Abstract
Bacteria of the genus Lysobacter are considered to be facultative predators that use a feeding strategy similar to that of myxobacteria. Experimental data supporting this assumption, however, are scarce. Therefore, the predatory activities of three Lysobacter species were tested in the prey spot plate assay and in the lawn predation assay, which are commonly used to analyze myxobacterial predation. Surprisingly, only one of the tested Lysobacter species showed predatory behavior in the two assays. This result suggested that not all Lysobacter strains are predatory or, alternatively, that the assays were not appropriate for determining the predatory potential of this bacterial group. To differentiate between the two scenarios, predation was tested in a CFU-based bioassay. For this purpose, defined numbers of Lysobacter cells were mixed together with potential prey bacteria featuring phenotypic markers, such as distinctive pigmentation or antibiotic resistance. After 24 h, cocultivated cells were streaked out on agar plates and sizes of bacterial populations were individually determined by counting the respective colonies. Using the CFU-based predation assay, we observed that Lysobacter spp. strongly antagonized other bacteria under nutrient-deficient conditions. Simultaneously, the Lysobacter population was increasing, which together with the killing of the cocultured bacteria indicated predation. Variation of the predator/prey ratio revealed that all three Lysobacter species tested needed to outnumber their prey for efficient predation, suggesting that they exclusively practiced group predation. In summary, the CFU-based predation assay not only enabled the quantification of prey killing and consumption by Lysobacter spp. but also provided insights into their mode of predation.
INTRODUCTION
In nature, microorganisms do not occur as isolated living entities. Instead, they exist in complex communities of multiple species that interact with each other (1). While some of these interactions are beneficial for the partners involved, others tend to be parasitic or even competitive (2). A commonly encountered negative interaction among microorganisms is predation, which is considered an important evolutionary force that shapes microbial biodiversity (3). Predatory behavior can be observed in many taxonomically unrelated groups of bacteria, encompassing both obligate and facultative predators (4–6). The latter are capable of preying on other organisms but can also survive by utilizing nonliving nutrient sources (6). Predatory bacteria show an enormous diversity of feeding strategies (7). At present, four basic predatory lifestyles are known, i.e., “wolf pack” or group predation, epibiotic attachment, direct cytoplasmic invasion, and periplasmic invasion (8). It is, however, difficult to categorize predatory bacteria based on their hunting behaviors, since clear distinctions between the aforementioned strategies are often not possible.
Among the most thoroughly investigated facultative predators are myxobacteria. Although they are individually capable of penetrating and digesting prey microcolonies (9), myxobacteria are generally assumed to hunt collectively (7). Group predation requires a quorum of cells as well as gliding motility, which allows myxobacteria to actively seek their prey (10, 11). Another commonly observed feature is the concerted release of cell wall-degrading enzymes and antibiotics (12–15). Few bioassays are available for investigating predatory interactions among bacteria. Myxobacterial predation is typically analyzed on agar plates. For this purpose, myxobacteria are inoculated onto a spot or lawn of prey organisms in order to monitor the emergence of lysis or swarming (16–19). A variation of this methodology involves the recovery and enumeration of surviving prey cells after transferring to agar media, which exclusively suppress myxobacterial growth (13, 20).
Bacteria of the genus Lysobacter share many properties with myxobacteria. Both groups feature a high G+C content in their DNA, the ability to glide on solid surfaces, and the secretion of lytic enzymes (10, 12, 21, 22). Prior to the introduction of phylogenetic markers, these commonalities caused some confusion concerning the taxonomic placement of isolates with the aforementioned features. As a consequence, many Lysobacter strains were originally falsely classified as myxobacteria (22). This also led to some ambiguities with regard to predatory behavior of the two bacterial groups. In general, Lysobacter spp. are assumed to practice group predation (4, 8), even though there was also evidence for epibiotic feeding (23, 24). In a study by Shilo (23), a concentrated suspension of a Lysobacter sp. (originally assigned as Myxobacter FP-1) was added to a cyanobacterial culture. After incubation, the mixed cultures were examined under the microscope, and lysis of cyanobacteria was observed after attachment of Lysobacter. In contrast, the hypothesis of wolf pack feeding was supported mainly by the observation that many Lysobacter strains are potent antibiotic producers (25).
Since its discovery by Christensen and Cook (21), the genus Lysobacter has been expanded from 4 to 30 species (www.bacterio.net/lysobacter.html). Several studies of the newly discovered species focused on the release of lytic enzymes and the production of antimicrobial compounds (see, e.g., references 26, 27, and 28) without providing direct evidence for their involvement in predatory interactions. To the best of our knowledge, there is only one recent study which investigated bacterial predation of Lysobacter. Lueders et al. quantified the incorporation of biomass carbon into a soil microbial food web (29). For this purpose, agricultural soil was inoculated with 13C-labeled Escherichia coli, which was expected to serve as prey for predatory bacteria. Preliminary data confirmed this assumption, and 16S rRNA sequencing indicated that the predators included bacteria of the genus Lysobacter (29).
Taken together, experimental data on the predatory activity of Lysobacter spp. or evidence for their predatory strategy are scarce. To fill this gap, this study aimed at evaluating the predatory potentials of three different species from this genus. The CFU-based assay confirmed that all Lysobacter spp. tested were able to feed on other bacteria, although killing efficiencies varied across prey types. Additionally, we obtained evidence that Lysobacter bacteria hunt exclusively in groups, which is in stark contrast to myxobacterial predation.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
Since predatory performance is known to be strongly affected by the type of prey (19), nine taxonomically distinct strains were chosen as prey bacteria. Agrobacterium tumefaciens and Ralstonia solanacearum are well-known soilborne plant pathogens (30), and their potential eradication by a predatory bacterium could be of agricultural interest. The actinobacterium Rhodococcus rhodochrous is frequently used as a soil inoculant (31). Moreover, its close relationship to Rhodococcus fascians made it an interesting model organism to evaluate potential effects of Lysobacter spp. against a Gram-positive plant pathogen (32). Bacillus subtilis, Pseudomonas fluorescens, and Chromobacterium pseudoviolaceum represent widely distributed soil bacteria. C. pseudoviolaceum is also known to produce a violet pigment (33), which allows easy identification on agar plates. Escherichia coli, although not being a typical soil inhabitant, was selected because of its common use in predation assays (13, 16, 19). Likewise, Micrococcus luteus and Lactococcus lactis had been reported as suitable prey organisms before (6, 34). Strains used in this study were obtained from the German Strain Collection of Microorganisms and Cell Cultures (DSMZ) or the Jena Microbial Resource Collection (JMRC). Ralstonia solanacearum GMI1000 was kindly provided by C. Allen (Department of Plant Pathology, University of Wisconsin—Madison, USA). Strain numbers and cultivation conditions are provided in Table 1.
TABLE 1.
Bacterial strains and cultivation conditions used
| Species | Strain | Growth mediuma | Growth temp (°C) |
|---|---|---|---|
| Predators | |||
| Lysobacter capsici | DSM 19286 | LB | 30 |
| Lysobacter enzymogenes | DSM 2043 | LB | 30 |
| Lysobacter oryzae | DSM 21044 | LB | 30 |
| Myxococcus fulvus | ST035975 | MD1 | 30 |
| Prey | |||
| Agrobacterium tumefaciens | DSM 5172 | LB | 30 |
| Bacillus subtilis | DSM 347 | LB | 30 |
| Chromobacterium pseudoviolaceum | DSM 23279 | LB | 30 |
| Escherichia coli | DSM 18039 | LB | 37 |
| Lactococcus lactis | DSM 20069 | SM17 | 30 |
| Micrococcus luteus | DSM 14234 | LB | 30 |
| Pseudomonas fluorescens | DSM 11532 | LB | 30 |
| Ralstonia solanacearum | GMI1000 | NB | 30 |
| Rhodococcus rhodochrous | DSM 43334 | LB | 30 |
LB, Luria broth; NB, nutrient broth; SM17, M17 medium (Sigma) with 0.5% sucrose.
Correlation of optical densities with viable cell count data.
For every bacterial strain, the statistical relationship between CFU and optical density at 600 nm (OD600) was determined (35). For this, bacteria were grown in the appropriate growth medium until they reached early stationary phase. At this time, cells were harvested by centrifugation (2,400 × g, 5 min). The cell pellet was suspended in phosphate-buffered saline (PBS) buffer (0.8% NaCl, 0.02% KCl, 0.144% Na2HPO4, 0.024% KH2PO4, pH 7.6). Serial dilutions of these suspensions with defined OD600 values were streaked out on a suitable agar medium. Following 4 days of incubation at 30°C, the number of CFU were determined and plotted against the respective optical densities (see Fig. S1 to S3 in the supplemental material).
Generation of antibiotic-resistant prey bacteria.
A pJET1.2-derived vector featuring a chloramphenicol resistance gene in its multiple-cloning site was introduced into E. coli by electroporation, yielding E. coli/pJET1.2-Cm. A. tumefaciens, P. fluorescens, and R. solanacearum were transformed with pBHR1 (Mobitec), generating the respective chloramphenicol-resistant strains. B. subtilis and L. lactis were transformed with pNZ8048 (36) to endow these bacteria with chloramphenicol resistance. Electrocompetent cells were prepared following previously described protocols (37–39).
Prey spot plate assay.
Prey bacteria were grown on LB agar medium at 30°C for 2 days, except for E. coli, which was cultured for 1 day at 37°C. The resulting cell lawn was collected with a sterile spatula and suspended in PBS buffer to yield a final concentration of 107 cells ml−1. Assay plates consisted of WAT agar (0.1% CaCl2 · 2H2O, 1.5% agar, pH 7.2) which had been spotted with 150 μl of freshly prepared prey suspensions. Each prey spot was inoculated with a single predator colony (Lysobacter capsici, 4.5 × 107 cells ml−1; Lysobacter oryzae, 6 × 107 cells ml−1; Lysobacter enzymogenes, 1 × 107 cells ml−1; and Myxococcus fulvus, 5 × 104 cells ml−1). For this purpose, the predatory bacteria had previously been grown on LB or MD1 agar (0.3% Casitone, 0.7% CaCl2 · 2H2O, 0.2% MgSO4 · 7H2O, 1.5% agar) for 5 days at 30°C. The assay plates were incubated at 30°C for 10 days. Lysis of prey spots was monitored during the incubation period. A prey spot without any added predator colony served as a negative control. The experiment was conducted in three biological replicates. The diameter of the lysis area was measured on days 1 and 10.
Lawn predation assay.
R. rhodochrous, C. pseudoviolaceum, B. subtilis, and M. luteus were cultivated in 5 ml LB medium for 2 days on a rotary shaker (220 rpm) at 30°C, whereas E. coli was incubated for 1 day at 37°C. After centrifugation (1,200 × g, 4°C, 5 min) and removal of the supernatant, the cell pellet was washed twice and suspended in TPM buffer (1 M 1.0% Tris-HCl, 0.1 M KH2PO4, 0.8 M 1.0% MgSO4, pH 7.6) to yield a final concentration of 1010 cells ml−1. Five hundred microliters of this suspension was evenly spread on a TPM agar plate (TPM buffer with 1.5% agar). Predatory bacteria were precultured in glass tubes containing 5 ml LB medium at 220 rpm for 2 days, except for M. fulvus, which was grown in 25 ml MD1 medium at 150 rpm for 5 days. The prey-covered TPM agar plates were individually spotted with 10 μl of Lysobacter and M. fulvus cell suspensions, which were adjusted to 5 × 107 cells ml−1. In subsequent experiments, the concentrations of the Lysobacter suspensions were increased to 1.2 × 109 cells ml−1 for L. capsici, 1.6 × 109 cells ml−1 for L. oryzae, and 1 × 109 cells ml−1 for L. enzymogenes. As a control, predator suspensions were spotted on TPM agar plates without prey bacteria. The experiment was replicated three times. The diameter of the predator swarm was measured on days 1 and 10.
CFU-based predation assay.
Glycerol stock cultures of the test bacteria were used to inoculate LB agar plates unless otherwise stated (Table 1). Agar cultures were incubated at 30°C until the appearance of first colonies. From every predator culture, six colonies were randomly selected and subcultured in glass tubes containing 5 ml LB medium at 220 rpm for 2 days, while M. fulvus was cultured in 25 ml MD1 medium at 150 rpm for 5 days. In parallel, a prey colony was selected and individually cultured in 10 ml of appropriate medium at 220 rpm for 2 days. After cultivation, 2 ml of each bacterial culture was harvested and centrifuged (1,200 × g, 4°C, 5 min). The supernatant was removed, and the cell pellet was washed three times with 2 ml of PBS buffer and then resuspended in 1.6 ml of nutrient-free PBS buffer. From these suspensions, 370-μl aliquots of prey (cell concentration adjusted to 1 × 106 cells ml−1) and predator (adjusted to 1 × 108 cells ml−1) were mixed in a 2-ml tube. The predator control sample contained 370 μl of predator cells and the same volume of PBS buffer, and the prey control sample contained 370 μl of prey mixed with 370 μl of PBS buffer. Control experiments included only monocultures of predator or prey. Every experiment was replicated six times. All cultures were incubated at 30°C and 220 rpm for 24 h. After cultivation, serial dilutions of cocultures and monocultures ranging from 10−3 to 10−5 were prepared by mixing with PBS buffer and were individually spread on nutrient-rich agar plates (Table 1). The CFU number was determined (see Fig. S4 in the supplemental material).
When using antibiotic resistance as a selection marker, prey bacteria harboring resistance plasmids were pregrown in medium supplemented with chloramphenicol (25 μg ml−1). The antibiotic was removed by washing with PBS buffer prior to the addition of the predator suspension. Cocultivation of predator and prey cells was carried out without any antibiotics added. Control experiments confirmed that no significant loss of resistance plasmids occurred during this period (see Fig. S5 in the supplemental material). After 24 h, cocultures were spread either on nutrient-rich agar containing 25 μg ml−1 chloramphenicol for counting the prey population or on LB agar supplemented with 50 μg ml−1 kanamycin for quantifying the number of Lysobacter colonies. After incubation at 30°C for 3 to 5 days, prey and predatory colonies were counted and compared to the numbers of colonies in the control plates. Every experiment was repeated two times.
Frequency dependence of predatory efficiency.
To determine whether the ability of Lysobacter spp. to effectively lyse its prey depends on the predator/prey ratio (PPR), the CFU-based assay was conducted by varying the initial ratio between predator and prey. For this, the number of prey cells (i.e., B. subtilis) was held constant (2 × 107 cells ml−1), while the number of predator cells (i.e., Lysobacter spp.) was varied ranging from 2 × 107 to 2 × 1010 cells ml−1.
Contact dependence of predatory behavior.
Lysobacter strains were grown in LB medium to an OD600 of 4. After centrifugation (2,400 × g, 5 min), the cell pellet was washed with PBS buffer and directly mixed with the prey bacterium B. subtilis as described previously. Alternatively, the recovered Lysobacter cells were propagated in PBS buffer for 24 h to mimic starvation conditions. Supernatants of starved (PBS) and nonstarved (LB) cultures were filter sterilized and diluted according to the predator cell suspension. Aliquots (370 μl) of these filtrates were mixed with 370 μl of prey suspension (adjusted to 1 × 106 cells ml−1). Control experiments included B. subtilis suspensions treated with 370 μl of LB medium or PBS buffer. All cocultures and monocultures of B. subtilis were incubated at 30°C and 220 rpm for 24 h. After cultivation, serial dilutions ranging from 10−3 to 10−5 were prepared by mixing with PBS buffer and were individually spread on nutrient-rich agar plates. The CFU number was determined.
Evaluation of the predation efficiency.
To quantify predatory activity, both the killing efficiency (e) and the utilization of prey (u) were determined for each experiment. The two parameters were calculated using the following formulas: e = (CFU of control prey − CFU of surviving prey)/CFU of control prey × 100 and u = (CFU of predator with prey − CFU of control predator)/CFU of control predator × 100.
Statistical analyses.
The data obtained from the prey spot plate assay and lawn predation assay were analyzed using a paired-sample t test and nonparametric statistical tests. The Mann-Whitney U test and Wilcoxon median tests were applied to statistically analyze the CFU-based predation assay. All statistical analyses were performed using the SPSS software (version 22.0; IBM, USA).
RESULTS
Prey spot plate assay.
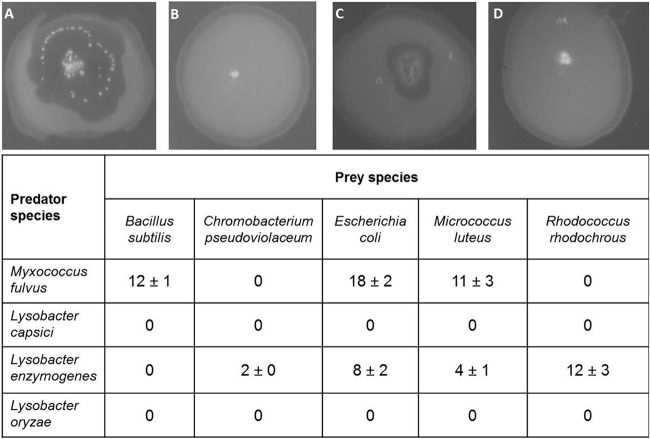
The predatory activity of three selected Lysobacter species was initially investigated using the prey spot plate assay. Since this bioassay was originally developed to isolate myxobacteria (40), Myxococcus fulvus was included as a positive control. After 10 days of incubation, the tested M. fulvus strain had produced lysis zones within spots of E. coli, B. subtilis, and M. luteus. However, no lysis was observed on plates covered with C. pseudoviolaceum and R. rhodochrous (Fig. 1). From the three Lysobacter strains tested, only L. enzymogenes exhibited lytic activity, whereas L. capsici and L. oryzae appeared to have no effect on any prey organism. L. enzymogenes was most active against R. rhodochrous. Moderate lytic activity could be observed against E. coli and M. luteus, while there was weak activity against C. pseudoviolaceum and no activity against B. subtilis. Although L. enzymogenes and M. fulvus were both found to attack E. coli and M. luteus, it appeared that the two prey strains were more susceptible to the myxobacterium. Thus, of the three Lysobacter strains tested, only one showed clear signs of predation using this assay.
FIG 1.
Effect of predators on different prey bacteria as determined by the prey spot plate assay. The table shows the mean (± 95% confidence interval [n = 3]) diameter of the lysis zone (in millimeters). Images depict spots of E. coli that have been coinoculated with a single colony of M. fulvus (A), L. capsici (B), L. enzymogenes (C), or L. oryzae (D) after 10 days of incubation.
Lawn predation assay.
The predatory performance of myxobacteria is often correlated with their ability to swarm on prey-covered plates (10). Similar to the occurrence of lysis plaques in the prey spot plate assay, the swarming rate of a predator is prey specific (41). Subjecting the same four species of predators to this test indicated that M. fulvus exhibited the fastest swarm expansion on E. coli, while it was significantly lower on M. luteus and B. subtilis (Fig. 2). Interestingly, the myxobacterium failed completely to swarm on plates covered with R. rhodochrous and C. pseudoviolaceum, as indicated by the fact that no significant swarm expansion was observed after 10 days of incubation relative to the first day (paired-sample t test, P < 0.05 [n = 3]) (see Table S1 in the supplemental material). These results suggested that C. pseudoviolaceum and R. rhodochrous are unsuitable as prey for M. fulvus. Since bacteria belonging to the genus Lysobacter are considered to have gliding motility (21, 42), we expected them to display swarming behavior in the lawn predation assay, similar to that of M. fulvus. Again, however, only L. enzymogenes showed some moderate predatory activity. Significant predatory swarming was observed on R. rhodochrous and E. coli plates, although the effects were less pronounced than in case of M. fulvus (Fig. 2). In contrast, L. capsici and L. oryzae did not exhibit swarming behavior on the selected prey bacteria under the experimental conditions used. Thus, again, only one of the three Lysobacter species analyzed showed moderate signs of predation.
FIG 2.
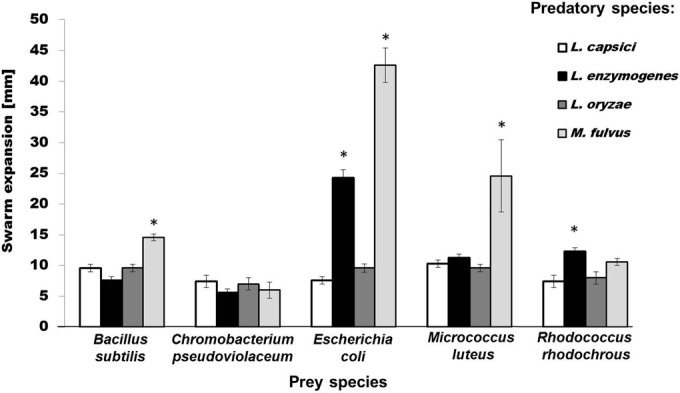
Effect of predators on different prey bacteria as determined by the lawn predation assay. Shown is the mean swarm expansion (± 95% confidence interval [n = 3]) (in millimeters) of four species of predatory bacteria. Paired t test: *, P < 0.05 between day 1 and day 10. All other comparisons were not significant (P > 0.05).
CFU-based predation assay.
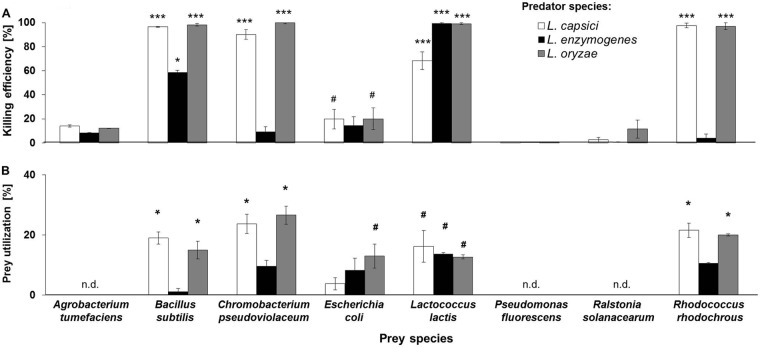
A prerequisite for the simultaneous determination of predator and prey populations from a mixed culture is the ability to phenotypically distinguish both partners. C. pseudoviolaceum and R. rhodochrous were initially selected as prey bacteria, because they form intensively colored colonies which can be easily differentiated from those of Lysobacter spp. To extend the application range of the assay, differences in antibiotic resistance also were used to discriminate predator and prey. Previous antibiotic susceptibility tests had revealed that all tested Lysobacter spp. were resistant to kanamycin (up to a concentration of 100 μg ml−1), whereas they shared sensitivity to chloramphenicol (25 μg ml−1). Therefore, chloramphenicol resistance genes were introduced into the kanamycin-sensitive prey bacteria A. tumefaciens, B. subtilis, E. coli, L. lactis, P. fluorescens, and R. solanacearum. Subsequently, the testing was carried out with the phenotypically labeled prey organisms. For evaluating the killing efficiency (e), the number of surviving prey bacteria from cocultures was compared with that from prey monocultures after incubation on LB-chloramphenicol plates. Likewise, LB-kanamycin plates were used to determine the prey utilization (u). The latter parameter quantified Lysobacter's consumption of prey and was calculated by comparing the number of Lysobacter CFU when grown in monoculture to the number of Lysobacter CFU achieved in coculture with its prey.
In the CFU-based predation assay, L. capsici and L. oryzae preyed on all Gram-positive bacteria tested, namely, B. subtilis, L. lactis, and R. rhodochrous (Fig. 3A; see Table S2 in the supplemental material). L. enzymogenes was not active against R. rhodochrous, but it was found to negatively affect populations of B. subtilis and L. lactis. Besides the species-dependent prey utilization, we also observed quantitative differences in prey consumption. All three Lysobacter strains were found to significantly reduce the CFU number of B. subtilis and L. lactis (Fig. 3A; see Table S2 in the supplemental material). In contrast to L. capsici and L. oryzae, L. enzymogenes did not completely eradicate the B. subtilis population. L. capsici exhibited a comparatively reduced killing efficiency against L. lactis (Mann-Whitney U test, P < 0.05 [n = 6]) (Fig. 3A). Overall, Gram-negative bacteria appeared to be more resistant toward Lysobacter predation than Gram-positive bacteria. The only exception was C. pseudoviolaceum, which turned out to be a preferred prey organism for both L. capsici and L. oryzae. While some weak predatory activity was also observed against E. coli (at least in the case of L. capsici and L. oryzae), the growth of A. tumefaciens, R. solanacearum, or P. fluorescens remained unaffected (Mann-Whitney U test, P < 0.05 [n = 6]) (Fig. 3A). It is noteworthy that L. enzymogenes failed to prey on all tested Gram-negative bacteria. Despite the limited prey range, it became obvious that both Gram-positive and Gram-negative bacteria can be killed by Lysobacter spp., although e and u values differed significantly depending on the prey species tested.
FIG 3.
Effect of predators on different prey bacteria and vice versa as determined by the CFU-based predation assay. (A) Mean killing efficiency (ē; ± 95% confidence interval) of all three Lysobacter spp. tested against different species of prey bacteria (percent). Asterisks denote significant differences between the number of prey CFU of the control group (i.e., monocultures) and samples containing both predator and prey (i.e., cocultures) (Mann-Whitney U test: ***, P < 0.001 *, P < 0.05; #, P < 0.07; df = 2). (B) Mean prey utilization (ū; ± 95% confidence interval) of all three Lysobacter spp. tested against different species of prey bacteria (percent). Asterisks denote significant differences in the prey utilization when comparing control groups consisting exclusively of predators with samples containing both predators and prey (Wilcoxon test: *, P < 0.05; #, P < 0.07). n.d., prey species for which u was not determined.
Prey utilization was assessed for prey strains that were susceptible to Lysobacter predation. Consistent with the observed killing efficiencies against Gram-positive bacteria, the populations of L. capsici and L. oryzae increased significantly in the presence of B. subtilis, R. rhodochrous, and L. lactis (Fig. 3B; see Table S2 in the supplemental material). Growth of L. enzymogenes, however, increased only when L. lactis was provided as a food source. The prey utilization of C. pseudoviolaceum by L. capsici was 23.7% ± 0.2% (mean ± 95% confidence interval) and that by L. oryzae was 26.6% ± 0.1%, indicating that both species benefitted equally from the presence of C. pseudoviolaceum (Wilcoxon test, P < 0.05 [n = 6]) (Fig. 3B). Similar observations were made when E. coli was used as prey. Under these conditions, only the growth of L. oryzae increased detectably (Fig. 3B).
For comparative purposes, Myxococcus fulvus also was included in this analysis and tested using the CFU-based predation assay. B. subtilis and E. coli were selected as prey, because they were susceptible to M. fulvus predation in both the prey spot predation assay and the lawn predation assay. In addition, C. pseudoviolaceum and R. rhodochrous were included as prey organisms, since the CFU-based predation assay had already revealed a larger prey spectrum for the tested Lysobacter spp. than was initially detected with the prey spot plate assay and the lawn predation assay. Since M. fulvus is a slow-growing bacterium, prey reduction was assessed after 24 h and 48 h of cocultivation. Surprisingly, M. fulvus completely failed to reduce the number of prey relative to that in control cultures (see Fig. S6 in the supplemental material).
Frequency dependence of predatory efficiency.
Outnumbering prey is an important feature of wolf pack predation (8). To investigate whether the predatory performance of Lysobacter depended on the predator/prey ratio (PPR), the CFU-based predation assay was repeated, but this time the number of prey cells (i.e., B. subtilis) was held constant, while initial numbers of predator cells were varied. When predator populations outnumbered the prey by 1,000:1 or 100:1, killing efficiency was very high, and an almost complete eradication of prey populations was observed (Fig. 4). Lowering the PPR to 10:1, however, led to a loss of killing efficiency for L. oryzae and L. enzymogenes, whereas L. capsici retained effective predation (e = 93.8% ± 4.0%). At a PPR of 1:1, no significant prey reduction was detectable for any of the three Lysobacter spp. tested. Thus, this experiment confirmed that the predatory success of the tested predators critically depended on their frequency relative to the number of prey bacteria.
FIG 4.
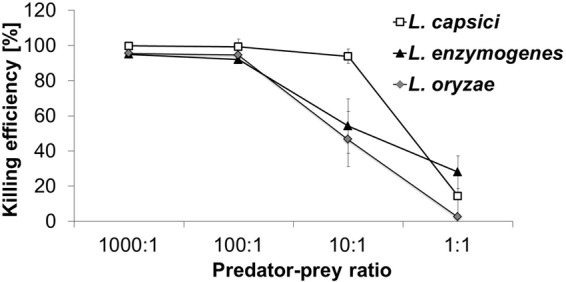
Frequency dependence of predatory efficiency. Shown are different predator/prey ratios versus the mean killing efficiency (ē; ± 95% confidence interval) of each predatory species. The number of B. subtilis CFU was held constant in all experiments.
Contact dependence of predatory behavior.
Finally, we set out to clarify whether the predatory activity of Lysobacter depends on physical proximity to its prey or whether it is mediated exclusively by extracellular factors, such as lytic enzymes or antibiotics. For this purpose, the killing of B. subtilis by Lysobacter spp. was compared to the effect of cell-free Lysobacter culture supernatants. Surprisingly, none of the tested supernatants affected the growth of B. subtilis (Fig. 5). The outcome of this experiment was the same irrespective of whether the supernatants originated from Lysobacter cultures grown under nutrient-rich or nutrient-deficient conditions. This suggests that cell contact is likely important for the lysis of prey by Lysobacter spp., thus corroborating previous observations (23, 43). Nevertheless, this conclusion does not exclude an involvement of degradative enzymes or antibiotics, whose production might be induced by the presence of prey.
FIG 5.
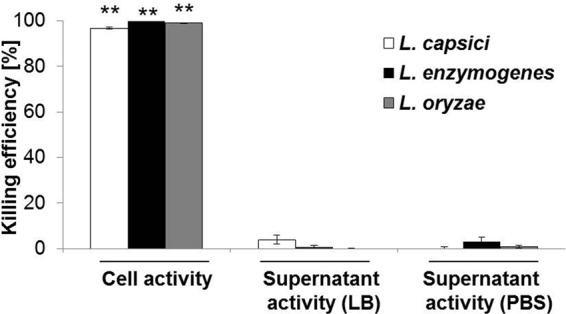
Contact dependence of predatory behavior. The killing of the prey bacterium B. subtilis by Lysobacter cells and culture supernatants was analyzed in the CFU-based predation assay. Asterisks denote significant differences between the number of prey CFU of the control group and samples containing both predator and prey (Mann-Whitney U test: **, P < 0.01; df = 2).
DISCUSSION
Although predatory behaviors pervade the entire bacterial realm, research in this area has focused on few taxonomic groups. In this context, especially facultative predators such as myxobacteria (7) as well as the obligate predator Bdellovibrio bacteriovorus (4, 5) have received the main attention. Assuming that Lysobacter spp. use a feeding strategy similar to that of myxobacteria (22), the predatory activity of three selected Lysobacter spp. was evaluated using two predation assays that had been previously established for myxobacteria (16, 17). In these assays, however, neither L. capsici nor L. oryzae displayed any lytic activity against the prey species tested. Also, L. enzymogenes showed only a relatively weak predatory activity compared to that of the myxobacterium M. fulvus. These results indicated that either (i) a different assay was needed for assessing their predatory activity or (ii) L. capsici and L. oryzae are not predatory bacteria.
To differentiate between these two possibilities, the selected Lysobacter strains were subjected to a CFU-based predation assay. Similar assays have been previously described to analyze the obligate bacterial predator Bdellovibrio by counting the numbers of plaques on plates after cocultivation with prey bacteria in liquid medium (44) as well as for quantifying the predatory efficiency of nonobligate myxobacterial predators (20). The initial setup of this assay required a cocultivation with prey bacteria that could be phenotypically distinguished from the predators using, for example, the pigmentation of their colonies. Subsequently, antibiotic resistance was used as an alternative labeling strategy, significantly extending the number of potential prey bacteria. Aside from measuring the killing of prey, the CFU-based predation assay enabled the simultaneous monitoring of growth of predators and prey. In this way, it was possible to exclude the possibility that the decline of prey resulted from competitive interactions (i.e., killing without feeding).
The CFU-based predation assay confirmed that the selected Lysobacter spp. were effectively feeding on C. pseudoviolaceum, R. rhodochrous, B. subtilis, and, to a reduced extent, also E. coli and L. lactis during 24 h of cocultivation. It is thus evident that Lysobacter can prey on both Gram-positive and Gram-negative bacteria. Since Lysobacter spp. often exhibit inhibitory effects against phytopathogenic fungi, they represent promising biocontrol agents (45, 46). It is noteworthy, however, that the three Lysobacter strains used in this study did not show any activity against the two phytopathogenic bacteria A. tumefaciens and R. solanacearum under the experimental conditions tested.
Further analyses revealed that to achieve high killing efficiencies, all Lysobacter strains required a numerical superiority over their prey, although they differed in their optimal PPRs. Overall, this suggested that the Lysobacter strains were restricted to group predation. This means that individual Lysobacter cells must work together to successfully kill their prey, which could be mediated, for instance, by the cooperative secretion of hydrolytic enzymes or antibiotics. Chemical analyses of Lysobacter spp. already have illuminated their huge potential for the production of antimicrobial agents (47). Among the antibiotics reported are inhibitors of cell wall biosynthesis, such as cephabacins (48) and tripropeptins (49), as well as a number of compounds which target the bacterial membrane (50–52). The strains used in this study are not yet known as antibiotic producers, although the biosynthesis of such compounds seems likely in light of previous investigations (26). In contrast to the case for myxobacteria (13), however, a clear causal link between antibiotic production and predation is still missing for Lysobacter spp.
In the CFU-based predation assay, the number of Lysobacter cells was more than 10 times higher than that of prey populations. Furthermore, both predator and prey were continuously mixed during the 24 h of cocultivation, contributing to a homogeneous distribution of diffusible lytic factors. On the basis of these findings, we hypothesize that the quorum of Lysobacter cells used was likely below the critical threshold in the prey spot plate and the lawn predation assays, and therefore predatory behavior was not observed. This result is consistent with earlier studies which showed that L. enzymogenes was unable to lyse cyanobacteria when the predator inoculum was less than 106 cells ml−1 (4). In case of Myxococcus, however, a much smaller predator concentration (i.e., a predator/prey ratio of 1:1) was sufficient to induce prey lysis (53). This finding is further corroborated by the results of the prey spot plate assay, in which M. fulvus was more efficient in lysing the prey organisms than L. enzymogenes despite a smaller initial inoculum (Fig. 1).
Some studies suggested that myxobacteria are single-cell hunters rather than wolf pack predators and that close proximity to the corresponding prey cells might be essential for them to penetrate and lyse prey colonies (7, 9). The M. fulvus strain tested here failed to exhibit predatory activity in the CFU-based predation assay. A possible explanation could be that myxobacterial cells do not just require close proximity to their prey but instead must establish physical contact with their prey for an extended period to promote lysis of prey cells. This condition seems to preclude effective lysis in the CFU-based predation assay, as the shaking of the liquid cocultures likely prevented effective predation.
L. capsici and L. oryzae exhibited no swarming behavior in this study, thereby limiting the use of the lawn predation and prey spot plate assays to analyze the predatory behavior of these bacteria. Obviously, Lysobacter spp. and myxobacteria do not show the same predation behavior. Nonetheless, further experiments are necessary to fully understand the predation mechanism used by these bacteria and to clarify the role of antibiotics in their predatory interactions. We expect the described CFU-based predation assay to facilitate studies on additional previously neglected predatory bacteria and assist in the quantitative evaluation of their predatory behavior.
Supplementary Material
ACKNOWLEDGMENTS
Research in the Secondary Metabolism of Predatory Bacteria and Experimental Ecology and Evolution research groups is supported by the DFG-funded graduate school Jena School for Microbial Communication (JSMC). Funding by the Volkswagen Foundation to Christian Kost is gratefully acknowledged.
We thank Franziska Müller von Klingspor for technical support, Ákos T. Kovács for providing plasmid pNZ8048, and Hirokazu Kage for providing R. solanacearum/pBHR1. Furthermore, we thank the three unnamed reviewers for their valuable input on improving the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01781-15.
REFERENCES
- 1.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 2.Lidicker WZ., Jr 1979. A clarification of interactions in ecological systems. Bioscience 29:475–477. doi: 10.2307/1307540. [DOI] [Google Scholar]
- 3.Abrams PA. 2000. The evolution of predator-prey interactions: theory and evidence. Annu Rev Ecol Syst 31:79–105. doi: 10.1146/annurev.ecolsys.31.1.79. [DOI] [Google Scholar]
- 4.Jurkevitch E, Davidov Y. 2007. Phylogenetic diversity and evolution of predatory prokaryotes, p 1–56. In Jurkevitch E. (ed), Predatory prokaryotes—biology, ecology, and evolution. Springer, Heidelberg, Germany. [Google Scholar]
- 5.Davidov Y, Jurkevitch E. 2004. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacterstarrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol 54:1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- 6.Casida LE. 1988. Nonobligate bacterial predation of bacteria in soil. Microb Ecol 15:1–8. doi: 10.1007/BF02012948. [DOI] [PubMed] [Google Scholar]
- 7.Berleman JE, Kirby JR. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev 33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin MO. 2002. Predatory prokaryotes: an emerging research opportunity. J Mol Microbiol Biotechnol 4:467–477. [PubMed] [Google Scholar]
- 9.McBride MJ, Zusman DR. 1996. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiol Lett 137:227–231. doi: 10.1111/j.1574-6968.1996.tb08110.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol 1:45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- 11.Hillesland KL, Lenski RE, Velicer GJ. 2007. Ecological variables affecting predatory success in Myxococcus xanthus. Microb Ecol 53:571–578. doi: 10.1007/s00248-006-9111-3. [DOI] [PubMed] [Google Scholar]
- 12.Rosenberg E, Varon M. 1984. Antibiotics and lytic enzymes, p 109–125. In Rosenberg E. (ed), Myxobacteria: development and cell interactions. Springer, New York, NY. [Google Scholar]
- 13.Xiao Y, Wei X, Ebright R, Wall D. 2011. Antibiotic production by Myxobacteria plays a role in predation. J Bacteriol 193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichenbach H, Höfle G. 1993. Biologically active secondary metabolites from myxobacteria. Biotech Adv 11:219–277. doi: 10.1016/0734-9750(93)90042-L. [DOI] [PubMed] [Google Scholar]
- 15.Schieferdecker S, König S, Weigel C, Dahse HM, Werz O, Nett M. 2014. Structure and biosynthetic assembly of gulmirecins, macrolide antibiotics from the predatory bacterium Pyxidicoccus fallax. Chem Eur J 20:15933–15940. doi: 10.1002/chem.201404291. [DOI] [PubMed] [Google Scholar]
- 16.Pham VD, Shebelut CW, Diodati ME, Bull CT, Singer M. 2005. Mutations affecting predation ability of the soil bacterium Myxococcus xanthus. Microbiology 151:1865–1874. doi: 10.1099/mic.0.27824-0. [DOI] [PubMed] [Google Scholar]
- 17.Berleman JE, Chumley T, Cheung P, Kirby JR. 2006. Rippling is a predatory behavior in Myxococcus xanthus. J Bacteriol 188:5888–5895. doi: 10.1128/JB.00559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berleman JE, Kirby JR. 2007. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J Bacteriol 189:5675–5682. doi: 10.1128/JB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan AD, MacLean RC, Hillesland KL, Velicer GJ. 2010. Comparative analysis of Myxococcus predation on soil bacteria. Appl Environ Microbiol 76:6920–6927. doi: 10.1128/AEM.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller S, Strack SN, Hoefler BC, Straight PD, Kearns DB, Kirby JR. 2014. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol 80:5603–5610. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen P, Cook FD. 1978. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol 28:367–393. doi: 10.1099/00207713-28-3-367. [DOI] [Google Scholar]
- 22.Reichenbach H. 2006. The genus Lysobacter, p 939–957. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer, Berlin, Germany. [Google Scholar]
- 23.Shilo M. 1970. Lysis of blue-green algae by myxobacter. J Bacteriol 104:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daft MJ, Stewart WDP. 1971. Bacterial pathogens of freshwater blue-green algae. New Phytol 70:819–829. doi: 10.1111/j.1469-8137.1971.tb02582.x. [DOI] [Google Scholar]
- 25.Nett M, König GM. 2007. The chemistry of gliding bacteria. Nat Prod Rep 24:1245–1261. doi: 10.1039/b612668p. [DOI] [PubMed] [Google Scholar]
- 26.Park JH, Kim R, Aslam Z, Jeon CO, Chung YR. 2008. Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int J Syst Evol Microbiol 58:387–392. doi: 10.1099/ijs.0.65290-0. [DOI] [PubMed] [Google Scholar]
- 27.Puopolo G, Raio A, Zoina A. 2010. Identification and characterization of Lysobacter capsici strain pg4: a new plant health-promoting rhizobacterium. J Plant Pathol 92:157–164. [Google Scholar]
- 28.Begunova EA, Stepnaya OA, Tsfasman IM, Kulaev IS. 2004. The effect of the extracellular bacteriolytic enzymes of Lysobacter sp. on Gram-negative bacteria. Mikrobiologiia 73:267–270. [PubMed] [Google Scholar]
- 29.Lueders T, Kindler R, Miltner A, Friedrich MW, Kaestner M. 2006. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl Environ Microbiol 72:5342–5348. doi: 10.1128/AEM.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínková L, Uhnáková B, Pátek M, Nešvera J, Kren V. 2009. Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177. doi: 10.1016/j.envint.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Goethals K, Vereecke D, Jaziri M, Montagu VM, Holsters M. 2001. Leafy gall formation by Rhodococcus fascians. Annu Rev Phytopathol 39:27–52. doi: 10.1146/annurev.phyto.39.1.27. [DOI] [PubMed] [Google Scholar]
- 33.Kämpfer P, Busse HJ, Scholz HC. 2009. Chromobacterium piscinae sp. nov. and Chromobacterium pseudoviolaceum sp. nov., from environmental samples. Int J Syst Evol Microbiol 59:2486–2490. doi: 10.1099/ijs.0.008888-0. [DOI] [PubMed] [Google Scholar]
- 34.Casida LE. 1980. Bacterial predators of Micrococcus luteus in soil. Appl Environ Microbiol 39:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madigan MT, Martinko JM. 2006. Brock biology of microorganisms, 11th ed Pearson Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- 36.Kuipers OP, Ruyter PG, Kleerebezem M, Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 37.Dower WJ. 1990. Electroporation of bacteria: a general approach to genetic transformation, p 275–296. In Setlow JK. (ed), Genetic engineering—principles and methods, vol 12 Plenum Publishing Corp., New York, NY. [DOI] [PubMed] [Google Scholar]
- 38.Xue G-P, Johnson JS, Dalrymple BP. 1999. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J Microbiol Methods 34:183–191. doi: 10.1016/S0167-7012(98)00087-6. [DOI] [Google Scholar]
- 39.Holo H, Nes IF. 1989. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh BN. 1947. Myxobacteria in soils and composts; their distribution, number and lytic action on bacteria. J Gen Microbiol 1:1–10. doi: 10.1099/00221287-1-1-1. [DOI] [PubMed] [Google Scholar]
- 41.Mendes-Soares SH, Velicer GJ. 2013. Decomposing predation: testing for parameters that correlate with predatory performance by a social bacterium. Microb Ecol 65:415–423. doi: 10.1007/s00248-012-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan RF, Holtman MA, Zylstra GJ, White JF, Kobayashi DY. 2003. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J Appl Microbiol 94:1079–1086. doi: 10.1046/j.1365-2672.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- 43.Jurkevitch E. 2007. Predatory behaviors in bacteria—diversity and transitions. Microbe 2:67–73. [Google Scholar]
- 44.Stolp H, Starr MP. 1963. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Jochum CC, Yu F, Zaleta-Rivera K, Du L, Harris SD, Yuen GY. 2008. An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology 98:695–701. doi: 10.1094/PHYTO-98-6-0695. [DOI] [PubMed] [Google Scholar]
- 46.Puopolo G, Giovannini O, Pertot I. 2014. Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiol Res 169:7–8. [DOI] [PubMed] [Google Scholar]
- 47.Xie Y, Wright S, Shen Y, Du L. 2012. Bioactive natural products from Lysobacter. Nat Prod Rep 29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brakhage AA, Al-Abdallah Q, Tüncher A, Spröte P. 2005. Evolution of β-lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry 66:1200–1210. doi: 10.1016/j.phytochem.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 49.Hashizume H, Sawa Harada S, Igarashi M, Adachi H, Nishimura Nomoto A. 2011. Tripropeptin C blocks the lipid cycle of cell wall biosynthesis by complex formation with undecaprenyl pyrophosphate. Antimicrob Agents Chemother 55:3821–3828. doi: 10.1128/AAC.00443-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonner DP, O'Sullivan J, Tanaka SK, Clark JM, Whitney RR. 1988. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. II. Biological properties. J Antibiot 41:1745–1751. [DOI] [PubMed] [Google Scholar]
- 51.Kato A, Nakaya S, Ohashi Y, Hirata H. 1997. WAP-8294A2, a novel anti-MRSA antibiotic produced by Lysobacter sp. J Am Chem Soc 119:6680–6681. doi: 10.1021/ja970895o. [DOI] [Google Scholar]
- 52.Hamamoto H, Urai M, Ishii K, Yasukawa J, Paudel A, Murai M, Kaji T, Kuranaga T, Hamase K, Katsu T, Su J, Adachi T, Uchida R, Tomoda H, Yamada M, Souma M, Kurihara H, Inoue M, Sekimizu K. 2015. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat Chem Biol 11:127–133. doi: 10.1038/nchembio.1710. [DOI] [PubMed] [Google Scholar]
- 53.Burnham JC, Collart SA, Melvin J. 1984. Myxococcal predation of the cyanobacterium Phormidium luridum in aqueous environments. Arch Microbiol 137:220–225. doi: 10.1007/BF00414547. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.