Abstract
Aggregation of misfolded protein in the endoplasmic reticulum (ER) induces a cellular protective response to ER stress, the unfolded protein response (UPR), which is mediated by a basic leucine zipper (bZIP) transcription factor, Hac1p/Xbp1. In this study, we identified and studied the molecular functions of a HAC1 homolog from the thermotolerant yeast Hansenula polymorpha (HpHAC1). We found that the HpHAC1 mRNA contains a nonconventional intron of 177 bp whose interaction with the 5′ untranslated region is responsible for the translational inhibition of the HpHAC1 mRNA. The H. polymorpha hac1-null (Hphac1Δ) mutant strain grew slowly, even under normal growth conditions, and was less thermotolerant than the wild-type (WT) strain. The mutant strain was also more sensitive to cell wall-perturbing agents and to the UPR-inducing agents dithiothreitol (DTT) and tunicamycin (TM). Using comparative transcriptome analysis of the WT and Hphac1Δ strains treated with DTT and TM, we identified HpHAC1-dependent core UPR targets, which included genes involved in protein secretion and processing, particularly those required for N-linked protein glycosylation. Notably, different glycosylation and processing patterns of the vacuolar glycoprotein carboxypeptidase Y were observed in the WT and Hphac1Δ strains. Moreover, overexpression of active HpHac1p significantly increased the N-linked glycosylation efficiency and TM resistance. Collectively, our results suggest that the function of HpHac1p is important not only for UPR induction but also for efficient glycosylation in H. polymorpha.
INTRODUCTION
The endoplasmic reticulum (ER) represents the central organelle of the eukaryotic cell in which the crucial steps of protein folding, modification, and selection for transport to other compartments take place. Perturbation of ER homeostasis through, for example, the accumulation of misfolded proteins or defects in the ER membrane leads to the activation of intracellular signaling pathways referred to as the unfolded protein response (UPR), ER-associated protein degradation (ERAD), and the calnexin cycle (CNX) for glycoprotein quality control (1). UPR induces multiple protective cellular events required for proper protein folding and misfolded protein degradation. The underlying regulatory mechanism and modes of action of UPR have been intensively studied in the traditional yeast Saccharomyces cerevisiae (2). UPR is mediated by a master transcription factor, Hac1p, in S. cerevisiae (ScHac1p) (3), and the synthesis of functionally active Hac1p is triggered via nonconventional splicing of the HAC1 mRNA, which is mediated by the endonuclease activity of the ER stress sensor, Ire1p (4). This unusual splicing of HAC1 mRNA induces the translation of HAC1 mRNA by eliminating base pairing between the 5′ untranslated region (UTR) and the splicing site (5). The newly synthesized Hac1p is then shuttled into the nucleus and functions as an active basic leucine zipper (bZIP) transcription factor during ER stress. Hac1p binds to a UPR element (UPRE) within the promoter regions of its target genes. The consensus motif of UPRE1, CANCNTG, is found in the promoters of the KAR2, PDI1, and FKB2 genes in S. cerevisiae (6).
The protein secretion pathways of yeast and filamentous fungi are of special interest to researchers that are developing industrial protein producers. Understanding the underlying mechanisms of UPR would provide solid knowledge on the regulation of cellular responses to protein secretion stress (7). In this aspect, the HAC1 homologs of non-Saccharomyces yeasts and filamentous fungi that are potential hosts for the industrial production of recombinant proteins have been identified and characterized. Unlike S. cerevisiae HAC1 mRNA, which contains an intron of 252 nucleotides (nt), the hac1/hacA genes encoding the key UPR transcription factors from the filamentous fungi Trichoderma reesei and Aspergillus nidulans contain introns that are 20 nt long (8). Similarly, Yarrowia lipolytica HAC1 mRNA harbors a short intron of 29 nt (9). In contrast, the intron of Pichia pastoris HAC1 mRNA is 322 nt, which is longer than the S. cerevisiae HAC1 mRNA intron (10). Despite the differences in intron length, the HAC1 mRNAs of these yeast and filamentous fungi undergo similar, unconventional splicing reactions to produce functional Hac1 proteins (8, 9, 11).
Hansenula polymorpha (syn., Pichia angusta and Ogataea angusta) is a thermotolerant methylotrophic yeast which can grow at temperatures up to 48°C and utilize methanol as a sole carbon and energy source. H. polymorpha has been a favorable model to study the mechanisms of genetic control of methanol metabolism and peroxisome biogenesis. In recent decades, H. polymorpha has also been used as a host for heterologous protein production (12). In particular, glycoengineered H. polymorpha strains have been developed to produce glycoproteins with human-compatible N-glycans on the basis of information on the host-specific structure and biosynthesis pathway of N-linked glycosylation in H. polymorpha (13–15). Likewise, a recent study on the identification and functional analysis of protein O-mannosyltransferases (PMTs) in H. polymorpha provided some unique features of PMT proteins in H. polymorpha (16). In this study, to obtain further details on the gene regulatory networks required for protein secretion and modification in H. polymorpha, we identified and characterized the HAC1 homolog of H. polymorpha (HpHAC1) via functional analysis and genome-wide gene expression profiling.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The H. polymorpha and S. cerevisiae strains used in this study are described in Table S1 in the supplemental material. The plasmids used in this study are listed in Table S2 in the supplemental material, and the primers used for the construction of strains and plasmids are listed in Table 1. Yeast cells were routinely grown in YPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose) or synthetic complete (SC) medium (0.67% yeast nitrogen base without amino acids, 2% glucose, dropout amino acid mixture supplemented with all required amino acids). Hphac1 and Hpire1 deletion mutants were constructed in the H. polymorpha DL1-L background by fusion PCR and in vivo DNA recombination (15), using the primer sets listed in Table 1. PCR fragments were introduced into yeast cells, and the transformants carrying the gene deletion as a result of in vivo DNA recombination were selected first by growth in minimal medium lacking leucine (SC-LEU) and then by PCR screening.
TABLE 1.
PCR primers used in this study
| Primer purpose and primera | Gene | Sequence (5′ → 3′) |
|---|---|---|
| H. polymorpha gene deletion | ||
| IRE1 NF | HpIRE1 | CCGGAATTGCTCCACTCC |
| IRE1 NR | HpIRE1 | CCTAAACACGCACGCCTCACGCCTGTCACTCTATTGAGAGC |
| IRE1 CF | HpIRE1 | CGTATTACCAAACCGCTTACGTACGTTGATGGACTTGCTCCGAGC |
| IRE1 CR | HpIRE1 | CTGTCCATCTGTAGACTCTGG |
| HAC1 NF | HpHAC1 | GCACAGTGTTGTATCAAACG |
| HAC1 NR | HpHAC1 | CCTAAACACGCACGCCTCACAGCTGTTGAGAGCAGTCAT |
| HAC1 CF | HpHAC1 | CGTATTACCAAACCGCTTACGTACGCTCTTTTAATAGCGTGCAT |
| HAC1 CR | HpHAC1 | CCAACGGTAAGAAATTCAAG |
| LEU2 NF | HpLEU2 | TTGGTGGAATCTACTTTGGT |
| LEU2 NR | HpLEU2 | GTGAGGCGTGCGTGTTTAGG |
| LEU2 CF | HpLEU2 | GTACGTAAGCGGTTTGGTAATACG |
| LEU2 CR | HpLEU2 | CAAACATGTTGTTGGTGACA |
| Expression vector construction | ||
| HpHAC1_HA_1F | HpHAC1 | GATGTTCCTGACTATGCGACTGCTCTCAACAGCTCT |
| HpHAC1_2B_SalI | HpHAC1 | TAGTCGACTCAAGACAAATAGTCGTCAAATTC |
| HpHAC1_HA_3F_HindIII | HpHAC1 | CAAAAGCTTATGTACCCATACGATGTTCCTGACTATGC |
| HpHAC1_4B_SalI | HpHAC1 | TAGTCGACCTTGTAGATGACATGTAGTG |
| HpHAC1p_9F_AscI | HpHAC1 | ATGGCGCGCCTGTGCATTCTTCTGTATACGG |
| HpHAC1p_10B_EcoRI | HpHAC1 | ACGAATTCTTTATTATGGGTATTTGTTTGAT |
| HpHAC1_11F_EcoRI | HpHAC1 | ACGAATTCATGACTGCTCTCAACAGC |
| HpHAC1PE_16B | HpHAC1 | GTTGTGAGACGATTTTTTCGATAACGATTTTGGCGAT |
| HpHAC1PE_17F | HpHAC1 | AAAAAATCGTCTCACAACCGCAAAAGAAAGGCGCAA |
| HpHAC1p fw BamHI | HpHAC1 | CGGGATCCACAAAGTAACGGAAGC |
| HpHAC1t rv BamHI | HpHAC1 | CGGGATCCTGTACACCAAATGAATTAAGAAAGC |
| ScACT1p_F_KpnI | ScACT1(p)b | GGGGTACCGCTTTGGACTCCACCAACGT |
| ScACT1p_B_BglII | ScACT1(p) | GAAGATCTTGTTAATTCAGTAAATTTTCGATC |
| ScHAC1p_F_KpnI | ScHAC1(p) | GGGGTACCAGAGCCACTATCATCGGCG |
| ScHAC1p_B_BglII | ScHAC1(p) | GAAGATCTAGTGGCGGTTGTTGTCGTAG |
| RT-PCR analysis | ||
| ACT1 F | HpACT1 | TCCAGGCTGTGCTGTCGTTG |
| ACT1 R | HpACT1 | ACCGGCCAAGTCGATTCTCA |
| ALG5 F | HpALG5 | TGGCTCCAGAGCACACATGG |
| ALG5 R | HpALG5 | CGTGTCCTTGATGGTGCGAA |
| ERO1 F | HpERO1 | ACACAGGCATCGGACAACCC |
| ERO1 R | HpERO1 | CAGAACGTTGCCGAGCTCCT |
| KAR2 F | HpKAR2 | ATTTTCCACGGCGGCTGATA |
| KAR2 R | HpKAR2 | TTGAGGGACGCCTCTTGGTG |
| LHS1 F | HpLHS1 | CAGTCTCGAGGCGTCGCTTT |
| LHS1 R | HpLHS1 | AACGTCCTCGAGCCACTCCA |
| MNN2 F | HpMNN2 | CCCAAACGTGGCTTGACTCC |
| MNN2 R | HpMNN2 | CCGGTCCTCCAGCTCCTTTT |
| OCR5 F | HpOCR5 | TGATCTACTGCGAGGGCGGT |
| OCR5 R | HpOCR5 | GCCCAGTTTTCGGCGTTATG |
| OST1 F | HpOST1 | ACAACTTCACCGTCGGCTGG |
| OST1 | HpOST1 | TTCCGGCAGGAAGAACGAGA |
| OST2 F | HpOST2 | CGTGGCGCAATTTGTGCTTA |
| OST2 R | HpOST2 | AGTCACCCAATGCGCGTTCT |
| PDI1 F | HpPDI1 | GGCGTCGAGATCACCGGATA |
| PDI1 R | HpPDI1 | ATGGCCAAACCGTCAACACC |
| SCJ1 F | HpSCJ1 | GCAATTTGGGCTATCGACGC |
| SCJ1 R | HpSCJ1 | GAAACGGAATTGTGCGCTCC |
| SWP1 F | HpSWP1 | GCGCTTTGGCAGAAAACCAG |
| SWP1 R | HpSWP1 | CAAAGCAACGACACCACCGA |
| WBP1 F | HpWBP1 | CGTGTGGAAAACTCGTCGCA |
| WBP1 R | HpWBP1 | AAGCTGTATCCCTCGCGCTG |
F, forward; R, reverse.
(p), the promoter of each gene.
Construction of HpHAC1 expression vectors.
The vectors pDUM2-HA-HAC1s and pDUM2-HA-HAC1u for expression of HpHac1p tagged at its N terminus by the epitope corresponding to amino acids 98 to 106 of human influenza virus hemagglutinin (HA) under the control of the MOX1 promoter were constructed as follows. The DNA fragment containing a spliced form of HpHAC1 (HpHAC1s) was amplified from the cDNA of dithiothreitol (DTT)-treated wild-type (WT) H. polymorpha cells with primers HpHAC1_HA_1F/HpHAC1_2B_SalI and subsequently subjected to a second round of PCR with primers HpHAC1_HA_3F_HindIII/HpHAC1_2B_SalI to add an HA sequence tag at the N terminus. The DNA fragment containing the unspliced form of HpHAC1 (HpHAC1u) was amplified from genomic DNA by using primers HpHAC1_HA_1F/HpHAC1_4B_SalI and subjected to a second round of PCR with primers HpHAC1_HA_3F_HindIII/HpHAC1_4B_SalI to add an HA sequence tag at the N terminus. The HA-HpHAC1s fragment (1,008 bp) and the HA-HpHAC1u fragment (1,185 bp) were digested with HindIII and SalI and cloned into the corresponding sites of the pDUM2-cmycYPS1N4 vector (S. A. Cheon, unpublished data), creating pDUM2-HA-HAC1s and pDUM2-HA-HAC1u, respectively. The vector pDUH-HA-HAC1s, expressing the N-terminally HA-tagged, spliced HpHac1p (HA-Hac1ps) under the control of its native promoter, was constructed as follows. The HpHAC1 promoter (759 bp), amplified with primers HpHAC1p_9F_AscI/HpHAC1p_10B_EcoRI from genomic DNA of DL1-L, and the HpHAC1s fragment (981 bp), amplified with primers HpHAC1_11F_EcoRI/HpHAC1_2B_SalI from pDUM2-HA-HAC1s, were digested with AscI/EcoRI and EcoRI/SalI, respectively, and inserted between the AscI and SalI sites of pDUM2-HA-HAC1u, resulting in pDUH-HA-HAC1s.
The expression vector for the N-terminally HA-tagged, spliced HpHac1 protein lacking the PEST domain, pDUH-HA-HAC1sΔP, was constructed as follows. To amplify the HpHAC1sΔPEST gene fragment lacking the putative PEST motif, the N-terminal fragment (458 bp) and the C-terminal fragment (431 bp) of HAC1s were amplified with primers HpHAC1_11F_EcoRI/HpHAC1PE_16B and HpHAC1PE_17F/HpHAC1_2B_SalI from pDUM2-HA-HAC1s, respectively, and then fused by PCR using primers HpHAC1_11F_EcoRI/HpHAC1_2B_SalI to generate the HpHAC1sΔPEST fragment (858 bp). Then the HpHAC1 promoter fragment (759 bp), obtained by AscI and SalI digestion of pDUH-HA-HAC1s, and the EcoRI/SalI-treated HpHAC1sΔPEST fragment (858 bp) were cloned between the AscI and SalI sites of pDUH-HA-HAC1u, resulting in pDUH-HA-HAC1sΔP. To integrate a single-copy vector into the HpHAC1 locus, pDUH-HA-HAC1s and pDUH-HA-HAC1sΔP were digested with NheI, which has a unique recognition site located in the HpHAC1 promoter, and transformed into the WT and Hphac1Δ strains.
The vectors pHZ-HA-HAC1u and pHZ-HA-HAC1s for expression of the N-terminally HA-tagged, unspliced and spliced HpHac1 proteins, respectively, under the control of the native HpHAC1 promoter and terminator were constructed as follows. The DNA fragment containing the HpHAC1 promoter (783 bp), the HpHAC1u open reading frame (972 bp), and the HpHAC1 terminator were amplified from the genomic DNA with primers HpHAC1p fw BamHI/HpHAC1t rv BamHI, digested with BamHI, and inserted between the BglII and BamHI sites of pHIMAZCH, resulting in pZAM519. Subsequently, the NheI/ClaI HA-HAC1u-treated fragment containing the 5′ UTR from pDUH-HA-HAC1u was subcloned between the NheI and ClaI sites of pZAM519, generating pHZ-HA-HAC1u. The vector pHZ-HA-HAC1s for expression of the N-terminally HA-tagged spliced HpHAC1s was constructed by replacing the XhoI/BamHI-treated HA-HAC1u fragment of pHZ-HA-HAC1u with the XhoI/BamHI-treated HA-HAC1s fragment from pDUH-HA-HAC1s. To integrate a single-copy vector into the HpHAC1 locus, the pHZ-HA-HAC1u and pHZ-HA-HAC1s vectors were digested with NheI, which has a unique recognition site located in the HpHAC1 promoter, and transformed into the WT and Hpire1Δ strains.
Western blot analysis.
To detect HA-tagged HpHac1p, total protein extracts were prepared by bead beating cells in lysis buffer (50 mM Tris [pH 7.6], 25 mM ammonium acetate, 1 mM EDTA) containing a protease inhibitor cocktail (Sigma, St. Louis, MO, USA) and 1 mM phenylmethylsulfonyl fluoride (PMSF). The cell extract was concentrated by trichloroacetic acid (TCA) precipitation. The dried TCA precipitates were resuspended in sample loading buffer containing 100 mM Tris (pH 11.0) and 3% sodium dodecyl sulfate (SDS) and heated at 100°C for 5 min. SDS-PAGE was performed on 4 to 20% gradient gels, immunoblotting was performed with an anti-HA antibody (Roche, Basel, Switzerland), and proteins were detected using an alkaline phosphatase substrate kit (Bio-Rad, CA, USA). To detect intracellular H. polymorpha carboxypeptidase Y (HpCPY) and S. cerevisiae CPY (ScCPY) in H. polymorpha, total protein extracts were prepared by bead beating cells in 50 mM sodium phosphate buffer (pH 7.5) containing protease inhibitor cocktails and 1 mM PMSF. After centrifugation at 14,240 × g for 10 min at 4°C, total cell lysates were obtained, and 30 μg of proteins was separated by SDS-PAGE for immunoblotting with anti-HpCPY antibody (13) or anti-ScCPY antibody (Invitrogen, Carlsbad, CA). The blots were detected using an ECL Advance Western blotting detection kit (GE Healthcare, Buckinghamshire, United Kingdom).
Transcriptome analysis using a cDNA microarray.
H. polymorpha DL1-L and the Hphac1Δ mutant were initially inoculated at an optical density at 600 nm (OD600) of 0.1 and incubated to an OD600 of 0.3. After treatment with either 5 mM DTT or 5 μg/ml tunicamycin (TM), samples were collected at the time points indicated below. Total RNA was isolated using the hot phenol method and purified using an RNeasy minikit (Qiagen, USA) according to the manufacturer's instructions. cDNA was synthesized from 10 to 20 μg of total RNA using a special RT-dT primer kit (Genisphere, Hatfield, PA, USA), which contained a specific capture sequence, and SuperScript reverse transcriptase (Invitrogen, Carlsbad, CA). The capture region of the synthesized cDNA was labeled with fluorescent dyes using a 3DNA Submicro EX expression array detection kit (Genisphere, Hatfield, PA, USA). After hybridization, the microarray slides (Gene Expression Omnibus [GEO] database accession number GPL4802) were washed with SSC buffer (0.15 M sodium chloride and 0.015 M sodium citrate) and then scanned with a ScanArray 5000 scanner (Packard, Billerica, CA). All arrays were analyzed using Quintet, an R-based unified cDNA microarray data analysis system with a graphical user interface (17). Genes with similar expression patterns across the time points were clustered using the K-means clustering program.
Accession numbers.
The microarray data have been deposited in the GEO database with the accession numbers GSE67084, GSE68528, and GSE68217. The DNA sequences of HpHAC1 and H. polymorpha IRE1 (HpIRE1) were deposited in GenBank under accession numbers DQ679915 and KP279465, respectively.
RESULTS
Identification and characterization of H. polymorpha HAC1.
The UPR, which is essential for proper protein folding and for degradation of misfolded proteins upon ER stress, is mediated by the master transcription factor Hac1p in S. cerevisiae (3). We identified an H. polymorpha homolog (HpHAC1) of S. cerevisiae HAC1 whose translated product, which contains an amino acid sequence similar to that of the DNA binding domain, exhibits a high degree of sequence identity with the DNA binding region of S. cerevisiae Hac1p (see Fig. S1A in the supplemental material). We also identified an H. polymorpha homolog of S. cerevisiae IRE1 (HpIRE1) whose translated product has a high degree of overall sequence identity (43.6%) with S. cerevisiae Ire1p (see Fig. S1B in the supplemental material). To determine whether HpHAC1 mRNA is spliced by HpIre1p, HpHAC1 mRNAs from cells treated with the UPR-inducing N-glycosylation inhibitor tunicamycin (TM) were analyzed by reverse transcription-PCR (RT-PCR). Upon TM treatment, most of the longer PCR product was converted into the shorter product in the WT strain (Fig. 1A, left), but the shorter PCR product was not detected in the Hpire1 null mutant (Hpire1Δ) strain, indicating that the TM-induced splicing of HpHAC1 mRNA is mediated by HpIre1p in H. polymorpha, as reported in S. cerevisiae (Fig. 1A, right). Sequencing of the two PCR products indicated that an internal stretch of 177 bp in the longer product, the unspliced HpHAC1 (HpHAC1u) mRNA, is absent in the shorter one, the spliced HpHAC1 (HpHAC1s) mRNA (Fig. 1B). The 5′ splicing site was located within the coding region of the HpHAC1 gene, and the stop codon in the unspliced form was cleaved out by splicing (Fig. 1B). Thus, after splicing, the C-terminal 7 amino acids of the unspliced HpHac1p (311 amino acids) are changed into a new sequence of 20 amino acids of the spliced HpHac1p (324 amino acids).
FIG 1.

Analysis of Ire1p-mediated H. polymorpha HAC1 splicing under UPR-induced conditions. (A) RT-PCR analysis of HpHAC1 splicing upon UPR induction by TM treatment in H. polymorpha DL1-L WT and Hpire1Δ mutant cells. (B) Diagram showing the binding site of the primers used for RT-PCR analysis. HpHAC1u, unspliced form of the HpHAC1 transcript; HpHAC1s, spliced form of the HpHAC1 transcript. (C) Structural organization of HpHac1p and ScHac1p translated after unconventional splicing of HAC1 mRNAs. Dark vertical lines, PEST domains; small grid lines, transcriptional activation domain (TAD); aa, amino acids.
Comparison of the amino acid sequences of the active HpHac1p and ScHac1p, which are generated after splicing, showed that the overall homology between the two yeast Hac1 proteins is quite low, with 8% identity and 15% similarity, but the homology between the basic motif and the leucine zipper domains is significantly higher with 68% identity and 77% similarity (Fig. 1C). Despite such low overall homology, all of the characteristic domains reported in the S. cerevisiae Hac1p, including the basic leucine zipper (bZIP; which is required for DNA binding), the PEST motif (which is involved in the ubiquitin-mediated degradation of Hac1p), and the transcriptional activation domain (TAD) (18), are also present in the H. polymorpha Hac1p.
We tested whether HpHac1p is a functional homolog of S. cerevisiae Hac1p. Two different forms of HpHAC1, HpHAC1u and HpHAC1s, were expressed under the control of the GAL10 promoter in the S. cerevisiae WT and hac1 deletion (Schac1Δ) strains. Expression of HpHAC1s severely inhibited the growth of both the WT and Schac1Δ strains (see Fig. S2A in the supplemental material), consistent with previous reports that the overexpression of endogenous or heterogeneous active Hac1 proteins severely reduces the rate of S. cerevisiae growth (19, 20). However, expression of the HpHAC1u form rescued the defective growth of the Schac1Δ mutant subjected to TM stress. We found that HpHAC1 mRNAs were spliced with a very low efficiency in S. cerevisiae upon TM treatment (data not shown). HpHac1p at such low levels might not be toxic to cell growth, but it might be functional enough to complement the defective growth of the Schac1Δ mutant. Indeed, the use of the S. cerevisiae ACT1 or HAC1 promoter to direct the less efficient expression of HpHAC1s was able to recover the growth defect of the Schac1Δ mutant in the presence of TM more efficiently than the use of the strong GAL10 promoter (see Fig. S2B in the supplemental material). Thus, these results strongly indicate that H. polymorpha Hac1p is a functional homolog of S. cerevisiae Hac1p.
Negative regulation of Hac1p expression by an unconventional intron in H. polymorpha.
As was observed previously for HAC1 orthologs found in other yeasts and filamentous fungi, the unconventional intron of the H. polymorpha HAC1 mRNA is predicted to have a secondary structure that forms two short stem-loops (Fig. 2A). The conserved splicing site CAG|CAG is present at both putative intron borders and is likely recognized by HpIre1p. Moreover, the intron contains the sequence 5′-TCAAACAAATACCCATAA-3′, which forms long-range base pairs with sequences in the 5′ UTR region. The base pairing between the HAC1 5′ UTR and its intron inhibits translation of the HAC1 mRNA, which is critical for activation of a plethora of UPR target genes, in S. cerevisiae (5).
FIG 2.
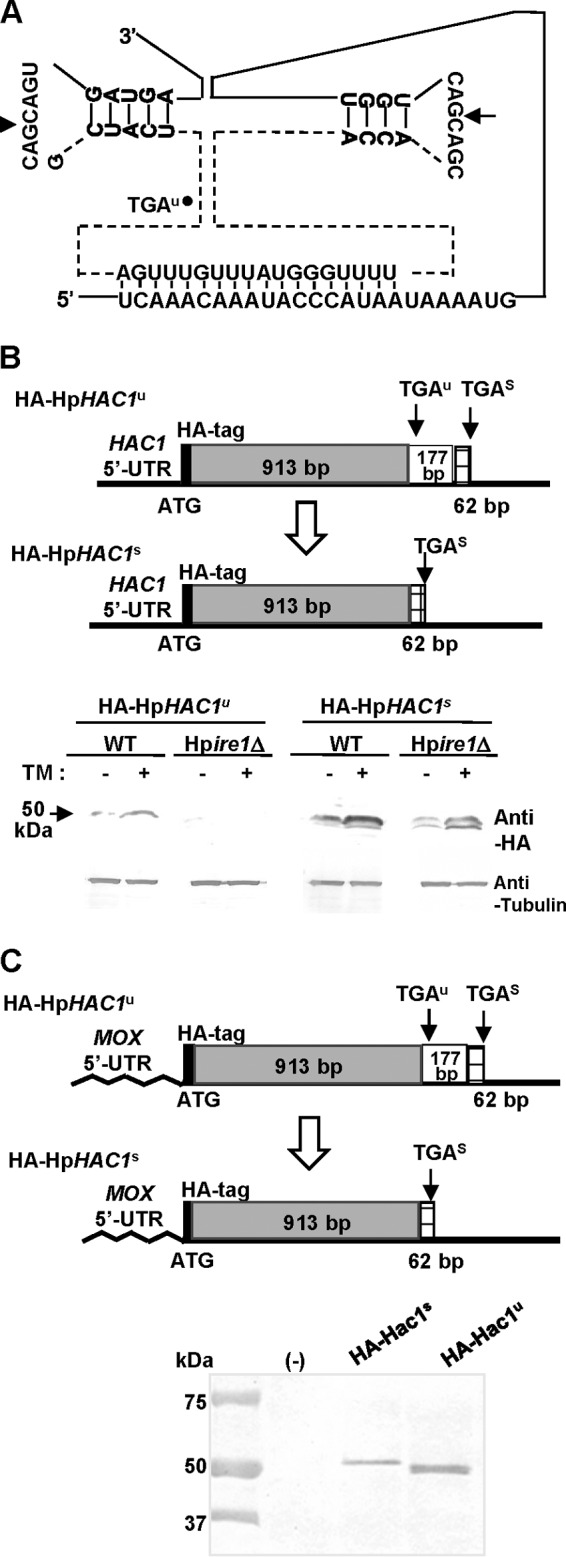
Analysis of HpHac1p expression in the H. polymorpha WT and ire1-disrupted strains. (A) Predicted secondary structure of the intron in the HpHAC1 mRNA loop and putative sites of cleavage by Ire1p. The HpHAC1 5′ UTR and intron can interact by base paring. (B, C) Western blot analysis of the N-terminally HA-tagged HpHac1u and HpHac1s proteins under the control of the native promoter (B) or under the control of the MOX promoter (C). The H. polymorpha cells were cultivated in YPD or YPM (1% Bacto yeast extract, 2% Bacto peptone, 2% methanol) at 37°C for 8 h, and 10 μg of total cell extracts was analyzed by immunoblotting with anti-HA antibody.
To determine whether this negative regulation of HAC1 expression at the translational level occurs in H. polymorpha, we constructed strains expressing an N-terminally HA-tagged, unspliced HpHAC1 protein (HA-HpHac1u) and spliced HpHAC1 protein (HA-HpHac1s) (Fig. 2B, top). While HA-HpHac1s was detected both in the WT and in the ire1-null (Hpire1Δ) mutant, HA-HpHac1u was expressed only in the WT and not in the Hpire1Δ mutant (Fig. 2B, bottom), suggesting that the unconventional intron of HpHAC1 mRNA inhibits its translation to prevent the synthesis of HpHac1p. To determine whether the translational inhibition is mediated by the interaction of the HpHAC1 intron with its 5′ UTR, we replaced the HpHAC1 5′ UTR with the HpMOX1 5′ UTR. We found that the HpMOX1 5′ UTR-HA-HpHAC1u construct was efficiently translated in yeast cells growing under normal conditions (Fig. 2C), indicating the involvement of the 5′ UTR in translational inhibition. As shown in Fig. 1B, the translation product of spliced HpHAC1 mRNA (HA-Hac1s), generated by using the stop codon at 975 bp, was found to be larger than the translation product of the unspliced HpHAC1 mRNA (HA-Hac1u), generated by using the stop codon at 936 bp. Altogether, these results strongly suggest that the base paring between the HpHAC1 intron and its 5′ UTR is responsible for the negative regulation of HpHAC1 expression at the translational step.
Roles of HpHac1 in growth under stress conditions.
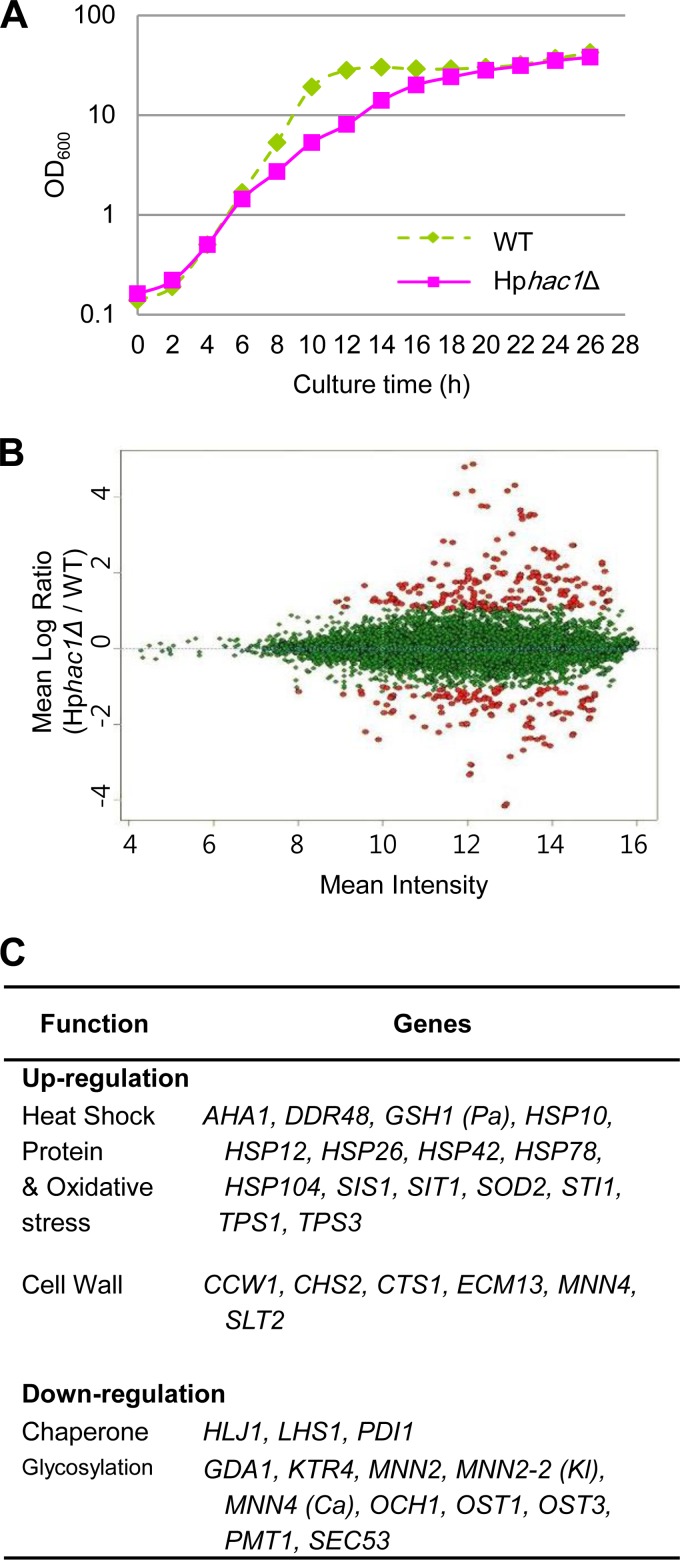
To investigate the physiological function of HpHAC1, we analyzed the growth phenotypes of a H. polymorpha hac1-null (Hphac1Δ) mutant. In contrast to S. cerevisiae hac1Δ, the Hphac1Δ mutant showed a slightly retarded growth even under normal growth conditions and a decreased thermal tolerance compared to that of the WT (Fig. 3A). Moreover, the Hphac1Δ strain did not show the inositol auxotrophic phenotype (Fig. 3B), which is interesting because a lack of Hac1p results in inositol auxotrophy in S. cerevisiae due to the involvement of UPR in lipid biogenesis (21). The growth of the Hphac1Δ strain was severely impaired in the presence of the UPR-inducing agents dithiothreitol (DTT) and TM. Intriguingly, the Hphac1Δ strain was also very sensitive to calcofluor white (CFW) and SDS, which did not affect the growth of the Schac1Δ strain (Fig. 3C). The growth defects caused by CFW and SDS were rescued by addition of 0.5 M KCl, an osmotic stabilizer (Fig. 3D), suggesting that the impaired growth of the Hphac1Δ strain may largely be due to the loss of cell wall integrity.
FIG 3.
Growth phenotype analyses of H. polymorpha hac1Δ strains. The results of thermotolerance analysis (A), inositol auxotrophy analysis (B), and analysis of sensitivity to cell wall perturbation (C, D) of the H. polymorpha WT [Hp(WT)] and Hphac1Δ strains are shown. The S. cerevisiae WT [Sc(WT)] and hac1Δ strains were spotted as controls. Yeast cells were grown to stationary phase in YPD medium, and a 10-fold dilution series was spotted onto a YPD plate at 30°C, 37°C, and 45°C or onto YPD plates containing 10 mM caffeine, 0.01% SDS, or 45 μg/ml CFW without or with the addition of 0.5 M KCl. Ino, inositol. (E) Effect of HpHAC1s and HpHAC1sΔPEST (HpHAC1s-ΔP) expression on ER stress resistance.
On the other hand, we determined whether the increased stability of HpHac1p makes H. polymorpha cells more resistant to TM. In S. cerevisiae, the Hac1p PEST motif mediates proteosomal degradation of the proteome, and thus, mutation of the PEST motif increases UPR-dependent transcription and resistance to ER stress (18). HpHac1p is predicted to contain two consecutive PEST motifs that are located between amino acids 148 and 177 (28 amino acids; PEST score, 13.86) and between amino acids 177 and 190 (12 amino acids; PEST score, 12.42). We constructed the spliced form of HpHAC1 lacking both PEST motifs (HpHAC1s-ΔPEST) and integrated it into the HpHAC1 locus in the WT and Hphac1Δ strains. We found that HpHAC1s-ΔPEST increased the level of resistance to TM compared to that achieved with HpHAC1s in both strains (Fig. 3E). This result confirms that HpHac1p is required for the growth of H. polymorpha under UPR-inducing conditions.
Identification of genes regulated by HpHac1p under normal growth conditions.
To investigate the differences in gene expression profiles between the WT and Hphac1Δ strains under normal growth condition, we analyzed the transcriptomes of the strains grown in YPD medium in the absence of UPR-inducing agents using microarray analysis. RNA samples were collected from H. polymorpha cells in early exponential growth phase because Hphac1Δ mutant cells grow slower in the late exponential phase than WT cells (Fig. 4A). The transcriptome analysis indicated that expression of a set of glycosylation and ER chaperone genes is downregulated in the Hphac1Δ mutant compared to their levels of expression in the WT (Fig. 4B and C), implying that the growth retardation of the Hphac1Δ mutant may be related to the low levels of expression of these genes even under normal growth conditions. The levels of expression of several genes involved in cell wall construction and remodeling, such as CHS2 (which encodes chitin synthase II), GAS1 (which encodes β-1,3-glucanosyltransferase), and CTS1 (which encodes extracellular endochitinase), were shown to be increased, reflecting the existence of compensatory mechanisms for defective cell wall integrity. The expression of a number of heat shock proteins (HSPs) was also upregulated, and this increased expression might facilitate protein folding in Hphac1Δ mutant cells with the decreased expression of ER chaperones.
FIG 4.
Transcriptome profile of the Hphac1 disruption mutant. (A) Growth of WT and Hphac1Δ mutant cells in YPD medium at 37°C. (B) Scatter plots reflecting the differential expression profiles of the WT and Hphac1Δ strains cultivated in YPD under normal condition. Total RNAs were obtained from WT strain DL1 and Hphac1Δ mutant cells grown to log phase (OD600, ∼0.5) in YPD medium and subjected to microarray analysis using a whole-genome microarray. Red spots, genes up- or downregulated more than 2-fold in the Hphac1 mutant compared to their expression in the WT strain; green spots, genes showing less than 2-fold differential expression in the WT and mutant strains. (C) Genes up- and downregulated in the Hphac1Δ mutant compared to their expression in the WT under normal growth conditions on YPD. The annotated genes were functionally categorized according to the Munich Information Centre for Protein Sequences (MIPS). Pa, Pichia angusta; Ca, Candida albicans; Kl, Kluyveromyces lactis.
Identification of HpHAC1-dependent core UPR genes upregulated in response to UPR induction.
To identify the HAC1-dependent core UPR genes that are commonly increased in response to both DTT and TM treatment, but only in the presence of HpHac1p, we carried out two types of time course transcriptome analyses. First, to identify genes whose expression is affected by DTT or TM, we compared the transcriptomes of WT cells grown in YPD medium containing either DTT or TM with the transcriptomes of WT cells grown in YPD medium only (Fig. 5A, left). DTT or TM treatment induced the expression of a number of genes associated with all steps of the secretion process but downregulated a number of genes involved in protein synthesis, transcription, and DNA synthesis (see Fig. S3B in the supplemental material). Interestingly, the UPR response to DTT treatment was rapid but transient, while the response to TM treatment was considerably slower but continued to increase over time (Fig. 3A). Most of the genes that were induced in response to DTT treatment are involved in the oxidative response, while most genes that were induced in response to TM treatment are involved in glycosylation. This suggests that the main stress caused by DTT is oxidative stress, the response to which was rapid and transient, and also suggests that TM treatment generates an N-glycosylation defect. The results confirm that these agents interfere with ER folding by different mechanisms: DTT, the strong reducing agent, prevents disulfide bond formation (22), whereas TM inhibits an early step of N-glycosylation by blocking the transfer of N-acetylglucosamine-1-phosphate from UDP-N-acetylglucosamine to dolichol phosphate in the synthesis of lipid-linked oligosaccharides (23). Second, to identify genes whose expression is dependent upon HpHac1p under UPR-induced conditions, we analyzed the differences in expression patterns between WT and Hphac1Δ mutant cells grown under DTT- or TM-induced UPR conditions (Fig. 5A, right) and selected genes that were downregulated more than 2-fold in the Hphac1Δ mutant compared to their levels of regulation in the WT. Thus, by combining the results of the two types of transcriptome analyses, we identified 50 HpHAC1-dependent core UPR genes whose expression is increased upon treatment with DTT and with TM, but only in the presence of HpHac1p (Fig. 5B).
FIG 5.
Identification of HpHAC1-dependent core UPR genes. (A) Schematic diagram of sample preparation for microarray analysis. (B) Venn diagrams illustrating the overlapped genes up- and downregulated in the WT and Hphac1Δ strains under conditions with TM and DTT treatment (left) and the functional categories of the core UPR target genes based on the Munich Information Centre for Protein Sequences (right). 1-1, amino acid metabolism; 1-2, nitrogen sulfur and selenium metabolism; 1-3, nucleotide/nucleoside/nucleobase metabolism; 1-4, phosphate metabolism; 1-5, C compound and carbohydrate metabolism; 2, energy; 3, cell cycle and DNA processing; 4, transcription; 6, protein fate; 7, protein with binding function or cofactor requirement; 9, cellular transport, transport facilitation, and transport routes; 14, unclassified proteins.
The HpHAC1-dependent core UPR genes include several gene families having specific functions in protein folding, processing, and transport (Table 2): genes playing a role in protein folding in the ER (KAR2, ERO1, SCJ1, LHS1, PDI1), genes required for protein N-glycosylation (ALG5, OST1, OST2, OST4, SWP1, WBP1, KTR1, OCR5, MNN4, MNN2), genes involved in protein transport from the ER to the Golgi apparatus (SEC66, SEC61, SEC31, SEC4, SEC53), and genes required for the reverse transport of vesicles from the Golgi apparatus to the ER (SEC21, SEC26, RET2). We validated the microarray data by quantitative RT-PCR (qRT-PCR) and confirmed the more apparent and sustained induction by TM treatment compared to the less and transient induction by DTT treatment (Fig. 3A and 6). A previous transcriptome study of the induction of the S. cerevisiae UPR by both DTT and TM in an IRE1- and HAC1-dependent manner reported that many aspects of secretory functions, including translocation, protein glycosylation, vesicular transport, vacuolar protein targeting, ER-associated degradation, lipid/inositol metabolism, and cell wall biosynthesis, were upregulated by the UPR (3). Compared to these S. cerevisiae UPR genes involved in several functions throughout the secretory pathway, the H. polymorpha UPR genes identified in this study were shown to be more enriched in the functional categories involved in protein glycosylation processes (see Table S3 in the supplemental material).
TABLE 2.
HpHAC1-dependent core UPR genes identified by transcriptome analysis and their functional category
| Functiona | H. polymorpha gene(s) |
|---|---|
| Cytoskeleton | ARP2, ARC35, BZZ1, CAP2, SLA2 |
| Glycosylation | ALG5, MNN2, MNN4,b KTR1, OST1, OST2, OST4, OCR5, SWP1, WBP1 |
| Lipid metabolism | INP51 |
| Protein degradation | CUE1, PNG1b |
| Protein folding | ERO1, KAR2, LHS1, PDI1, SCJ1 |
| Secretion | SEC4, SEC21, SEC26, SEC31, SEC53, SEC61, SEC66, RET2 |
| Signaling | DIG1, SCK1, SET3, SKS1 |
| Transport | FCY2, PMR1, YBR235W |
| Vacuolar | VMA10 |
| Other | BDH1, BUD7, CAF120, ERG6, HPC2, YDR100W, YHR140W, YHR112C, YIL057C, IPF3486,b SPAC13C5.04b |
The distribution of differentially expressed genes was made according to functional categories of the Munich Information Center for Protein Sequences (MIPS).
H. polymorpha open reading frames annotated by gene information for organisms other than S. cerevisiae.
FIG 6.

Quantitative real-time PCR analyses of HpHAC1-dependent core UPR genes. H. polymorpha DL1-L (WT) and Hphac1Δ mutant cells grown to log phase (OD600, ∼0.3) in YPD medium were transferred to YPD supplemented with 5 mM DTT or 5 μg/ml TM. RNA samples from the WT treated with TM (squares) or DTT (diamonds) and from the Hphac1Δ mutant treated with TM (triangles) or DTT (×) were prepared at the times indicated on the x axes (in minutes) and analyzed by qRT-PCR. The relative levels of induced expression are expressed as the ratio of the mRNA level of each gene to the mRNA level of HpACT1.
Modulation of protein glycosylation activity by HpHac1p in H. polymorpha.
Our transcriptome data showed that several genes involved in N-linked glycosylation are HpHAC1-dependent core UPR targets. To determine whether N-glycosylation is affected by HpHac1p, we analyzed the glycosylation pattern of a vacuolar glycoprotein, carboxypeptidase Y (CPY), in the WT and the Hphac1Δ mutant. H. polymorpha CPY (HpCPY) has three N-glycosylation sites (24), and the precursor HpCPY, with a calculated molecular weight (MW) of 60,793, is converted into the mature HpCPY with a predicted MW of 47,142 (Fig. S4A) by a process that is mediated by the vacuolar proteinase A HpPep4p (25). We detected diffuse protein bands with MWs higher than 60,000 in the intracellular extract of H. polymorpha WT cells, which suggests that the HpCPY protein is modified by N-linked glycosylation in the ER followed by hypermannosylation in the Golgi apparatus. We found that the sizes of the diffuse bands of HpCPY were significantly reduced in the Hphac1Δ mutant grown in YPD medium without TM (Fig. 7A), suggesting that overall N-glycosylation activity decreases in the Hphac1Δ mutant, even under normal growth conditions. We confirmed that peptide-N-glycosidase F treatment, which eliminates N-glycans from glycoproteins, generated HpCPY proteins with the same size of 47 kDa as major products both in the WT and in the Hphac1Δ mutant strain (see Fig. S4B in the supplemental material). This strongly supports the suggestion that the smaller size of diffuse bands of HpCPY in the Hphac1Δ mutant is mainly due to the lower efficiency of N-glycosylation and is not due to HpCPY degradation. This finding is consistent with the results from our transcriptome analysis (Fig. 4C), which indicated that the expression levels of many N-glycosylation genes were lower in the Hphac1Δ mutant than in the WT, even without UPR induction. Quite interestingly, upon TM treatment, the level of hyperglycosylated HpCPY decreased, while HpCPY of ∼47 kDa (designated X in Fig. 7A) accumulated in the WT (Fig. 7A). In contrast, the extent of HpCPY hyperglycosylation did not change significantly, even 3 h after TM treatment, and the ∼47-kDa HpCPY protein barely accumulated in the Hphac1Δ strain. The ∼47-kDa HpCPY protein appears to be an unglycosylated form of the mature CPY protein, on the basis of the fact that its migration pattern was almost identical to that of the HpCPY protein treated with peptide-N-glycosidase F (see Fig. S4B in the supplemental material). Although the detailed mechanism underlying this differential pattern of CPY processing in the WT and the Hphac1Δ mutant remains unknown, the results suggest the involvement of Hac1p in protein processing under TM-induced UPR conditions.
FIG 7.

Comparative analysis of CPY protein processing in the WT, Hphac1Δ mutant, and HpHAC1s-overexpressing strains. (A) Time course analysis of HpCPY processing after TM treatment. The H. polymorpha WT and Hphac1Δ mutant were grown in YPD to exponential phase (OD600 ∼0.5) before being treated with 5 μg/ml TM for 3 h. Western blot analysis was performed with total intracellular protein extracts using anti-HpCPY antibody. (B) Effect of HpHAC1s on N-glycosylation of ScCPY proteins. H. polymorpha DL1-LdU strains harboring the ScCPY expression vector pDLGAP-ScCPY with or without the HA-HpHAC1s expression vector pDUM2-HA-HAC1s were incubated in YPM medium containing 0.5% methanol for 12 h. Total cell extracts were analyzed by immunoblotting with anti-ScCPY antibody.
Next, we tested whether overexpression of HpHac1p can increase the N-glycosylation levels of heterologous glycoproteins in H. polymorpha. To do this, we compared the N-glycosylation patterns of the S. cerevisiae CPY (ScCPY), which has four N-glycosylation sites, in the WT with or without the HpHac1s protein (Fig. 7B). Quite interestingly, overexpression of the active HpHac1s protein shifted heterogeneously glycosylated ScCPY to forms that were more homogeneous and hyperglycosylated in WT cells than in the control cells without HpHac1s expression (Fig. 7B). To confirm the positive effect of HpHac1p on the modulation of glycosylation, we performed the same experiment employing the Hpalg3Δ mutant strain, which has a defect in the conversion step of Man5GlcNAc2-dolichyl pyrophosphate (Dol-PP) to Man6GlcNAc2-Dol-PP in the ER (15). Our previous study reported that the S. cerevisiae CPY protein was hypoglycosylated with a reduced extent of occupancy at N-glycosylation sites in the Hpalg3Δ mutant (13). Overexpression of the active HpHac1s protein increased the amount of hyperglycosylated ScCPY in Hpalg3Δ mutant cells compared to that in control cells without HpHAC1s expression (Fig. 7B). Collectively, these results demonstrate that increased expression of active H. polymorpha Hac1p enhances protein glycosylation activity in H. polymorpha.
DISCUSSION
Elucidating the mechanisms underlying UPR is of special interest to industrial mycologists because yeast and filamentous fungi overexpressing heterologous proteins experience ER stress caused by the accumulation of unfolded proteins and defects in the ER membrane. Moreover, recent studies strongly implicate UPR as a central regulator of fungal pathogenesis (26). The basic features of the Hac1-dependent UPR signaling pathway appear to be well conserved in most fungal species, although several recent reports have revealed unexpected variations, such as the lack of the Hac1 homolog in the fission yeast Schizosaccharomyces pombe (27) and the absence of the canonical Ire1-Hac1 UPR in Candida glabrata (28). Each of the HAC1 orthologs of Candida albicans, Cryptococcus neoformans, Aspergillus fumigatus, and Alternaria brassicicola has a short unconventional intron (19, 20, 29, and 56 nt, respectively) (29–32). In this study, we show that HpHAC1 mRNA contains a nonconventional intron of 177 bp which inhibits the expression of HpHAC1 at the translational level and that HpHac1p is essential for UPR induction as well as normal cell growth. Most importantly, we demonstrate that protein glycosylation is modulated by HpHac1p in H. polymorpha.
Like S. cerevisiae HAC1, which also contains a long intron, we showed that expression of HpHAC1 is regulated by base pairing of its intron with the complementary sequences in its 5′ UTR (Fig. 2). In contrast, the fungal HAC1 orthologs and mammalian XBP1, which contain short introns, use different regulatory mechanisms to control the expression level of Hac1/XBP1 homologs. Mammalian cells translate the unspliced XBP1 mRNA under unstressed conditions but rapidly degrade the resulting XBP1 protein by the proteasome (33). C. albicans, T. reesei, A. nidulans, and Aspergillus niger express HAC1 mRNAs with truncated 5′ UTRs in response to stress, and these are translated more efficiently (34). It would be interesting to study the evolutionary relationship between the size of the unconventional intron and the mechanism of regulation of HAC1 expression.
In S. cerevisiae, the classical UPR element UPRE1 was found in the promoters of several UPR target genes, such as KAR2/BiP, and was defined to have a semipalindromic 7-nucleotide consensus sequence, CAGNGTG (6). Further studies by computational genetic screens of S. cerevisiae UPR gene promoters led to the discovery of two additional novel UPREs, UPRE2 and UPRE3, which are necessary and sufficient for UPR activation of the promoters, and the additional role of Gcn4p, another bZIP transcription factor, in the activation UPR (35). We detected almost identical UPRE sequences, including the classical UPRE1 and the additional UPRE2 and UPRE3 motifs, in the promoter regions of the core HAC1 regulons of H. polymorpha (Table 3). It is possible that H. polymorpha Hac1p can bind to the same DNA elements present in the promoters of UPR target genes of S. cerevisiae, thus complementing the defects of S. cerevisiae hac1 mutants (see Fig. S1 in the supplemental material), despite the overall low degree of homology to ScHac1p. Several novel binding motifs were also identified in the promoters of H. polymorpha HAC1 regulons (data not shown), but the biological significance of these motifs has yet to be investigated.
TABLE 3.
Analysis of UPRE regulatory motif in HpHAC1-dependent UPR target promoters
| UPRE in S. cerevisiaea | Sequence | H. polymorpha gene(s) |
|---|---|---|
| UPRE1, classical UPRE (23/50) | CAGNGTG | ALG5, ARC35, CAF120, CUE1, DIG1, FCY2, HPC2, INP51, KAR2, LHS1, MNN2, OCR5, OST1, OST4, PMR1, SEC4, SEC31, SEC53, SEC61, SEC66, SKS1, WBP1, YBR235W |
| UPRE2, putative motif 1 (21/50) | CGTGTCGG | FCY2, HPC2, STE3 |
| ACGTGTCG | KAR2, SKS1 | |
| CGTGTCC | BUD7, HPC2, SEC4, SEC61, YBR235W | |
| TACGTG | ERG6, ERO1, HPC2, KAR2, KTR1, LHS1, OCR5, PDI1, SEC4, SEC21, SEC26, SLA2, SKS1, WBP1, YBR235W, YHR140W, YIL057C | |
| UPRE3, putative motif 8 (1/50) | AGTAGGAC | |
| AGGACAAC | KAR2 | |
| Other (16/50) | ARP2, BDH1, BZZ1, CAP2, MNN4, OST2, PNG1, RET2, SCJ1, SCK1, SWP1, VAM10, YHR112C, YDR100W, IPF3486, SPAC13C5.04 |
The UPRE sequences in S. cerevisiae are from the reference 35. Data in parentheses represent the number of genes with the UPRE/total number of HpHAC1-dependent UPR targets.
In an effort to enhance the protein secretion and folding capacity of yeast and fungal cells, several attempts have been made to manipulate the expression level of Hac1p. Hac1p overexpression increased the expression of a set of ER chaperones, improving the secretory production of some heterologous proteins (20) and the cell surface expression of bacterial esterase A in S. cerevisiae (36) and secreted, surface-displayed, and membrane proteins in P. pastoris (10). However, depending on the target protein, expression of Hac1p either decreased the amount of heterologous proteins or exerted only a modest to no effect on expression levels, suggesting that the effect of Hac1p overexpression needs to be evaluated on a case-by-case basis (10, 20, 37). We report here that overexpression of HpHac1p has a positive effect on the N-glycosylation pattern of a heterologous glycoprotein, S. cerevisiae CPY, generating more homogeneously glycosylated forms (Fig. 7B). Similarly, overexpression of Hac1p improved the processing of the signal peptide of the adenosine A2 receptor in P. pastoris (10). Thus, we suggest that the beneficial effects of increased Hac1p activity include enhanced protein processing, such as glycosylation activity and proteolytic cleavage. Further studies of the UPR induced by the expression of an aberrant secreted protein or a misfiled heterologous protein in H. polymorpha would generate more relevant information on the practical application of UPR to the development of host strains producing secretory recombinant proteins of industrial interest. We propose that the systematic manipulation of UPR mediated by Hac1p would facilitate the successful exploitation of yeast as an intelligent cell factory for the secretory production of correctly folded and processed recombinant proteins.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by grant no. NRF-2013M3A6A8073554 (Global Frontier Program for the Intelligent Synthetic Biology) from the National Research Foundation of Korea (NRF) and by grant no. 914007-4 (Strategic Initiative for Microbiomes in Agriculture and Food) from the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01440-15.
REFERENCES
- 1.Ron D, Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Cox JS, Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 3.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 4.Sidrauski C, Walter P. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90:1031–1039. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 5.Ruegsegger U, Leber JH, Walter P. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107:103–114. doi: 10.1016/S0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. 1998. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J Biol Chem 273:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- 7.Arvas M, Pakula T, Lanthaler K, Saloheimo M, Valkonen M, Suortti T, Robson G, Penttila M. 2006. Common features and interesting differences in transcriptional responses to secretion stress in the fungi Trichoderma reesei and Saccharomyces cerevisiae. BMC Genomics 7:32. doi: 10.1186/1471-2164-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saloheimo M, Valkonen M, Penttila M. 2003. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol Microbiol 47:1149–1161. doi: 10.1046/j.1365-2958.2003.03363.x. [DOI] [PubMed] [Google Scholar]
- 9.Oh MH, Cheon SA, Kang HA, Kim JY. 2010. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 27:443–452. doi: 10.1002/yea.1762. [DOI] [PubMed] [Google Scholar]
- 10.Guerfal M, Ryckaert S, Jacobs PP, Ameloot P, Van Craenenbroeck K, Derycke R, Callewaert N. 2010. The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb Cell Fact 9:49. doi: 10.1186/1475-2859-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whyteside G, Nor RM, Alcocer MJ, Archer DB. 2011. Activation of the unfolded protein response in Pichia pastoris requires splicing of a HAC1 mRNA intron and retention of the C-terminal tail of Hac1p. FEBS Lett 585:1037–1041. doi: 10.1016/j.febslet.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 12.Kang HA, Gellisen G. 2005. Hansenula polymorpha, p 111–142. In Gellissen G. (ed), Production of recombinant proteins: novel microbial and eukaryotic expression systems. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 13.Cheon SA, Kim H, Oh DB, Kwon O, Kang HA. 2012. Remodeling of the glycosylation pathway in the methylotrophic yeast Hansenula polymorpha to produce human hybrid-type N-glycans. J Microbiol 50:341–348. doi: 10.1007/s12275-012-2097-2. [DOI] [PubMed] [Google Scholar]
- 14.Kim MW, Kim EJ, Kim JY, Park JS, Oh DB, Shimma Y, Chiba Y, Jigami Y, Rhee SK, Kang HA. 2006. Functional characterization of the Hansenula polymorpha HOC1, OCH1, and OCR1 genes as members of the yeast OCH1 mannosyltransferase family involved in protein glycosylation. J Biol Chem 281:6261–6272. doi: 10.1074/jbc.M508507200. [DOI] [PubMed] [Google Scholar]
- 15.Oh DB, Park JS, Kim MW, Cheon SA, Kim EJ, Moon HY, Kwon O, Rhee SK, Kang HA. 2008. Glycoengineering of the methylotrophic yeast Hansenula polymorpha for the production of glycoproteins with trimannosyl core N-glycan by blocking core oligosaccharide assembly. Biotechnol J 3:659–668. doi: 10.1002/biot.200700252. [DOI] [PubMed] [Google Scholar]
- 16.Kim H, Moon HY, Lee DJ, Cheon SA, Yoo SJ, Park JN, Agaphonov MO, Oh DB, Kwon O, Kang HA. 2013. Functional and molecular characterization of novel Hansenula polymorpha genes, HpPMT5 and HpPMT6, encoding protein O-mannosyltransferases. Fungal Genet Biol 58-59:10–24. doi: 10.1016/j.fgb.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Choe JK, Chung TH, Park S, Cho HG, Hur CG. 2005. An open source microarray data analysis system with GUI: Quintet, p 392–401. In Slezak D, Yao J, Peters JM, Ziarko W, Hu X (ed.), Rough sets, fuzzy sets, data mining, and granular computing, part II. Lecture notes in computer science, vol 3642 Springer-Verlag, Berlin, Germany. doi: 10.1007/11548706_41. [DOI] [Google Scholar]
- 18.Pal B, Chan NC, Helfenbaum L, Tan K, Tansey WP, Gething MJ. 2007. SCFCdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae. Mol Biol Cell 18:426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawahara T, Yanagi H, Yura T, Mori K. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell 8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valkonen M, Penttila M, Saloheimo M. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 69:2065–2072. doi: 10.1128/AEM.69.4.2065-2072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang HJ, Jesch SA, Gaspar ML, Henry SA. 2004. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braakman I, Helenius J, Helenius A. 1992. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J 11:1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes G, Hansen WJ, Holcomb CL, Rine J. 1984. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol Cell Biol 4:2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellu AR, van der Klei IJ, Rechinger KB, Yavuz M, Veenhuis M, Kiel JA. 1999. Characterization of the Hansenula polymorpha CPY gene encoding carboxypeptidase Y. Yeast 15:181–189. [DOI] [PubMed] [Google Scholar]
- 25.Bae JH, Sohn JH, Rhee SK, Choi ES. 2005. Cloning and characterization of the Hansenula polymorpha PEP4 gene encoding proteinase A. Yeast 22:13–19. [DOI] [PubMed] [Google Scholar]
- 26.Krishnan K, Askew DS. 2014. Endoplasmic reticulum stress and fungal pathogenesis. Fungal Biol Rev 28:29–35. doi: 10.1016/j.fbr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragon T, Li H, Walter P. 2012. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife 1:e00048. doi: 10.7554/eLife.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki T, Nakayama H, Nagayoshi Y, Kakeya H, Kohno S. 2013. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata. PLoS Pathog 9:e1003160. doi: 10.1371/journal.ppat.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, Kang HA. 2011. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog 7:e1002177. doi: 10.1371/journal.ppat.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joubert A, Simoneau P, Campion C, Bataille-Simoneau N, Iacomi-Vasilescu B, Poupard P, Francois JM, Georgeault S, Sellier E, Guillemette T. 2011. Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol Microbiol 79:1305–1324. doi: 10.1111/j.1365-2958.2010.07522.x. [DOI] [PubMed] [Google Scholar]
- 31.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, White S, McCarthy JW, Latge JP, Feldmesser M, Rhodes JC, Askew DS. 2009. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog 5:e1000258. doi: 10.1371/journal.ppat.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, Brown AJ, Archer DB. 2008. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol 45:1235–1247. doi: 10.1016/j.fgb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Bernales S, Papa FR, Walter P. 2006. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 34.Mulder HJ, Saloheimo M, Penttila M, Madrid SM. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol Genet Genomics 271:130–140. doi: 10.1007/s00438-003-0965-5. [DOI] [PubMed] [Google Scholar]
- 35.Patil CK, Li H, Walter P. 2004. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol 2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breinig F, Diehl B, Rau S, Zimmer C, Schwab H, Schmitt MJ. 2006. Cell surface expression of bacterial esterase A by Saccharomyces cerevisiae and its enhancement by constitutive activation of the cellular unfolded protein response. Appl Environ Microbiol 72:7140–7147. doi: 10.1128/AEM.00503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasser B, Maurer M, Gach J, Kunert R, Mattanovich D. 2006. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol Bioeng 94:353–361. doi: 10.1002/bit.20851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.