Abstract
The symptoms of viral infections of fungi range from cryptic to severe, but there is little knowledge of the factors involved in this transition of fungal/viral interactions. Brown cap mushroom disease of the cultivated Agaricus bisporus is economically important and represents a model system to describe this transition. Differentially expressed transcript fragments between mushrooms showing the symptoms of brown cap mushroom disease and control white noninfected mushrooms have been identified and sequenced. Ten of these RNA fragments have been found to be upregulated over 1,000-fold between diseased and nondiseased tissue but are absent from the Agaricus bisporus genome sequence and hybridize to double-stranded RNAs extracted from diseased tissue. We hypothesize that these transcript fragments are viral and represent components of the disease-causing agent, a bipartite virus with similarities to the family Partitiviridae. The virus fragments were found at two distinct levels within infected mushrooms, at raised levels in infected, nonsymptomatic, white mushrooms and at much greater levels (3,500 to 87,000 times greater) in infected mushrooms exhibiting brown coloration. In addition, differential screening revealed 9 upregulated and 32 downregulated host Agaricus bisporus transcripts. Chromametric analysis was able to distinguish color differences between noninfected white mushrooms and white infected mushrooms at an early stage of mushroom growth. This method may be the basis for an “on-farm” disease detection assay.
INTRODUCTION
Mycoviruses infect most classes of fungi; however, these infections are generally symptomless and persistent. Studies to understand the transition of viral infection from cryptic (nonsymptomatic) to severe symptoms have largely been aimed toward improving the success of mycovirus biocontrol agents for phytopathogenic fungi such as the Cryphonectria parasitica hypovirus (1). However, when the agricultural crop is fungal, i.e., the cultivated mushroom, Agaricus bisporus, the transition of mycoviral infection from cryptic to severe is economically damaging due to losses in mushroom quality and yield. A. bisporus is a well-characterized fungus, and the emergence of symptomatic virus disease provides a model for studying virus-fungus interactions in this fruit body-forming fungus.
A. bisporus is the principal edible mushroom species under commercial cultivation with an annual global production of $4.7 billion (2). A disease of mushrooms first described in the 1990s, it continues to cause serious economic damage to the European mushroom industry (3). The disease is associated with the presence of double-stranded RNA (dsRNA) molecules, 26 dsRNAs have been size separated, and up to 16 have been found in a single sample (4). The disease exhibits a number of symptoms: mushroom cap browning, primordial growth inhibition or delay, and cap malformations. The collective name mushroom virus X (MVX) has been given to the disease prior to any detailed knowledge of specific dsRNAs or causative disease agents. However, the disease symptoms of MVX have been separated geographically, the brown symptom initially being more prevalent in Ireland, The Netherlands, and Belgium, while a primordial inhibition/delay symptom was more commonly recorded in Britain, strongly suggesting different causative agents responsible for the different symptoms (3). The symptom of brown coloration on white strains of mushrooms has serious economic consequences due to market rejection of brown mushrooms because of associations with poor quality damage, age, and/or disease. This report is aimed at identifying the causative agents of brown cap mushroom disease (BCMD) and understanding how host responses lead to the brown symptom development.
The coloration associated with BCMB is a light to dark brown generally uniform across the mushroom cap (Fig. 1), which is distinct from the discrete dark brown lesions associated with bacterial blotch disease caused by Pseudomonas tolaasii (5). The primordia of infected crops are white, the brown coloration developing during mushroom growth or after harvest between the farm and retailer. A significant proportion of mushrooms in a crop (up to 40%) can show the brown symptom, but more often a smaller number of mushrooms are brown, while the remainder, even within centimeters, are white (4) (Fig. 1). This observation of symptom expression localized to individual mushrooms but neighboring mushrooms apparently asymptomatic suggests that the development of disease symptoms is probably not influenced by the aerial environment. BCMD is distinct from La France disease, which is associated with the encapsidated virus, Agaricus bisporus virus 1 (ABV1), also known as La France isometric virus (LIV) (6, 7), since no particles or LIV immunological signals have been consistently observed in MVX mushroom samples (4).
FIG 1.
Pale brown mushroom emerging from an MVX-infected crop of A. bisporus and among white mushrooms.
Two of the 26 dsRNAs (from MVX-infected mushrooms) have been fully or partially sequenced; a 14.5-kbp molecule showed sequence similarity to an endornavirus (AbEV1) (8), and a 17-kbp molecule had similarities to Cryphonectria parasitica hypovirus (9). Two studies have reported an apparent association between the brown symptom with the presence of four or five currently unsequenced low-molecular-weight dsRNAs (0.65 to 2.3 kbp) (4, 9), respectively. Three of the 26 dsRNA molecules identified from MVX diseased A. bisporus mushrooms have also been recorded in apparently healthy mushroom crops (4).
Previous research to understand the causative agents within the MVX complex has centered largely on dsRNAs (4, 9). In this study, we used the suppression subtractive hybridization (SSH) technique to identify differentially expressed transcript fragments between BCMB-diseased and nondiseased mushrooms. The identified transcripts were found to be both host (Agaricus bisporus) and non-Agaricus in origin. We hypothesize the non-Agaricus transcript fragments are viral forming a single bipartite virus in the Partitiviridae which represents the disease-causing agent. The Partitiviridae are a family of dsRNA fungal and plant viruses with two or more genomic segments (10). We propose that this virus coexists within A. bisporus at two levels analogous to a transition in viral life-strategy from persistent to acute, moderate levels with no visible symptoms and high levels associated with brown symptom development, the viral level being determined during early mushroom development.
MATERIALS AND METHODS
Biological material.
Agaricus bisporus, strain A15 (Sylvan, Inc., United Kingdom), was grown on composted wheat straw according to commercial practice. Mushroom fruit bodies were harvested as closed-cups, developmental stage 2 (11), unless otherwise stated. Mushrooms for RNA analysis were frozen immediately under liquid nitrogen and stored at −80°C until required.
For suppression subtractive hybridization, A. bisporus mushrooms were grown as (i) BCMD infected and (ii) noninfected. (i) An A. bisporus crop was infected by horizontal transfer with an infected A. bisporus culture (MVX 4614) originally collected as diseased material (with brown coloration) from a commercial farm in Ireland and then grown at the Teagasc experimental mushroom unit at the Kinsealy Research Centre, Dublin, Ireland (12). (ii) A noninfected crop cultivated under nearly identical, disease-free conditions at the experimental mushroom unit at Warwick HRI, University of Warwick.
To screen and quantify the changes in transcript levels (by microarray and quantitative PCR) as a result of brown disease infection, four replicate brown infected mushroom (BIM) samples were collected from each of five commercial mushroom farms in Ireland and the United Kingdom during BCMD disease outbreaks and compared to 12 noninfected controls cultivated under nearly identical but disease-free conditions at Warwick HRI. These transcript levels were compared to white infected mushrooms (WIM) growing in close proximity to brown mushrooms in an infected crop grown at Kinsealy Research Centre (as described above).
To examine time course changes in gene expression and color development in an infected crop, an experimental crop was infected by horizontal transfer with an infected A. bisporus culture, MVX 4569 at the Kinsealy Research Centre. Small segments of pileus (cap) tissue were explanted from the growing fruit body at 24-h intervals over 4 days, such that the cap had five missing sections upon completion of the time course. Prior to this work it had not been possible to predict which mushrooms in a crop would become symptomatic and develop brown coloration. Therefore, more than 100 mushroom fruit bodies were explant sampled. All samples were labeled and frozen in liquid nitrogen. At the beginning of this experiment, the mushrooms were ∼20 mm in diameter at developmental stages 1 to 2, and after 4 days they had grown to ∼60 mm in diameter at developmental stages 5 to 6 (10). Prior to each explant removal during the time course, mushroom cap color (chromametric) data were recorded for all mushrooms being examined using the Konica Minolta Chroma Meter CR-300 (Minolta UK).
At the end of the sampling period, the chromametric data were analyzed, and the mushrooms with brown color (on day 4) as defined chromametrically with L values ≤83 and b values of ≥13 were designated “brown infected mushrooms” (BIM); a description of chromatographic parameters L and b is given in Results (“Time course analysis of color symptom development and transcript levels”). Infected samples that did not meet the chromametric criteria for brownness were inferred to be “white infected mushrooms” (WIM). Color development in the segments was compared to transcript levels (by quantitative reverse transcriptase PCR from extracted RNA) in the same segments in brown infected, white infected, and noninfected control mushrooms (grown at Warwick HRI). Three replicate mushrooms per infection-type (BIM, WIM, and control white noninfected) were color analyzed and explant sampled (five segments taken from each).
RNA isolation.
Double-stranded RNA was isolated using a phenol extraction and cellulose affinity protocol adapted from the Valverde extraction method (13). Double-stranded RNA was separated from phenol-extracted total RNA by selective binding to cellulose in the presence of 15% ethanol. Samples were treated with S1 nuclease and DNase 1 (Sigma) to eliminate residual DNA and ssRNA contamination. The presence of dsRNA molecules was confirmed by gel electrophoresis (see Fig. S1 in the supplemental material).
RNA for SSH, microarray, and quantitative reverse transcriptase PCR (RT-qPCR) was initially isolated from mushroom fruit bodies by the TRIzol extraction method (14) using TRI Reagent (Sigma-Aldrich). RNA integrity and quantity were confirmed by using a Bioanalyzer (Agilent Technologies) and a NanoDrop-1000 (Thermo Scientific), and 1 μg of RNA was used for RT-qPCR cDNA synthesis. mRNA was prepared according to the Qiagen Oligotex mRNA minikit protocol prior to analysis by suppression subtractive hybridization (SSH) and microarrays.
SSH.
Differentially expressed genes were selectively amplified by SSH, carried out as described by Eastwood et al. (15) and the manufacturer's instructions (Clontech Laboratories, Inc., California). RNA was extracted from A. bisporus fruit bodies in three pathological states: noninfected (NI), infected with BCMD and showing the brown coloration symptom (brown infected mushrooms [BIM]), and white mushrooms from the infected growing areas but displaying no apparent browning symptom (white infected mushrooms [WIM]). The presence of dsRNA molecules of all samples was determined before continuing SSH using the agarose gel electrophoresis described previously (4); no MVX-associated molecules were detected in the noninfected controls (see Fig. S1 in the supplemental material). The RNA samples were then enriched for poly(A)+ mRNA, reverse transcribed, and RsaI restricted, followed by two hybridization steps that enriched the differentially expressed sequences. Comparisons were made in both directions between noninfected and white infected mushrooms and between white infected mushrooms and brown infected mushrooms. The gene fragments identified by SSH were Sanger sequenced by the Genomic Centre Sequencing Service, Warwick HRI, University of Warwick (15). Transcript sequences identified during the course of these experiments were compared to known sequences available on open access and proprietary databases, putative gene function was recorded where possible. Comparisons were made with the A. bisporus genome sequence after its public release in May 2010 (16) to ascertain the A. bisporus or non-A. bisporus origin.
Expression analysis using microarrays.
The gene sequences identified from the SSH work were compared to known A. bisporus sequences, and the novel sequences were incorporated into the A. bisporus microarray design of Eastwood et al. (15) using eArray software (Agilent Technologies, Santa Clara, CA). The microarrays were manufactured by Agilent Technologies with 60-mer in situ-synthesized probes of 1,250 genes or transcripts and validated as described by Eastwood et al. (15). Each probe was replicated five times on each microarray. Further information on microarray design can be obtained via the GEO database (accession no. GSE70464).
RNA was isolated from 32 mushroom samples: 20 samples from 5 separate infected mushroom farms with crops displaying the brown symptom (4 replicate samples per farm) and 12 samples from a noninfected crop grown at Warwick HRI. The RNA was further purified as per Eastwood et al. (15) and converted by random priming into cRNA via cDNA, fluorescently labeled with cyanine-3, and hybridized to the array. To determine the amount of hybridization to each probe, arrays were laser scanned, and the resulting images were analyzed using commercially available image analysis software (15). Comparisons between treatments were made and analyzed statistically using GeneSpring software (Agilent Technologies). RNA integrity and quality were tested prior to use by electrophoretic separation using Bioanalyzer (Agilent Technologies).
For statistical analysis and normalization, the data from all 32 microarrays were combined. Outliers were identified based on the range across the five replicate probes per microarray as observations being >2.0 (on the log scale), when the difference between the maximum and/or minimum value and the median value was >0.95. Mean values for each probe were calculated across the replicate spots on each array. These mean values were then analyzed using a MicroArray analysis of variance (MAANOVA) approach in R (http://cran.rproject.org/web/packages/maanova/index.html). The MAANOVA mixed model R-package was used to analyze the response for each gene separately, allowing for differences between slides and between arrays within slide (there being eight identical arrays per slide) and assessing for differences between the six different sources of material (five infected farms and the control noninfected mushrooms). Individual F-tests were used to assess the overall differences in gene expression between the noninfected control samples and the brown infected samples, between each infected farm samples and the control, and among the BCMD-infected farm samples.
RT-qPCR.
Gene expression levels of the samples used for microarray analysis were quantified by quantitative reverse transcriptase PCR (RT-qPCR). cDNA synthesis reactions were performed with the ThermoScript RT-PCR system (Invitrogen) using 50 ng of random hexamers per reaction, according to the manufacturer's instructions. PCR primers (see Table S1 in the supplemental material) were designed using Primer Express (Applied Biosystems) to specifically amplify sections of transcripts based on their sequence established by Sanger sequencing (above). These primers were then optimized using SYBR Green I as a fluorophore for detection and an ABI 7900HT thermocycler (Applied Biosystems). Briefly, PCRs included 1 μM concentrations of forward and reverse primers, 20 ng of cDNA, 7.5 μl of 2× SYBR green PCR mix (Applied Biosystems), and the following cycling conditions: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. In addition, a dissociation curve (95°C for 15 s, 60°C for 15 s, and 2% ramp rate to 95°C) was carried out for all reactions to ensure spurious products were absent in the reactions. Standard curves were determined and assessed for linearity and efficiency to ensure accurate quantification. The transcript levels of four A. bisporus genes—beta-tubulin, actin, pyruvate dehydrogenase, and succinate dehydrogenase—were shown not to vary between treatments, and a geometric mean of their expression levels was used as an internal standard to normalize the transcript levels of 10 non-Agaricus sequences identified from microarray analysis.
For the time course experiment, RT-qPCR was performed on the RNA extracted from explant segments of three replicate mushrooms (per sampling day) each of brown infected mushrooms, white infected mushrooms, and noninfected mushrooms. The RT-qPCR time course data were log transformed and statistically analyzed by analysis of variance in a “split-plot” design.
Chromametric analysis.
Three color measurements were taken per day of each mushroom from the brown infected, white infected, and noninfected treatments (three replicate mushrooms per treatment) over a time course (day 0 to day 4) using a ChromaMeter CR-300 (as described above). The instrument was calibrated prior to use with a CR-A43 white calibration plate. The color data were statistically analyzed by analysis of variance.
Northern hybridization analysis.
DsRNAs were extracted from different disease samples, and the extracts were pooled (to maximize the number of dsRNAs) and loaded onto multiple lanes of gels in ∼300-ng aliquots per lane and with the appropriate markers in separate lanes. A 900-ng aliquot of dsRNA was applied to a single well to act as a visual reference for identification of hybridized dsRNA bands on the corresponding membranes (ethidium bromide stained as described previously [4]). Double-stranded RNAs were fractionated by prolonged low-voltage gel electrophoresis in 0.8% agarose (4). The separated dsRNA molecules were transferred to Hybond-N+ blotting membranes (GE Healthcare) overnight by capillary action according to standard protocols (17). After electrophoresis and prior to hybridization, gels were photographed to indicate dsRNA migration from the origin.
The membranes were cut into separate identical lanes and each lane of membrane was hybridized overnight with 25 ng of dsRNA-specific probe labeled with [α-32P]dCTPs in the presence of 50% formamide solution containing 5× SSPE (0.75 M sodium chloride, 50 mM sodium dihydrogen orthophosphate, 5 mM EDTA), 0.5% sodium dodecyl sulfate, 5× Denhardt's solution, and 100 μg of heat-denatured salmon sperm ml−1 at 42°C overnight. The probes were produced by PCR amplification to the 16 non-Agaricus gene sequences (presumed to be viral) identified by SSH and microarray analysis; the PCR products were analyzed by gel electrophoresis for specific, single-product amplification, and the DNA fragments of the appropriate predicted length were excised directly from the gel and purified. A Rediprime II random prime labeling system (GE Healthcare) was used to incorporate radiolabel into the PCR product.
Membrane hybridization was detected autoradiographically by exposure to blue sensitive autoradiographic film (Kodak) at −70°C for a minimum of 24 h. Autoradiographs were laser scanned densitometrically to record the intensities of hybridized bands using a Personal Densitometer SI v.4.0 (Molecular Dynamics) at a pixel size setting of 100 μm.
Accession numbers.
Newly determined sequence data were deposited in GenBank under accession numbers LN867086 to LN867098 and in the GEO database under accession number GSE70464.
RESULTS
Identification of putative differentially expressed transcripts associated with BCMD disease.
Suppression subtractive hybridization generated cDNAs in the size range of 40 to 703 bp for putative differentially expressed transcripts associated with BCMD disease. These transcript fragments were sequenced and compared, and a total of 197 unique sequences were identified. Probes were designed to the sequences and incorporated onto the A. bisporus custom microarray design.
Screening putative BCMB-associated transcripts by microarray analysis.
Transcript levels were compared between brown infected mushrooms and noninfected mushrooms by microarray analysis (microarray data GEO accession no. GSE70464). Two criteria were applied to identify transcripts with different levels between the two infection states: statistical analysis by MAANOVA (P < 0.012) and fold changes of >2 at four or five farms. This analysis resulted in the identification of 25 transcripts with increased levels and 32 transcripts with decreased levels between noninfected and brown infected states (Tables 1 and 2). Sixteen of the 25 transcripts with increased levels were found to be non-Agaricus in origin, i.e., no homology with the A. bisporus genome sequence (16), and had between 4.5 and 4,000-fold higher transcript levels in brown infected compared to control noninfected mushrooms (Tables 1 and 3). Fourteen sequences longer than 100 bp were submitted to publicly available databases with accession numbers LN867086 to LN867098, the remaining two sequences of <100 bp are shown in Table S2 in the supplemental material. Ten of the non-Agaricus transcripts with the highest fold changes (150- to 4,020-fold infected/noninfected) were selected for further investigation. Table 1 shows the 9 host Agaricus bisporus transcripts with increased levels (infected/noninfected). These code for genes involved in nitrogen/amino acid metabolism, DNA binding/transcription regulation and intracellular trafficking/secretion/vesicular transport. The transcripts with the highest fold changes and putatively identified functions had homologies to centrosome-microtubule-binding protein (7.7-fold) and to DNA-directed RNA polymerase and delta-1-pyrroline-5-carboxylate (both 4-fold).
TABLE 1.
Agaricus bisporus genes with significantly upregulated expression in MVX-infected mushroomsa
| KOG class and putative gene function | Accession no. | Avg fold change in expression |
|---|---|---|
| Nitrogen/amino acid metabolism | ||
| Serine proteinase | AJ344211.1 | 1.4 |
| Delta-1-pyrroline-5-carboxylate dehydrogenase (PruA) | X95584.1 | 4.0 |
| Alpha-aminoadipate reductase | XM_006463916.1 | 1.7 |
| DNA binding/transcription regulation | ||
| DNA-directed RNA polymerase | XM_006457975.1 | 4.0 |
| Hypothetical protein with C2H2 zinc finger domain (DNA-directed RNA polymerase Mortierella verticillata 7e10−6) | XM_006459481.1 | 2.9 |
| Intracellular trafficking, secretion, and vesicular transport | ||
| Centrosome-microtubule-binding domain protein | XM_006462296.1 | 7.7 |
| Dynamin-like GTPase | XM_006461407.1 | 1.5 |
| Unknown function (present in A. bisporus genome sequence) | ||
| Hypothetical protein 1 | XM_006456447.1 | 7.5 |
| Hypothetical protein 2 | XM_006455011.1 | 2.2 |
| Putative virus sequences (not present in A. bisporus genome sequence) | ||
| A. bisporus microarray transcript ID 2127 | LN867096 | 4,019.2 |
| A. bisporus microarray transcript ID 2135 | LN867098 | 3,702.2 |
| A. bisporus virus X genomic RNA band VX3BMF (microarray ID 2121) | AJ421988.1 | 3,466 |
| A. bisporus microarray transcript ID 2120 | LN867091 | 2,787.4 |
| A. bisporus microarray transcript ID 2125 | See Table S2 in the supplemental material | 2,767.6 |
| A. bisporus microarray transcript ID 2008 | LN867086 | 2,649 |
| A. bisporus microarray transcript ID 2126 | See Table S2 in the supplemental material | 2,550.4 |
| A. bisporus microarray transcript ID 2019 | LN867088 | 1,727.4 |
| A. bisporus microarray transcript ID 2122 | LN867095 | 1,125 |
| A. bisporus microarray transcript ID 2129 | LN867090 | 148.4 |
| A. bisporus microarray transcript ID 2029 | LN867097 | 16.0 |
| A. bisporus microarray transcript ID 2123 | LN867092 | 12.3 |
| A. bisporus microarray transcript ID 2020 | LN867089 | 7.1 |
| A. bisporus microarray transcript ID 2011 | LN867093 | 6.3 |
| A. bisporus microarray transcript ID 2118 | LN867094 | 4.8 |
| A. bisporus microarray transcript ID 2015 | LN867087 | 4.5 |
TABLE 2.
Agaricus bisporus genes with significantly downregulated expression in MVX-infected mushrooms
| KOG class and putative gene function | Accession no. | Avg fold change in expression |
|---|---|---|
| Translation, ribosomal structure, and biogenesis | ||
| 60S ribosomal protein L6E | XM_006460202.1 | 3.0 |
| 60S ribosomal protein L2 | XM_006459690.1 | 2.3 |
| 40S ribosomal protein S8 | XM_006461143.1 | 2.2 |
| 30S ribosomal protein S28e | XM_006454373.1 | 1.5 |
| Exoribonuclease | XM_006456512.1 | 1.5 |
| Amino acid metabolism/synthesis | ||
| Dihydroxy-acid dehydratase | XM_006462514.1 | 5.6 |
| Glutamine synthase | XM_006458078.1 | 3.5 |
| Phosphoribosylformimino-5-aminoimidazole carboxamide ribonucleotide (ProFAR) isomerase | XM_006460035.1 | 1.3 |
| Protein processing and turnover | ||
| PRA1 family protein | XM_006460744.1 | 2.5 |
| O-Fucosyltransferase domain | XM_006460837.1 | 1.5 |
| 20S proteasome endopeptidase | XM_006453886.1 | 1.4 |
| 20S proteasome Sβ3 | XM_006454065.1 | 1.3 |
| Aminopeptidase | XM_006458368.1 | 1.2 |
| Intracellular trafficking, secretion, and vesicular transport | ||
| Rab GTPase | XM_006458558.1 | 3.1 |
| SHR3 chaperone domain protein | XM_006458113.1 | 2.5 |
| Intracellular architecture | ||
| Hypothetical protein with NUDE motif | XM_006454367.1 | 3.1 |
| Cofilin-like actin-depolymerising factor domain protein | XM_006462466.1 | 1.8 |
| Cell surface/membrane architecture | ||
| CFEM domain membrane protein | XM_006458728.1 | 4.5 |
| Hypothetical protein with hydrophobic surface binding protein A domain | XM_006464087.1 | 4.2 |
| GDP-mannose 4,6 dehydratase | XM_006455145.1 | 4.0 |
| Galactose-binding lectin | XM_006455186.1 | 2.7 |
| Cell signaling | ||
| Ste20p-like protein kinase (serine/threonine protein kinase) | XM_006461835.1 | 1.2 |
| Nucleic acid binding/metabolism | ||
| Hypothetical protein with RNA recognition motif domain | XM_006463787.1 | 3.6 |
| Glutaredoxin (DNA synthesis) | XM_006460655.1 | 2.4 |
| DNA helicase TIP49 | XM_006454015.1 | 1.6 |
| Cellular metabolic processes | ||
| Fatty acid synthase | XM_006459000.1 | 3.5 |
| 4-aminobenzoate hydroxylase | XM_006456196.1 | 2.1 |
| Heparinize II/III-like protein | XM_006454602.1 | 1.7 |
| Metallo-dependant hydrolase/carbohydrate esterase 9 family | XM_006461088.1 | 1.3 |
| Transposable element | ||
| Putative transposable element | XM_006456594.1 | 1.9 |
| Hypothetical proteins (present in the A. bisporus genome) | ||
| Hypothetical protein 1 | XM_006454602.1 | 4.0 |
| Hypothetical protein 2 | XM_006463391.1 | 1.3 |
TABLE 3.
Fold change differences of non-Agaricus transcripts with significantly higher levels in infected mushrooms compared to noninfected mushrooms and with a fold change of >2a
| Microarray transcript ID | Microarray analysis (BIM vs control) | RT-qPCR analysis (fold change) |
|
|---|---|---|---|
| BIM vs control | BIM vs WIM | ||
| 2127 | 4,019 | 662,368 | 35,921 |
| 2135 | 3,702 | 65,873 | 16,621 |
| 2121 | 3,466 | 207,805 | 3,707 |
| 2120 | 2,787 | 618,604 | 31,640 |
| 2125 | 2,768 | 1,916,670 | 17,172 |
| 2008 | 2,649 | 2,106,633 | 11,525 |
| 2126 | 2,550 | 65,873 | 16,621 |
| 2019 | 1,727 | 1,427,255 | 87,515 |
| 2122 | 1,125 | 262,186 | 11,746 |
| 2129 | 148 | 265,049 | 3,468 |
| 2029 | 16.1 | nm | nm |
| 2123 | 12.3 | nm | nm |
| 2020 | 7.1 | nm | nm |
| 2011 | 6.3 | nm | nm |
| 2118 | 4.8 | nm | nm |
| 2015 | 4.5 | nm | nm |
Fold change differences were determined by microarray analysis and RT-qPCR. BIM, brown infected mushrooms; control, noninfected white mushrooms; WIM, white infected mushrooms; nm, not measured.
In contrast, all 32 downregulated transcripts code for A. bisporus genes. These are shown in Table 2 and have functions involved in protein synthesis/processing and turnover, membrane function and architecture, and nucleic acid binding metabolic functions. The transcripts with the most reduced levels (noninfected compared to brown infected) had putative functions associated with branched-chain amino acid synthesis (dihydroxy acid dehydratase) and cell surface/membrane architecture. Sequences found of non-Agaricus origin are shown in Table S2 in the supplemental material. Database homology searches revealed no significant similarity to any known sequence other than transcript 2121, which showed identical similarity to the A. bisporus virus X genomic RNA band VX3BMF (accession no. AJ421988).
RT-qPCR of highly expressed transcripts.
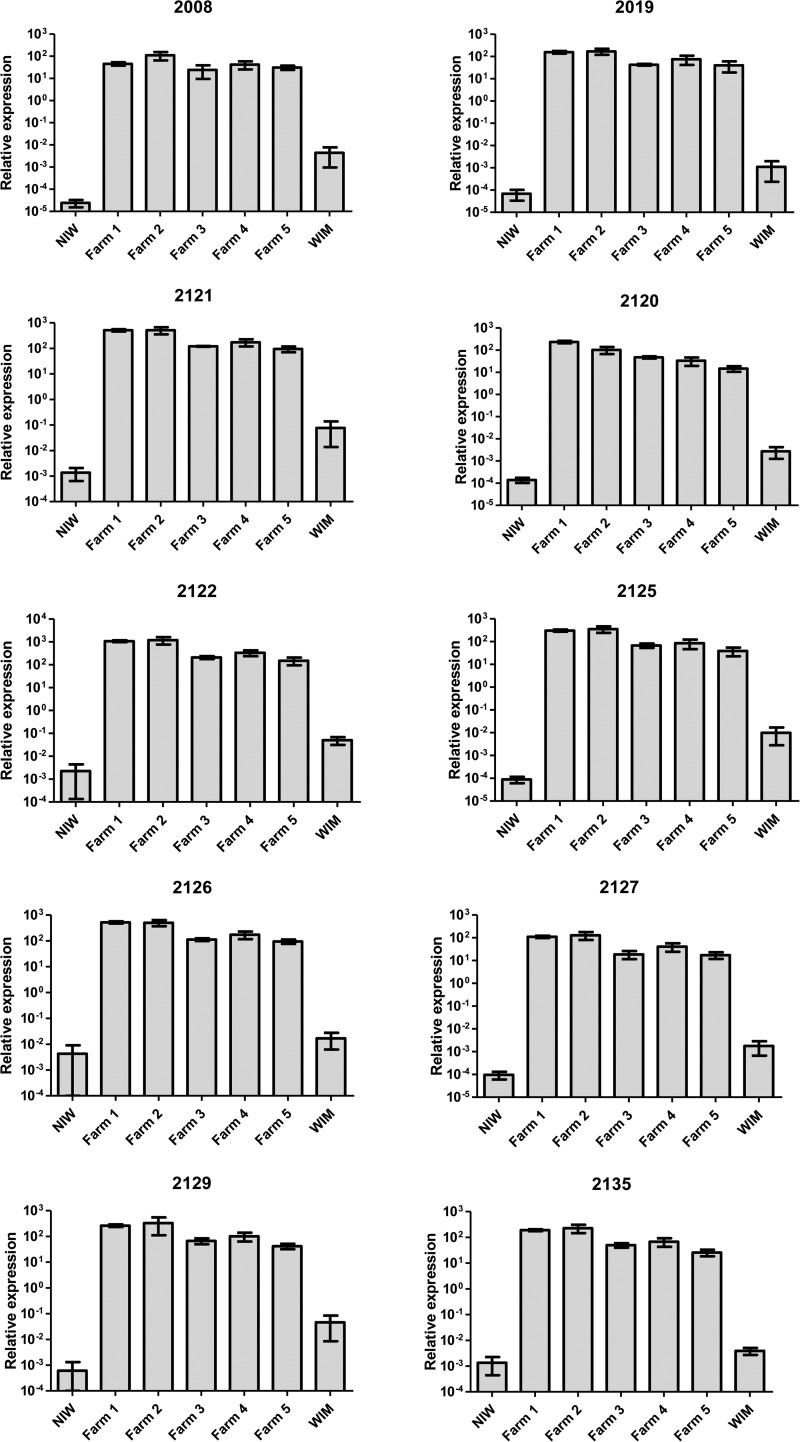
RT-qPCR analysis of the 10 most upregulated non-Agaricus transcripts showed significantly higher levels in brown infected and white infected mushrooms compared to noninfected mushrooms from the same extracted samples used for microarray analysis (P < 0.05) (Table 3 and Fig. 2). However, the fold change measured by RT-qPCR was much higher (>105 greater) than that measured by microarray. The huge fold change (brown infected/control) may indicate that these transcripts were absent from the noninfected mushrooms and that the microarray signals from noninfected mushrooms may represent nonspecific hybridizations or saturation of the array (Table 3). The level of the 10 transcripts in nonsymptomatic, white infected mushrooms (sampled in close proximity to brown infected mushrooms) were also significantly greater compared than in noninfected control mushrooms (P < 0.05) (Table 3 and Fig. 2). The levels of virus fragments were found at two distinct ranges: at increased levels in infected white mushrooms and between 374 and 12,000 times greater in infected brown mushrooms both compared to noninfected controls.
FIG 2.
RT-qPCR analysis of putative dsRNA fragments from brown mushrooms sampled from five different farms and noninfected white (NIW) negative control and white infected mushrooms (WIM). The relative expressions were calculated against the geometric mean of four housekeeping genes (β-tubulin, actin, pyruvate dehydrogenase, and succinate dehydrogenase). The A. bisporus microarray transcript fragment number is shown above each graph. Error bars indicate the standard errors.
Time course analysis of color symptom development and transcript levels.
Transcript levels of the 10 non-Agaricus transcripts and mushroom cap color were measured over a 4-day time course during mushroom growth and symptom development in (i) infected mushrooms developing a brown coloration, (ii) infected mushrooms that remained white, and (iii) control noninfected mushrooms.
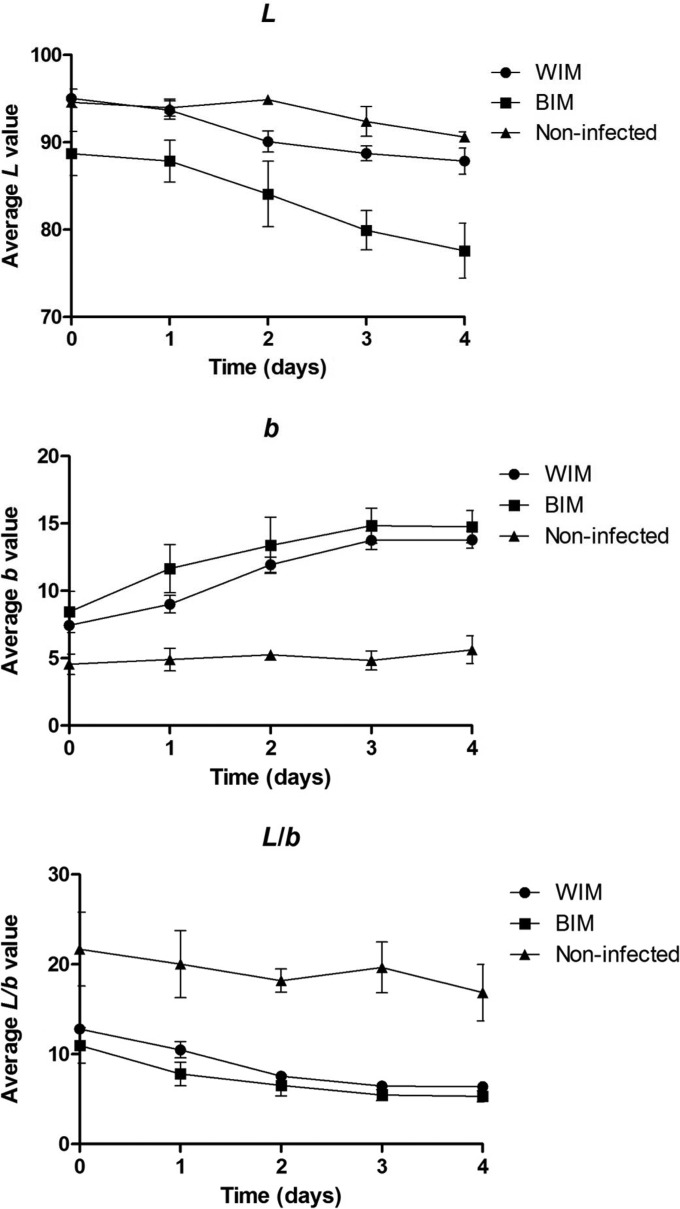
Three chromametric values were measured: L, a, and b. The L value represents the “whiteness to darkness” on a monochromatic scale; the a value represents the “redness,” and the b value represents the “yellowness.” The average L color value of the mushrooms decreased significantly (P < 0.05) for all three treatments over time (i.e., more discolored) (Fig. 3a). The L value of the infected mushrooms (which later developed to brown infected) was already significantly lower than the other two treatments at day 0, even though this was not apparent to the human eye. No significant difference was measured in L value between white infected and noninfected controls at day 0 and 1. Analyses of the a color value did not reveal consistent significant differences over time and between the samples tested (data not shown). The b values (indicating yellowness) of the brown infected and white infected mushrooms did not differ significantly from each other but were significantly different (P < 0.05) from the control noninfected mushrooms at day 0 and thereafter (more yellow) (Fig. 3b).
FIG 3.
Mushroom color over a time course (4 days). The average L and b values and the L/b ratios of brown infected mushrooms (BIM), white infected mushrooms (WIM), and noninfected mushrooms are depicted. Error bars indicate the least significant differences (5%).
When two color parameters were combined in a ratio, L/b, both white infected and brown infected mushrooms had significantly different L/b ratios (P < 0.05) compared to the noninfected controls from day 0 to day 4 (Fig. 3c). This combined ratio, L/b, can therefore identify infected mushrooms that will develop brown coloration before detection by the human eye. This also implies that the biochemistry responsible for the infection-related color difference was already active during early fruit body development.
Transcript levels of the 10 non-Agaricus transcripts were measured by RT-qPCR in brown infected, white infected, and noninfected mushrooms did not change significantly over the 4 days (see Fig. S2 in the supplemental material), with the exception of transcript fragment number 2008, which had significantly higher levels on days 2 to 4 than on day 0 (P < 0.05). The levels of transcripts from the brown infected and white infected mushrooms were significantly higher than those from the control noninfected mushrooms by 4 orders of magnitude.
Hybridization of non-Agaricus transcripts to existing dsRNA profiles.
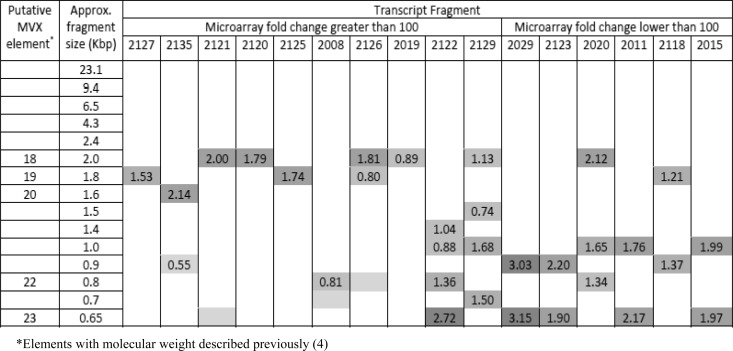
Northern hybridization analyses were conducted using probes designed to the 16 non-Agaricus transcripts. All probes hybridized to low-molecular-weight dsRNAs of size 2.0 kbp or smaller (Fig. 4) there was no evidence of hybridization to larger molecules present in the samples. Four probes (transcript fragment numbers 2019, 2120, 2125, and 2127) hybridized strongly to a single dsRNA fragment. These hybridization signals included the four low-molecular-weight dsRNAs previously associated with the brown symptom (0.6, 0.8, 1.8 and 2.0 kbp). In general, the non-Agaricus transcripts which showed the greatest expression by microarray analysis hybridized to fewer and larger dsRNA elements (Fig. 4).
FIG 4.
Schematic representation of Northern hybridizations of transcript fragments to dsRNAs and scanning densitometry analysis. dsRNA was extracted from mushrooms with the browning symptom and separated by gel electrophoresis, and Northern hybridizations were conducted using PCR-amplified fragments for each of the 16 differentially regulated non-Agaricus transcript identified. Approximate fragment sizes were calculated using a molecular weight marker and measuring the distance migrated from the well (densitometry measurements of >0.5 are included shown for each hybridization, with the relative signal intensity indicated by darker shading). Transcripts are ranked in order of increasing level of expression as measured by microarray hybridization. The transcript fragment indicates a A. bisporus microarray transcript reference.
DISCUSSION
Strong evidence (microarray and RT-qPCR) has been presented that transcript fragments identified by SSH are viral in origin. These transcripts were at much greater levels in infected than noninfected mushrooms, hybridized to dsRNAs, and are not coded by the Agaricus bisporus genome (16). The transcript levels were so low in noninfected mushrooms (105 lower than in infected mushrooms) that the signals probably represent nonspecific hybridizations on the microarray and therefore are absent in noninfected mushrooms.
The non-Agaricus transcript fragments hybridized to low-molecular-weight dsRNA bands (≤2 kbp; Fig. 4) and provided no evidence that these are fragments of larger RNA molecules. Seven of the sixteen probes hybridized to the 2.0- or 1.8-kbp band. Twelve of the sixteen probes hybridized to two or more elements, possibly indicating close sequence similarity between the viral elements. The identification of transcripts hybridizing to dsRNAs with sizes 0.65, 0.7, 0.8, 0.9, 1.0, 1.4, 1.5, 1.6, 1.8, 1.9, and 2.0 kbp is consistent with previously associations made between the browning symptom and small dsRNAs of 0.6 to 2.0 kbp (from mushrooms in the United Kingdom) (4) and 0.6 to 2.3 kbp (from mushrooms in The Netherlands) (9). The increased numbers of transcripts identified in this study may reflect the increased sensitivity of detection by autoradiography.
We hypothesize from the hybridization results that the functional infective unit of BCMD is the 2.0 and 1.8-kbp dsRNAs. This cooccurring pair is sufficient size to contain the coding potential required for independent existence. The RNA-dependent RNA polymerase (RDRP) of mycoviruses has a coding size range of ∼2 kbp. The RNA elements with sizes lower than 1.8 kbp are possibly satellite dsRNAs and may not have coding function because of length restrictions (18). Satellites are capable of modulating disease expression, however (19). A virus of this inferred genome size and organization mirrors the characteristics of the bisegmented genomes of Partitiviridae members, e.g., Rhizoctonia solani virus 717 (20) and Pleurotus ostreatus virus 1 (21), both mycoviruses of basidiomycete fungi. The Partitiviridae have a dsRNA genome divided into two segments, infect both fungi and plants, and have a persistent life-strategy being generally asymptomatic (14, 22). However, the two segments of a Partitiviridae code for an RDRP enzyme and a coat protein. It is uncertain whether this disease is associated with a coat protein since viral particles have not been identified in mushrooms with BCMD (4).
Viral transcripts have been quantified by RT-qPCR at two levels: moderate levels (102- to 103-fold increase above noninfected controls) in nonsymptomatic mushrooms and very high levels (104- to 106-fold increase) in highly symptomatic (brown colored) mushrooms (Table 3). There appears to be a step change in viral activity analogous to the transition in viral life strategy from persistent to acute proposed by Villarreal et al. (23) for animal viruses and Roossinck (22) for plant viruses. The two levels of A. bisporus viral transcripts were found in brown infected and white infected mushroom fruit bodies connected by a mycelial network. However, the transition from moderate to high levels appears to be determined at an early stage of fruit body development and thus associated with the phase change from vegetative to reproductive growth and the subsequent differentiation and cell division. Persistent viruses are generally asymptomatic, do not move from cell to cell, are present at low titers and transmitted vertically, and when found in plants, belong in the Partitiviridae family and the Endonavirus genus, two viral classifications which also infect fungi. Acute viruses accumulate to high levels, move from cell to cell via generally horizontal transmission, and flourish in monoculture (22). Cultivated mushrooms are grown in monoculture and may contain the viral populations from wild A. bisporus since the organism has only relatively recently been taken into cultivation (about 400 years) and since then largely propagated vegetatively. The proposed potential for transition in viral life strategy from persistent to acute could have been stimulated by a change in the growing technology of A. bisporus known as phase 3 composting. This process involves bulk A. bisporus mycelial growth in 100-tonne batches, followed by cell breakage by machine (to facilitate compost loading/unloading for transport), cell mixing, and increased cell division and hyphal fusions (anastomoses). This breakage and mixing of cells may have increased the potential for viral elements to spread through the mycelial network. Phase 3 composting was adopted by the mushroom industry at about the time when the disease was first described.
The A. bisporus transcript with the largest increase in expression levels (brown infected compared to control) has homology to centrosome-microtubule-binding domain protein, exhibiting a 7.7-fold increase. It is interesting that the centrosome (i.e., the fungal equivalent is the spindle pole body) has been reported to bind RNA and is part of the cytoskeleton which can interact with viral proteins and the endoplasmic reticulum forming a viral replication complex (24, 25). The 4-fold increase in expression of DNA-directed RNA polymerase is difficult to explain in response to increased transcription of an RNA virus, other than perhaps reflecting a change in host transcriptional patterns. The increased expression of delta-1-pyrroline-5-carboxylate dehydrogenase transcripts may indicate a redistribution of nutritional resources in cap tissue as suggested by Wagemaker et al. (26).
Of the 26 genes of A. bisporus to be involved in melanin biosynthesis (27) and the formation of brown tissue only one, 4-aminobenzoate hydroxylase, had changed expression levels, but with a decrease (brown infected compared to control). However, the increased serine protease transcript level in brown infected mushrooms is consistent with its ability to activate tyrosinase (28, 29); however, the degree of upregulation is small (1.4-fold). Further studies on enzyme activity and chemical analyses in brown infected mushrooms are needed to determine which enzymes are responsible for color formation. The downregulated genes show a general decease in cellular biosynthesis of macromolecules consistent with the appropriation of the cellular functions for viral replication particularly protein synthesis and nitrogen metabolism were targeted. The downregulation of a broad range of genes associated with cellular biosynthesis and growth, e.g., ribosomal proteins, also corresponds with the observed 24-h delay in primordium formation associated with A. bisporus MVX infection (4).
The use of a sensitive and objective technology to measure reflected color (the Minolta meter) has shown that a color difference can be detected between white infected mushrooms (which appear to be white to the human eye) and white noninfected mushrooms (Fig. 3). This instrumentation could be used by the mushroom growing industry to detect the disease earlier in a crop before any coloration becomes apparent and the use of the combined color parameter ratio L/b makes this detection more sensitive. The color data also demonstrated that the symptom of diseased mushrooms brown or “off-white” (i.e., white to the human eye but with a low L/b ratio measured by the Minolta meter) was determined early in the development of the mushroom and correlating with the two levels of viral transcripts. Mushroom primordia growing in close proximity may have either high viral transcript loads and become brown mushrooms or low viral transcript loads and become off-white mushrooms (Fig. 1). This difference in biology is localized to individual mushroom fruit bodies and thus not easily transmissible at a later growth stage. However, the observation that the brown coloration is uniform across a mushroom cap suggests that the signal/changed biology was at the primordial stage when cell division is highly active.
Transcript fragments from mushrooms with BMCD hybridized to dsRNAs and had sequences not present in the Agaricus bisporus genome and thus are presumed to be viral. We hypothesize that the transcripts may represent a single bi- or multipartite virus in the family Partitiviridae, although this is based on the similar sizes of the RNAs without sequence comparisons. The viral transcript levels were found at two broad ranges and there appears to be a step change to the higher level at an early developmental stage resulting in expression of the brown tissue symptom. This may be an example of change in viral life strategy from persistent to acute caused by factors currently unknown. The sequence information from the viral transcripts could form the basis of an RT-qPCR-based test to detect the level of viral presence in mycelium. Preliminary evidence has demonstrated that virus can be quantified in the mycelium of an infected A. bisporus crop (30). Future work should involve full sequencing of the viral transcripts and a study of the factors that determine viral transcript levels and disease symptom expression.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge research funding from Teagasc (grant HDHG 5515) and The Walsh Fellowship, Teagasc, Ireland (project 2006082).
We thank Robert Coutts, Imperial College London, for useful discussion.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01093-15.
REFERENCES
- 1.Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384. doi: 10.1146/annurev-phyto-080508-081932. [DOI] [PubMed] [Google Scholar]
- 2.Sonnenberg ASM, Baars JJP, Hendrickx PM, Lavrijssen B, Gao W, Weijn A, Mes JJ. 2011. Breeding and strain protection in the button mushroom Agaricus bisporus, p 7–15. In Savoie J-M, Foulongue-Oriol M, Largeteau M, Barroso G (ed), Proceedings of the 7th International Conference on Mushroom Biology and Mushroom Products, 4–7 October 2011, Arcachon, France INRA, UR1264, Mycology and Food Safety, Bordeaux, France. [Google Scholar]
- 3.Burton KS, Green J, Baker A, Eastwood DC, Grogan HM. 2011. Mushroom virus X: the identification of brown cap mushroom virus and a new highly sensitive diagnostic test for phase III compost, p 466–473. In Savoie J-M, Foulongue-Oriol M, Largeteau M, Barroso G (ed), Proceedings of the 7th International Conference on Mushroom Biology and Mushroom Products, 4–7 October 2011, Arcachon, France INRA, UR1264, Mycology and Food Safety, Bordeaux, France. [Google Scholar]
- 4.Grogan HM, Adie BAT, Gaze RH, Challen MP, Mills PR. 2003. Double-stranded RNA elements associated with the MVX disease of Agaricus bisporus. Mycol Res 107:147–154. doi: 10.1017/S0953756203007202. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher JT, Gaze RH. 2008. Mushroom pest and disease handbook. Mansion Publishing, London, United Kingdom. [Google Scholar]
- 6.Goodin MM, Schlagnhaufer B, Romaine CP. 1992. Encapsidation of La France disease specific dsRNAs in 36-nm isometric particles. Phytopathology 82:285 290. doi: 10.1094/Phyto-82-285. [DOI] [Google Scholar]
- 7.van der Lende TR, Harmsent MC, Wessels JGH. 1994. Double-stranded RNAs and proteins associated with the 34 nm virus particles of the cultivated mushroom Agaricus bisporus. J Gen Virol 75:2533–2536. doi: 10.1099/0022-1317-75-9-2533. [DOI] [PubMed] [Google Scholar]
- 8.Maffettone E. 2007. Characterization of a novel virus associated with the MVX disease of Agaricus bisporus. Ph.D. thesis Cranfield University, Bedford, United Kingdom. [Google Scholar]
- 9.Sonnenberg ASM, Lavrijssen B. 2004. Browning and the presence of viral double-stranded RNA in Dutch mushrooms, p 541–546. In Romaine CP, Keil CB, Rinker DL, Royse DJ (ed), Science and cultivation of edible and medicinal fungi. Penn State University, State College, PA. [Google Scholar]
- 10.Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N. 2014. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188:128–141. doi: 10.1016/j.virusres.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Hammond JBW, Nichols R. 1975. Changes in respiration and soluble carbohydrates during the post-harvest storage of mushrooms (Agaricus bisporus). J Sci Food Agr 26:835–842. doi: 10.1002/jsfa.2740260615. [DOI] [Google Scholar]
- 12.Grogan HM, Tomprefa N, Mulcahy J, Holcroft S, Gaze RH. 2004. Transmission of mushroom virus X disease in crops, p 489–497. In Romaine CP, Keil CB, Royse DJ (ed), Mushroom science XVI: science and cultivation of edible and medicinal fungi. Penn State University, State College, PA. [Google Scholar]
- 13.Valverde RA, Nameth ST, Jordan R. 1990. Analysis of double-stranded RNA for plant virus diagnosis. Plant Dis 74:255–258. [Google Scholar]
- 14.Chomczynski P. 1993. TRIZOL™: a new reagent for optimal single-step isolation of RNA. Biotechniques 15:532–537. [PubMed] [Google Scholar]
- 15.Eastwood DC, Herman B, Noble R, Dobrovin-Pennington A, Sreenivasaprasad S, Burton KS. 2013. Environmental regulation of reproductive phase change in Agaricus bisporus by 1-octen-3-ol, temperature and CO2. Fungal Genet Biol 55:54–66. doi: 10.1016/j.fgb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Deakin G, Doddapaneni H, Floudas D, Grimwood J, Hildén K, Kües U, Labutti KM, Lapidus A, Lindquist EA, Lucas SM, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wösten HA, Xu J, Eastwood DC, Foster GD, Sonnenberg AS, Cullen D, de Vries RP, Lundell T, Hibbett DS, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F. 2012. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci U S A 109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 18.Xu P, Roossinck MJ. 2011. Plant virus satellites. John Wiley & Sons, Ltd, Chichester, United Kingdom. [Google Scholar]
- 19.Kaper JM, Tousignant ME. 1994. Viral satellites: parasitic nucleic acids capable of modulating disease expression. Endeavour 8:194–200. [DOI] [PubMed] [Google Scholar]
- 20.Strauss EE, Lakshman DK, Tavantzis SM. 2000. Molecular characterization of a partitivirus from the basidiomycete Rhizoctonia solani. J Gen Virol 81:549–555. [DOI] [PubMed] [Google Scholar]
- 21.Lim WS, Jeong JH, Jeong RD, Yoo YB, Yie SW, Kim KH. 2005. Complete nucleotide sequence and genome organization of a dsRNA partitivirus infecting Pleurotus ostreatus. Virus Res 108:111–119. doi: 10.1016/j.virusres.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Roossinck MJ. 2010. Lifestyles of plant viruses. Trans R Soc B 365:1899–1905. doi: 10.1098/rstb.2010.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villarreal LP, Defilippis VR, Gottlieb KA. 2000. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology 272:1–6. doi: 10.1006/viro.2000.0381. [DOI] [PubMed] [Google Scholar]
- 24.Marshall WF. 2009. Centriole evolution. Curr Opin Cell Biol 21:14–19. doi: 10.1016/j.ceb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferralli J, Ashby J, Fasler M, Boyko V, Heinlein M. 2006. Disruption of microtubule organization and centrosome function by expression of tobacco mosaic virus movement protein. J Virol 80:5807–5821. doi: 10.1128/JVI.00254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagemaker MJM, Eastwood DC, Welagen J, Van Der Drift C, Jetten MSM, Burton KS, van Griensven LJLD, Op Den Camp HJM. 2007. The role of ornithine aminotransferase in fruiting body formation of the mushroom Agaricus bisporus. Mycol Res 111:909–918. doi: 10.1016/j.mycres.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Weijn A, Bastiaan-Net S, Wichers HJ, Mes JJ. 2013. Melanin biosynthesis pathway in Agaricus bisporus mushrooms. Fungal Genet Biol 55:42–53. doi: 10.1016/j.fgb.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Burton KS, Noble R. 1993. The influence of flush number, bruising and storage temperature on mushroom quality. Postharvest Biol Technol 5:39–47. [Google Scholar]
- 29.Espín JC, van Leeuwen J, Wichers HJ. 1999. Kinetic study of the activation process of a latent mushroom (Agaricus bisporus) tyrosinase by serine proteinases. J Agric Food Chem 47:3509–3517. doi: 10.1021/jf9813539. [DOI] [PubMed] [Google Scholar]
- 30.Burton KS, Baker A, Eastwood DC, Grogan H. 2010. Developing an accurate, quantitative and predictive test for Mushroom Virus X. HDC report M51. Horticulture Development Company, Kenilworth, United Kingdom: http://www.hdc.org.uk/sites/default/files/research_papers/M51%20Final%20Report.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.