and in freshwater samples belonged to Proteobacteria. Using this method, the authors were able to determine that human and swine feces were the sources of fecal pollution in the river. Cao and colleagues evaluated terminal restriction fragment length polymorphism (TRFLP), phylogenetic microarray, and next-generation sequencing (Illumina) data for 64 blind samples, from single or dual sources, that were generated from 12 host groups. They determined that all three methods were able to identify correctly the dominant sources of host groups for 95% of the blind samples (22).
The aim of the current study was to assess toolbox approaches using host-associated molecular markers and bacterial 16S rRNA gene community analyses to identify the sources of fecal pollution in environmental waters in Brisbane, Australia. Fecal samples were collected from six host groups and surface water from the Brisbane River. Bacterial communities were sequenced using the Illumina MiSeq platform. In addition, the water samples were tested for the presence of bird-, cattle-, horse-, and human-associated molecular markers to provide multiple lines of evidence regarding the presence of fecal pollution from specific sources.
MATERIALS AND METHODS
Water sampling.
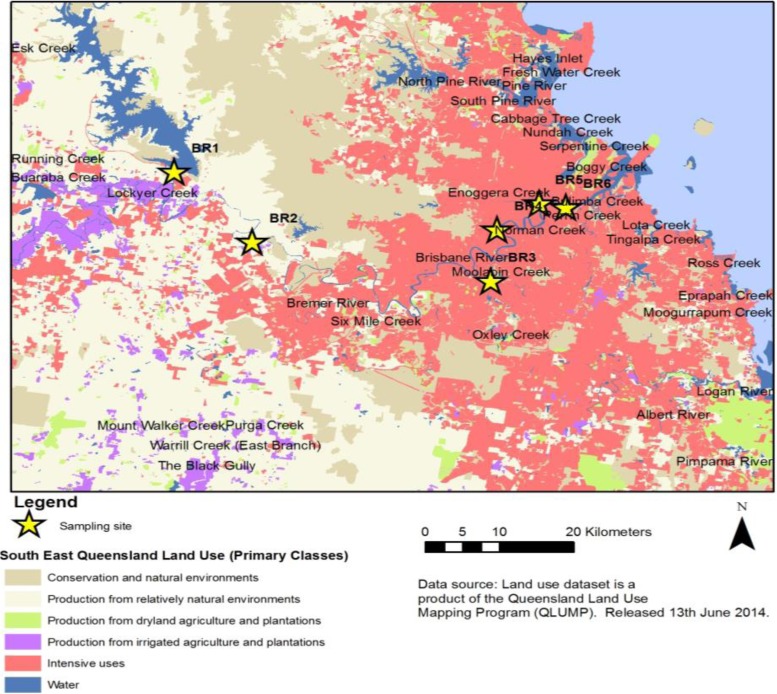
In December 2013, water samples were collected from six sites (designated BR1 to BR6) along the Brisbane River in Queensland, Australia (Fig. 1). Each site was sampled in triplicate at one event, giving a total of 18 samples. Sampling site descriptions and GPS coordinates are provided in Table S1 in the supplemental material. Site BR1 is located on the upper reaches of the Brisbane River. This site receives overflow of water from the Wivenhoe Reservoir. Site BR2 is located in a periurban nonsewered catchment. Site BR3 is at a major tributary of the Brisbane River and is tidally influenced. The catchment where site BR3 is located has residential and industrial developments and is serviced by a wastewater treatment plant (WWTP). Site BR4 is located in a highly urban area and also is tidally influenced. This site receives urban runoff through a stormwater drain. Sites BR5 and BR6 are located on the lower reaches of the river, in highly urbanized areas. A 10-liter water sample was collected from each site, in sterile carboy containers, 30 cm below the water surface. The water samples were transported on ice to the laboratory and were processed within 6 to 8 h.
FIG 1.
Map of the Brisbane River, showing sampling sites.
Enumeration of fecal indicator bacteria.
The membrane filtration method was used for isolation and enumeration of fecal indicator bacteria (FIB). Serial dilutions of water samples were made in sterile MilliQ water and were filtered through nitrocellulose membranes (pore size, 0.45 μm; diameter, 47 mm; Millipore, Tokyo, Japan). Dilutions were placed on modified membrane-thermotolerant Escherichia coli (mTEC) agar (Difco, Detroit, MI) and membrane-Enterococcus indoxyl-d-glucoside (mEI) agar (Difco) for the isolation of E. coli and Enterococcus spp., respectively. Modified mTEC agar plates were incubated at 35°C for 2 h to recover stressed cells, followed by incubation at 44°C for 22 h (23), while mEI agar plates were incubated at 41°C for 48 h (24).
Biomass collection from water samples.
A 2-liter subsample of the 10-liter sample from each site was filtered through a nitrocellulose membrane (pore size, 0.45 μm; diameter, 47 mm; Millipore). In cases of membrane clogging due to turbidity, multiple membranes were used. The membrane (or membranes) was immediately transferred to a 15-ml sterile tube containing phosphate-buffered saline (Sigma-Aldrich, St. Louis, MO). The sample tube was shaken with a vortex mixer for 5 min to detach the microbial biomass from the membrane, followed by centrifugation at 4,500 × g for 15 min at 4°C to yield a pellet (25).
Host group sampling.
Fecal and wastewater samples were collected from six host groups. Individual cat fecal samples (n = 14) were collected from the veterinary hospital located at the University of Queensland, Gatton Campus, on two separate occasions. Composite cattle wastewater samples (mixtures of urine and feces; n = 12) were collected from two abattoirs located on the outskirts of Brisbane on three separate occasions. Individual dog fecal samples (n = 14) were collected from a dog park and a veterinary hospital on two separate occasions. Individual horse fecal samples (n = 14) were collected from a horse racecourse on one occasion. Individual kangaroo fecal samples (n = 14) were collected from the Lone Pine Koala Sanctuary. Composite human wastewater samples (n = 24) were collected from the primary influent of four WWTPs, serving 50,000 to 500,000 people in Brisbane and the Sunshine Coast region, on six separate occasions. All fecal samples were collected from the fresh defecation of each animal (cat, dog, horse, and kangaroo). All samples were transported on ice to the laboratory, stored at 4°C, and processed within 6 h.
Biomass collection from cattle and human wastewater samples.
The composite cattle and human wastewater samples were concentrated using an Amicon Ultra-15 centrifugal filter device (molecular weight cutoff value, 30,000; Millipore). In brief, 10 ml of a wastewater sample was added to the Amicon device and centrifuged at 4,000 × g for 10 min. Approximately 180 to 200 μl of concentrated sample was collected from the filter device sample reservoir by using a pipette (26). The concentrated sample was stored at −20°C for a maximum of 24 h prior to DNA extraction.
DNA extraction.
DNA was extracted from the pellet obtained from each water sample by using a Mo Bio PowerSoil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA). A DNeasy blood and tissue kit (Qiagen, Valencia, CA) was used to extract DNA from the concentrated cattle and human wastewater samples. A QIAamp stool DNA kit (Qiagen) was used to extract DNA directly from 100 to 220 mg of fresh individual animal fecal samples. The extracted DNA samples were quantified using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technology, Wilmington, DE).
Real-time PCR assays.
The previously published primer sequences and amplification conditions for real-time PCR assays that were used in this study are shown in Table S2 in the supplemental material. Each DNA sample was amplified using a previously published universal bacterial real-time PCR assay to confirm the presence of DNA (27). The universal bacterial real-time PCR amplification was performed in 20-μl reaction mixtures using SsoFast EvaGreen supermix (Bio-Rad Laboratories, Richmond, CA). The universal bacterial real-time PCR assay mixtures contained 10 μl of supermix, 300 nM each primer, and 2 μl of template DNA. Sketa22 real-time PCR assays were undertaken to assess the potential presence of PCR inhibitors in the DNA samples extracted from fecal/wastewater and water samples, according to previously published methods (28, 29). Sketa22 real-time PCR assays were performed in 25-μl reaction mixtures using iQ supermix (Bio-Rad Laboratories). The real-time PCR assay mixtures contained 12.5 μl of supermix, 300 nM each primer, 400 nM probe, 2 μl of template DNA, and 10 pg (2 μl) of Oncorhynchus keta DNA.
The HF183 (30, 31), GFD (32), and horse (33) real-time PCR assays were performed in 20-μl reaction mixtures using SsoFast EvaGreen supermix (Bio-Rad Laboratories). The real-time PCR mixtures contained 10 μl of supermix, 300 nM each primer (for HF183), 100 nM each primer (for GFD), or 300 nM each primer (for horse), and 2 μl of template DNA. To separate the specific product from nonspecific products, including primer dimers, melting curve analysis was performed for each real-time PCR run. During melting curve analysis, the temperature was increased from 65°C to 95°C in 0.5°C increments. The CowM3 (34) real-time PCR mixtures contained 12.5 μl of iQ supermix (Bio-Rad Laboratories), 400 nM each primer, 80 nM probe, and 5 μl of template DNA. All real-time PCR assays were performed in triplicate. For each real-time PCR assay, triplicate positive and negative (sterile water) controls were included.
PCR and Illumina MiSeq sequencing.
The V5 and V6 regions of the 16S rRNA gene were amplified using the primer set described previously (35) (see Table S2 in the supplemental material). Amplicons from each sample were pooled in equal amounts. All samples were sequenced (paired-end sequenced at a length of 300 nucleotides [nt] in each direction) by the University of Minnesota Genomics Center (Minneapolis, MN), using version 3 chemistry on the MiSeq platform.
Sequence data analysis.
Sequence processing was performed using mothur software (version 1.33.3) (36). Sequences were trimmed to 150 nt and paired-end joined using fastq-join (37). Quality trimming was performed to remove sequences with average quality scores of <35 over a window of 50 nt, homopolymers of >8 nt, ambiguous bases, or mismatches to primer sequences. High-quality sequences were aligned against the SILVA database (version 115) (38). Sequences were further quality trimmed by using a 2% precluster error (39, 40) and chimera removal using UCHIME (41). Assignment of OTUs was performed at 97% identity using the furthest-neighbor algorithm. Taxonomic assignments were made against the Ribosomal Database Project database (version 9) (42). Samples were grouped by environmental water or animal host groups. For comparisons, groups were normalized to include 11 samples per group, each randomly subsampled to 25,000 sequence reads (275,000 sequence reads per group). Rarefaction curves based on the numbers of OTUs for water samples and host groups are shown in Fig. S1 to S7 in the supplemental material. For determination of the percentages of sequence reads and OTUs (97% sequence similarity) unique to each group, no normalization was performed.
The Bayesian classifier software SourceTracker was used to assign sources of fecal pollution in water samples (43). SourceTracker calculated the probability that an OTU present in the bacterial community of environmental water samples (the sink) came from host groups (the source). In this study, all samples with rarefication to 100, 1,000, and 10,000 sequence reads, using default settings at α values of 0.001, were used in SourceTracker when possible. If fewer sequences were available for a sample, then all sequences for the sample were used. In contrast to the method used by Shanks and colleagues (44), all source groups were included in SourceTracker runs at the different rarefaction depths, and SourceTracker assignments of source contributions are reported. Details regarding the mathematical modeling for source-specific OTU assignments are reported elsewhere (43).
Statistical analyses.
Alpha diversity indices, including sample coverage, number of OTUs observed, Shannon diversity index (45), and abundance-based coverage estimate (ACE) (46), were calculated using mothur. These diversity indices were chosen to provide both parametric and nonparametric estimates of diversity. For comparison of community structures, Bray-Curtis dissimilarity (47) was calculated. Differences in phylogenetic composition and abundance-weighted phylogenetic composition among sample groups were evaluated using unweighted and weighted UniFrac metrics (48), respectively, in mothur. Similarly, differences in beta diversity were evaluated using analysis of similarity (ANOSIM) (49), and the significance of group clustering was evaluated in mothur using analysis of molecular variance (AMOVA) (50). Spearman rank correlation and binary logistic regression analyses were performed to determine the correlations between microbial water quality parameters, using SPSS statistical software (version 19.2; IBM, Armonk, NY). All statistics were evaluated with α values of 0.05.
Sequence data accession number.
Raw sequence data from MiSeq sequencing were received as fastq files and were submitted to the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA257794.
RESULTS
Bacterial community diversity and composition.
The coverage of the sample groups (275,000 sequence reads per group, with 11 independent samples subsampled to 25,000 sequence reads) ranged from 97 to 100%, with an average of 96% ± 1.2% among all samples. Cat and dog fecal samples and cattle wastewater samples had significantly lower Shannon diversity index values, observed richness (Sobs) values, and estimated richness values based on the ACE index than did human wastewater samples, horse and kangaroo fecal samples, and Brisbane River water samples (P < 0.05) (Table 1). The Brisbane River water samples had the highest richness values based on the ACE index. Notably, cattle wastewater samples showed considerably greater variability in both richness and diversity than did the other samples tested (Table 1). Conversely, bacterial communities in fecal samples from pets (cats and dogs) and kangaroos showed comparatively little variation in richness, based on both observed richness and ACE index values, but these groups also had the lowest richness values, compared to human wastewater samples, horse fecal samples, and Brisbane River water samples.
TABLE 1.
Shannon diversity indices, observed richness values, and abundance-based coverage estimates of richness for sample groups
| Sample group | Mean ± SDa |
||
|---|---|---|---|
| Shannon diversity index | Sobs | ACE index | |
| Brisbane River water | 5.00 ± 0.24 A | 1,725 ± 172 AC | 4,024 ± 1,035 A |
| Human wastewater | 4.70 ± 0.40 A | 1,443 ± 324 A | 3,102 ± 710 E |
| Cat feces | 3.15 ± 0.38 B | 215 ± 26 B | 318 ± 55 BCF |
| Cattle wastewater | 2.07 ± 2.00 B | 374 ± 729 B | 557 ± 861 BCF |
| Dog feces | 2.13 ± 0.75 B | 145 ± 26 B | 222 ± 69 BCF |
| Horse feces | 5.80 ± 0.73 A | 1,972 ± 341 C | 2,881 ± 776 E |
| Kangaroo feces | 5.15 ± 0.93 A | 1,348 ± 168 A | 1,816 ± 170 D |
Values followed by the same capital letter were not significantly different at an α of 0.05 via Tukey's post hoc test.
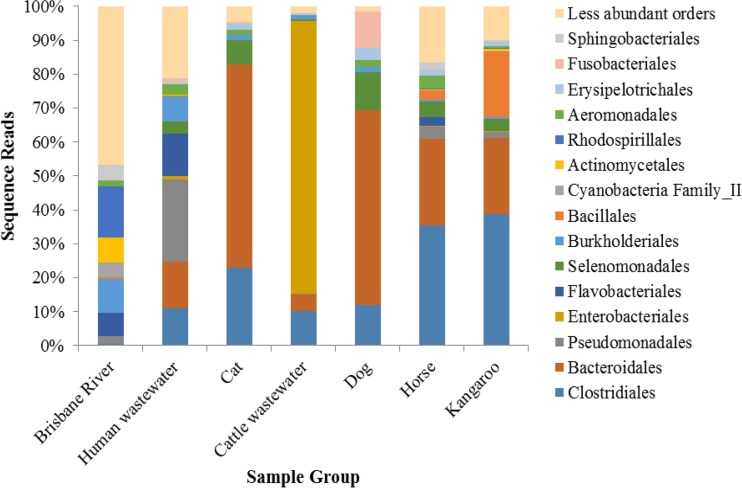
Among fecal samples, communities were predominantly composed of Clostridiales, Bacteroidales, and Enterobacteriales (Fig. 2). The WWTP samples showed low relative abundances of Enterobacteriales but higher relative abundances of Pseudomonadales. Orders that were abundant in fecal samples accounted for only ∼50% of sequence reads for the Brisbane River water samples, which included predominantly Rhodospirillales (15% of reads), Burkholderiales (10%), an unclassified order of Cyanobacteria (8%), Actinomycetales (8%), Flavobacteriales (7%), and Alteromonadales (6%). All other orders accounted for fewer than 5% of reads in the Brisbane River samples. Distributions of the 15 most abundant orders among 11 individual samples from the Brisbane River and host groups are shown in Fig. S8 to S14 in the supplemental material.
FIG 2.
Distribution of the 15 most abundant orders among samples (11 samples per group, with sequence numbers normalized). A total of 148 orders were identified among all samples.
Variations in host-associated communities.
In order to assess maximum variations in community compositions, the percentages of sequence reads and OTUs that were associated with each host group were calculated using all samples, without normalization or subsampling (Table 2). Host-associated communities were defined as communities that were unique to a particular host group. The horse and kangaroo host groups were composed of 59% and 40% host-associated communities, respectively, and represented the majority of species richness in these samples. Human and cattle wastewater communities showed approximately 10% and 8% host association, respectively, accounting for about 50% of the species richness, while cat and dog communities were highly cosmopolitan. Of particular note, the predominant host-associated OTUs (those unique to a host group) among human wastewater, cat fecal, cattle wastewater, horse fecal, and kangaroo fecal samples belonged to Clostridiales (24 to 47%). Bacteroidales were also predominant in cattle wastewater, horse fecal, and kangaroo fecal samples (32 to 37%). In contrast, cat fecal samples were dominated by Coriobacteriales (23%) and Erysipelotrichales (19%), whereas dog fecal samples were dominated by Anaeroplasmatales (46%).
TABLE 2.
Distributions of host-associated communities and operational taxonomic units among fecal sources, without normalization or subsampling
| Sample group | No. of samples in group | No. of sequence readsa | Host-associated communitiesb (%) | Host-associated OTUsc (%) | Predominant unique ordersd (%) |
|---|---|---|---|---|---|
| Human wastewater | 24 | 1,367,124 | 10 | 62 | Clostridiales (24), Bacteroidales (11), Selenomonadales (8), Verrucomicrobiales (8), Burkholderiales (6) |
| Cat feces | 14 | 1,150,863 | 0.9 | 23 | Clostridiales (47), Coriobacteriales (23), Erysipelotrichales (19), Desulfovibrionales (6) |
| Cattle wastewater | 12 | 1,342,864 | 8 | 49 | Clostridiales (38), Bacteroidales (37) |
| Dog feces | 14 | 921,392 | 0.4 | 19 | Anaeroplasmatales (46), Coriobacteriales (14), Fusobacteriales (10), Clostridiales (10), Bacteroidales (6), Erysipelotrichales (5) |
| Horse feces | 14 | 770,531 | 59 | 75 | Clostridiales (33), Bacteroidales (32), Verrucomicrobiales subdivision 5 (8) |
| Kangaroo feces | 14 | 934,609 | 40 | 62 | Clostridiales (38), Bacteroidales (36), Verrucomicrobiales (7), Selenomonadales (7) |
Total number of high-quality sequences for the group.
Percentage of communities associated with the host group.
Percentage of host-associated OTUs among the OTUs present in the group.
Orders accounting for >5% of host-associated sequence reads.
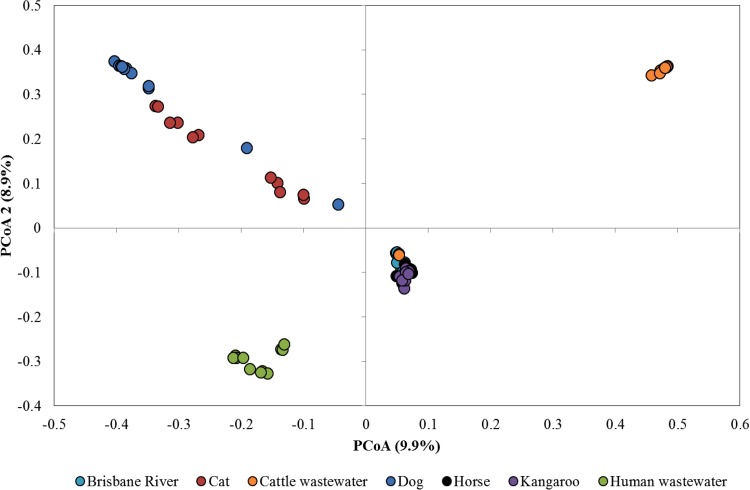
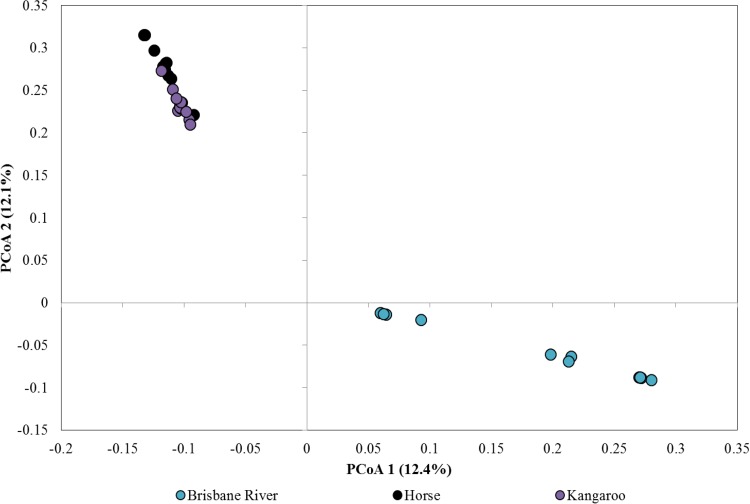
Weighted UniFrac distances were significantly different among all host groups (P ≤ 0.004), indicating differences in abundance-weighted phylogenetic community structures. However, unweighted UniFrac distances differed significantly only among Brisbane River water, kangaroo fecal, and human wastewater samples (P ≤ 0.027). Unweighted phylogenetic structures did not differ significantly among most host groups and Brisbane River samples (P ≥ 0.063). Ordination of samples via principal-coordinate analysis (PCoA) indicated that samples tended to group by source (Fig. 3). Apparent overlap of Brisbane River water, horse fecal, and kangaroo fecal samples when all samples were plotted was further resolved (Fig. 4). Brisbane River water samples, however, as well as horse and kangaroo fecal samples, were still highly similar. Nevertheless, clustering of samples by source was supported by AMOVA (P < 0.001).
FIG 3.
Principal-coordinate analysis (PCoA) of all source-specific samples and water samples (11 samples subsampled to 25,000 sequences). A total of 99 axes were required to explain total variance.
FIG 4.
Principal-coordinate analysis (PCoA) of samples clustered in the center of Fig. 3. A total of 55 axes were required to explain total variance.
Microbial water quality parameters and sources of fecal pollution in the Brisbane River.
Among the 18 water samples analyzed from the six sites, all samples yielded culturable E. coli and Enterococcus spp. The concentrations of E. coli and Enterococcus spp. in water samples ranged from 12 to 480 CFU per 100 ml of water and from 3 to 600 CFU per 100 ml of water, respectively (Table 3). The concentrations of E. coli and Enterococcus spp. were much greater in water samples from sites BR3 and BR4 than from the other sites. In contrast, the percentages of Enterobacteriales were relatively low among communities characterized using 16S rRNA gene sequencing, with this order accounting for 0.004 to 0.032% of sequence reads. The percentages of Enterobacteriales were not correlated with the concentrations of either E. coli or Enterococcus spp. (Spearman r values of 0.492 and 0.445, with P values of 0.124 and 0.169, respectively). Among the host-associated molecular markers tested, the bird-associated GFD marker was more prevalent than the others. Among the 18 samples tested, 15 (83%), 6 (33%), 3 (17%), and 3 (17%) were PCR positive for the GFD, HF183, CowM3, and horse markers, respectively (Table 3). The host specificity and host sensitivity values for the GFD, HF183, CowM3, and horse markers are shown in Table S3 in the supplemental material.
TABLE 3.
Sampling sites, numbers (ranges) of fecal indicator bacteria, and real-time PCR positive/negative results for host-associated molecular markers
| Sampling sitea | No. of FIB (CFU/100 ml) |
Detection of molecular markersb |
||||
|---|---|---|---|---|---|---|
| E. coli | Enterococcus spp. | CowM3 | HF183 | GFD | Horse | |
| BR1 | 12–18 | 3–6 | − − − | − − − | + + + | − − − |
| BR2 | 51–80 | 19–25 | + + + | − − − | − − − | − − − |
| BR3 | 242–340 | 510–600 | − − − | + + + | + + + | − − − |
| BR4 | 395–480 | 522–580 | − − − | + + + | + + + | − − − |
| BR5 | 121–160 | 174–208 | − − − | − − − | + + + | + + − |
| BR6 | 70–103 | 150–182 | − − − | − − − | + + + | − − − |
Triplicate samples from each site were tested.
−, negative for the marker; +, positive for the marker. The result for each of the triplicate samples tested is shown.
The potential sources of fecal pollution in water samples from the Brisbane River, analyzed using DNA sequencing, were determined using SourceTracker (Table 4). Brisbane River water samples tended to show relatively small contributions from WWTPs (up to 13% at rarefaction to 100 reads and ≤4% at ≥1,000 reads). Contributions from other animal sources were rarely detected and were very small (≤3% of sequence reads). Notably, rarefaction depth had a significant effect on host group assignments for Brisbane River samples. Among river samples, contributions attributed to horses and WWTPs were significantly greater at 100 sequence reads than at either 1,000 or 10,000 reads (P values of 0.001 and 0.006 to 0.007, respectively).
TABLE 4.
Percentages of source assignments determined using SourceTracker with rarefaction to 100, 1,000, and 10,000 sequence reads
| Sampling site | Rarefaction size (no. of sequence reads) | % of samples assigned to indicated sourcea |
|||||
|---|---|---|---|---|---|---|---|
| Cat | Cattle | Dog | Horse | Kangaroo | WWTPs | ||
| BR1 | 100 | — | — | 0–1 | 0–1 | 0–1 | 2–12 |
| 1,000 | — | — | — | — | — | 0–4 | |
| 10,000 | — | — | — | — | — | 0–3 | |
| BR2 | 100 | 0–1 | 0–1 | 0–1 | 0–1 | — | 1–6 |
| 1,000 | — | — | — | — | — | 1–1 | |
| 10,000 | — | — | — | — | — | 1–1 | |
| BR3 | 100 | — | — | — | — | — | 1–13 |
| 1,000 | — | — | — | — | — | 0–2 | |
| 10,000 | — | — | — | — | — | 0–2 | |
| BR4b | 100c | 0–1 | 0–1 | 0–2 | 0–1 | — | 1–2 |
| 1,000c | — | — | — | — | — | 0–1 | |
| BR5 | 100 | 0–1 | — | — | 0–1 | — | 0–2 |
| 1,000 | — | — | — | — | — | 0–1 | |
| 10,000 | — | — | — | — | — | 0–1 | |
| BR6 | 100 | — | — | — | 0–1 | 0–1 | 2–7 |
| 1,000 | — | — | — | — | — | 2–3 | |
| 10,000 | — | — | — | — | — | 1–2 | |
—, could not be assigned to a source.
No replicate had 10,000 sequence reads for analysis.
One replicate was excluded from analysis due to a paucity of sequence reads.
Correlations between microbial water quality parameters.
Among all water samples collected, the concentrations of E. coli were significantly and positively correlated with those of Enterococcus spp. by Spearman rank correlation (r = 0.910, P < 0.001). Binary logistic modeling relating the concentrations of E. coli and Enterococcus spp. to real-time PCR detection of host-associated molecular markers for all samples was performed. The regression models for the CowM3 and horse markers were not significant (χ2 = 5.20, P = 0.074). The model for the HF183 marker was significant (χ2 = 11.03, P = 0.001) and explained 40.4% of the variations in detection (Nagelkerke R2). Similarly, the model for the GFD marker was significant (χ2 = 25.30, P < 0.001) and explained 67.9% of the variance; the analysis revealed that samples with elevated concentrations of E. coli (P = 0.008), but not Enterococcus spp. (P = 0.108), were 82 times more likely to test positive for this marker.
Due to differences in detection and extents of source assignments, correlations and binary logistic models were calculated for Brisbane River samples at each rarefaction depth using only SourceTracker data for samples with sufficient sequence reads for rarefaction. Correlation analysis revealed a significant negative correlation (r = −0.496, P = 0.043) between E. coli concentrations and percentages of sequence reads assigned to sewage for Brisbane River samples. Spearman correlations were not significant. Binary logistic models did not significantly explain the variations in detection of the CowM3 (χ2 ≤ 10.83, P ≥ 0.146), HF183 (χ2 ≤ 11.52, P ≥ 0.103), GFD (χ2 ≤ 10.83, P ≥ 0.146), or horse (χ2 ≤ 10.83, P ≥ 0.146) marker.
DISCUSSION
Microbial community analysis can be used to obtain information about different bacterial florae that are unique to animal feces and environments such as soils and water (51, 52). The recent advancement and reduced costs of high-throughput sequencing have accelerated the analysis of microbial communities of animal feces and environmental water samples (53). However, only a few studies have investigated the potential application of next-generation sequencing for MST (21, 22, 54–57). In this study, we developed a library of bacterial sequences from six host groups as a proof of concept. The sequences in the library were then compared to the sequences obtained from an urban river in order to assess the utility of bacterial community analysis for MST field studies. The V5-to-V6 region of the 16S rRNA gene was selected as the target, because this region was shown previously to provide richness estimates comparable to those from full-length 16S rRNA gene analysis. Furthermore, Illumina gene sequencing offers significantly more reads with improved quality than other next-generation sequencing platforms (58, 59).
The Shannon diversity indices for cat fecal samples in this study were greater than those in a previous study, but those for dog fecal samples were similar (60). Among the host groups, human wastewater samples had significantly greater observed richness, Shannon diversity indices, and ACE richness than did cat and dog fecal samples. Cattle wastewater samples also showed considerably greater variability in both diversity and richness than did fecal samples from other host groups. This could be due to the fact that the cattle wastewater samples were mixtures of urine and feces and were collected from wastewater drains potentially containing bacterial populations from environmental sources in addition to those of fecal origin. The higher-diversity indices for human wastewater samples could be attributed to wastewater-associated bacterial communities as well as human fecal bacteria from a large population releasing wastewater into the WWTP stream (57). The Shannon diversity index for horse fecal samples was similar to the value reported in previous studies that investigated the 16S rRNA gene in horse fecal samples using pyrosequencing (61–63). Diversity indices can vary greatly among studies, which can be attributed to factors such as variable numbers of reads used in the analyses, different OTU similarity cutoff values, the target regions sequenced, and the methods used, in addition to variations due to the DNA extraction kits used in the studies (64).
Direct comparisons of taxonomic orders for individual fecal samples from cat, dog, horse, and kangaroo host groups showed that the bacterial communities were predominantly composed of Clostridiales and Bacteroidales. In contrast, the Enterobacteriales order was highly prevalent in composite cattle wastewater samples. This is not in agreement with a previous study that reported that Firmicutes was the most abundant phylum across all communities in dairy cows (65). Such a discrepancy could be due to the fact that we collected cattle wastewater samples from wastewater drains in abattoirs, whereas Pitta and colleagues (65) collected ruminant contents from the stomachs of dairy cows. It is possible that portions of microbial communities in the drains came from other animals, such as pigs and sheep, which were also present in the abattoirs. In addition, the possibility of regrowth of Enterobacteriales in the drains cannot be ruled out. Since the cattle wastewater samples were composite samples, it is also possible that the mixtures of feces and urine from large numbers of cattle contributed to the greater abundance of Enterobacteriales. In contrast, Brisbane River water samples were dominated by Rhodospirillales, Burkholderiales, Actinomycetales, and Flavobacteriales. The presence of these taxonomic orders in surface waters has been reported previously (66–68).
Host-associated communities were greatest in horse fecal samples, followed by kangaroo fecal and human wastewater samples. The percentages of host-associated OTUs among the host groups ranged from 19 to 75%. Among the human wastewater samples, these OTUs were classified among Clostridiales, Bacteroidales, Selenomonadales, Verrucomicrobiales, and Burkholderiales. However, OTUs belonging to Clostridiales and Bacteroidales were also identified in the other host groups. Therefore, it appears that the use of a single order for source identification may not be suitable, although unique OTUs exist among host groups. Similar observations were also reported by Unno and colleagues (21). However, differences in DNA extraction procedures, different microbial decay rates, and temporal variations in the bacterial community might have artificially resulted in differences in the observed community structures. Therefore, the conclusions presented here should be interpreted cautiously and as preliminary only. Further studies are needed in order to identify novel host-associated markers based on species-specific sequence differences, which may allow verification of the relative contributions of known host groups.
Evaluation of fecal contamination at high taxonomic resolution (orders) will likely not result in robust assessments of pollution, as several host groups may possess OTUs classified in the same taxa, as described above. Thus, for determination of fecal pollution using sequence analysis, we employed a previously developed OTU-based Bayesian classification approach, SourceTracker (43), to improve the accuracy of source assignments. However, due to differences between the host and the environment, not all host-associated species were likely to persist and be detected in water samples.
Application of SourceTracker indicated that all water samples collected from the Brisbane River had human wastewater signatures representing 1 to 13% of the bacterial community analyzed. Similar results were reported in a previous study that investigated the presence of human wastewater in water samples from harbor, river, and storm water outfall sites in Milwaukee, Wisconsin, and Lake Michigan (57). In Milwaukee, the environmental water samples demonstrated 0.2 to 12.2% human wastewater signatures during dry and wet weather periods. Small numbers of samples were occasionally classified as being polluted with feces from animals. However, the contributions from animals were small. Such low-level fecal pollution could be attributed to the fact that the water samples were collected during dry weather conditions, when the river flow was reduced. In addition, the SourceTracker software estimated the probability of the presence of the OTUs in environmental samples derived from the host groups. Large variations in the host community or small sample sizes for the host community tend to decrease the chances of assignment of an OTU to its source (57). At present, it is not known what constitutes a representative library. In this study, the numbers of samples included in the host groups ranged from 12 to 24. It is possible that these samples did not capture the variations in the bacterial communities, as required for effective source identification. It has been reported that large variations in bacterial communities may occur among human fecal samples (69, 70). A large library composed of more fecal samples might improve the ability to identify the sources of fecal pollution in environmental waters. Notably, the rarefaction depth used for SourceTracker assignments influenced the percentages of source contributions to the water samples; greater rarefaction depth tended to reduce the percentages of source-associated OTUs. The influences of reference library size and rarefaction depth on the accuracy of SourceTracker assignments have not yet been carefully evaluated. However, the results presented herein suggest that these parameters may be of critical importance. Therefore, SourceTracker assignments were evaluated in the context of established and well-validated host-associated molecular markers (9).
In this study, source contributions determined via sequence library analysis versus host-associated molecular markers in the same river water sample showed variable agreement. For example, water samples from site BR2 were positive for the CowM3 marker and were classified as being polluted with cattle feces using low-rarefaction sequence analysis. Samples from only two sites (BR3 and BR4) were positive for the HF183 marker, whereas all six sites (BR1 to BR6) were classified as being polluted with human wastewater using sequence analysis. Discrepancies between results obtained using sequence analysis and HF183 detection may be due in part to bias in the taxa amplified using different primer sets. For example, the HF183 target occurs in the V2 hypervariable region and may amplify species or strains of Bacteroidales not amplified using the V5/V6 primer set. Most of the water samples from the Brisbane River were positive for the GFD marker, but the results could not be compared to the sequence analysis results because our sequence library did not contain any sequences from avian hosts. We attempted to sequence a number of bird fecal samples, but the samples did not pass the quality control check at the sequencing center. The results obtained using host-associated markers should be interpreted with care, as HF183, CowM3, and GFD were detected in fecal samples from nontarget host groups in Brisbane, Australia (3, 71) (see Table S3 in the supplemental material). Taken together, our data clearly suggest the presence of human wastewater pollution in the tested water samples. A recent study also reported the presence of chronic sewage pollution in the Brisbane River, using a toolbox of host-associated molecular markers (72).
Concentrations of E. coli and Enterococcus spp. were significantly related to detection of the human wastewater-associated HF183 marker but were poorly related to the CowM3 marker and reads assigned to sewage. These results suggest that, while some of these markers may be useful indicators of sewage pollution, they may fail to indicate human health risks from nonhuman sources. The lack of a relationship between FIB, host-associated molecular markers, and community structure likely reflects the differences in methodologies, with FIB analysis providing data on the concentrations of viable E. coli or Enterococcus spp., host-associated molecular marker detection providing information on the presence or absence of host-associated fecal pollution, and community sequence analysis providing data on the relative abundance of pollution. Discrepancies between these methods have been noted previously (56). Host-associated molecular markers represent a single taxonomically narrow group of target microorganisms and thus are suitable for detection or quantification of specific sources of fecal pollution, but they fail to provide a comprehensive assessment of source contributions (73). However, comparisons of sequence libraries created from larger numbers of samples from various hosts may ultimately allow for more comprehensive assessment of source contributions by identifying multiple sources of fecal pollution in a single sample. This could be advantageous for scenarios in which the pollution sources are not known or in which mixed sources of pollution affect waterways.
In this study, we show that bacterial community structure and molecular marker analyses can be combined to identify potential sources of fecal pollution in an urban river. The bacterial community-based approach is an emerging technique that will require further validation in order to determine optimal library size, sample type (individual samples versus composite samples), and sequencing depth, similar to the original library-dependent MST methodologies developed in the late 1990s (9). Nonetheless, this study is a proof of concept that has provided multiple lines of evidence; based on the results obtained, we recommend using sequence-based analysis to obtain information on the multiple sources of fecal pollution (where possible) and then quantifying host-associated molecular markers of interest for the effective management of environmental water quality. One limitation of the community-based approach is that it takes time to develop a library, and it is yet not known what constitutes a representative library. At present, very little information is available on the geographical and temporal stability of the bacterial sequences in host groups. The analysis of large data sets can be time-consuming and remains a significant challenge, although several open-source tools, such as mothur, QIIME, and SourceTracker, have been developed by genomic centers and are efficient, flexible, and simple to use.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded as part of the CSIRO Capability Development Theme.
We thank the University of Minnesota Genomics Center for DNA sequencing. Our sincere thanks go to Stephen Cook for supplying the GIS map.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02032-15.
REFERENCES
- 1.Tallon P, Magajna B, Lofranco C, Leung KT. 2005. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Pollut 166:139–166. doi: 10.1007/s11270-005-7905-4. [DOI] [Google Scholar]
- 2.Griffith JF, Cao YP, McGee CD, Weisberg SB. 2009. Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res 43:4900–4907. doi: 10.1016/j.watres.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed W, Sritharan T, Palmer A, Sidhu JPS, Toze S. 2013. Evaluation of bovine feces-associated microbial source tracking markers and their correlations with fecal indicators and zoonotic pathogens. Appl Environ Microbiol 79:2682–2691. doi: 10.1128/AEM.03234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McQuaig S, Griffith J, Harwood VJ. 2012. The association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl Environ Microbiol 78:6423–6432. doi: 10.1128/AEM.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badgley BD, Thomas FI, Harwood VJ. 2011. Quantifying environmental reservoirs of fecal indicator bacteria associated with sediment and submerged aquatic vegetation. Environ Microbiol 13:932–942. doi: 10.1111/j.1462-2920.2010.02397.x. [DOI] [PubMed] [Google Scholar]
- 7.Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl Environ Microbiol 72:612–621. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field KG, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 9.Harwood VJ, Staley C, Badgely BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 10.Scott TM, Rose JB, Jenkins TM, Farrah SR, Lukasik J. 2002. Microbial source tracking: current methodology and future directions. Appl Environ Microbiol 68:5796–5803. doi: 10.1128/AEM.68.12.5796-5803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoeckel DM, Harwood VJ. 2007. Performance, design, and analysis in microbial source tracking studies. Appl Environ Microbiol 73:2405–2415. doi: 10.1128/AEM.02473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon DM, Bauer S, Johnson JR. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513–1522. [DOI] [PubMed] [Google Scholar]
- 13.Hartel PG, Summer JD, Hill JL, Collins JV, Entry JA, Segars WI. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J Environ Qual 31:1273–1278. doi: 10.2134/jeq2002.1273. [DOI] [PubMed] [Google Scholar]
- 14.Harwood VJ, Wiggins BA, Hagedorn C, Ellender RD, Gooch J, Kern J, Samadpour M, Chapman ACH, Robinson BJ. 2003. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study. J Water Health 1:153–166. [PubMed] [Google Scholar]
- 15.Ahmed W, Sidhu JPS, Toze S. 2012. Evaluation of the nifH gene marker of Methanobrevibacter smithii for the detection of sewage pollution in environmental waters. Environ Sci Technol 46:543–550. doi: 10.1021/es203372u. [DOI] [PubMed] [Google Scholar]
- 16.Mauffret A, Caparis MP, Gourmelon M. 2012. Relevance of Bacteroidales and F-specific RNA bacteriophages for efficient fecal contamination tracking at the level of a catchment in France. Appl Environ Microbiol 78:5143–5152. doi: 10.1128/AEM.00315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noble RT, Griffith JF, Blackwood AD, Fuhrman JA, Gregory JB, Hernandez X, Liang X, Bera AA, Schiff K. 2006. Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Appl Environ Microbiol 72:1604–1612. doi: 10.1128/AEM.72.2.1604-1612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y, Van De Werfhorst LC, Sercu B, Murray JLS, Holden PA. 2011. Application of an integrated community analysis approach for microbial source tracking in a coastal creek. Environ Sci Technol 45:7195–7201. doi: 10.1021/es201118r. [DOI] [PubMed] [Google Scholar]
- 19.Dubinsky EA, Esmaili L, Hulls JR, Cao Y, Griffith JF, Anderson GL. 2012. Application of phylogenetic microarray analysis to discriminate sources of fecal pollution. Environ Sci Technol 46:4340–4347. doi: 10.1021/es2040366. [DOI] [PubMed] [Google Scholar]
- 20.Staley C, Unno T, Gould TJ, Jarvis B, Phillips J, Cotner JB, Sadowsky MJ. 2013. Application of Illumina next-generation sequencing to characterize the bacterial community of the upper Mississippi River. J Appl Microbiol 115:1147–1158. doi: 10.1111/jam.12323. [DOI] [PubMed] [Google Scholar]
- 21.Unno T, Jang J, Han D, Kim JH, Sadowsky MJ, Kim O-S, Chun J, Hur H-G. 2010. Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Environ Sci Technol 44:7777–7782. doi: 10.1021/es101500z. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Van De Werfhorst LC, Dubinsky EA, Badgely BD, Sadowsky MJ, Anderson GL, Griffith JF, Holden PA. 2013. Evaluation of molecular community analysis methods for discerning fecal sources and human waste. Water Res 47:6862–6872. doi: 10.1016/j.watres.2013.02.061. [DOI] [PubMed] [Google Scholar]
- 23.Office of Water, Environmental Protection Agency. 1997. Method 1600: membrane filter test method for enterococci in water. EPA/821/R-97/004. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 24.Office of Water, Environmental Protection Agency. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane thermotolerant Escherichia coli agar (modified mTEC). EPA/821/R-02/023. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 25.Ahmed W, Sawant S, Huygens F, Goonetilleke A, Gardner T. 2009. Prevalence and occurrence of zoonotic bacterial pathogens in surface waters determined by quantitative PCR. Water Res 43:4918–4928. doi: 10.1016/j.watres.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed W, Goonetilleke A, Gardner T. 2010. Human and bovine adenoviruses for the detection of source-specific fecal pollution in coastal waters in Australia. Water Res 44:4662–4673. doi: 10.1016/j.watres.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Boon N, Top EM, Verstraete W, Siciliano SD. 2003. Bioaugmentation as a tool to predict the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl Environ Microbiol 69:1511–1520. doi: 10.1128/AEM.69.3.1511-1520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed W, Harwood VJ, Gyawali P, Sidhu JPS, Toze S. 2015. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl Environ Microbiol doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haugland A, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res 39:559–568. doi: 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic markers with real-time PCR for assessment of human fecal pollution in freshwater. Environ Microbiol 7:249–259. doi: 10.1111/j.1462-2920.2004.00702.x. [DOI] [PubMed] [Google Scholar]
- 32.Green HC, Dick LK, Gilpin B, Samadpour M, Field KG. 2012. Genetic markers for rapid PCR-based identification of gull, Canada goose, duck, and chicken fecal contamination in water. Appl Environ Microbiol 78:503–510. doi: 10.1128/AEM.05734-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dick LK, Bernhard AE, Brodeur TJ, Santo Domingo JW, Simpson JM, Walters SP, Field KG. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl Environ Microbiol 71:3184–3191. doi: 10.1128/AEM.71.6.3184-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanks OC, Atikovic E, Blackwood AD, Lu J, Noble RT, Santo Domingo J, Seifring S, Sivaganesan M, Haugland RA. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl Environ Microbiol 74:745–752. doi: 10.1128/AEM.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claesson MJ, Wang QO, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinforma J 7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 38.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 41.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. 2013. Comparison of microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL. [Google Scholar]
- 46.Chao A, Lee S-M. 1992. Estimating the number of classes via sample coverage. J Am Stat Assoc 87:210–217. doi: 10.1080/01621459.1992.10475194. [DOI] [Google Scholar]
- 47.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 48.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 50.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JE, Lee S, Sung J, Ko G. 2011. Analysis of human and animal fecal microbiota for microbial source tracking. ISME J 5:362–365. doi: 10.1038/ismej.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas T, Gilbert J, Meyer F. 2012. Metagenomics: a guide from sampling to data analysis. Microb Inform Exp 2:3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong JY, Park HD, Lee KH, Weon HY, Ka JO. 2011. Microbial community analysis and identification of alternative host-specific fecal indicators in fecal and river water samples using pyrosequencing. J Microbiol 49:585–594. doi: 10.1007/s12275-011-0530-6. [DOI] [PubMed] [Google Scholar]
- 55.McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML. 2010. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12:378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neave M, Luter H, Padovan M, Townsend S, Schobben X, Gibb K. 2014. Multiple approaches to microbial source tracking in tropical northern Australia. Microbiologyopen 3:860–874. doi: 10.1002/mbo3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL. 2013. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb Ecol 65:1011–1023. doi: 10.1007/s00248-013-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. 2009. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75:5227–5236. doi: 10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacteria and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 61.Dougal K, de La Fuente G, Harris PA, Gridwood SE, Pinloche E, Newbold CJ. 2013. Identification of a core bacterial community within the large intestine of the horse. PLoS One 8:e77660. doi: 10.1371/journal.pone.0077660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shepherd ML, Swecker WS, Jensen RV, Ponder MA. 2012. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V5 gene amplicons. FEMS Microbiol Lett 326:62–68. doi: 10.1111/j.1574-6968.2011.02434.x. [DOI] [PubMed] [Google Scholar]
- 63.Steelman SM, Chowdhary BP, Dowd S, Suchodolski J, Janečka JE. 2012. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet Res 8:231. doi: 10.1186/1746-6148-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gihring TM, Green SJ, Schadt CW. 2012. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- 65.Pitta DW, Kumar S, Vecchiarelli B, Shirley DJ, Bittinger K, Baker LD, Ferguson JD, Thomsen N. 2014. Temporal dynamics in the ruminal microbiome of dairy cows during the transition period. J Anim Sci 92:4014–4022. doi: 10.2527/jas.2014-7621. [DOI] [PubMed] [Google Scholar]
- 66.Ponce-Terashima R, Moskey AM, Reis MG, McLellan SL, Blanton RE. 2014. Sources and distribution of surface water fecal contamination and prevalence of schistosomiasis in a Brazilian village. PLoS Negl Trop Dis 8:e3186. doi: 10.1371/journal.pntd.0003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. 2014. Bacterial community structure is indicative of chemical inputs in the upper Mississippi River. Front Microbiol 5:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. 2015. Species sorting and seasonal dynamics primarily shape bacterial communities in the upper Mississippi River. Sci Total Environ 505:435–445. doi: 10.1016/j.scitotenv.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 69.Huse SM, Ye Z, Zhou Y, Fodor A. 2012. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporasp JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmed W, Masters N, Toze S. 2012. Consistency in the host specificity and host sensitivity of the Bacteroides HF183 marker for sewage pollution tracking. Lett Appl Microbiol 55:283–289. doi: 10.1111/j.1472-765X.2012.03291.x. [DOI] [PubMed] [Google Scholar]
- 72.Sidhu JPS, Hodgers L, Ahmed W, Chong NM, Toze S. 2012. Prevalence of human pathogens and indicators in stormwater run-off in Brisbane, Australia. Water Res 46:6652–6660. doi: 10.1016/j.watres.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 73.McLellan SL, Boehm AB, Shanks OC. 2012. Marine and freshwater fecal indicators and source identification, p 199–235. In Meyers RA. (ed), Encyclopedia of sustainability science and technology. Springer, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.