Abstract
The identification of core genes involved in the biosynthesis of saxitoxin (STX) offers a great opportunity to detect toxic algae associated with paralytic shellfish toxins (PST). In the Yellow Sea (YS) in China, both toxic and nontoxic Alexandrium species are present, which makes it a difficult issue to specifically monitor PST-producing toxic algae. In this study, a quantitative PCR (qPCR) assay targeting sxtA4, a domain in the sxt gene cluster that encodes a unique enzyme involved in STX biosynthesis, was applied to analyze samples collected from the YS in spring of 2012. The abundance of two toxic species within the Alexandrium tamarense species complex, i.e., A. fundyense and A. pacificum, was also determined with TaqMan-based qPCR assays, and PSTs in net-concentrated phytoplankton samples were analyzed with high-performance liquid chromatography coupled with a fluorescence detector. It was found that the distribution of the sxtA4 gene in the YS was consistent with the toxic algae and PSTs, and the quantitation results of sxtA4 correlated well with the abundance of the two toxic species (r = 0.857). These results suggested that the two toxic species were major PST producers during the sampling season and that sxtA-based qPCR is a promising method to detect toxic algae associated with PSTs in the YS. The correlation between PST levels and sxtA-based qPCR results, however, was less significant (r = 0.552), implying that sxtA-based qPCR is not accurate enough to reflect the toxicity of PST-producing toxic algae. The combination of an sxtA-based qPCR assay and chemical means might be a promising method for monitoring toxic algal blooms.
INTRODUCTION
Saxitoxin (STX) and its analogues, commonly known as paralytic shellfish toxins (PSTs), are potent neurotoxic alkaloids (1) synthesized by marine dinoflagellates in the genera Alexandrium, Gymnodinium, and Pyrodinium (2–4) and some cyanobacteria in freshwater (5–7). PSTs ingested by humans via shellfish vectors can reversibly bind to voltage-gated Na+ channels and inhibit the flow of sodium ions (1, 8), which leads to paralytic poisoning symptoms, including neurological numbness, tingling and burning of the lips and skin, ataxia, and fever. Severe poisoning may lead to a loss of muscular coordination and respiratory distress, which can be fatal (9).
The biosynthetic pathway for STX, a compound with a complex chemical structure, remained a mystery for a long time prior to the identification of the STX synthesis genes in several cyanobacterial species (10). The identification and characterization of this set of core genes involved in STX synthesis provided the possibility of distinguishing the toxic potential of incipient blooms. A variety of genes related to toxin synthesis in cyanobacteria have been applied successfully not only in the detection, differentiation, and quantification of toxic cyanobacteria but also in studies on the regulation of toxin biosynthesis (7, 11–14). In contrast, the genetic basis for STX production in dinoflagellates remains elusive, due to the huge size of the haploid genome, which is up to 60 times the size of that of humans; it consists of a considerable number of unknown genes and a high frequency of repeats. However, the precursor incorporation patterns and stereochemistries of PSTs should be identical in cyanobacteria and dinoflagellates (15). Recently, the cyanobacterial sxt gene homologs, consisting of four domains (sxtA1 to sxtA4), were also demonstrated in dinoflagellates (16). The domain sxtA4 in the sxt gene cluster, which encodes the unique enzyme putatively involved in the sxtA pathway for STX synthesis in marine dinoflagellates, has been adapted to develop a saxitoxin-specific quantitative PCR (qPCR) assay (17). This assay has been used to assess the toxic potential of blooms in Australia and has shown promise as an accurate, fast, and cost-effective means of quantifying the potential for STX production in marine phytoplankton samples. It will also be useful for biological oceanographic studies and monitoring of toxic algal blooms.
The Yellow Sea (YS) in China has many important aquaculture zones in which PSTs have been frequently detected in shellfish samples (18–20). Several PST-producing species in the genus Alexandrium, such as A. tamarense (Lebour) Balech, A. catenella (Whedon & Kofoid) Balech, and A. minutum Halim, have been identified in parallel with nontoxic species, like A. affine (Inoue & Fukuyo) Balech, A. andersonii Balech, and A. leei Balech (21). A. tamarense (Lebour) Balech, A. catenella (Whedon & Kofoid) Balech, and another morphologically defined species, A. fundyense Balech, together constitute the A. tamarense species complex, which can be classified into different ribotypes/groups (groups I to V) based on the sequences of rRNA genes and internal transcribed spacer (ITS) regions (22–24). In the YS, cells of both groups I and IV from the species complex have been detected (18, 21). Recently, the nomenclature of the species complex was formally revised, and species names were assigned for the 5 groups as A. fundyense (group I), A. mediterraneum (group II), A. tamarense (group III), A. pacificum (group IV), and A. australiense (group V) (25). The morphologically derived name A. catenella was rejected. In this paper, the new nomenclature of the species complex is adopted, and A. fundyense and A. pacificum are used to represent groups I and IV of the species complex previously identified as A. tamarense and A. catenella, respectively, by their morphological features.
Due to the similarities in the morphological features of many Alexandrium species, particularly those species within the A. tamarense species complex, the traditional morphological approach is not accurate enough for distinguishing toxic and nontoxic species. The detection of a gene specific for STX synthesis, therefore, is a better choice to monitor the blooms of PST-producing algae and to understand the potential impacts of those toxic algal blooms. In this study, the sxtA-based qPCR assay was applied to analyze the field samples collected from the YS in parallel with two TaqMan-based qPCR assays for the quantification of A. fundyense and A. pacificum, and the PST content in net-concentrated phytoplankton samples was determined with high-performance liquid chromatography (HPLC) at the same time. The results of the qPCR assays and HPLC were compared to explore the potential of using an STX synthesis-related gene in monitoring PST-producing algal blooms in the YS.
MATERIALS AND METHODS
Cruises and sample collection.
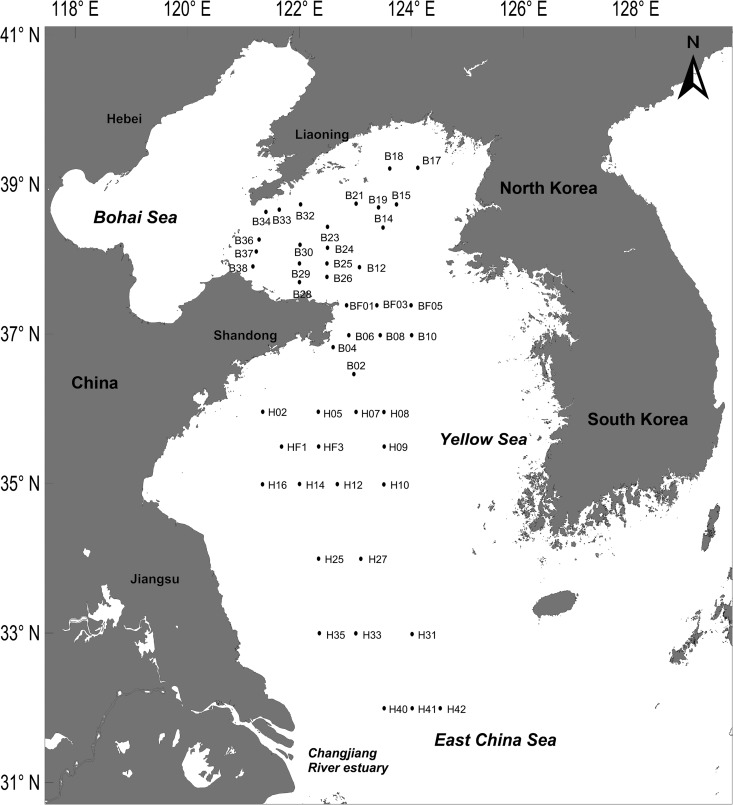
Altogether, 52 samples collected from the YS (121.0°E to 124.5°E, 32.0°N to 39.0°N) in a cruise organized by the National Natural Science Foundation of China (NSFC) from 12 to 20 May 2012 (Fig. 1) were analyzed in this study.
FIG 1.
Investigation area and sampling sites (labeled dots) in the Yellow Sea and East China Sea.
About 1,000 liters of surface seawater was collected with a submersible pump (the exact volume was calculated with the flow rate and sampling time), and phytoplankton were concentrated using a net with mesh size of 20 μm (T3 Monocron HM 200). The concentrated cells were flushed into a flask with membrane-filtered (pore size, 0.45 μm) seawater, and the final volume was made up to 500 ml. After the cells and seawater were mixed, 100 ml of the net-concentrated sample was filtered with a glass fiber membrane (Whatman; grade GF/C, 47-mm diameter) under low pressure (vacuum, <50 kPa) for a determination of the PSTs. Another 50 ml of net-concentrated sample was filtered with a 25-mm-diameter nylon gauze (mesh size, 10 μm) for qPCR assays. The membranes were put into cryogenic tubes and stored in a refrigerator at −20°C until analysis.
qPCR assays. (i) DNA extraction for qPCR assays.
DNA extraction was performed according to the method of Hosoi-Tanabe and Sako (26). The frozen samples collected on the nylon mesh were thawed at room temperature, and the cells were immediately rinsed with 1 ml of Tris-EDTA (TE) buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0]) and added to a centrifugation tube. After centrifugation at 3,300 × g for 1 min (3-16K centrifuge; Sigma, Germany), the supernatant in the centrifugation tube was removed. The cell pellet was resuspended with 400 μl of TE buffer and boiled at 100°C. Next, 400 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added. The mixture was shaken at room temperature for 1 min. After centrifugation at 14,324 × g for 3 min at 4°C, 300 μl of the supernatant was transferred to a new tube. Next, 15 μl of 3 M sodium acetate (pH 5.2) and 400 μl of 100% ethanol (−20°C) were added. After centrifugation at 14,324 × g for 3 min at 4°C, the DNA pellet was rinsed with 70% ethanol, dried, and dissolved in 20 μl of TE buffer for the qPCR assays.
(ii) qPCR assay for sxtA4.
A qPCR assay for sxtA4 using SYBR green developed by Murray et al. (17) was adopted for use in this study. The qPCR assay was carried out using a primer pair, sxtA4F (5′-CTGAGCAAGGCGTTCAATTC-3′) and sxtA4R (5′-TACAGATMGGCCCTGTGARC-3′), which was designed based on the consensus sequence of the sxtA4 domain from 9 strains of the STX-producing species A. catenella, A. tamarense, A. minutum, A. fundyense, and Gymnodinium catenatum. The PCR product was a 125-bp fragment. The primers were synthesized by TaKaRa Biotechnology Co., Ltd., Dalian, China.
The qPCR assay was performed in triplicate in an 8-well plate format in a CFX96 Touch real-time PCR detection system (Bio-Rad, CA, USA). The fluorescence threshold was set by the analytical software for the real-time PCR detection system (Bio-Rad). The final volume for PCR was 20 μl, containing 10 μl of SYBR Premix Ex Taq II (2×) (product no. DRR081; TaKaRa Biotechnology Co., Ltd., Dalian, China), 6.8 μl of double-distilled water (ddH2O), 1.6 μl of the template, and 0.8 μl of 10 μM each primer. The qPCR assays were performed under the following cycling conditions, as suggested by the reagent protocol: initial denaturation at 95°C for 10 s and 39 cycles of 15 s of denaturation at 95°C and 30 s of annealing/extension at 60°C. Melting curve analysis was performed at the end of each cycle, and selected PCR products were sequenced to confirm the specificity of PCR amplification.
To test the specificity of the qPCR assay for sxtA4, 8 strains of STX-producing Alexandrium species, 3 strains of nontoxic Alexandrium species, and a species of diatom (Table 1) were analyzed with the qPCR assay. These cultures were maintained at 20°C with f/2-Si medium (the diatom was cultured with f/2 medium). The light intensity was 56 microeinsteins m−2 s−1 on a 14-h light/10-h dark cycle. Also, 20,000 cells were counted under light microscopy (Eclipse E100; Nikon, Japan) and collected from four PST-producing species, i.e., A. minutum (strain Am-LYG), A. pacificum (strains ACDH and ATHK), A. fundyense (strain ATLY), and the nontoxic species A. affine (strain ASCH) during the exponential-growth phase. DNA was extracted and amplified with the qPCR assay in triplicate to test whether there was a significant difference in the copy numbers of the sxtA4 domain among the STX-producing species. TE buffer for dissolving the DNA pellet was used as a negative control in this experiment and during the analysis of field samples.
TABLE 1.
List of microalgal species used in the experiment
| Current species name (group) | Morphological species name | GenBank accession no. (LSU rRNA gene D1-D2 region) | Strain | PSTs detected | Source | Presence of sxtA4 qPCR product |
|---|---|---|---|---|---|---|
| A. pacificum (IV) | A. tamarense | DQ176650 | ATCI02 | Yes | South China Sea | + |
| A. pacificum (IV) | A. tamarense | DQ176651 | ATCI03 | Yes | South China Sea | + |
| A. pacificum (IV) | A. tamarense | DQ176649 | AT5-3 | Yes | South China Sea | + |
| A. pacificum (IV) | A. tamarense | DQ176652 | ATHK | Yes | South China Sea | + |
| A. pacificum (IV) | A. catenella | DQ176647 | ACDH | Yes | East China Sea | + |
| A. tamarense (III) | A. tamarense | DQ176655 | AT-6 | No | Europe | − |
| A. fundyense (I) | A. tamarense | ATLY | Yes | Yellow Sea | + | |
| A. minutum | A. minutum | DQ176657 | AM | Yes | East China Sea | + |
| A. minutum | A. minutum | Am-LYG | Yes | Yellow Sea | + | |
| A. affine | A. affine | ASCH | No | Yellow Sea | − | |
| A. affine | A. affine | DQ176654 | AC-1 | No | South China Sea | − |
| Skeletonema costatum | S. costatum | SC-1 | No | Jiaozhou Bay of Yellow Sea | − |
To assess the relative abundances of the sxtA4 gene in field samples, a calibration curve was established with the DNA sample prepared from a representative toxic species, A. pacificum (strain ACDH). With this calibration curve, the threshold cycle (CT) values of the field samples can be converted into the cell number of this representative toxic species, which will be used in a direct comparison with the abundance of the toxic A. tamarense species complex (here, A. fundyense and A. pacificum). To prepare the calibration curve, a DNA sample was extracted from 200,000 cells of A. pacificum (strain ACDH) at the exponential-growth phase and diluted at 50% over 3 orders of magnitude to prepare a set of standard solutions (n = 8). Triplicate standard solutions were analyzed with the qPCR assay to establish the calibration curve.
(iii) TaqMan-based qPCR assays for A. fundyense and A. pacificum.
The quantification of A. fundyense and A. pacificum organisms in the YS was previously conducted with two TaqMan-based qPCR assays (27), according to the protocols of Hosoi-Tanabe and Sako (26) and Gao et al. (28), with some modifications. The primer pairs used for A. fundyense were AtI-F (5′-GCTTGGTGGGAGTGTTGCAC-3′) and AtI-R (5′-TAAGTCCAAGGAAGGAAGCATC-3′), and the TaqMan probe was AtI-P (5′-FAM-AGAGCTTTGGGCTGTGGGTGTA-TAMRA-3′) (FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine). For A. pacificum (group IV of the A. tamarense species complex), the primer pairs used were AtIV-F (5′-CCTCAGTGAGATTGTAGTGC-3′) and AtIV-R (5′-GTGCAAAGGTAATCAAATGTCC-3′), and the TaqMan probe was AtIV-P (5′-FAM-ATGGGTTTTGGCTGCAAGTGCA-TAMRA-3′). The primers and probes were synthesized by TaKaRa Biotechnology Co., Ltd., Dalian, China.
The qPCRs were performed in a strip of eight 0.2-ml thin-wall tubes (product no. PCR-02-C; Axygen Scientific, Inc., CA, USA) with a CFX96 Touch real-time PCR detection system (Bio-Rad, CA, USA). Each 25-μl qPCR mixture contained 12.5 μl of 2× Premix Ex Taq (probe qPCR) (product no. DR390A; TaKaRa Biotechnology Co., Ltd., Dalian, China), 8.5 μl of ddH2O, 2 μl of the template, 0.5 μM each primer, and 1 μM TaqMan Probe. The qPCR assays were performed in triplicate, with the following protocol: initial denaturation at 95°C for 30 s and then 40 cycles of 5 s of denaturation at 95°C and 30 s of annealing/extension at 60°C.
The efficiencies of the two qPCR assays for A. fundyense and A. pacificum were 100% and 92%, respectively. The field samples were quantified with the calibration curves prepared with two strains representing A. fundyense (ATLY) and A. pacificum (ACDH) (27).
Analysis of paralytic shellfish toxins with HPLC.
PSTs in the phytoplankton samples were analyzed by HPLC using a postcolumn oxidation method (29), with some modifications (18).
(i) Apparatus and reagents.
PSTs were analyzed with the Alliance HPLC system (Waters Corporation, Milford, MA, USA), including an e2695 separation module, a postcolumn reaction module, and a 2475 fluorescence detector (FLD). The system was run under the software Empower 2.
All chemical reagents required for toxin analysis were of HPLC or analytical grade. The water used for toxin analysis was Milli-Q water prepared with a Millipore Simplicity water purification system (EMD Millipore Corporation, Billerica, MA, USA). PST standards, including those for gonyautoxin 1 and 4 (GTX1 and GTX4, respectively), gonyautoxin 2 and 3 (GTX2 and GTX3, respectively), gonyautoxin 5 (GTX5), decarbomylgonyautoxin 2 and 3 (dcGTX2 and dcGTX3, respectively), saxitoxin (STX), neosaxitoxin (neoSTX), decarbamoylsaxitoxin (dcSTX), and N-sulfocarbamoyl toxin 1 and 2 (C1 and C2, respectively), were purchased from the Institute for Marine Bioscience, National Research Council, Canada.
(ii) Sample extraction and cleanup of paralytic shellfish toxins.
Glass fiber membranes were cut into small pieces and put into a 0.05 M acetic acid solution. The mixture was treated with a probe sonicator (Scientz Biotechnology Co., Ltd., Ningbo, China) in an ice bath for five cycles of 20 s at 200 W before being allowed to stand for another 40 s. The mixture was then centrifuged at 6,654 × g for 5 min, and 1 ml of supernatant was collected and filtered through a 0.22-μm-pore-size syringe filter prior to analysis with HPLC.
(iii) HPLC analysis of paralytic shellfish toxins.
Briefly, the carbamate toxins GTX1 to GTX4, STX, and neoSTX, decarbamoyl toxins dcGTX2, dcGTX3, and dcSTX, and N-sulfocarbamoyl toxin GTX5 were analyzed with an Agilent Zorbax Bonus-RP column (150-mm length by 4.6-mm inner diameter [i.d.], 3.5-μm particle size). A gradient method was applied using mobile phase A (5.5 mM ammonium phosphate buffer [pH 7.10] containing 11 mM sodium heptanesulfonate as an ion pair reagent) and mobile phase B (16.5 mM ammonium phosphate buffer [pH 7.10] containing 11 mM sodium heptanesulfonate and 11.5% acetonitrile). Toxins were eluted isocratically with mobile phase A for 10.4 min and then mobile phase B for 10.6 min, followed by mobile phase A again for the remaining 7 min. The flow rate of both mobile phases was 0.8 ml min−1.
N-Sulfocarbamoyl toxins C1 and C2 were analyzed with a Phenomenex Synergi Hydro-RP column (150-mm length, 4.6-mm i.d., 4-μm particle size). The method used two mobile phases (mobile phase A, 1% NH4OH solution containing 3 mM tetrabutyl ammonium phosphate [pH 5.8]; mobile phase B, 1% NH4OH solution containing 3 mM tetrabutyl ammonium phosphate and 4% methyl cyanide [MeCN] [pH 5.8]). Toxins were eluted with mobile phase A for 8 min, mobile phase B from 8.1 to 9.1 min, and mobile phase A again for another 6.9 min. The flow rate of both mobile phases was 0.8 ml min−1.
The oxidant solution and acid solution were the same as those described in reference 29. The flow rate of both the oxidant and acid solutions was 0.4 ml min−1. The temperature for the postcolumn reaction was maintained at 85°C. To detect PSTs, the excitation wavelength was 330 nm, and the emission wavelength was 390 nm.
The limits of detection (LODs) (signal-to-noise ratio [S/N] = 3) and limits of quantification (LOQs) (S/N = 10) of the HPLC method were 9.61 pg and 32.0 pg for C1, 18.0 pg and 59.9 pg for C2, 21.7 pg and 72.3 pg for GTX4, 32.2 pg and 107 pg for GTX1, 5.45 pg and 18.2 pg for dcGTX3, 41.1 pg and 137 pg for GTX5, 8.84 pg and 29.5 pg for dcGTX2, 1.77 pg and 5.9 pg for GTX3, 4.33 pg and 14.4 pg for GTX2, 28.7 pg and 95.8 pg for neoSTX, 19.9 pg and 66.3 pg for dcSTX, and 11.6 pg and 38.5 pg for STX.
Statistics.
The significance of the difference in CT values among the toxic strains was analyzed with Duncan's multiple-range test using SPSS 16.0. The linear correlation was analyzed with Pearson correlation coefficients by SPSS 16.0, and the linear-regression analysis used SigmaPlot 12.0.
RESULTS
sxtA-based qPCR assay in the Yellow Sea.
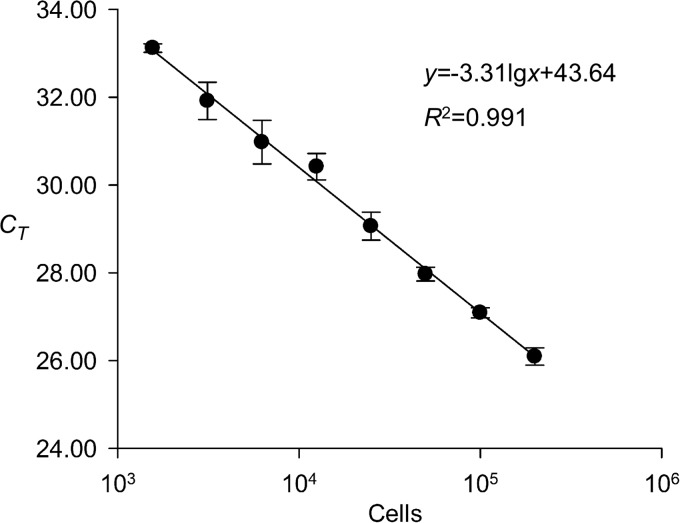
The specificity of the sxtA-based qPCR assay for the detection of PST-producing microalgae was examined with several species within the genus Alexandrium commonly present in the coastal waters of China. All PST-producing species, i.e., A. fundyense (strain ATLY), A. pacificum (strains ACDH, ATHK, ATCI02, ATCI03, and AT5-3), and A. minutum (strains AM and Am-LYG), showed positive responses. Negative responses were observed in all nontoxic Alexandrium species or strains, i.e., A. affine (strains ASCH and AC-1) and A. tamarense (strain AT-6 from European coastal waters), and the diatom Skeletonema costatum. The specificity of the primer set could be verified by the single peak in the melting curve (data not shown), and the PCR efficiency was 100.5%, according to the slope of the standard curve (Fig. 2). The detection limit for this sxtA-based qPCR assay, expressed as the number of A. pacificum cells (strain ACDH), was 150 cells, and the assay exhibited a linear response in a wide range, from 150 cells to 200,000 cells. In accordance with the protocol for field sample preparation described above under “Cruises and sample collection” and “qPCR assays” (1/10 of the 500-ml phytoplankton sample concentrated from 1,000 liters of seawater was used for DNA preparation in the qPCR analysis, and 1/10 of the 20-μl DNA sample was used as the template for qPCRs), the detection limit for the qPCR assay was about 15 cells liter−1.
FIG 2.
Standard curve of the sxtA-based qPCR assay (error bars indicate standard deviations of measured CT values).
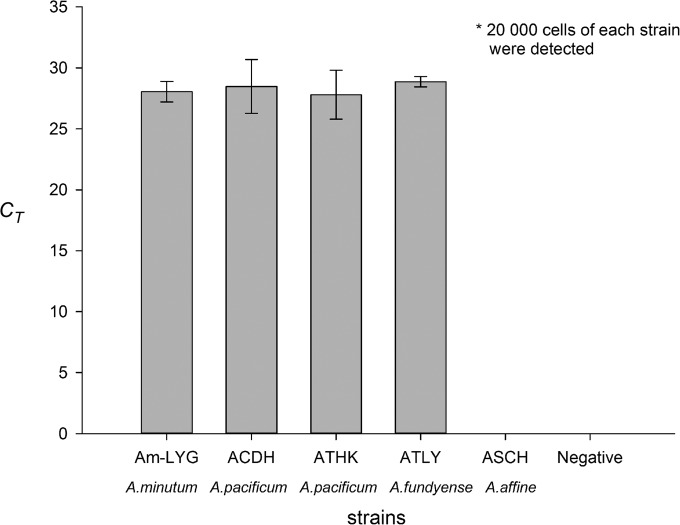
For the four strains of PST-producing Alexandrium species isolated from the coastal waters of China, i.e., A. minutum (AM-LYG), A. pacificum (ACDH and ATHK), and A. fundyense (ATLY), there was no significant difference in CT values (P > 0.05, n = 3) among the DNA samples prepared from the same number of cells (Fig. 3), based on Duncan's multiple-range test.
FIG 3.
Comparison of CT values for the DNA samples prepared from the same number of Alexandrium cells using the sxtA-based qPCR assay (error bars indicate standard deviations of measured CT values).
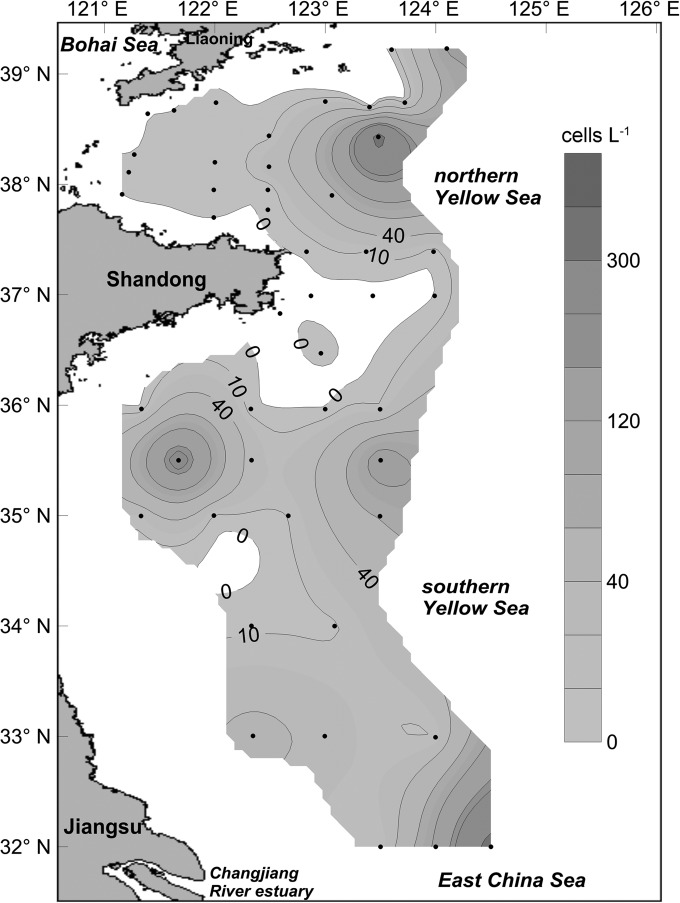
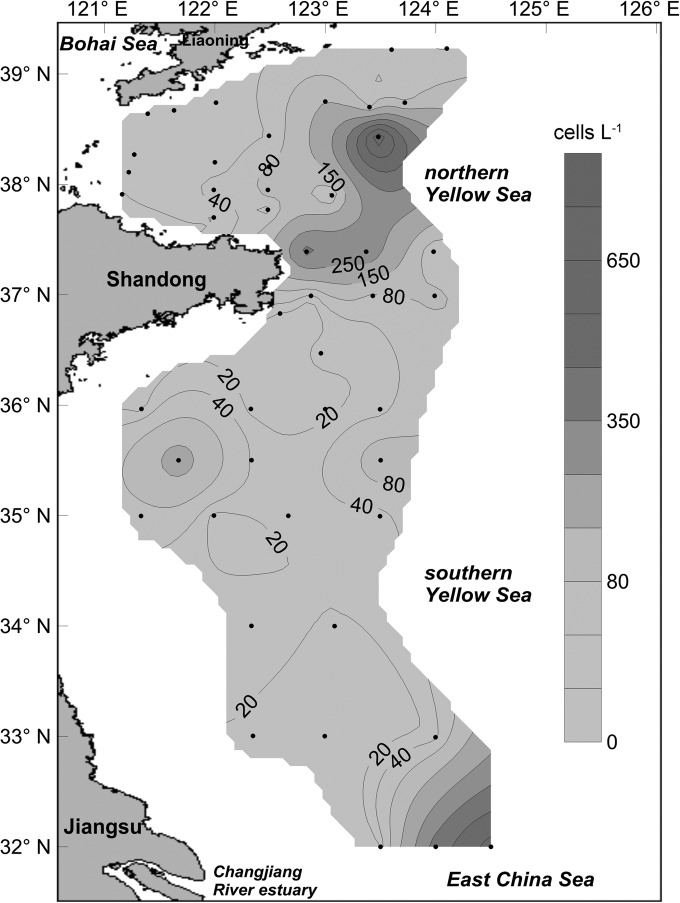
Using the calibration curve prepared with the DNA samples extracted from toxic A. pacificum (strain ACDH), the relative abundances of the sxtA4 gene in the YS were calculated and expressed as the cell number of this representative toxic species. The distribution pattern and abundances of this representative toxic species in the YS are shown in Fig. 4. It can be seen that four patches of PST-producing toxic algae were present in the YS. The maximum cell density was 343 cells liter−1, which appeared in the northern YS near sampling site B14. Another patch with similar cell density (338 cells liter−1) appeared in the joint area (sampling site H42) between the YS and East China Sea (ECS), which is close to the Chang Jiang River estuary. The remaining two patches, one of which appeared in the coastal waters of Shandong province (sampling site HF1) and the other at the central area of the southern YS (sampling site H09), had relatively lower cell densities of 240 and 150 cells liter−1, respectively.
FIG 4.
Distribution and abundance of PST-producing toxic cells representing the relative abundance of the sxtA4 domain in the Yellow Sea, as determined by the sxtA-based qPCR (cells liter−1).
qPCR assay of A. fundyense and A. pacificum in the Yellow Sea.
Two TaqMan-based qPCR assays were applied to quantify A. fundyense and A. pacificum in the YS. The distribution of the two toxic species as a whole representing the toxic A. tamarense species complex in the YS is shown in Fig. 5. Similarly, four patches of toxic A. tamarense species complex were found in the YS during the investigation. The highest cell density was 731 cells liter−1 at sampling site B14, located in the northern YS. Another patch with a relatively lower cell density (maximum, 590 cells liter−1) was present in the joint area between the YS and ECS adjacent to the Chang Jiang River estuary. The remaining two patches, with maximum cell densities of 198 cells liter−1 and 117 cells liter−1, were present in the coastal waters near Shandong province and the central area of the southern YS.
FIG 5.
Distribution and abundance of toxic A. tamarense species complex (A. fundyense and A. pacificum) in the Yellow Sea, as determined by two species-specific qPCR assays (cells liter−1).
Determination of paralytic shellfish toxins in the Yellow Sea.
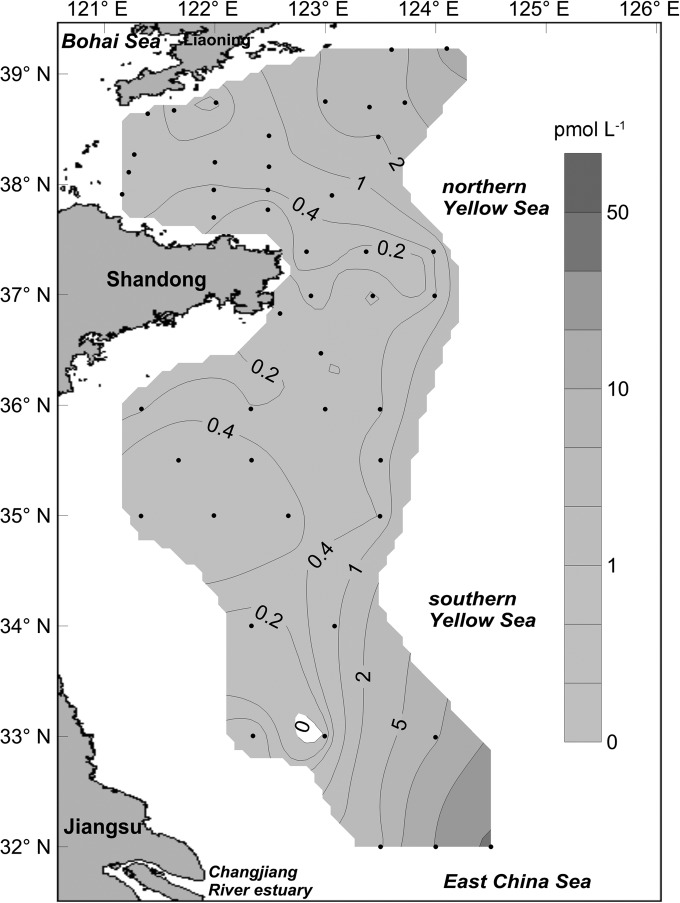
PSTs in net-concentrated phytoplankton samples collected from YS were analyzed with HPLC coupled with FLD, and the results were interpreted as the toxin level (total amount of PSTs in 1 liter of seawater) from a comparison of the abundance of toxic algae and the relative abundance of the sxtA4 gene. The distribution of PSTs in the YS is shown in Fig. 6. A high level of PSTs (45.3 pmol liter−1) was found in the joint area between the YS and ECS close to the Chang Jiang River estuary. In the northern YS, the PST level was much lower, and the maximum level of PSTs was 11.7 pmol liter−1 (at sampling site B15). In the coastal water near Shandong province, the PST level was even lower, and the maximum value was only about 1.01 pmol liter−1. The toxin profiles of PSTs in the samples collected from these areas were quite similar and were characterized by a high proportion of low-potency N-sulfocarbamoyl toxins (C1/C2) with trace amounts of GTX2, GTX3, and GTX4.
FIG 6.
Distribution and toxin level (picomoles liter−1) of paralytic shellfish toxins in the Yellow Sea, as determined by high-performance liquid chromatography coupled with a fluorescence detector.
Relationship between the results of qPCR assays and PST analysis.
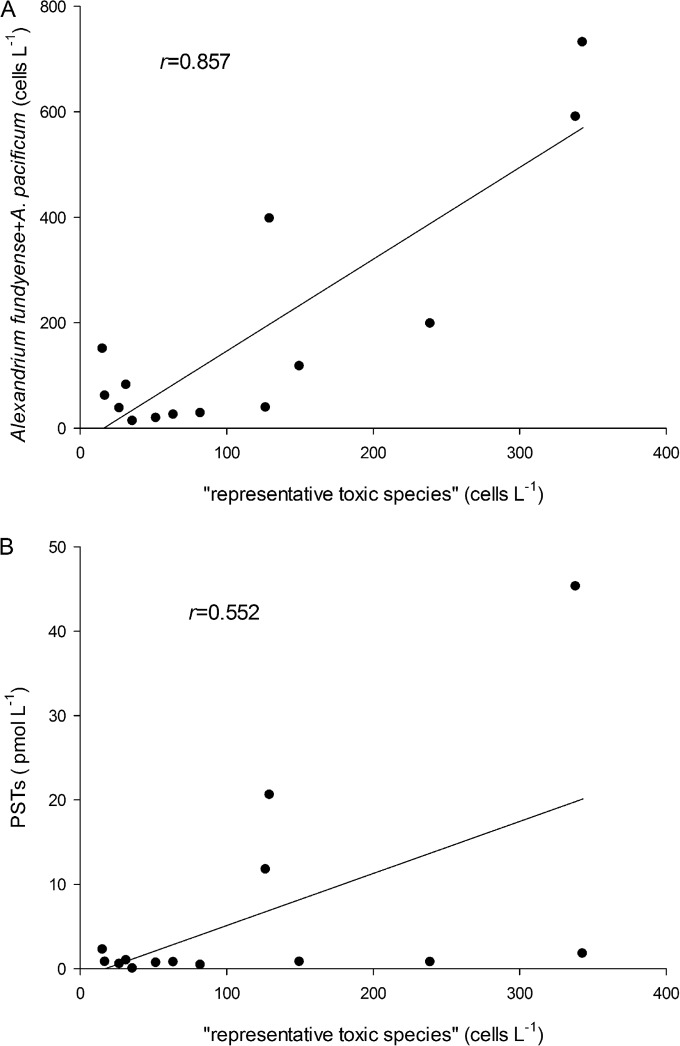
The relationship between the cell density of the representative toxic species and that of toxic A. tamarense species complex in the YS were analyzed using data above the theoretical detection limit of the sxtA4 qPCR assay. There was a significant positive correlation (Fig. 7A) (r = 0.857; P < 0.05) between them, although the cell densities of the toxic A. tamarense species complex quantified with TaqMan-based qPCR assays were generally higher than those from the sxtA4 qPCR assay. The correlation between PST level and the abundance of representative toxic species showed a much lower r value and was only weakly significant (Fig. 7B) (r = 0.552; P < 0.05).
FIG 7.
Correlation between sxtA4 quantitation results (expressed as the abundance of representative toxic species) and the abundance of toxic A. tamarense species complex (A) and PSTs levels (B).
DISCUSSION
Feasibility of the sxtA-based qPCR assay for detection of PST-producing toxic algae.
Monitoring of toxic algae has previously relied on morphological identification or molecular detection of specific species (4, 30). However, neither of those methods is directly related to toxin synthesis. The detection of unique genes related to toxin synthesis, which can cover all toxin-producing organisms rather than a single species, can provide a comprehensive and effective means to assess the toxic potential of the incipient toxic blooms. Such genes have been applied successfully in the detection or monitoring of toxic cyanobacterial species in freshwater environments. For example, a variety of mcy genes have been used as valid markers for the detection of hepatotoxic Anabaena, Microcystis, and Planktothrix species (7). Mixed polyketide synthase/nonribosomal peptide synthesis (PKS/NRPS) and ndaF genes have been applied to monitor blooms of Nodularia species (11). The gene rpoC1 has been used to differentiate Cylindrospermopsis raciborskii from Anabaena bergii and Aphanizomenon ovalisporum (12). Recently, multiplex qPCR assays were developed to detect several different genes related to toxin biosynthesis in cyanobacteria (13, 31), one of which included sxt (13).
With the identification of genes associated with STX production in dinoflagellates, a SYBR green qPCR assay targeting the sxtA4 domain of the core gene sxtA was developed (17). This qPCR assay has been applied to study toxic algal blooms associated with PSTs in the sea and demonstrated positive correlations with the results of a long-subunit (LSU) qPCR assay and microscopic observations. In addition, a sensitive probe-based qPCR assay was developed and applied to analyze mRNA transcripts of the sxtA gene, the results of which showed a good correlation with PSTs (32). However, there are still some concerns about the feasibility of using an sxtA-based qPCR assay for monitoring toxic algal blooms. One reason is that most genes in dinoflagellate genomes have multiple copies, whose numbers vary from those of isolates (28, 33, 34). Also, the uniqueness of the sxtA4 domain in PST-producing species still needs to be validated in different waters. It was reported that the sxtA1 and sxtA4 domains were present in two nontoxic strains of A. australiense (A. tamarense species complex group V) (17), Therefore, the feasibility of this sxtA-based qPCR assay still needs to be tested further with more PST-producing toxic species and requires validation in different regions around the world.
In this study, we tested the specificity of this sxtA-based qPCR assay for the detection of PST-producing microalgae using several species in the genus Alexandrium commonly present in the YS. The specificity of the qPCR assay to detect toxic algae associated with PSTs was further confirmed. Although the sxtA4 domain has been detected in nontoxic strains of A. australiense (17), there is no report of this species in the YS so far. The CT values for DNA samples prepared from the same number of cells of four toxic strains belonging to A. fundyense, A. pacificum, and A. minutum exhibited no significant differences in our study, suggesting that the number of copies of the sxtA4 domain was relatively constant among these PST-producing species in the YS (Fig. 3). This is in accordance with the findings of a previous study (17). Therefore, the application of this sxtA-based qPCR assay to study PST-producing toxic algae in the YS should be feasible.
Potential of sxtA-based qPCR assay for monitoring toxic algal bloom in the Yellow Sea.
Alexandrium spp. have been widely reported in the coastal waters of China, and blooms of Alexandrium spp. have been reported in the northern YS, East China Sea, and South China Sea (35, 36). In the YS, many species in the genus Alexandrium, such as A. fundyense and A. pacificum of the toxic A. tamarense species complex (reported previously as either A. tamarense or A. catenella), A. minutum, and the nontoxic A. affine, A. andersonii, and A. leei species, have been identified through cyst germination, morphological observation, and also single-cell sequencing (21). In addition, cyst beds of Alexandrium species, with abundance as high as 3,788 cysts/g sediment (dry weight), have been reported in the YS (37–39), where PSTs have been frequently detected in scallops (19, 20, 40). The coexistence of multiple toxic and nontoxic Alexandrium species makes it difficult to monitor every toxic species in this region. The qPCR assay based on the unique toxin synthetic gene employed in this study has a great advantage in indicating the potential existence of PSTs and their producers.
The sxtA-based qPCR assay was applied to analyze PST-producing microalgae in the YS in this study, and we also performed a comprehensive investigation on the toxic A. tamarense species complex, PST-related gene, and PSTs in the YS. The distribution pattern of a PST synthesis-related gene in the YS, as indicated by the representative toxic species, is consistent with the distribution of toxic algae (A. fundyense and A. pacificum) and PSTs. Based on these findings, it was strongly suggested that toxic A. fundyense and A. pacificum should be the major PST producers in the YS. This opinion was also supported by the toxin profile of field samples, which was similar to those of strains of A. fundyense and A. pacificum established from the YS and ECS, which predominantly produce C1/C2 congeners (21, 35). The coincidence of the distribution pattern in the representative toxic species, toxic A. tamarense species complex, and PSTs fully supported the sxtA-based qPCR assay as a reliable method to study PST-producing algae in the YS.
In this study, the quantitation results of the toxic A. tamarense species complex correlated well with the abundance of the representative toxic species derived from the sxtA-based qPCR assay (r = 0.857), which is similar to the studies performed in Australia (17). The abundance of A. fundyense and A. pacificum combined, however, was generally higher than that of the representative toxic species (slope, 1.74). This is probably due to the overestimation of the abundance of A. fundyense and A. pacificum organisms caused by the variance of DNA extraction efficiency and LSU rRNA gene copy number (28, 33). In contrast, the correlation between the quantitation results of the sxtA-based qPCR assay and PST levels in the phytoplankton samples was less significant (r = 0.552). This is not surprising, since the intracellular toxins, as secondary metabolites, are affected by many environmental factors, like temperature, salinity, light, and availability of nutrients (41–44). Toxin production in dinoflagellates has been supposed to be an adaptation that evolved to offset the disadvantage of low-nutrient affinity (42, 45–47), and phosphorus deficiency has been shown to increase significantly the cellular quota of PSTs in toxic A. tamarense species complex in comparison to that of nutrient-sufficient or N-deficient cells (41, 42, 45–48). In addition, the depression of essential reactions for STX biosynthesis might block toxin production and result in a loss of toxicity (49). Another issue that may influence the correlation between the quantitation results of sxtA4 and PST levels is the difference in toxin content between A. fundyense and A. pacificum. Gu et al. (21) determined the content of PSTs in six strains of A. fundyense isolated from the northern YS and found that the cellular toxin quota ranged from 1.1 to 5.0 fmol cell−1, which is significantly lower than that for A. pacificum (50–52). According to our previous analysis, A. fundyense was distributed mainly in the YS north to 34°N, while A. pacificum was confined in the joint area between YS and ECS close to the Chang Jiang River estuary (28). This will also result in the relatively poor correlation between the sxtA4 quantitation results and PST levels.
The less significant correlation between the sxtA4 quantitation results and PST levels in this study suggested that the combination of molecular biological approaches and chemical measurements of toxins might be an appropriate method for monitoring toxic algal blooms. Most of the monitoring programs on phycotoxins to date have focused on toxins in shellfish, which reflect the historic exposure to toxic algae. The detection of PSTs and PST-producing algae in seawater might reflect the current toxicity level and thus facilitate an early warning of shellfish contamination. Moreover, it provides the possibility of tracing the origin of toxins in the field and promotes better understanding in ecological studies.
There are many important mariculture zones in the YS, and PSTs have been detected from the shellfish samples from time to time (18–20). However, there is little knowledge so far on the annual bloom dynamics of PST-producing toxic algae in the YS. The results of this study confirmed the feasibility of the sxtA-based qPCR assay in monitoring PST-producing toxic algae in the YS in spring, and its feasibility still needs to be further tested in other seasons in the future.
Conclusions.
In this study, an sxtA-based qPCR assay was applied to study toxic algae associated with PSTs in the YS. The qPCR assay showed high specificity for detecting PST-producing microalgae, and the quantitation results of sxtA4 represented well the abundance and distribution of the toxic A. tamarense species complex (A. fundyense and A. pacificum) in the YS during the sampling season. However, the correlation between PST levels and sxtA-based qPCR results was less significant (r = 0.552), implying that sxtA-based qPCR is not accurate enough to reflect the toxicity of blooms associated with PSTs. The combination of sxtA-based qPCR assay and chemical means might be an appropriate method for monitoring toxic algal blooms.
ACKNOWLEDGMENTS
This study was supported by project 41176100 of the National Natural Science Foundation of China (NSFC), the Strategic Priority Research Program (grant XDA11020304) of the Chinese Academy of Sciences (CAS), the Public Science and Technology Research Funds Project of Ocean (grant Y32323101H) from the State Ocean Administration (SOA), and project U1406403 jointly supported by Shandong Province and the NSFC. The sampling process in this study was supported by the open cruise organized by the NSFC.
We thank Gu Haifeng from the Third Institute of Oceanography, SOA, who offered strains ATLY and ASCH for use in this study.
REFERENCES
- 1.Wiese M, D'Agostino PM, Mihali TK, Moffitt MC, Neilan BA. 2010. Neurotoxic alkaloids: saxitoxin and its analogs. Mar Drugs 8:2185–2211. doi: 10.3390/md8072185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Usup G, Kulis DM, Anderson DM. 1994. Growth and toxin production of the toxic dinoflagellate Pyrodinium bahamense var. compressum in laboratory cultures. Nat Toxins 2:254–262. doi: 10.1002/nt.2620020503. [DOI] [PubMed] [Google Scholar]
- 3.Oshima Y, Blackburn SI, Hallegraeff GM. 1993. Comparative-study on paralytic shellfish toxin profiles of the dinoflagellate Gymnodinium catenatum from 3 different countries. Mar Biol 116:471–476. doi: 10.1007/BF00350064. [DOI] [Google Scholar]
- 4.Anderson DM, Alpermann TJ, Cembella AD, Collos Y, Masseret E, Montresor M. 2012. The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14:10–35. doi: 10.1016/j.hal.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alam M, Ikawa M, Sasner JJ Jr, Sawyer P. 1973. Purification of Aphanizomenon flosaquae toxin and its chemical and physiological properties. Toxicon 11:65–72. doi: 10.1016/0041-0101(73)90154-2. [DOI] [PubMed] [Google Scholar]
- 6.Araoz R, Molgo J, de Marsac NT. 2010. Neurotoxic cyanobacterial toxins. Toxicon 56(5):813–828. doi: 10.1016/j.toxicon.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 7.Pearson LA, Neilan BA. 2008. The molecular genetics of cyanobacterial toxicity as a basis for monitoring water quality and public health risk. Curr Opin Biotech 19:281–288. doi: 10.1016/j.copbio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Akimoto T, Masuda A, Yotsu-Yamashita M, Hirokawa T, Nagasawa K. 2013. Synthesis of saxitoxin derivatives bearing guanidine and urea groups at C13 and evaluation of their inhibitory activity on voltage-gated sodium channels. Org Biomol Chem 11:6642–6649. doi: 10.1039/c3ob41398e. [DOI] [PubMed] [Google Scholar]
- 9.Shumway SE. 1990. A review of the effects of algal blooms on shellfish and aquaculture. J World Aquac Soc 21:65–104. doi: 10.1111/j.1749-7345.1990.tb00529.x. [DOI] [Google Scholar]
- 10.Murray SA, Mihali TK, Neilan BA. 2011. Extraordinary conservation, gene loss, and positive selection in the evolution of an ancient neurotoxin. Mol Biol Evol 28:1173–1182. doi: 10.1093/molbev/msq295. [DOI] [PubMed] [Google Scholar]
- 11.Koskenniemi K, Lyra C, Rajaniemi-Wacklin P, Jokela J, Sivonen K. 2007. Quantitative real-time PCR detection of toxic Nodularia cyanobacteria in the Baltic Sea. Appl Environ Microbiol 73:2173–2179. doi: 10.1128/AEM.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fergusson KM, Saint CP. 2003. Multiplex PCR assay for Cylindrospermopsis raciborskii and cylindrospermopsin-producing cyanobacteria. Environ Toxicol 18:120–125. doi: 10.1002/tox.10108. [DOI] [PubMed] [Google Scholar]
- 13.Al-Tebrineh J, Pearson LA, Yasar SA, Neilan BA. 2012. A multiplex qPCR targeting hepato- and neurotoxigenic cyanobacteria of global significance. Harmful Algae 15:19–25. doi: 10.1016/j.hal.2011.11.001. [DOI] [Google Scholar]
- 14.Mejean A, Ploux O. 2013. A genomic view of secondary metabolite production in cyanobacteria. Adv Bot Res 65:189–234. doi: 10.1016/B978-0-12-394313-2.00006-8. [DOI] [Google Scholar]
- 15.Shimizu Y. 1996. Microalgal metabolites: a new perspective. Annu Rev Microbiol 50:431–465. doi: 10.1146/annurev.micro.50.1.431. [DOI] [PubMed] [Google Scholar]
- 16.Stüken A, Orr RJ, Kellmann R, Murray SA, Neilan BA, Jakobsen KS. 2011. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS One 6:e20096. doi: 10.1371/journal.pone.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray SA, Wiese M, Stüken A, Brett S, Kellmann R, Hallegraeff G, Neilan BA. 2011. sxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl Environ Microbiol 77:7050–7057. doi: 10.1128/AEM.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JH, Yu RC, Gao Y, Kong FZ, Wang YF, Zhang QC, Kang ZJ, Yan T, Zhou MJ. 2013. Tracing the origin of paralytic shellfish toxins in scallop Patinopecten yessoensis in the northern Yellow Sea. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 30:1933–1945. doi: 10.1080/19440049.2013.838644. [DOI] [PubMed] [Google Scholar]
- 19.Jiang TJ, Jiang T. 2007. Detection and analysis of PSP toxins in shellfish in coastal areas of China. Oceanol Limnol Sin 38:36–41. (In Chinese with English abstract.) [Google Scholar]
- 20.Kong FZ, Xu ZJ, Li QL, Zhang AJ, Zhang HL. 2005. Research on distribution of shellfish toxin and harmful red tide planktons in the Bohai Sea and the Yellow Sea, p 27–34. In Red Tide Study, Prevention and Control Committee of China (ed), Red tide studies, prevention and control in China. Ocean Press, Beijing, China: (In Chinese with English abstract.) [Google Scholar]
- 21.Gu HF, Zeng N, Liu TT, Yang WD, Müller A, Krock B. 2013. Morphology, toxicity, and phylogeny of Alexandrium (Dinophyceae) species along the coast of China. Harmful Algae 27:68–81. doi: 10.1016/j.hal.2013.05.008. [DOI] [Google Scholar]
- 22.Scholin CA, Hallegraeff GM, Anderson DM. 1995. Molecular evolution of the Alexandrium tamarense ‘species complex' (Dinophyceae): dispersal in the North American and West Pacific regions. Phycologia 34:472–485. doi: 10.2216/i0031-8884-34-6-472.1. [DOI] [Google Scholar]
- 23.John U, Fensome RA, Medlin LK. 2003. The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense “species complex” (Dinophyceae). Mol Biol Evol 20:1015–1027. doi: 10.1093/molbev/msg105. [DOI] [PubMed] [Google Scholar]
- 24.Lilly EL, Halanych KM, Anderson DM. 2007. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae). J Phycol 43:1329–1338. doi: 10.1111/j.1529-8817.2007.00420.x. [DOI] [Google Scholar]
- 25.John U, Litaker RW, Montresor M, Murray S, Brosnahan ML, Anderson DM. 2014. Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: the introduction of five species with emphasis on molecular-based (rDNA) classification. Protist 165:779–804. doi: 10.1016/j.protis.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosoi-Tanabe S, Sako Y. 2005. Species-specific detection and quantification of toxic marine dinoflagellates Alexandrium tamarense and A. catenella by real-time PCR assay. Mar Biotechnol 7:506–514. doi: 10.1007/s10126-004-4128-4. [DOI] [PubMed] [Google Scholar]
- 27.Gao Y, Yu RC, Chen JH, Zhang QC, Kong FZ, Zhou MJ. 2015. Distribution of Alexandrium fundyense and A. pacificum (Dinophyceae) in the Yellow Sea and Bohai Sea. Mar Pollut Bull 96:210–219. doi: 10.1016/j.marpolbul.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Yu RC, Zhang QC, Zhou MJ. 2013. Application of qPCR method in detection of Alexandrium tamarense species complex in China. Acta Sci Circum 33:2256–2263. (In Chinese with English abstract.) [Google Scholar]
- 29.Van de Riet JM, Gibbs RS, Chou FW, Muggah PM, Rourke WA, Burns G, Thomas K, Quilliam MA. 2009. Liquid chromatographic post-column oxidation method for analysis of paralytic shellfish toxins in mussels, clams, scallops, and oysters: single-laboratory validation. J AOAC Int 92:1690–1704. [PubMed] [Google Scholar]
- 30.Godhe A, Cusack C, Pedersen J, Andersen P, Anderson DM, Bresnan E, Cembella A, Dahl E, Diercks S, Elbrächter M, Edler L, Galluzzi L, Gescher C, Gladstone M, Karlson B, Kulis D, LeGresley M, Lindahl O, Marin R, McDermott G, Medlin LK, Naustvoli L-J, Penna A, Töbe K. 2007. Intercalibration of classical and molecular techniques for identification of Alexandrium fundyense (Dinophyceae) and estimation of cell densities. Harmful Algae 6:56–72. doi: 10.1016/j.hal.2006.06.002. [DOI] [Google Scholar]
- 31.Ngwa FF, Madramootoo CA, Jabaji S. 2014. Development and application of a multiplex qPCR technique to detect multiple microcystin-producing cyanobacterial genera in a Canadian freshwater lake. J Appl Phycol 26:1675–1687. doi: 10.1007/s10811-013-0199-9. [DOI] [Google Scholar]
- 32.Stüken A, Dittami SM, Eikrem W, McNamee S, Campbell K, Jakobsen KS, Edvardsen B. 2013. Novel hydrolysis-probe based qPCR assay to detect saxitoxin transcripts of dinoflagellates in environmental samples. Harmful Algae 28:108–117. doi: 10.1016/j.hal.2013.06.003. [DOI] [Google Scholar]
- 33.Galluzzi L, Bertozzini E, Penna A, Perini F, Garcés E, Magnani M. 2010. Analysis of rRNA gene content in the Mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: implications for the quantitative real-time PCR-based monitoring methods. J Appl Phycol 22:1–9. doi: 10.1007/s10811-009-9411-3. [DOI] [Google Scholar]
- 34.Hariganeya N, Tanimoto Y, Yamaguchi H, Nishimura T, Tawong W, Sakanari H, Yoshimatsu T, Sato S, Preston CM, Adachi M. 2013. Quantitative PCR method for enumeration of cells of cryptic species of the toxic marine dinoflagellate Ostreopsis spp. in coastal waters of Japan. PLoS One 8:e57627. doi: 10.1371/journal.pone.0057627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou C, Ye RM, Zheng JW, Luo ZH, Gu HF, Yang WD, Li HY, Liu JS. 2014. Molecular phylogeny and PSP toxin profile of the Alexandrium tamarense species complex along the coast of China. Mar Pollut Bull 89:209–219. [DOI] [PubMed] [Google Scholar]
- 36.Yu RC, Gao Y, Luo X, Song JJ, Zhou MJ. 2014. Toxic algae and phycotoxins in the coastal waters of China, p 92–106. In Sun S, Adrianov AV, Lutaenko KA, Sun XX (ed), Marine biodiversity and ecosystem dynamics of the Northwest Pacific Ocean. Science Press, Beijing, China. [Google Scholar]
- 37.Cho HJ, Matsuoka K. 2001. Distribution of dinoflagellate cysts in surface sediments from the Yellow Sea and East China Sea. Mar Micropaleontol 42:103–123. doi: 10.1016/S0377-8398(01)00016-0. [DOI] [Google Scholar]
- 38.Pan J, Li RX, Li Y, Sun P. 2010. Distribution of dinoflagellate cysts in surface sediments from the southern Yellow Sea in autumn. Adv Mar Sci 28:41–49. (In Chinese with English abstract.) [Google Scholar]
- 39.Shi YJ, Liu DY, Shao HB, Di BP, Dong ZJ, Wang YJ. 2011. Distribution of dinoflagellate cysts in the surface sediments from the northern Yellow Sea, China. Mar Sci Bull 30:320–327. (In Chinese with English abstract.) [Google Scholar]
- 40.Li A, Ma J, Cao J, Wang Q, Yu RC, Thomas K, Quilliam MA. 2012. Analysis of paralytic shellfish toxins and their metabolites in shellfish from the North Yellow Sea of China. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 29:1455–1464. doi: 10.1080/19440049.2012.699005. [DOI] [PubMed] [Google Scholar]
- 41.Graneli E, Flynn K. 2006. Chemical and physical factors influencing toxin content, p 229–241. In Graneli E, Turner JT (ed), Ecology of harmful algae, vol 189 Springer, Berlin, Germany. [Google Scholar]
- 42.John EH, Flynn KJ. 2002. Modelling changes in paralytic shellfish toxin content of dinoflagellates in response to nitrogen and phosphorus supply. Mar Ecol Prog Ser 225:147–160. doi: 10.3354/meps225147. [DOI] [Google Scholar]
- 43.Maestrini SY, Bechemin C, Grzebyk D, Hummert C. 2000. Phosphorus limitation might promote more toxin content in the marine invader dinoflagellate Alexandrium minutum. Plankton Biol Ecol 47:7–11. [Google Scholar]
- 44.Lim PT, Leaw CP, Kobiyama A, Ogata T. 2010. Growth and toxin production of tropical Alexandrium minutum Halim (Dinophyceae) under various nitrogen to phosphorus ratios. J Appl Phycol 22:203–210. doi: 10.1007/s10811-009-9443-8. [DOI] [Google Scholar]
- 45.Boyer GL, Sullivan JJ, Andersen RJ, Harrison PJ, Taylor FJR. 1987. Effects of nutrient limitation on toxin production and composition in the marine dinoflagellate Protogonyaulax tamarensis. Mar Biol 96:123–128. doi: 10.1007/BF00394845. [DOI] [Google Scholar]
- 46.Frangópulos M, Guisande C, deBlas E, Maneiro I. 2004. Toxin production and competitive abilities under phosphorus limitation of Alexandrium species. Harmful Algae 3:131–139. doi: 10.1016/S1568-9883(03)00061-1. [DOI] [Google Scholar]
- 47.Anderson DM, Kulis DM, Sullivan JJ, Hall S, Lee C. 1990. Dynamics and physiology of saxitoxin production by the dinoflagellates Alexandrium spp. Mar Biol 104:511–524. doi: 10.1007/BF01314358. [DOI] [Google Scholar]
- 48.Murata A, Leong SCY, Nagashima Y, Taguchi S. 2006. Nitrogen: phosphorus supply ratio may control the protein and total toxin of dinoflagellate Alexandrium tamarense. Toxicon 48:683–689. doi: 10.1016/j.toxicon.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Zhang SF, Lin L, Wang DZ. 2014. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its nontoxic mutant. Mar Drug 12:5698–5718. doi: 10.3390/md12115698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li TS, Yu RC, Zhou MJ. 2011. Short-term effects of different nitrogen substrates on growth and toxin production of dinoflagellate Alexandrium catenella Balech (strain ACDH). Harmful Algae 12:46–54. doi: 10.1016/j.hal.2011.08.011. [DOI] [Google Scholar]
- 51.Wang DZ, Jiang TJ, Hsieh DPH. 2005. Toxin composition variations in cultures of Alexandrium species isolated from the coastal waters of southern China. Harmful Algae 4:109–121. doi: 10.1016/j.hal.2003.12.003. [DOI] [Google Scholar]
- 52.Wang DZ, Hsieh DP. 2005. Growth and toxin production in batch cultures of a marine dinoflagellate Alexandrium tamarense HK9301 isolated from the South China Sea. Harmful Algae 4:401–410. doi: 10.1016/j.hal.2004.07.002. [DOI] [Google Scholar]