Abstract
Staphylococcal enterotoxins (SEs) produced by Staphylococcus aureus have superantigenic and emetic activities, which cause toxic shock syndrome and staphylococcal food poisoning, respectively. Our previous study demonstrated that the sequence of SET has a low level of similarity to the sequences of other SEs and exhibits atypical bioactivities. Hence, we further explored whether there is an additional SET-related gene in S. aureus strains. One SET-like gene was found in the genome of S. aureus isolates that originated from a case of food poisoning, a human nasal swab, and a case of bovine mastitis. The deduced amino acid sequence of the SET-like gene showed 32% identity with the amino acid sequence of SET. The SET-like gene product was designated SElY. In the food poisoning and nasal swab isolates, mRNA encoding SElY was highly expressed in the early log phase of cultivation, whereas a high level of expression of this mRNA was found in the bovine mastitis isolate at the early stationary phase. To estimate whether SElY has both superantigenic and emetic activities, recombinant SElY was prepared. Cell proliferation and cytokine production were examined to assess the superantigenic activity of SElY. SElY exhibited superantigenic activity in human peripheral blood mononuclear cells but not in mouse splenocytes. In addition, SElY exhibited emetic activity in house musk shrews after intraperitoneal and oral administration. However, the stability of SElY against heating and pepsin and trypsin digestion was different from that of SET and SEA. From these results, we identified SElY to be a novel staphylococcal emetic toxin.
INTRODUCTION
Staphylococcus aureus produces a variety of exotoxins, including staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin 1 (TSST-1) (1). These toxins are superantigens, which have the ability to stimulate a large repertoire of the Vβ elements of T cell receptor (TCR)-bearing T cells. SEs are also emetic toxins causing staphylococcal food poisoning in humans, although the mechanism of SE-induced emesis has not been entirely verified (2). Due to these properties, SEs are assumed to be a menace to public health. Five major serological types (SEA to SEE) have been characterized (2), and new types of SE-related toxins (SEG to SEI, SElJ, SEK to SET, SElU, SElV, and SElX) have recently been reported (2–8). Moreover, superantigen-related genes, such as staphylococcal superantigen-like protein (SSL) genes (ssl1 to ssl26), were discovered during determination of the complete genome sequences of several S. aureus strains (9–11). It has been recognized that the superantigen genes are associated with mobile genetic elements (MGEs), such as pathogenicity islands, prophages, or plasmids (5, 11–14). This fact implies that these superantigen genes move among S. aureus strains by horizontal transfer and that such MGEs play an important role in the evolution of S. aureus as a pathogen.
We have reported that SET shows mitogenicity to human T cells and requires major histocompatibility complex (MHC) class II molecules for stimulation, although a relatively large amount of SET is required to stimulate T cells compared with the amount of SEA that is required (5). Moreover, the emetic activity of SET is slightly different from that of typical SEs (5). SET requires a prolonged incubation time for the induction of emesis, and the long duration of the emetic response has been shown in the primate model. Additionally, as revealed by phylogenetic analysis, SET is included in the streptococcal pyrogenic exotoxin K (SpeK), SpeL, and SpeM group encoded by the streptococcal genome instead of being closely related to other SEs (5). These data indicate that SET has a low level of similarity to other SEs and shows atypical bioactivities. We then explored additional SET-related genes in S. aureus strains. One SET-like gene was found in S. aureus isolates. In the study described here, we identified and characterized this newly recognized SE toxin, designated SElY.
MATERIALS AND METHODS
Bacterial strains and DNA isolation.
Total DNA of S. aureus was purified using a QIAamp DNA minikit (Qiagen, Tokyo, Japan). The bacterial strains used in this study are shown in Table 1. S. aureus was grown in Trypticase soy broth (Nissui Pharmaceutical Co., Tokyo, Japan) at 37°C with aeration for total DNA isolation. Escherichia coli strains were grown in LB broth (Sigma-Aldrich, St. Louis, MO) containing 100 μg/ml of ampicillin (Wako Pure Chemicals, Osaka, Japan) for plasmid isolation. E. coli plasmid DNA was purified with a QIAprep spin miniprep kit (Qiagen) following the manufacturer's instructions.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | SE genotype | Source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| 11689 | sea seb sek seq sely | Food poisoning |
| IVM54 | seb sek seq sely | Nasal colonization |
| AV3008 | seg sei sem sen seo sely | Bovine mastitis |
| E. coli BL21 | SE negative | Stratagene |
| Plasmids | ||
| pGEX 6P-1 | Apr, GST fusion expression vector | Pharmacia |
| pKYX1 | Apr, pGEX 6P-1 with sely | Current study |
Quantitative PCR.
For RNA isolation, the S. aureus strains in Table 1 were grown in 10 ml of brain heart infusion broth (Difco Laboratories, Detroit, MI) supplemented with 1% yeast extract (Difco) in a 60-ml flask at 37°C with aeration (5). The bacterial cultures were centrifuged at 2,000 × g for 10 min at 4°C. After the supernatant was discarded, the pellet were stored at 4°C within 12 h. Total RNA was extracted from S. aureus cultures using a Fast RNA Pro kit (MP-Biomedicals, Santa Ana, CA) according to the manufacturer's instructions. Bacterial cells were broken by use of a bead shocker. To degrade contaminating genomic DNA, purified total RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI) according to the manufacturer's instructions. Annealing of total RNA with a random hexamer primer (12 mM) was performed for 10 min at 20°C. cDNA was synthesized by reverse transcription for 30 min at 55°C using a Transcriptor first-strand cDNA synthesis kit (Roche, Basel, Switzerland), followed by enzyme inactivation (5 min at 85°C). The expression level of each gene was analyzed by PCR using a Bio-Rad CFX96 Touch quantitative PCR system (Bio-Rad, Tokyo, Japan). The PCR mixture (final volume, 20 μl) consisted of 10 μl of SsoAdvanced SYBR green supermix (Bio-Rad), 2 μl of each primer (final concentration, 0.5 μM), 2 μl of ultrapure H2O, and 4 μl of a 1/10 dilution of cDNA. The primer sequences are shown in Table 2. gyrB was adopted as an internal control (15, 16). Cycling conditions were as follows: 60 s at 95°C, followed by 40 rounds of 95°C for 15 s, 62°C (gyrB) or 60°C (sely) for 15 s, and 72°C for 30 s. To determine the dissociation of PCR products, melting curve analysis was performed at temperatures of between 70°C and 95°C. The PCR efficiency of each primer pair was determined by the dilution series method using cDNA as a template. Analysis of the data from the quantitative PCR was performed with CFX manager software (version 3.0). Data are expressed as mean values from duplicate experiments. Statistical analysis was performed using analysis of variance (ANOVA) followed by the Tukey test. A P value of less than 0.05 was considered statistically significant. The normality of the data was confirmed using the Shapiro-Wilk normality test.
TABLE 2.
Nucleotide sequences and predicted sizes of PCR products
| Primer use and gene | Primer | Oligonucleotide sequence (5′-3′) | PCR product size (bp) |
|---|---|---|---|
| Real-time PCR | |||
| gyrB | gyrBF | AGGTCTTGGAGAAATGAATG | 113 |
| gyrBR | CAAATGTTTGGTCCGCTT | ||
| sely | SElYF | CAATGTACGGACAGTGCTCTACAA | 189 |
| SElYR | TGACCGTTAACAAACAAGTTCATTC | ||
| Cloning, sely | SElYGSTF | CCCCGGATCCAAAACAACTGGATTGATTA | 599 |
| SElYGSTR | CCCCGTCGACCTATTTCATATAAATATCT |
Cloning of SE-related genes and expression of recombinant proteins.
The cloning and preparation of recombinant SEA and SET have been described elsewhere (5, 17). To construct the recombinant SElY expression plasmid, PCR primers including the BamHI and EcoRI sites were designed to amplify a fragment of the sely gene corresponding to the mature form of SElY (Table 2). The N-terminal signal peptide sequence of SElY was predicted using the online signal peptide prediction software SignalP (http://www.cbs.dtu.dk/services/SignalP) (18). The sely gene fragment from S. aureus AV3008 was amplified by PCR using KOD Plus DNA polymerase (Toyobo, Osaka, Japan), and the PCR products were digested with BamHI and EcoRI. The fragment of the sely gene was then cloned into the pGEX6P-1 glutathione S-transferase (GST) fusion expression vector, and the vector containing the clone was designated pKYX. The nucleotide sequence was verified by use of an ABI3100 Avant DNA sequencer (Applied Biosystems, Foster City, CA). Expression, purification of the GST-fused recombinant protein, and cleavage and removal of the GST tag from SElY were performed by the methods described previously (3, 5). The resulting recombinant SElY had 5 additional amino acid residues, GPLGS, at the N terminus. Multiple-sequence alignments and the construction of the phylogenetic tree of the SEs and SSLs, including SElY, were performed using ClustalW software (19).
Animals.
Adult (age, 2 to 8 months) house musk shrews (Suncus murinus) and specific-pathogen-free C57/BL6 mice were purchased from Clea Japan, Tokyo, Japan. Both sets of animals were kept at 22 to 25°C in a room lit for 12 h from 8:00 a.m. to 8:00 p.m. All animal experiments described in this paper were conducted in accordance with the Animal Research Ethics Committee, Hirosaki University Graduate School of Medicine, and followed the guidelines for animal experimentation of Hirosaki University.
Assays of superantigenic activity.
Recombinant SEA, SElY, and SET were treated with a ProteoSpin endotoxin removal maxikit (Norgen Biotek, Thorold, Ontario, Canada). Two types of superantigenic assays were conducted. First, the mitogenic activity of SElY was determined using human peripheral blood mononuclear cells (PBMCs) and mouse splenocytes. Human PBMCs were obtained from three healthy donors and processed by Ficoll-Paque Plus (GE Healthcare Japan, Tokyo, Japan) density gradient centrifugation. We obtained informed consent for the study from all healthy donors. This study was performed with the approval of the local ethical committee of the Hirosaki University Graduate School of Medicine. Splenocytes were isolated from C57/BL6 mice. The PBMCs and mouse splenocytes were incubated for 72 h in 96-well round-bottomed tissue culture plates (AGC Techno Glass Co., Shizuoka, Japan) with different concentrations of SEA, SElY, or SET and then assayed for the uptake of [3H]thymidine (PerkinElmer Japan Co., Ltd., Kanagawa, Japan). Data (in counts per minute) are presented as the means ± standard errors of triplicate determinations, as previously described (20).
Second, gamma interferon (IFN-γ) and interleukin-2 (IL-2) production in mouse splenocytes and human PBMCs stimulated by SEs was also measured to assess superantigenic activity. Briefly, splenocytes isolated from C57/BL6 mice and human PBMCs were stimulated with 0.01 to 10 μg/ml of purified SEA, SElY, and SET. The cells were incubated at 37°C for 72 h in a humidified 5% CO2 atmosphere. The culture supernatants were harvested for IFN-γ and IL-2 assays. Production of mouse IFN-γ was determined by a sandwich enzyme-linked immunosorbent assay (ELISA) as described elsewhere (21). The production of mouse IL-2 and human IFN-γ was determined by use of a mouse IL-2 DuoSet ELISA development kit (R&D Systems, Minneapolis, MN) and a human IFN-γ immunoassay kit (Biosource International, Inc., Camarillo, CA), respectively, according to the manufacturers' instructions. Data (in picograms per milliliter) are presented as the means ± standard deviations of triplicate determinations, as previously described (22).
Emesis assay.
The emesis assay was performed using house musk shrews and the method described previously (5, 22). Purified SEA, SElY, and SET were diluted in phosphate-buffered saline (PBS). House musk shrews were administered each toxin (100 or 500 μg/animal) or PBS (as a control) intraperitoneally (200 μl) or perorally (1 ml). The animals were observed for emesis for 3 h after administration. The incubation period for the first vomiting episode and the number of vomiting episodes were recorded. A total of four to seven animals were used in two experiments.
Assays of SEs stability.
Three types of protein stability assays were performed as reported previously (23). Bovine serum albumin (BSA; Sigma, St. Louis, MO) was used as a protein control. To study the stability of SEs against heat treatment, 500 μl of the toxin at 100 μg/ml in PBS was added to each 1.5-ml microtube with a cap and then placed into a heat block maintained at 100°C. At desired time intervals ranging from 30 min to 12 h, tubes were removed from the heat block, immediately put into an ice bath for 5 min to cool down, and then placed at −20°C.
Purified SEA, SElY, SET, or BSA was incubated in the presence of pepsin (Wako) to assess the potential stability of SElY in the gastrointestinal environment. Each protein at a concentration of 100 μg/ml was incubated with pepsin (100 μg/ml) in a final volume of 500 μl of 0.1 M sodium acetate buffer (pH 4.5) at 37°C (23). After incubation for the desired periods of time, tubes were removed. The digestion was terminated by treatment at 95°C for 5 min, and the tubes were immediately cooled down and stored at −20°C.
SEA, SElY, SET, or BSA was incubated in the presence of trypsin (Becton Dickinson and Company, Sparks, MD) to assess the stability of the toxins in the intestinal tract under the conditions described previously (24, 25). Each protein at a concentration of 100 μg/ml was incubated with trypsin (50 μg/ml in 0.01 M Tris-HCl, pH 8.0) in a final volume of 500 μl at 37°C. After incubation for the desired periods of time, tubes were removed, treated at 95°C for 5 min, immediately cooled down, and stored at −20°C.
Twenty microliters of treated samples was mixed in SDS-PAGE sample buffer. The extent of degradation was assessed by SDS-PAGE. Controls were BSA or toxin proteins without heating or proteolysis. After electrophoresis, each gel was stained with Coomassie brilliant blue.
Nucleotide sequence accession number.
The GenBank/DDBJ accession number for the SElY nucleotide and amino acid sequence data is AB924045.
RESULTS
Identification of a SET-like gene in S. aureus strains.
A BLAST search of the translated nucleotide database was performed using a protein query to detect a SET-like gene(s) from the S. aureus complete genome sequence. One SET-like gene was identified in three S. aureus strains isolated from a case of bovine mastitis (isolate RF122) and community-associated methicillin-resistant S. aureus infections (isolates 11819-97 and M013). In each isolate, the SET-like gene existed in non-MGEs. It was located on the S. aureus chromosome in the 56′ region. The deduced amino acid sequence of the SET-like gene has 32% sequence identity with the SET amino acid sequence. The SET-like gene encoded a polypeptide 221 amino acids in length, and the mature form of the SET-like hypothetical protein was predicted to have a molecular weight of 22,529. The SET-like gene was presumed to be an SE-related gene and was designated sely; its corresponding protein was designated SElY. Phylogenetic analysis showed that SElY belongs to the SET/SE-related toxin subgroup, and the subgroup is related to a group consisting of SElX, TSST-1, and SSLs (Fig. 1). As shown in Table 1, the sely gene was also found in our S. aureus isolates that originated from a case of food poisoning (isolate 11689), a human nasal swab (isolate IVM54), and a case of bovine mastitis (isolate AV3008).
FIG 1.
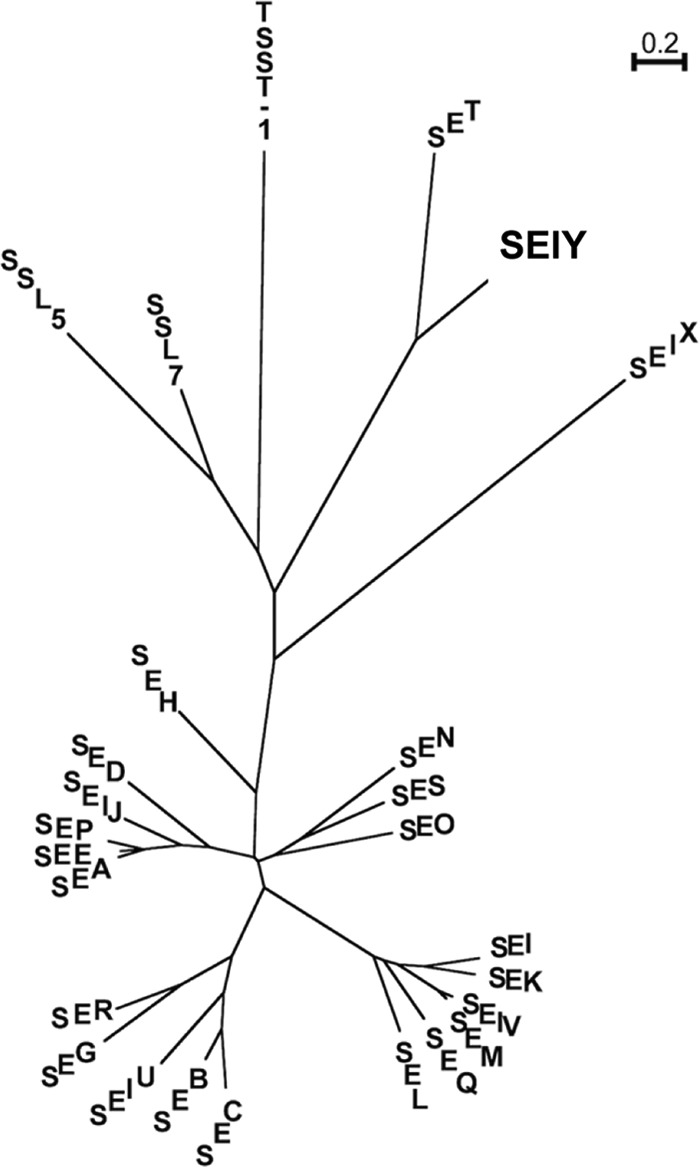
Phylogenetic tree of staphylococcal enterotoxins, including SElY and TSST-1. This phylogenetic tree was constructed using ClustalW. The tree shows that SElY belongs to the SET/SElY subgroup, and that subgroup is related to a subgroup consisting of SElX and TSST-1.
Transcription of sely in S. aureus isolates.
We determined the level of transcription of the sely gene in our three S. aureus isolates using a housekeeping gene, gyrB, as a reference. Total RNA isolated from S. aureus was subjected to real-time quantitative PCR analysis for sely mRNA expression at various time points throughout growth. The growth curves of the S. aureus isolates and the levels of expression of the sely gene in these isolates at each point are shown in Fig. 2. All PCR efficiencies were between 1.90 and 2.10, and all r2 values for the calibration curve were above 0.98. The level of sely gene expression in each isolate was shown as the ratio of the copy number of sely to that of gyrB in purified total RNA. sely mRNA was transcribed in all growth phases of all three S. aureus isolates. In food poisoning isolate 11689 and nasal swab isolate IVM54, the sely gene was highly expressed in the early log phase (ratios of sely expression to gyrB expression, 0.374 and 0.387, respectively). In contrast, AV3008, an isolate from a case of bovine mastitis, showed a lower level of sely expression throughout growth than the other two isolates, and sely mRNA reached a maximum level at 9 h after inoculation.
FIG 2.
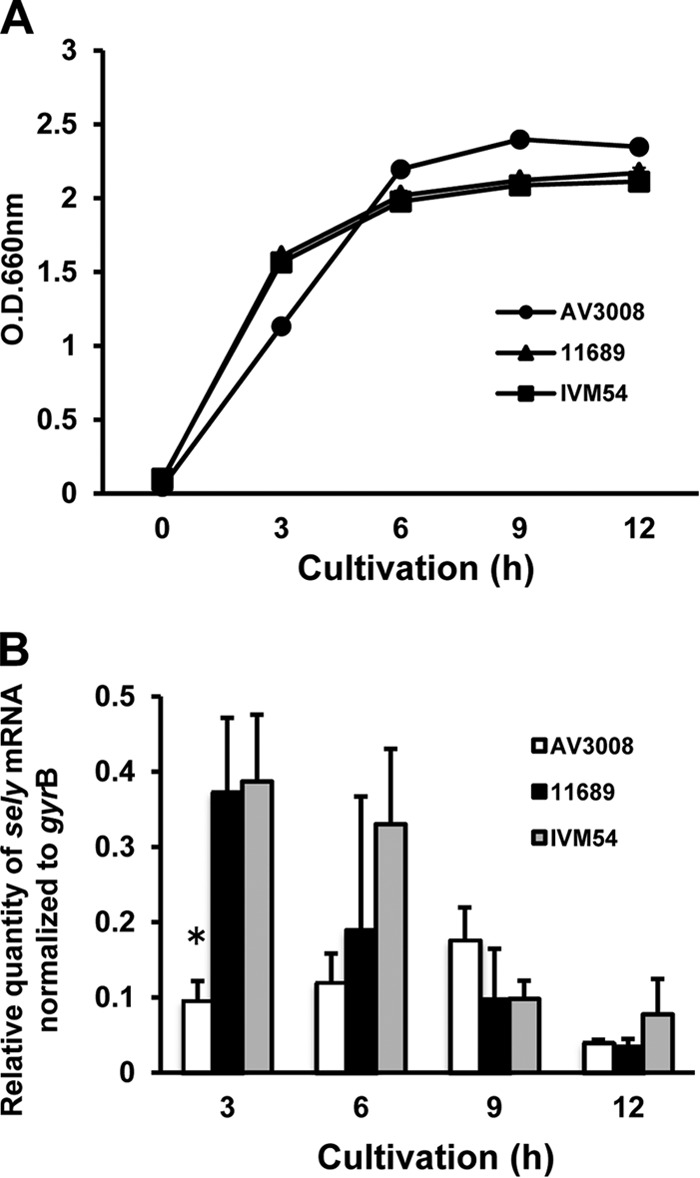
Detection of sely mRNA by real-time quantitative PCR. (A) The growth of the S. aureus isolates was determined by measurement of the optical density (O.D.) at 660 nm. Total RNA was prepared from S. aureus cells collected at 3, 6, 9, and 12 h after inoculation. (B) Expression of sely mRNA was determined by normalization to gyrB mRNA expression. Data are expressed as the mean ± standard deviation for four samples. These data were reproducible in three experiments. Statistical analysis was performed using ANOVA followed by the Tukey test. A P value of less than 0.05 was considered statistically significant. *, P < 0.05.
SElY exhibits superantigenic activity in human PBMCs.
To assess the superantigenic activity of SElY, lymphocyte proliferation and cytokine production were measured. First, SElY was tested for mitogenic activity with mouse splenocytes, and that activity was compared with the mitogenic activities of SEA and SET. Representative results from three experiments are shown in Fig. 3A. SEA strongly induced the lymphocyte proliferation of mouse splenocytes at a minimum SEA concentration of 100 pg/ml, whereas the mitogenic activity of SET was very weak in comparison with that of SEA. However, the induction of lymphocyte proliferation of mouse splenocytes by 10 μg/ml of SElY could not be detected. Second, IFN-γ and IL-2 production from mouse splenocytes was determined. Similar to the findings for lymphocyte proliferation, SEA strongly stimulated the production of both cytokines, whereas SET exhibited an extremely low level of activity. Furthermore, SElY also failed to induce IFN-γ and IL-2 production even when a concentration of up to 10 μg/ml was used. These results suggest that SElY may not exhibit superantigenic activity in mouse splenocytes. Next, we examined the superantigenic activity of SElY using human PBMCs. SEA strongly induced human PBMC proliferation, as was found for mouse splenocytes. Interestingly, SElY could induce human PBMC proliferation, comparable to the findings for SET. The minimum concentration required to induce PBMC proliferation was 100 pg/ml for both SElY and SET. SElY required a concentration 2 orders of magnitude higher than that of SEA to achieve substantial lymphocyte proliferation (Fig. 4A). Furthermore, the IFN-γ titer in human PBMCs stimulated by SElY was measured and compared with that in human PBMCs stimulated by SEA and SET. SElY induced IFN-γ production as well as SEA and SET did. These results demonstrate that SElY exhibits superantigenic activity in human PBMCs.
FIG 3.
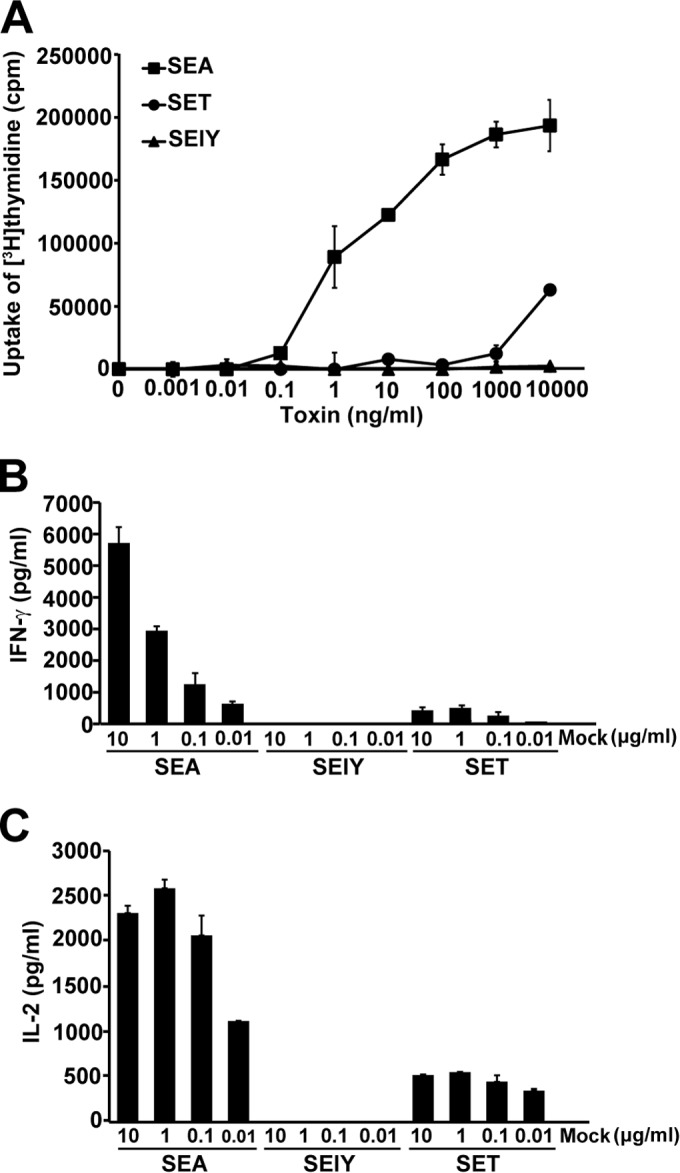
Superantigenic activity of SElY in mouse splenocytes. Mouse splenocytes were incubated with various concentrations of SEA, SET, and SElY for 72 h. (A) The uptake of [3H]thymidine was determined to assess lymphocyte proliferation. (B and C) The production of IFN-γ (Β) and IL-2 (C) was determined by sandwich ELISAs. Mock, mock treatment with no toxin. Each bar represents the mean ± standard deviation for triplicate wells from a representative experiment. These data were reproducible in three experiments.
FIG 4.
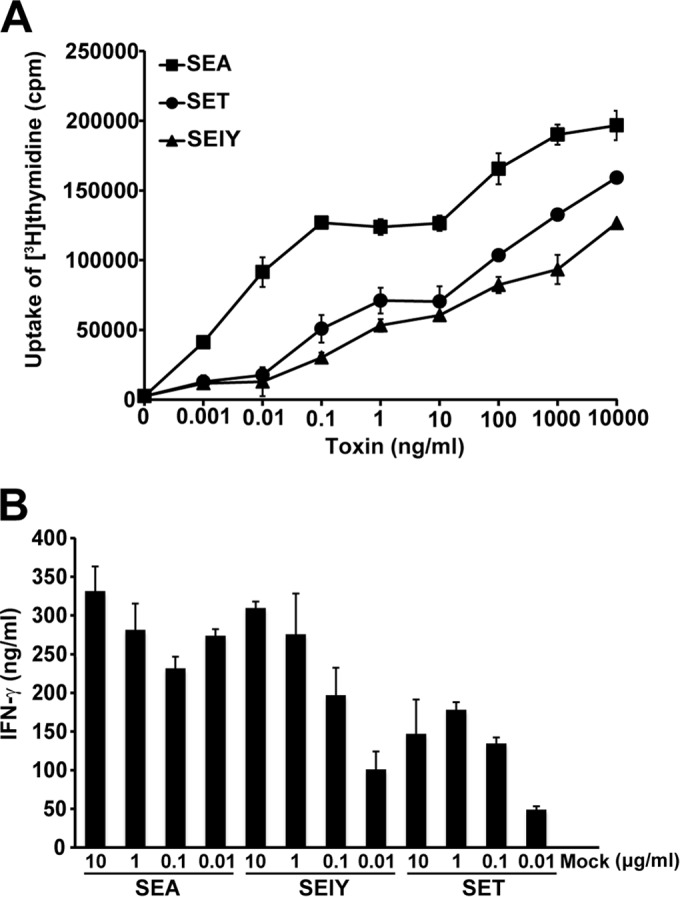
Superantigenic activity of SElY in human PBMCs. Human PBMCs were incubated with various concentrations of SEA, SET, and SElY for 72 h. (A) The uptake of [3H]thymidine was determined to assess lymphocyte proliferation. (Β) The production of IFN-γ was determined by sandwich ELISAs. Each bar represents the mean ± standard deviation for triplicate wells from a representative experiment. These data were reproducible in three experiments.
SElY exhibited emetic activity in house musk shrews.
SElY was examined for emetic activity using house musk shrews. This animal has been recognized to be the animal model that should be used to confirm the emetic activity of staphylococcal enterotoxins. SEA at a dose of 100 μg/animal induced an emetic response in 7 of 7 house musk shrews within 37 to 120 min after intraperitoneal administration (Table 3). Although no house musk shrew vomited after receiving a dose of 100 μg SElY per animal by intraperitoneal injection, SElY at a dose of 500 μg/animal by intraperitoneal injection was able to induce an emetic response in 4 of 7 house musk shrews. In addition, SElY at a dose of 500 μg/animal by oral administration induced an emetic response in 3 of 7 house musk shrews (Table 3). The emetic response to SElY was observed within 10 to 150 min and 51 to 100 min after intraperitoneal and oral administration, respectively.
TABLE 3.
Emetic activity of SElY in house musk shrews
| Toxin and dosea (μg/animal) | No. of shrews |
Incubation period (min) observed in each shrew | No. of emetic episodes | |
|---|---|---|---|---|
| Testedb | That vomited | |||
| SEA, 100 i.p. | 7 | 7 | 37, 51, 95, 98, 104, 108, 120 | 11, 5, 11, 14, 6, 2, 6 |
| SElY | ||||
| 100 i.p. | 4 | 0 | ||
| 500 i.p. | 7 | 4 | 10, 13, 17, 150 | 12, 7, 4, 3 |
| 500 p.o. | 7 | 3 | 51, 94, 100 | 2, 2, 9 |
| PBS | 7 | 0 | ||
i.p., intraperitoneally; p.o., orally.
The total number of shrews from two experiments is indicated.
SElY is unstable in response to heat treatment and digestive enzymes.
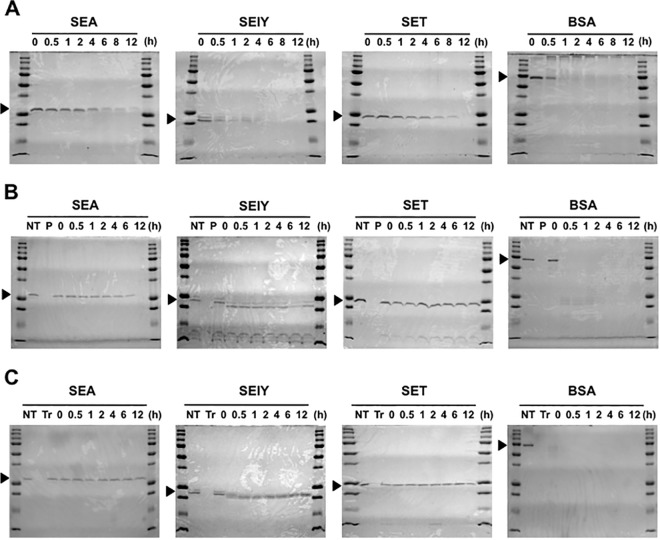
We examined whether SElY resists heat and pepsin and trypsin digestion, which are related to food preparation and gastrointestinal conditions. After heat treatment, the samples were analyzed by SDS-PAGE (Fig. 5A). The band for BSA was immediately reduced at 30 min and disappeared at 1 h. The band for SElY was also immediately reduced at 30 min. A faint band for SElY could be detected at up to 2 h of heat treatment. After that, the band for SElY totally disappeared. In contrast, the bands for SEA and SET were still observed at up to 8 to 12 h after heat treatment. For proteolytic analysis, SElY was degraded at 30 min of incubation with pepsin treatment. The degraded SElY band had a molecular weight slightly lower than its actual molecular weight. This small protein band was detectable until 12 h after pepsin treatment (Fig. 5B). SEA and SET were more resistant to pepsin digestion, and each showed a single band corresponding to its authentic size. In contrast, BSA was rapidly digested by pepsin treatment (Fig. 5B). We further investigated the stability of SEA, SET, and SElY in response to trypsin digestion. Trypsin-treated SElY appeared as smaller bands in the SDS-polyacrylamide gels, as did pepsin-treated SElY. These bands were not further degraded up to 12 h after digestion (Fig. 5C). In contrast, trypsin-treated SEA and SET each appeared as a single band corresponding to its actual size, and the protein bands were clearly observed until 12 h after treatment (Fig. 5C). These results indicate that SEA and SET have high degrees of stability against heating and the digestive enzymes pepsin and trypsin. In contrast, SElY is unstable in response to pepsin, trypsin, or heat treatment.
FIG 5.
Effects of heat and digestive enzyme treatments on SEs. (A) SEs and BSA were maintained at 100°C. After the heat treatment, each sample was analyzed by SDS-PAGE. (B) Each protein (100 μg/ml) was incubated with pepsin (10 μg/ml) and analyzed by SDS-PAGE. (C) Each protein (100 μg/ml) was incubated with trypsin (100 μg/ml) and analyzed by SDS-PAGE. NT, no treatment; P, pepsin; Tr, trypsin.
DISCUSSION
We identified and characterized a new staphylococcal emetic toxin that is transcribed by S. aureus strains. The International Nomenclature Committee for Staphylococcal Superantigens recommends that only toxins that induce emesis after oral administration in primates be designated SEs and that other related toxins that either lack emetic properties or are unexamined in primates should be designated staphylococcal enterotoxin-like (SEl) superantigens (26). In addition, SE-related toxins that lack both superantigenic and emetic activities should be renamed SSLs (26). The properties of the new emetic toxin described in the present study were matched with SEls according to the definition of SEls because SElY has superantigenic activity in human PBMCs and an emetic response is shown in the house musk shrew model.
Our study revealed that SElY at a dose of 500 μg/animal induced an emetic response in the house musk shrew model. Although this activity was lower than that of SEA, it seems to be stronger than that of SEC2, SEH, and SET, for which a minimum dose of 1,000 μg/animal is required to induce an emetic response in house musk shrews (5, 22).
Our present results indicate that SElY, which shows homology to SET, acts as a superantigen for human lymphocytes but not mouse lymphocytes because SElY completely failed to induce lymphocyte proliferation and cytokine production from mouse splenocytes (Fig. 3 and 4). The nonresponsiveness of mouse splenocytes to SElY is presumed, as SElY was not capable of binding to TCR Vβ elements and/or MHC class II molecules in the mouse. We previously reported that SET showed superantigenic activity lower than that of known SEs, and a Vβ specificity for the activation of SET on T cells was not found (5). In this study, the proliferation of and cytokine production by SET-stimulated lymphocytes were also remarkably lower than those of SEA (Fig. 3 and 4).
S. aureus strain 11689 was isolated from a food poisoning outbreak. This strain carries other SE genes (sea, seb, sek, and seq) as well as sely. In this study, we demonstrated that expression of sely mRNA in strain 11689 is significantly increased in the early log phase. Therefore, SElY is assumed to be involved in human food poisoning, even though an epidemiological analysis will be necessary to reveal the prevalence of the sely gene in S. aureus. The present study revealed that the sely gene exists not only in a food poisoning isolate but also in isolates from cases of infectious disease, such as bovine mastitis and community-associated methicillin-resistant S. aureus infection.
SElY has emetic and superantigenic activities like other SEs. However, SElY was unstable in response to digestive enzymes and heat treatment. This property is different from that of other SEs because SEs are well-known to resist heating and digestive enzymes (23). From a phylogenetic tree analysis, SElY was found to be closely related to TSST-1 and SSLs rather than to other SEs. Furthermore, the gene encoding SElY exists not only in food poisoning isolates but also in isolates from cases of S. aureus infection. Therefore, SEY may have a novel function(s) and may contribute to the pathogenesis of S. aureus infection.
In conclusion, the present study has demonstrated the biological characteristics of newly identified SElY. This toxin has superantigenic activity in human PBMCs and acts as an emetic toxin in house musk shrews. Therefore, it is presumed that SElY probably acts as an emetic toxin in primates.
ACKNOWLEDGMENTS
This study was supported in part by JSPS KAKENHI grant numbers 20580338 (to K.O.) and 23390100 (to A.N.) and a Sasakawa scientific research grant from The Japan Science Society.
REFERENCES
- 1.Dinges MM, Orwin PM, Schlievert PM. 2000. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13:16–34. doi: 10.1128/CMR.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCormick JK, Yarwood JM, Schlievert PM. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol 55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 3.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect Immun 71:6088–6094. doi: 10.1128/IAI.71.10.6088-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, Gougeon M, Lina G, Vandenesch F. 2006. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect Immun 74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono HK, Omoe K, Imanishi K, Iwakabe Y, Hu DL, Kato H, Saito N, Nakane A, Uchiyama T, Shinagawa K. 2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect Immun 76:4999–5005. doi: 10.1128/IAI.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, Merriman JA, Guinane CM, Park JY, Bohach GA, Schlievert PM, Morrison WI, Fitzgerald JR. 2011. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog 7:e1002271. doi: 10.1371/journal.ppat.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munson SH, Tremaine MT, Betley MJ, Welch RA. 1998. Identification and characterization of staphylococcal enterotoxin type G and I from Staphylococcus aureus. Infect Immun 66:3337–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su YC, Wong ACL. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol 61:1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RJ, Ward JM, Henderson B, Poole S, O'Hara BP, Wilson M, Nair SP. 2000. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect Immun 68:4407–4415. doi: 10.1128/IAI.68.8.4407-4415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda M, Ohata T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian JQ, Ito T, Kanamori M, Matumura H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 11.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 12.Betley MJ, Mekalanos JJ. 1985. Staphylococcal enterotoxin A is encoded by phage. Science 229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 13.Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol 166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 14.Sato'o Y, Omoe K, Ono HK, Nakane A, Hu DL. 2013. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol Immunol 57:91–99. doi: 10.1111/1348-0421.12007. [DOI] [PubMed] [Google Scholar]
- 15.Duquenne M, Fleurot I, Aigle M, Darrigo C, Borezée-Durant E, Derzelle S, Bouix M, Deperrois-Lafarge V, Delacroix-Buchet A. 2010. Tool for quantification of staphylococcal enterotoxin gene expression in cheese. Appl Environ Microbiol 76:1367–1374. doi: 10.1128/AEM.01736-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joost I, Blass D, Burian M, Georke C, Wolz C, von Müller L, Becker K, Preissner K, Herrmann M, Bischoff M. 2009. Transcription analysis of the extracellular adherence protein from Staphylococcus aureus in authentic human infection and in vitro. J Infect Dis 199:1471–1478. doi: 10.1086/598484. [DOI] [PubMed] [Google Scholar]
- 17.Ono HK, Nishizawa M, Yamamoto Y, Hu DL, Nakane A, Shinagawa K, Omoe K. 2012. Submucosal mast cells in the gastrointestinal tract are a target of staphylococcal enterotoxin type A. FEMS Immunol Med Microbiol 64:392–402. doi: 10.1111/j.1574-695X.2011.00924.x. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DJ, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchiyama T, Miyoshi-Akiyama T, Kato H, Fujimaki W, Imanishi K, Yan XJ. 1993. Superantigenic properties of a novel mitogenic substance produced by Yersinia pseudotuberculosis isolated from patients manifesting acute and systemic symptoms. J Immunol 151:4407–4413. [PubMed] [Google Scholar]
- 21.Nakane A, Numata A, Minagawa T. 1992. Endogenous tumor necrosis factor, interleukin-6, and gamma interferon levels during Listeria monocytogenes infection in mice. Infect Immun 60:523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu DL, Omoe K, Shimoda Y, Nakane A, Shinagawa K. 2003. Induction of emetic response to staphylococcal enterotoxins in the house musk shrew (Suncus murinus). Infect Immun 71:567–570. doi: 10.1128/IAI.71.1.567-570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S-J, Hu D-L, Maina EK, Shinagawa K, Omoe K, Nakane A. 2011. Superantigenic activity of toxic shock syndrome toxin-1 is resistant to heating and digestive enzymes. J Appl Microbiol 110:729–736. doi: 10.1111/j.1365-2672.2010.04927.x. [DOI] [PubMed] [Google Scholar]
- 24.Chi YI, Sadler I, Jablonski LM, Callantine SD, Deobald CF, Stauffacher CV, Bohach GA. 2002. Zinc-mediated dimerization and its effect on activity and conformation of staphylococcal enterotoxin type C. J Biol Chem 277:22839–22846. doi: 10.1074/jbc.M201932200. [DOI] [PubMed] [Google Scholar]
- 25.Hovde CJ, Marr JC, Hoffmann ML, Hackett SP, Chi YI, Crum KK, Stevens DL, Stauffacher CV, Bohach GA. 1994. Investigation of the role of the disulphide bond in the activity and structure of staphylococcal enterotoxin C1. Mol Microbiol 13:897–909. doi: 10.1111/j.1365-2958.1994.tb00481.x. [DOI] [PubMed] [Google Scholar]
- 26.Lina G, Bohach GA, Nair SP, Hiramatsu K, Jouvin-Marche E, Mariuzza R. 2004. Standard nomenclature for the superantigens expressed by Staphylococcus. J Infect Dis 189:2334–2336. doi: 10.1086/420852. [DOI] [PubMed] [Google Scholar]