Abstract
Massive blooms of toxic cyanobacteria frequently occur in the central Baltic Sea during the summer. In the surface scum, cyanobacterial cells are exposed to high light (HL) intensity, high oxygen partial pressure and other stresses. To mimic these conditions, cultures of Nodularia spumigena CCY9414, which is a strain isolated from a cyanobacterial summer bloom in the Baltic Sea, were incubated at a HL intensity of 1200 μmol photons m−2 s−1 or a combination of HL and increased oxygen partial pressure. Using differential RNA sequencing, we compared the global primary transcriptomes of control and stressed cells. The combination of oxidative and light stresses induced the expression of twofold more genes compared with HL stress alone. In addition to the induction of known stress-responsive genes, such as psbA, ocp and sodB, Nodularia cells activated the expression of genes coding for many previously unknown light- and oxidative stress-related proteins. In addition, the expression of non-protein-coding RNAs was found to be stimulated by these stresses. Among them was an antisense RNA to the phycocyanin-encoding mRNA cpcBAC and the trans-encoded regulator of photosystem I, PsrR1. The large genome capacity allowed Nodularia to harbor more copies of stress-relevant genes such as psbA and small chlorophyll-binding protein genes, combined with the coordinated induction of these and many additional genes for stress acclimation. Our data provide a first insight on how N. spumigena became adapted to conditions relevant for a cyanobacterial bloom in the Baltic Sea.
Introduction
Cyanobacteria have an important role as primary producers in the global carbon cycle. In the central oceans, picoplanktonic cyanobacteria of the genera Synechococcus and Prochlorococcus represent the main carbon fixers (Scanlan et al., 2009). In addition, cyanobacterial species capable of fixing atmospheric N2 contribute substantial amounts of combined nitrogen to the marine food web (Zehr, 2011). Cyanobacteria also represent an important part of the phytoplankton communities in coastal, brackish and freshwater ecosystems. Many of these cyanobacteria can also produce bioactive compounds, which may have allelopathic activities against other phytoplankton organisms (Paz-Yepes et al., 2013) and often exhibit toxic effects on animals, including humans (Pearson et al., 2010). Mass developments (‘blooms') of toxic cyanobacteria in coastal waters or freshwater systems have a dramatic impact on the use of these water bodies for recreational or drinking water purposes. It has been predicted that the expected future increases in global temperatures and atmospheric CO2 concentrations will lead to the even further stimulation of toxic cyanobacterial blooms (Paerl and Huisman, 2008; Gehringer and Wannicke, 2014).
The Baltic Sea represents a brackish water ecosystem. Massive blooms of toxic cyanobacteria occur in its central regions almost every summer. These blooms are often dominated by N. spumigena, which is a filamentous cyanobacterium that produces a cocktail of bioactive compounds including the hepatotoxin nodularin (Fewer et al., 2013; Mazur-Marzec et al., 2013). N. spumigena is capable of N2 fixation in specialized cells that are termed heterocysts (Sivonen et al., 2007; Ploug et al., 2011). However, the molecular basis for bloom formation is not completely understood. It is thought that excess phosphorus combined with very low nitrogen concentrations favor the growth and bloom formation of N2-fixing cyanobacteria during the summer season (Sellner, 1997). This phenomenon particularly occurs under stably stratified warm water conditions. Then, gas vesicles that provide buoyancy to N. spumigena and related cyanobacteria lead to the formation of large surface scums in the absence of mixing.
Hence, Nodularia seems to have a selective advantage in the virtually nitrogen-free conditions of the Baltic Sea, which occur during the summer as a diazotrophic cyanobacterium that has adapted to brackish waters (Möke et al., 2013). The cyanobacterial filaments in the surface layer are exposed to extreme environmental conditions, such as high light (HL), high oxygen partial pressure and low nutrients, including iron and CO2. The acclimation of cyanobacteria to such conditions can be analyzed by transcriptomic techniques. Gene expression changes under HL and oxidative stress have been reported for unicellular, non-diazotrophic model cyanobacteria, such as Synechocystis sp. PCC 6803 (from here: Synechocystis) and Synechococcus sp. PCC 7002 (Hihara et al., 2001; Los et al., 2008; Ludwig and Bryant, 2012; Kopf et al., 2014), but not for representatives of bloom-forming marine strains. Recently, small non-protein-coding RNAs (ncRNAs) have been identified in cyanobacteria that are also regulated by environmental factors (Sakurai et al., 2012; Georg et al., 2014).
Investigations with model cyanobacteria showed that the D1 protein of photosystem 2 (PS2) represents the prime target for inactivation via HL-induced oxidative stress. In almost all cyanobacteria, the D1 protein is encoded by a small gene family that comprises D1 protein forms that are preferentially used under different light and oxygen stress conditions (reviewed by Mulo et al., 2012). Moreover, a multitude of protective measures interact to prevent extensive PS2 damage. These strategies include the non-photochemical quenching achieved via the orange carotenoid protein (Kirilovsky and Kerfeld, 2012), the protective actions of special flavodiiron proteins (Allahverdiyeva et al., 2013) and the activation of photorespiration under very low CO2 and high O2 partial pressure (Hackenberg et al., 2009; Allahverdiyeva et al., 2011).
In general, longer exposure to combined HL and oxidative stress is very unfavorable for oxygenic phototrophs and can only be tolerated by specialized organisms, such as bloom-forming cyanobacteria. Only recently, the first nearly complete genome sequence (Genbank accession number CP007203) of a cyanobacterium isolated from a Baltic Sea summer bloom has become available (Voß et al., 2013). These data serve as a foundation for molecular studies of bloom formation using N. spumigena strain CCY9414 (hereafter referred to as Nodularia CCY9414) as a model. Here we applied the differential RNA sequencing approach (Sharma et al., 2010), which provides comprehensive information on active transcriptional start sites. We analyzed global transcriptomic changes in Nodularia CCY9414 after exposure to HL and oxidative stress, which are characteristic for the situation in a surface scum, to identify specific adaptations to the bloom-forming lifestyle in the Nodularia CCY9414 genome. Our hypothesis was that those genes are not necessarily only expressed under these conditions in the bloom-forming cyanobacterium, but could be identified via comparison with data from Synechocystis, because they should not be present or differently regulated in cells of the non-blooming model cyanobacterium.
Materials and methods
Strain and culture conditions
N. spumigena strain CCY9414 was obtained from the Culture Collection Yerseke (CCY) at the Royal Netherlands Institute for Sea Research. It was isolated from surface waters of the Baltic Sea, east of the island of Bornholm (Hayes and Barker, 1997). Standard cultivation of axenic Nodularia CCY9414 cells was performed in cell culture bottles under N2-fixing conditions with nitrate-free ASN III medium (Rippka et al., 1979) supplemented with NaCl (the standard medium contained 12.5 g NaCl per liter of ASN III). The cultures were mixed daily and left to grow for 2 weeks under a 16/8 h light/dark cycle (40 μmol photons m−2 s−1 of light) at 20 °C. Cells of the glucose-tolerant unicellular model strain Synechocystis sp. PCC 6803 were grown in CO2-supplemented BG11 medium under standard conditions, as described previously (Hackenberg et al., 2009).
For the stress experiments, Nodularia CCY9414 cells cultivated under the above mentioned standard conditions (that is, N2-fixing cells in ASN III containing 12.5 g l−1 NaCl) were used, and one aliquot was collected immediately (control). The second aliquot of cells was exposed to 1 200 μmol photons m−2 s−1 of HL (the light source was a Leica Pradovit P150 slide projector, Leica, Braunschweig, Germany) without gassing (HL stress). The third aliquot of cells was exposed to increased oxygen partial pressure via the streaming of air enriched with 40% oxygen (GMS 600 2CH, QCAL gas mixing unit, QCAL, Oberostendorf, Germany) through the cell suspension at the control light intensity of 40 μmol photons m−2 s−1 (oxygen stress, +O2). The fourth aliquot was exposed to HL conditions of 1200 μmol photons m−2 s−1 and was additionally gassed by a stream of 40% oxygen (combined oxygen/HL stress, HL+O2). Samples were collected at defined time intervals (0.5, 3 and 8 h) after the onset of the stress conditions.
Photosynthetic activity
The photosynthetic activity of Nodularia CCY9414 and Synechocystis was measured as O2 production using Hansatech oxygen electrodes (Oxygraph, Norfolk, UK). A total of 4 ml of cell suspension containing ∼3 μg of chlorophyll a per ml was used for measurements at 20 °C and 30 °C corresponding to the growth temperatures of Nodularia and Synechocystis, respectively. The cells were exposed for 5 min to light of defined photon flux rates (25–1400 μmol photons m−2 s−1). Respiratory oxygen consumption was measured for 5 min in the dark, in the cells exposed to the lowest light intensity. Chlorophyll a was extracted from 4 ml of cells with 90% acetone and quantified according to Arnon (1949). The soluble amino acid concentrations were determined as described by Hagemann et al. (2005).
RNA isolation and RT-PCR
Aliquots of 50 ml of cells obtained from the stress experiments were collected by quick filtration through sterile glass fiber filters (Whatman GF/F) that were immediately frozen in liquid nitrogen and stored at −80 °C. Nodularia CCY9414 total RNA was isolated using a Total RNA Isolation Kit for plants (Macherey-Nagel, Düren, Germany) as described previously (Voß et al., 2013). Expression of the selected genes was analyzed using reverse-transcription PCR (RT-PCR). The RevertAid cDNA Synthesis Kit (MBI Fermentas, St. Leon-Rot, Germany) was used to generate cDNA from 1 μg of DNA-free RNA, using random hexamers as primers. The cDNA amounts were normalized to the constitutively expressed rnpA gene (ribonuclease P, NSP_30860). The PCR products were separated in 1% agarose gels and stained with ethidium bromide. The gene-specific primers are listed in Supplementary Table S1.
RNA sequencing
For transcriptome analysis, cDNA libraries were constructed (vertis Biotechnologie AG, Freising, Germany) and analyzed on an Illumina HiSeq 2000 sequencer as previously described (Mitschke et al., 2011a, 2011b). In brief, total RNA was enriched for primary transcripts by treatment with Terminator-5′-phosphate-dependent exonuclease (Epicentre, Madison, WI, USA). Then, 5′ppp-RNA was cleaved enzymatically using tobacco acid pyrophosphatase. The ‘de-capped' RNA was ligated to an RNA linker and first-strand cDNA synthesis was initiated by random priming. Second-strand cDNA synthesis was primed with a biotinylated antisense 5′-Solexa primer, after which cDNA fragments were bound to streptavidin beads. The bead-bound cDNA was blunted and 3′-ligated to a Solexa adapter. The cDNA fragments were amplified by 22 cycles of PCR. For Illumina HiSeq 2000 (San Diego, CA, USA) analysis (100- bp read lengths), cDNAs in the size range of 200–500 bp were eluted from a preparative agarose gel. Similar numbers of reads were obtained for each cDNA library, 41 519 905 reads from the control cells, 38 851 231 reads from the HL-stressed cells and 39 503 192 reads from the combined HL+O2 stress conditions. For normalization, the two stress libraries were scaled to the same library size as the control. Transcriptional start sites (TSSs) were predicted as previously described (Voß et al., 2013), but with a higher read threshold of 620 normalized counts. The classification of the TSSs into gTSSs, iTSSs, aTSSs and nTSSs was carried out as previously described allowing a maximal distance of 200 nt to a downstream gene for the classification as gTSS (Mitschke et al., 2011b).
For the detection of differentially expressed TSSs, the raw counts from all TSS positions were analyzed with the Analysis of Sequence Counts (ASC) approach (Wu et al., 2010). Genes were considered to be differentially expressed, when a fold change >2 or <0.5 was detected between a stress condition and the control, with a probability of at least 0.95. The data were deposited into the NCBI Sequence Read Archive under the following accession numbers: control, SRR696127; O2+HL, SRR1571498; and HL, SRR1572178.
Results and discussion
Photosynthetic activity under high irradiances
To test the hypothesis that bloom-forming cyanobacteria are better adapted than established model cyanobacteria to environmental stresses such as HL that are characteristic for cells in the surface scum, we compared the photosynthesis at different light intensities (0–1400 μmol photons m−2 s−1) of Nodularia CCY9414 with that of Synechocystis. The oxygen evolution rate of Nodularia CCY9414 was significantly higher than that of Synechocystis from 50 μmol photons m−2 s−1 upwards at 20 °C and from 100 μmol photons m−2 s−1 upwards at 30 °C (Supplementary Figure S1a). At both temperatures, Nodularia CCY9414 exhibited the greatest difference from Synechocystis at the highest measured light intensity. The results of this experiment supported the hypothesis that Nodularia CCY9414 has a better capability for using HL intensities for photosynthesis.
To gain insight into the superior light resistance of bloom-forming cyanobacteria, we analyzed Nodularia CCY9414 under HL stress, oxidative stress induced by doubling the amount of oxygen partial pressure (+O2) and under combined light and oxidative stress (HL+O2) conditions. These stress conditions stimulated photorespiratory activity in Nodularia CCY9414 as indicated by the increased glycine/serine ratio (Supplementary Figure S1b), which may be considered as a proxy for increased photorespiration (Allaverdiyeva et al., 2011). In the cells exposed to the +O2 stress, the highest ratios were detected after 0.5 or 3 h of stress, after which the levels began to decrease. We interpret these findings as an indication of the acclimation of Nodularia CCY9414 to the longer-term stress conditions. An increased photorespiratory flux has also been reported for cells of the bloom-forming freshwater cyanobacterium Microcystis aeruginosa under HL stress, whereas Synechocystis cells do not exhibit such behavior (Meissner et al., 2014).
Stress-related accumulation of selected transcripts
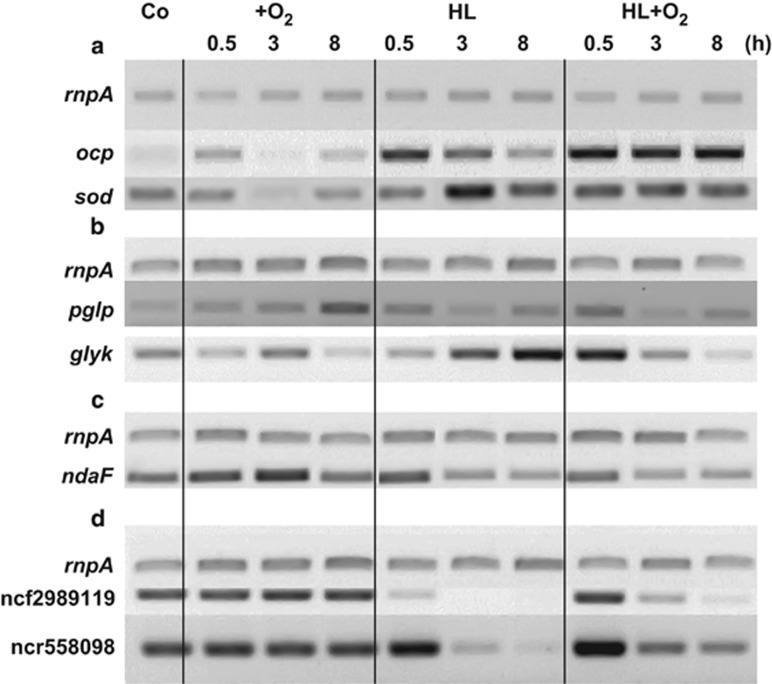
RNA isolated from Nodularia CCY9414 cells exposed to a time course of the studied stress conditions was used for an initial qualitative characterization of the stress response by RT-PCR (Figure 1). Transcript levels of the genes sod and ocp, which are known to be induced following exposure to HL and +O2 stress in several cyanobacteria (Hihara et al., 2001), increased under all of the selected stress treatments. Under the combined HL+O2 stress, ocp showed a higher level of induction compared with sod, but the expression of the two genes remained elevated during the entire stress period. Under HL, ocp expression was immediately enhanced and then exhibited a declining trend following longer periods of HL exposure, whereas sod transcript accumulation peaked at 3 h. The exposure of Nodularia to the single +O2 stress condition caused only small changes on ocp and sod expression (Figure 1a).
Figure 1.
Semi-quantitative analysis of the expression of selected genes in cells of N. spumigena CCY9414 by RT-PCR. Shown are RT-PCR results after exposure to an enhanced oxygen concentration (+O2; 40%), HL (1200 μmol photons m−2 s−1) or both stresses combined (HL+O2) for a period of 0.5, 3 or 8 h. RNase P mRNA (rnpA) was amplified for control. Expression of these genes in control cells (Co) is shown in the first lane. (a) Transcript accumulation for the orange carotenoid protein (ocp) and the superoxide dismutase subunit B (sodB) as markers for oxidative stress. (b) Transcript accumulation for the phosphoglycolate phosphatase (glgp) and glycerate 3-kinase (glyk) as markers for photorespiration. (c) Transcript accumulation for the nodularin synthase gene F (ndaF) as marker for the toxin synthesis. (d) Transcript accumulation for selected ncRNAs.
Moreover, we analyzed the expression of genes coding for selected enzymes of photorespiratory metabolism, including pglp, which codes for the entrance enzyme 2-phosphoglycolate phosphatase, and glyk, which codes for the last enzyme in the pathway, glycerate 3-kinase (Eisenhut et al., 2008). In particular, the glyk gene showed increased expression under the HL and HL+O2 stresses (Figure 1b), whereas pglp induction was less obvious. Finally, we analyzed the expression of ndaF, which encodes one subunit of nodularin synthase (Moffitt and Neilan, 2004). Its expression was found to be stimulated after oxidative and HL stresses (Figure 1c), corresponding with the increased amount of toxins that have been reported in cyanobacterial communities under similar conditions (Gehringer and Wannicke, 2014).
Comparative transcriptome analysis
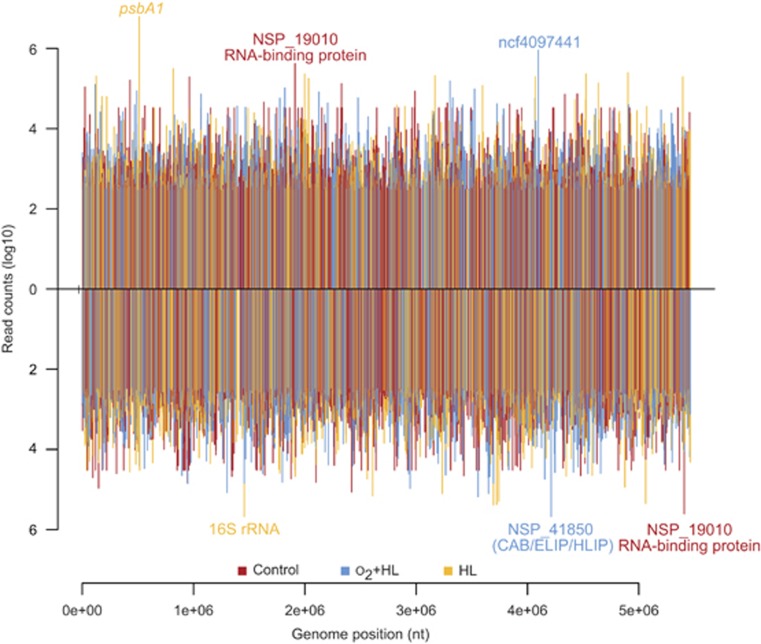
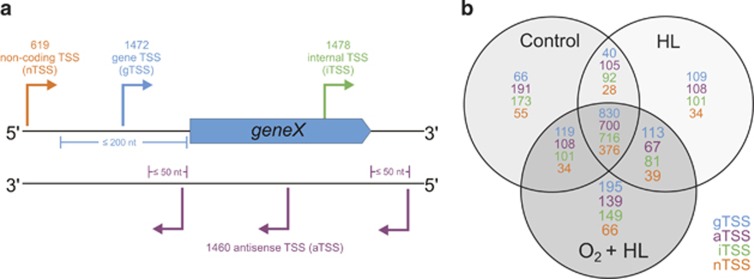
Initial RT-PCR analyses indicated that HL and especially the HL+O2 stress, resulted in the expected cellular response. Therefore, we compared gene expression in non-stressed cells (control) and cells exposed to the studied stress conditions by the transcriptome-wide mapping of TSSs. A total of 38 851 231 sequence reads were obtained for the HL-stressed sample and 39 503 192 reads for the sample exposed to HL+O2 (Supplementary Table S2). These data were compared with a total of 41 519 905 sequence reads from control cells. To exclude very weak or possible false-positive TSSs from this comparison, we chose a more stringent TSS threshold of 620 reads, reducing the number of TSSs from the initial 6519 (Voß et al., 2013) to 3734 for the control cells. However, we also identified 849 and 746 previously unknown additional TSSs that were active after 3 h HL and 0.5 h HL+O2, respectively. Among them, 300 TSSs were stimulated by both stresses, altogether leading to a total of 5029 TSSs in this study (Supplementary Table S3). The activation of a high number of additional TSSs under various stress conditions has also been found for Synechocystis (Kopf et al., 2014). The highest expression levels in Nodularia CCY9414 (measured as absolute read counts) were found for the 16S rRNA operons, several ribosomal protein genes, the stress-induced genes psbA and NSP_41850, which codes for a chlorophyll-binding protein (CAB)/ELIP/HL-inducible protein family, as well as the ncRNA ncf4097441 (Figure 2). The TSSs were classified as gTSSs (protein-coding mRNAs), iTSSs (TSSs within protein-coding regions), aTSSs (for antisense RNAs) and nTSSs (for ncRNAs within intergenic regions) (Table 1, Figure 3 and Supplementary Table S2).
Figure 2.
Global overview of all TSS with >620 reads along the chromosome draft sequence (upper part, forward strand; lower part, reverse strand). The TSSs are color-coded according to the respective condition in which they were maximal and selected examples are annotated. For details of all mapped TSSs, see Supplementary Table S3.
Table 1. Overview of total numbers of the TSS mapping (relative proportion in brackets in %) via the differential RNAseq approach.
| Control | HL+O2 stress | HL stress | Total | |
|---|---|---|---|---|
| gTSS | 1055 (28.25) | 1257 (32.79) | 1092 (30.06) | 1472 (29.27) |
| aTSS | 1104 (29.57) | 1014 (26.45) | 1022 (28.13) | 1460 (29.03) |
| iTSS | 1084 (29.03) | 1049 (27.37) | 1055 (29.04) | 1478 (29.39) |
| nTSS | 491 (13.15) | 513 (13.38) | 464 (12.77) | 619 (12.31) |
| Sum | 3734 | 3833 | 3633 | 5029 |
Abbreviations: HL, high light; RNAseq, RNA sequencing; TSS, transcription start site.
Figure 3.
Definition and specificities of the identified TSSs. (a) Details of annotation and classification of 5029 TSS into 1472 putative gTSS giving rise to mRNA, 1460 aTSS producing asRNA, 1478 iTSS for internal sense transcripts and 619 nTSS for candidate ncRNAs. aTSS was classified as gTSS when the TSS was located 1–200 nt upstream and in the same orientation as an open reading frame (ORF). TSSs located antisense to an annotated gene or within ≤50 bp 5′ or 3′ to it on the reverse strand were classified as aTSS. TSSs positioned within an annotated gene were classified as iTSS and all remaining TSSs as nTSS from which ncRNAs originate. (b) Overlaps and specificities of the different types of TSSs in the three compared conditions.
Recently, the activities of thousands of TSSs have been studied in the marine filamentous cyanobacterium Trichodesmium erythraeum, in which 26.7% of the mapped 6080 TSSs active under standard growth conditions were nTSSs (Pfreundt et al., 2014). However, the 12.4% nTSSs in Nodularia CCY9414 observed in this study is similar to what has been previously reported in other cyanobacteria, for example, 12.1% in Synechocystis (Kopf et al., 2014) and 10.3% in Anabaena PCC7120 (Mitschke et al., 2011b). Thus, the composition of the Nodularia CCY9414 primary transcriptome resembles that of other cyanobacteria, whereas that of the Trichodesmium transcriptome differs.
The full list of 5029 identified TSSs together with their expression levels are presented in Supplementary Table S3. We have provided a visualization of all mapped reads alongside the genome, including all identified TSSs, ncRNAs and asRNAs, as Supplementary Data 1, which is searchable with Nodularia CCY9414 locus tags and gene names.
Our data set allowed for the identification of strongly up- and downregulated TSSs under the HL and combined HL+O2 stress conditions (Supplementary Tables S4 and S5). Using a 3-fold change in expression as threshold, 30 gTSSs showed increased transcription under both stresses (Table 2). A much higher number of gTSSs (105) responded specifically to the combined application of the HL+O2 stress (Table 3), whereas only 37 gTSSs are specifically regulated by HL alone (Table 4). These changes were statistically robust as revealed by the empirical Bayes analysis of sequence counts (Wu et al., 2010). More genes were likely undergoing increased transcription, particularly those situated downstream in operons; however, our method only detected the 5′-ends of the primary transcripts. Orthologs for 18 out of the 29 genes listed in Table 2 have also been shown to be HL-induced in the model cyanobacterium Synechocystis (comparisons were performed using the CyanoEXpress database: http://193.136.227.181/cgi-bin/gx2?n=environmental; Hernandez-Prieto and Futschik, 2012). These similarities indicate that our stress conditions and time points were carefully selected and allowed for a reliable characterization of the transcriptome associated with stress acclimation in Nodularia CCY9414.
Table 2. Protein-coding genes whose TSS showed a more than threefold increase in the number of reads.
| Protein in Nodularia CCY9414 | Absolute read counts (control) | FC HL+O2/control | ASC DE probability O2+HL/control | FC HL/control | ASC DE probability HL/control | Annotation | ORF in 6803 (e-value) |
|---|---|---|---|---|---|---|---|
| NSP_44260 | 1 | 555 | 1.0 | 542.5 | 1.0 | Hypothetical protein | Slr0270 (e−34) |
| NSP_9140 | 47 | 83.7 | 1.0 | 54.5 | 1.0 | Processing peptidase-like protein | Sll0055 (e−163) |
| NSP_27800 | 293 | 57.6 | 1.0 | 5.2 | 1.0 | CAB/ELIP/HLIP superfamily protein | Ssr2595HL (e−29) |
| NSP_44180 | 725 | 38 | 1.0 | 19.3 | 1.0 | NADH dehydrogenase I subunit 4. Involved in cyclic electron flow | Slr1291HL (0.0) |
| NSP_4910 | 1175 | 26.3 | 1.0 | 10.9 | 1.0 | Ubiquinone biosynthesis monooxygenase UbiB, or ABC1-domain containing ABC transporter | Sll0005HL (0.0) |
| NSP_48910 | 988 | 4.1 | 1.0 | 23.8 | 1.0 | CAB/ELIP/HLIP superfamily protein | Ssl1633HL (e−14) |
| NSP_32430 | 7791 | 19.6 | 1.0 | 9.2 | 1.0 | Orange carotenoid protein | Slr1963HL (e−130) |
| NSP_46960 | 189 | 14.9 | 1.0 | 19.3 | 1.0 | Zn-dependent proteases | Sll0528HL (e−137) |
| NSP_9460 | 409 | 14.2 | 1.0 | 8 | 1.0 | Phycoerythrin linker protein CpeS, or, chromophore lyase CpcS | Slr2049 (e−10) |
| NSP_14420 | 2282 | 7.6 | 1.0 | 14.2 | 1.0 | Photosystem II protein D1 (psbA3 gene) | Slr1311 (0.0) Sll1867HL (0.0) |
| NSP_5400 | 2170 | 12 | 1.0 | 5.8 | 1.0 | Recombination protein RecR | None |
| NSP_39950 | 396 | 11.8 | 1.0 | 6.1 | 1.0 | Solanesyl diphosphate synthase | Slr0611HL (e−125) |
| NSP_51570 | 135 | 10.2 | 1.0 | 9 | 1.0 | 2-Octaprenyl-6-methoxyphenol hydroxylase | Slr1300 (e−146) |
| NSP_44510 | 557 | 7.5 | 1.0 | 9.8 | 1.0 | Adenosine deaminase | None |
| NSP_3210 | 249 | 4.6 | 1.0 | 9 | 1.0 | Hypothetical protein | Slr0217 (e−42) |
| NSP_41850 | 54170 | 8.7 | 1.0 | 5.8 | 1.0 | CAB/ELIP/HLIP superfamily protein | Ssr2595HL (e−25) |
| NSP_42110 | 4661 | 8.1 | 1.0 | 6.8 | 1.0 | Beta-carotene ketolase/hydroxylase, or, fatty acid desaturase | Sll1468 (e−05) |
| NSP_33570 | 2709 | 7 | 1.0 | 5.5 | 1.0 | Photosystem II protein D1 (psbA4 gene) | Slr1311 (0.0) Sll1867HL (0.0) |
| NSP_9820 | 696 | 6.4 | 1.0 | 3.5 | 1.0 | DNA-binding, ferritin-like protein Dps (oxidative damage) | Slr1894 (e−09) |
| NSP_17520 | 6085 | 3.1 | 1.0 | 6.4 | 1.0 | CAB/ELIP/HLIP superfamily protein | Ssl1633HL (e−16) |
| NSP_46010 | 22114 | 5.4 | 1.0 | 6.2 | 1.0 | CAB/ELIP/HLIP superfamily protein | Ssl1633HL (e−15) |
| NSP_32830 | 789 | 3.5 | 1.0 | 5.4 | 1.0 | GTP cyclohydrolase I | Slr0426HL (e−100) |
| NSP_9990 | 499 | 4.8 | 1.0 | 5.3 | 1.0 | Peptidoglucan-binding protein | None |
| NSP_48320 | 696 | 3.3 | 1.0 | 5.3 | 1.0 | CopG family transcriptional regulator | Ssr5117 (e−26) |
| NSP_7660 | 347 | 5.2 | 1.0 | 4.8 | 1.0 | Chaperone protein DnaJ | Sll0897HL (e−177) |
| NSP_29690 | 677 | 4.5 | 1.0 | 4.5 | 1.0 | Hypothetical protein; possible iron–sulfur cluster-binding protein | Sll1697HL (e−166) |
| NSP_31650 | 442 | 3.5 | 1.0 | 4.3 | 1.0 | Superoxide dismutase [Fe] | Slr1516HL (e−88) |
| NSP_8400 | 427 | 4.1 | 1.0 | 3.4 | 1.0 | Hypothetical protein | None |
| NSP_1700 | 607 | 3.2 | 1.0 | 4.1 | 1.0 | Beta-carotene hydroxylase | Sll1468HL (e−148) |
| NSP_46010 | 445 | 3.4 | 1.0 | 3.3 | 1.0 | CAB/ELIP/HLIP superfamily protein | Ssl1633HL (e−15) |
Abbreviations: ASC, analysis of sequence counts; DE, differentially expressed; ELIP, early light induced protein; FC, fold change; HL, high light; HLIP, HL-inducible protein; ORF, open reading frame; TSS, transcription start site.
Protein-coding genes whose TSS showed a more than threefold increase in the number of reads under HL stress for 3 h as well as under combined oxidative and HL (HL+O2) stress for 0.5 h (minimum number of 620 reads at both conditions).
For comparison, the orthologs in Synechocystis are given and indicated by the suffix HL when these genes were HL-induced in Synechocystis microarray data sets (CyanoEXpress 1.2 database; Hernandez-Prieto and Futschik, 2012). For the complete list of TSSs see Supplementary Table S3.
Table 3. Protein coding genes whose TSS showed a more than threefold increase in the number of reads.
| Protein in Nodularia CCY9414 | Absolute read counts (control) | FC HL+O2/control | ASC DE probability O2+HL/control | FC HL/control | ASC DE probability HL/control | Annotation | ORF in 6803 (e-value) |
|---|---|---|---|---|---|---|---|
| NSP_50790 | 1 | 1334 | 1.0 | 1 | 0.0 | Glycosyl transferase, group 1 | Sll0045 (e−30) |
| NSP_43710 | 20 | 50.3 | 1.0 | 0.1 | 0.0 | LSU ribosomal protein L5p (L11e) | Sll1808 (e−108) |
| NSP_41430 | 96 | 18.9 | 1.0 | 0.9 | 0.0 | 4Fe–4S ferredoxin, nitrogenase-associated | Sll0741 (2e−08) |
| NSP_16900 | 109 | 13.3 | 1.0 | 0.9 | 0.0 | Hypothetical protein | None |
| NSP_24490 | 89 | 13.1 | 1.0 | 1.1 | 0.0 | Phytoene synthase | Slr1255 (2e−135) |
| NSP_14160 | 338 | 12.7 | 1.0 | 0.6 | 0.0 | Cytochrome b559 α-Chain (PsbE) | Ssr3451 (2e−45) |
| NSP_3790 | 142 | 10.2 | 1.0 | 2 | 0.61 | Photosystem I subunit IX (PsaJ) | Sml0008 (2e−04) |
| NSP_6400 | 149 | 9.8 | 1.0 | 2.9 | 1.0 | NifU-like protein | Ssl2667 (e−43) |
| NSP_18580 | 409 | 9.3 | 1.0 | 0.5 | 0.0 | General secretion pathway protein H, Type IV pilin PilA | Sll1694HL (e−32) |
| NSP_2690 | 384 | 8.8 | 1.0 | 0.2 | 0.0 | Hypothetical protein | Slr1900 (e−49) |
| NSP_22750 | 299 | 7.9 | 1.0 | 0 | 0.0 | WD repeat-containing protein | Slr8038 (3e−39) |
| NSP_47150 | 203 | 7.4 | 1.0 | 1 | 0.0 | Hypothetical protein | None |
| NSP_6930 | 206 | 7.3 | 1.0 | 1.2 | 0.0 | Hypothetical protein | None |
| NSP_9450 | 1264 | 6.9 | 1.0 | 2.9 | 1.0 | RND efflux membrane fusion protein | Sll0141 (e−83) |
| NSP_45100 | 200 | 6.8 | 1.0 | 1.3 | 0.0 | Phycoerythrin linker protein CpeS homolog | Slr2049 (e−59) |
| NSP_38040 | 217 | 6.6 | 1.0 | 2.4 | 1.0 | Hypothetical protein | None |
| NSP_48690 | 450 | 6.3 | 1.0 | 1.5 | 0.0 | Protein CP12, regulation of Calvin cycle | Ssl3364 (2e−21) |
| NSP_49270 | 178 | 6.1 | 1.0 | 2.6 | 1.0 | WD40 repeat | Sll0877 (2e−77) |
| NSP_19890 | 226 | 5.9 | 1.0 | 1.1 | 0.0 | Ferric uptake regulation protein | Sll1937 (2e−06) |
| NSP_33730 | 341 | 5.6 | 1.0 | 2.1 | 0.94 | Dihydroxy-acid dehydratase | Slr0452 (0.0) |
| NSP_14650 | 306 | 5.5 | 1.0 | 0.1 | 0.0 | Major facilitator family transporter | Sll1154 (e−112) |
| NSP_10860 | 578 | 5.4 | 1.0 | 0.7 | 0.0 | Carbohydrate-selective porin. OprB family | Sll0772 (e−98) |
| NSP_36640 | 195 | 5.4 | 1.0 | 0.4 | 0.0 | Hypothetical protein | None |
| NSP_31680 | 704 | 5.4 | 1.0 | 2.5 | 1.0 | PHP family metal-dependent phosphoesterase | Sll0549 (e−73) |
| NSP_34350 | 677 | 5.3 | 1.0 | 1.1 | 0.0 | Hypothetical protein | Slr2073 (3e−49) |
| NSP_19030 | 308 | 5.3 | 1.0 | 2.5 | 1.0 | Hypothetical protein | Slr1391 (e−16) |
| NSP_15460 | 1702 | 5.3 | 1.0 | 0.6 | 0.0 | ATPase involved in DNA repair | None |
| NSP_700 | 368 | 5.1 | 1.0 | 0.7 | 0.0 | Uracil–DNA glycosylase, family 4 | None |
| NSP_42260 | 614 | 5 | 1.0 | 1.6 | 0.0 | Chorismate synthase | Sll1747 (0.0) |
| NSP_48530 | 992 | 4.9 | 1.0 | 1.1 | 0.0 | Cell division trigger factor (EC 5.2.1.8) | Sll0533HL (e−125) |
| NSP_52140 | 387 | 4.8 | 1.0 | 0.5 | 0.0 | Sigma54 homolog | Ssr0657 (3e−32) |
| NSP_33420 | 320 | 4.8 | 1.0 | 0.9 | 0.0 | Hypothetical protein | Slr0975 (6e−117) |
| NSP_24390 | 434 | 4.7 | 1.0 | 1.1 | 0.0 | NADH dehydrogenase I subunit 4, PS1 cyclic electron flow | Slr1291HL (0.0) |
| NSP_50790 | 1 | 4.7 | 1.0 | 2.4 | NA | Glycosyl transferase, group 1 | Sll0045 (2e−30) |
| NSP_40430 | 248 | 4.6 | 1.0 | 0.7 | 0.0 | Protein of unknown function DUF820 | Slr1613 (3e−44) |
| NSP_42360 | 322 | 4.6 | 1.0 | 0.6 | 0.0 | Nucleoside triphosphate pyrophosphohydrolase MazG | Sll1005HL (e−121) |
| NSP_15410 | 281 | 4.5 | 1.0 | 0.7 | 0.0 | ATPase associated with various activities or gas vesicle protein GvpN | Slr1416 (e−05) |
| NSP_35650 | 417 | 4.5 | 1.0 | 0.9 | 0.0 | Hypothetical protein | None |
| NSP_14220 | 4891 | 4.4 | 1.0 | 1.1 | 0.0 | Transposase. IS605 OrfB | None |
| NSP_25450 | 638 | 4.4 | 1.0 | 1 | 0.0 | Hypothetical protein | None |
| NSP_48520 | 453 | 4.3 | 1.0 | 1.2 | 0.0 | ATP-dependent Clp protease proteolytic subunit (EC 3.4.21.92) | Sll0534 (e−125) |
| NSP_1470 | 356 | 4.3 | 1.0 | 2.2 | 0.99 | Hypothetical protein | None |
| NSP_27850 | 1945 | 4.2 | 1.0 | 0.3 | 0.0 | TldD protein, part of proposed TldE/TldD proteolytic complex | Slr1322 (0.0) |
| NSP_35830 | 777 | 4.2 | 1.0 | 1.5 | 0.0 | Hemolysin-like | Slr0483 (e−38) |
| NSP_42990 | 333 | 4.2 | 1.0 | 2.6 | 1.0 | Nuclease subunit of the excinuclease complex | Slr1035 (e−12) |
| NSP_10200 | 1257 | 4.1 | 1.0 | 1.1 | 0.0 | Hypothetical protein | Slr0575 (e−78) |
| NSP_47200 | 6886 | 4.1 | 1.0 | 0.5 | 0.0 | Hypothetical protein | Sll0749 (e−38) |
| NSP_39370 | 265 | 4 | 1.0 | 1.2 | 0.0 | Dihydrolipoamide dehydrogenase | Slr1096 (0.0) |
| NSP_10630 | 444 | 4 | 1.0 | 2.2 | 1.0 | ATP-dependent peptidase S16 | Sll0195 (e−117) |
| NSP_17370 | 303 | 4 | 1.0 | 2 | 0.92 | ATP-dependent Clp protease proteolytic subunit (EC 3.4.21.92) | Slr0164 (e−99) |
| NSP_15490 | 549 | 3.9 | 1.0 | 1 | 0.0 | Cell division control protein FtsH | Slr0228HL (e−58) |
| NSP_17490 | 2512 | 3.9 | 1.0 | 2.1 | 1.0 | Hypothetical protein | Sll1483HL (e−43) |
| NSP_48980 | 1335 | 3.9 | 1.0 | 0.8 | 0.0 | Hypothetical protein | None |
| NSP_46280 | 1582 | 3.9 | 1.0 | 1.2 | 0.0 | 3-oxoacyl-[acyl-carrier protein] reductase (EC 1.1.1.100) | Sll0330 (e−17) |
| NSP_16440 | 414 | 3.9 | 1.0 | 1.8 | 0.13 | Hypothetical protein | None |
| NSP_33860 | 721 | 3.9 | 1.0 | 1.1 | 0.0 | Putative protein | Slr1378 (e−75) |
| NSP_830 | 283 | 3.8 | 1.0 | 0.9 | 0.0 | Octaprenyl-diphosphate synthase (EC 2.5.1.-)/Geranylgeranyl pyrophosphate synthetase | Slr0739 (e−156) |
| NSP_51180 | 279 | 3.8 | 1.0 | 0.7 | 0.0 | Hypothetical protein | None |
| NSP_4850 | 268 | 3.8 | 1.0 | 0.2 | 0.0 | EsV-1-176 | Slr0645 (2e−94) |
| NSP_53130 | 640 | 3.7 | 1.0 | 0.5 | 0.0 | Translation IF3 | Slr0974 (e−26) |
| NSP_50960 | 510 | 3.7 | 1.0 | 1.4 | 0.0 | Hypothetical protein | None |
| NSP_1350 | 1083 | 3.7 | 1.0 | 0.7 | 0.0 | Hypothetical Protein | Slr0476HL (9e−15) |
| NSP_38540 | 1663 | 3.7 | 1.0 | 0.4 | 0.0 | Photosystem II protein PsbK | Sml0005 (e−13) |
| NSP_3140 | 1342 | 3.7 | 1.0 | 0.5 | 0.0 | Hypothetical protein | None |
| NSP_50680 | 446 | 3.6 | 1.0 | 1.5 | 0.0 | Glycogen phosphorylase (EC 2.4.1.1) | Slr1367 (e−18) |
| NSP_25450 | 638 | 3.6 | 1.0 | 2.2 | 0.0 | Hypothetical protein | None |
| NSP_51790 | 945 | 3.6 | 1.0 | 1.4 | 0.0 | Cytochrome b6-f complex iron-sulfur subunit PetC1 (Rieske) | Sll1316 (e−98) |
| NSP_36900 | 814 | 3.6 | 1.0 | 1 | 0.0 | Peptidoglycan-binding domain 1 | None |
| NSP_30660 | 517 | 3.6 | 1.0 | 1.9 | 0.59 | Hypothetical protein | None |
| NSP_51310 | 1074 | 3.6 | 1.0 | 1.4 | 0.0 | Ton-B like periplasmic protein | None |
| NSP_19230 | 4411 | 3.5 | 1.0 | 1.1 | 0.0 | Hypothetical protein | Slr2070 (e−61) |
| NSP_51330 | 368 | 3.5 | 1.0 | 0.2 | 0.0 | Phage shock protein A | Sll0617 (e−85) |
| NSP_46110 | 555 | 3.5 | 1.0 | 1.2 | 0.0 | NADH dehydrogenase (EC 1.6.99.3), NdhD | Slr1743 (e−170) |
| NSP_52960 | 1906 | 3.5 | 1.0 | 0.9 | 0.0 | Shikimate kinase I (EC 2.7.1.71) | Sll1660 (e−70) |
| NSP_30840 | 381 | 3.5 | 1.0 | 1.5 | 0.0 | NAD (P) transhydrogenase alpha subunit (EC 1.6.1.2) | Slr1239 (e−107) |
| NSP_52090 | 313 | 3.5 | 1.0 | 0.9 | 0.0 | Two-component response regulator | Slr1909 (e−109) |
| NSP_25460 | 964 | 3.5 | 1.0 | 1.7 | 0.0 | Oxidoreductase, Gfo/Idh/MocA family | Slr0338 (0.0) |
| NSP_15680 | 3773 | 3.4 | 1.0 | 1.1 | 0.0 | Hypothetical protein | Slr2025 (e−37) |
| NSP_51030 | 335 | 3.4 | 1.0 | 2 | 0.71 | Hypothetical protein | None |
| NSP_43620 | 299 | 3.4 | 1.0 | 0.5 | 0.0 | Large SU ribosomal protein L36p | Sml0006 (e−17) |
| NSP_41520 | 335 | 3.3 | 1.0 | 2.1 | 0.96 | Photosystem I P700 chlorophyll a apoprotein subunit Ia (PsaA) | Slr1834HL (0.0) |
| NSP_16880 | 962 | 3.3 | 1.0 | 1.8 | 0.11 | Sorbitol dehydrogenase (EC 1.1.1.14) | Sll0990 (e−13) |
| NSP_40730 | 1758 | 3.3 | 1.0 | 1.7 | 0.0 | DNA methylase N-4/N-6 | None |
| NSP_26680 | 734 | 3.3 | 1.0 | 2.2 | 1.0 | Phage integrase | None |
| NSP_33380 | 1633 | 3.3 | 1.0 | 1.7 | 0.0 | Chaperone protein DnaK | Sll1932 (0.0) |
| NSP_7750 | 464 | 3.3 | 1.0 | 0.3 | 0.0 | Iron (III)-traNSP_ort ATP-binding protein SfuC | Slr0354 (e−101) |
| NSP_9910 | 2965 | 3.3 | 1.0 | 1.3 | 0.0 | Hypothetical protein | None |
| NSP_38190 | 375 | 3.3 | 1.0 | 2 | 0.88 | Similar to Ymc | None |
| NSP_19230 | 4411 | 3.2 | 1.0 | 1 | 0.0 | Hypothetical protein | Slr2070 (2e−67) |
| NSP_22130 | 7582 | 3.2 | 1.0 | 1.1 | 0.0 | Hypothetical protein | None |
| NSP_300 | 403 | 3.2 | 1.0 | 2.1 | 0.98 | 3-Polyprenyl-4-hydroxybenzoate carboxy-lyase | Sll0936 (0.0) |
| NSP_19990 | 736 | 3.2 | 1.0 | 1.6 | 0.0 | ADP-heptose–lipooligosaccharide heptosyltransferase II (EC 2.4.1.-) | Slr0606 (e−101) |
| NSP_24440 | 328 | 3.2 | 1.0 | 2.3 | 1.0 | Hypothetical protein | None |
| NSP_37380 | 514 | 3.1 | 1.0 | 0.1 | 0.0 | ATP synthase β-chain (EC 3.6.3.14) | Slr1329HL (0.0) |
| NSP_45180 | 2481 | 3.1 | 1.0 | 2.7 | 1.0 | Carboxyl-terminal processing protease | Slr0008 (e−171) |
| NSP_1410 | 1125 | 3.1 | 1.0 | 1.2 | 0.93 | Ribulose-phosphate 3-epimerase (EC 5.1.3.1) | Sll0807 (e−144) |
| NSP_12600 | 483 | 3.1 | 1.0 | 0.6 | 0.0 | Hypothetical protein | Sll1381 (3e−53) |
| NSP_50770 | 1649 | 3.1 | 1.0 | 0.8 | 0.0 | RNA-binding protein | Slr0193 (e−48) |
| NSP_890 | 591 | 3.1 | 1.0 | 1.5 | 0.0 | Flavodoxin reductases (ferredoxin–NADPH reductases) family 1 | Sll0765 (e−110) |
| NSP_50880 | 1046 | 3 | 1.0 | 0.6 | 0.0 | Phage integrase | None |
| NSP_42140 | 351 | 3 | 1.0 | 0.3 | 0.0 | Amino acid adenylation protein NdaA involved in nodularin synthesis | None |
| NSP_6650 | 927 | 3 | 1.0 | 0.9 | 0.0 | Serine/threonine kinase | Sll1380 (e−23) |
| NSP_29410 | 734 | 3 | 1.0 | 0.7 | 0.0 | Hypothetical protein | Sll1142 (e−82) |
| NSP_1450 | 1317 | 3 | 1.0 | 0.3 | 0.0 | Acetolactate synthase large subunit (EC 2.2.1.6) | Slr2088 (0.0) |
| NSP_2440 | 424 | 3 | 1.0 | 0.8 | 0.0 | Hypothetical protein | Sll1022 (e−82) |
Abbreviations: ASC, analysis of sequence counts; DE, differentially expressed; ELIP, early light induced protein; FC, fold change; HL, high light; IF3, initiation factor 3; LSU, large subunit; ORF, open reading frame; RND, resistance-nodulation-cell division; WD, Trp-Asp.
Protein coding genes whose TSS showed a more than threefold increase in the number of reads specifically under combined HL and oxygen (HL+O2) stress (minimum number of 620 reads at combined HL+O2 stress).
For comparison, the expression levels after HL stress in Nodularia CCY9414 as well as the orthologs in Synechocystis are given. Genes were indicated by the suffix HL, when they were HL-induced in Synechocystis according to microarray data sets (CyanoEXpress 1.2 database; Hernandez-Prieto and Futschik, 2012). For the complete list of TSSs see Supplementary Table S3.
Table 4. Protein coding genes whose TSS showed a more than threefold increase in the number of reads specifically under high light (HL) stress (minimum number of 1000 reads at HL stress).
| Protein in Nodularia CCY9414 | Absolute read counts (Control) | FC HL+O2/ control | ASC DE probability O2+HL/control | FC HL/ control | ASC DE probability HL/control | Annotation | ORF in 6803 (e-value) |
|---|---|---|---|---|---|---|---|
| NSP_3660 | 1 | 0.5 | 0.0 | 502.5 | 1.0 | ATP-dependent DNA helicase RecQ | Slr1536 (4e−69) |
| NSP_44910 | 22592 | 2.1 | 1.0 | 10.2 | 1.0 | Phycobilisome degradation protein NblA | Ssl0453 (e−04) |
| NSP_16590 | 901 | 0.3 | 0.0 | 10.0 | 1.0 | (Manganese) ABC transporter. ATP-binding protein | Sll0489 (e−09) |
| NSP_31420 | 533 | 2.4 | 1.0 | 6.2 | 1.0 | Ribose-phosphate pyrophosphokinase | Sll0469 (0.0) |
| NSP_45620 | 202 | 2.05 | 0.65 | 5.72 | 1.0 | NADH dehydrogenase subunit 2 | Sll0223HL (0.0) |
| NSP_10590 | 1417 | 2.8 | 1.0 | 5.7 | 1.0 | Translation elongation factor G | Sll1098 (0.0) |
| NSP_31650 | 2630 | 2.77 | 1.0 | 5.45 | 1.0 | Superoxide dismutase [Fe] (EC 1.15.1.1) | Slr1516 (5e−87) |
| NSP_36400 | 213 | 1.39 | 0.0 | 4.92 | 1.0 | COG2931: RTX toxins and related Ca2+-binding proteins | None |
| NSP_26350 | 409 | 2.3 | 1.0 | 4.8 | 1.0 | Ribosomal protein L11 methyltransferase | Sll1909 (e−135) |
| NSP_46680 | 220 | 0.38 | 0.0 | 4.62 | 1.0 | Hypothetical protein | Slr5058 (9e−79) |
| NSP_450 | 7516 | 1.2 | 0.0 | 4.4 | 1.0 | Alkyl hydroperoxide reductase subunit Cell wall binding protein | None |
| NSP_600 | 311 | 0.3 | 0.0 | 4.3 | 1.0 | Probable iron binding protein, HesB_IscA_SufA family | Slr1417 (1e−61) |
| NSP_4830 | 384 | 1.7 | 0.0 | 4.2 | 1.0 | Hypothetical protein | None |
| NSP_3830 | 284 | 2.4 | 1.0 | 4.2 | 1.0 | Enolase (EC 4.2.1.11) | Slr0752 (0.0) |
| NSP_53110 | 2651 | 2.8 | 1.0 | 4.0 | 1.0 | Hypothetical protein | None |
| NSP_36850 | 51014 | 2.2 | 1.0 | 3.9 | 1.0 | Hypothetical protein | Slr1676HL (e−59) |
| NSP_16910 | 382 | 1.4 | 0.0 | 3.9 | 1.0 | Serine/threonine kinase | Slr0599 (e−67) |
| NSP_39220 | 494 | 0.8 | 0.0 | 3.7 | 1.0 | Putative anti-sigma factor antagonist | Ssr1600 (e−50) |
| NSP_11290 | 6147 | 2.2 | 1.0 | 3.7 | 1.0 | Photosystem II protein D2 (PsbD) | Slr0927 (0.0) |
| NSP_4280 | 797 | 1.3 | 0.0 | 3.5 | 1.0 | Two-component system, regulatory protein | Slr1042 (5e−67) |
| NSP_3570 | 548 | 2.5 | 1.0 | 3.4 | 1.0 | Sulfur acceptor protein SufE iron-sulfur cluster assembly | Sll1151 (e−173) |
| NSP_6070 | 775 | 1.6 | 0.0 | 3.3 | 1.0 | Hypothetical protein | Sll2002 (e−115) |
| NSP_9380 | 1437 | 2.7 | 1.0 | 3.3 | 1.0 | LSU ribosomal protein L11p (L12e) | Sll1743HL (5e−84) |
| NSP_44900 | 17518 | 1.44 | 0.0 | 3.29 | 1.0 | Hypothetical protein | None |
| NSP_39280 | 379 | 1.9 | 0.27 | 3.3 | 1.0 | tRNA:m (5)U-54 MTase gid | Sll0204 (0.0) |
| NSP_44740 | 351 | 1.6 | 0.0 | 3.3 | 1.0 | Omega-3 fatty acid desaturase | Sll1441HL (e−178) |
| NSP_640 | 937 | 1.4 | 0.0 | 3.3 | 1.0 | Cell division inhibitor | Slr1223 (e−130) |
| NSP_15880 | 437 | 2.1 | 0.93 | 3.2 | 1.0 | Acetyltransferase (EC 2.3.1.-) | None |
| NSP_21560 | 811 | 0.4 | 0.0 | 3.2 | 1.0 | Putative multicomponent Na+/H+ antiporter subunit C | Slr2006HL (e−49) |
| NSP_19170 | 786 | 1.3 | 0.0 | 3.2 | 1.0 | Hypothetical protein | Slr0806 (e−138) |
| NSP_7020 | 1081 | 2.0 | 0.75 | 3.1 | 1.0 | Hypothetical protein | None |
| NSP_32830 | 344 | 2.9 | 1.0 | 3.1 | 1.0 | GTP cyclohydrolase I (EC 3.5.4.16) | Slr0426HL (1e−98) |
| NSP_7400 | 814 | 2.6 | 1.0 | 3.1 | 1.0 | SAM-dependent methyltransferase | None |
| NSP_1020 | 472 | 1.0 | 0.0 | 3.1 | 1.0 | Kynurenine 3-monooxygenase (EC 1.14.13.9) | None |
| NSP_15330 | 728 | 2.4 | 1.0 | 3.0 | 1.0 | NADP-dependent malic enzyme (EC 1.1.1.40) | Slr0721 (0.0) |
| NSP_42850 | 570 | 0.9 | 0.0 | 3.0 | 1.0 | Phenylalanyl-tRNA synthetase α-chain (EC 6.1.1.20) | Sll0454 (0.0) |
Abbreviations: ASC, analysis of sequence counts; DE, differentially expressed; FC, fold change; HL, high light; LSU, large subunit; RTX, repeats in toxin; SAM, S-adenosylmethionine.
For comparison, the expression levels after combined HL+O2 in Nodularia CCY9414 as well as the orthologs in Synechocystis (indicated, if present, as in Table 2) are given. For the complete list of TSSs see Supplementary Table S3.
Many of the genes listed in Table 2 code for proteins that have important roles in HL and oxidative stress acclimation. Among them, the D1 protein of PS2 encoded by the psbA gene is known to be the primary target of these stresses (Mulo et al., 2012). Special D1 protein forms are expressed in model cyanobacteria under stress conditions, such as HL (Garczarek et al., 2008) or anoxia (Summerfield et al., 2008; Sicora et al., 2009). In Nodularia CCY9414, the psbA gene family consists of four members. All four genes (psbA1-4) are transcribed from single TSSs as monocistronic transcripts. One of these genes (psbA1, NSP_5370) represented >90% of all reads associated with the four psbA genes but was only weakly stimulated by the HL and HL+O2 stress conditions. Similarly, the psbA2 gene (NSP_35290) was relatively strongly but constitutively expressed. In contrast, the psbA3 and psbA4 genes (NSP_14420 and NSP_33570) were induced by both stress conditions (5- to 14-fold). Interestingly, there are no specialized forms of D1 proteins in Nodularia CCY9414 and the proteins encoded by psbA1-4 have identical amino acid sequences. Moreover, the D1 protein in Nodularia CCY9414 exhibits sequence characteristics that set it apart from its orthologs in model cyanobacteria. It's sequence is 10% divergent from the D1 proteins encoded by psbA2/3 in Synechocystis. It is tempting to speculate that the D1 protein encoded by psbA1-4 share an amino acid sequence optimally adapted to HL conditions. We conclude that Nodularia CCY9414 uses a unique strategy to cope with HL-induced damage to PS2 and the D1 protein, because it expresses a presumably HL-adapted D1 protein form from multiple gene copies, some of which are induced under stress conditions serving as backups.
In addition to psbA, sod and ocp, five genes coding for small CABs, one gene coding for the NDH complex and one coding for the chaperone DnaJ were prominently expressed (Table 2). CABs are binding proteins for chlorophyll a and carotenoids (Funk and Vermaas, 1999). These proteins are rapidly induced by light stress (also called HL-inducible proteins) and have been suggested to confer protection to the photosynthetic apparatus against oxidative damage (reviewed in Engelken et al., 2012). Nodularia CCY9414 possesses nine such genes plus a gene coding for a ferrochelatase–CAB hybrid protein compared with the four CAB genes and one encoding a ferrochelatase–CAB hybrid that have been described in Synechocystis. A Synechocystis mutant lacking all four of these HL-inducible proteins has been reported, which is sensitive to HL (He et al., 2001; Xu et al., 2004). The presence of twice as many CAB genes in Nodularia CCY9414 compared with Synechocystis suggests that these proteins may have important roles in conferring greatly increased resilience to HL stress. An increased number of CAB genes have also been reported in HL-adapted compared with low-light-adapted Prochlorococcus strains (Scanlan et al., 2009).
Nodularia CCY9414 exhibited the activated expression of many additional genes (for example, coding for adenosine deaminase, which is a peptidoglycan-binding protein) that did not have homologs in the smaller genome of the model cyanobacterium Synechocystis or were not previously known to be regulated by HL or HL+O2 stress (Table 2). The overlap with the already known stress-induced genes became much smaller when those genes were considered, which were particularly induced either by the HL or the HL+O2 stress (orthologs for only 20% and 8.6% of the genes listed in Tables 3 and 4, respectively, are also HL-induced in Synechocystis). Among these genes, many code for hypothetical proteins, that is, their functions have not yet been described in model organisms. It is not unlikely that some of these proteins are involved in stress acclimation, contributing to the bloom-forming capability of Nodularia CCY9414 in a yet unknown manner. Another group of genes particularly induced following exposure to the combined HL+O2 stress included those coding for proteins that function in DNA structure maintenance or modification, such as uracil–DNA glycosylase, DNA methylase and nuclease. Interestingly, genes coding for two phage integrases also showed higher transcript levels in the stressed Nodularia CCY9414 cells. Cyanophages have been reported to be crucial factors that determine the fates of toxic cyanobacterial blooms (Yoshida-Takashima et al., 2012). Finally, the TSS of the ndaA gene, which is the first gene in the operon coding for the huge nodularin synthetase complex (Moffitt and Neilan, 2004), showed threefold higher expression (corresponding to higher amounts of ndaF in our RT-PCR experiments, see Figure 1c), indicating an increase in nodularin synthesis under bloom-forming conditions. The crucial role of toxins in the HL resistance of bloom-forming cyanobacteria has been shown for the freshwater cyanobacterium M. aeruginosa that accumulates the hepatotoxin microcystin, which is similar to nodularin. Microcystin seems to have an important role in HL resistance due to its binding to the main carboxylating enzyme Rubisco (Zilliges et al., 2011). In addition, the increased expression of genes coding for a number of proteins that are involved in enzymatic reactions as well as transport processes (for example, iron (tonB-like) and manganese transporters) was also observed. These changes indicate that stress situations resembling bloom-forming conditions induce complex metabolic reorganization.
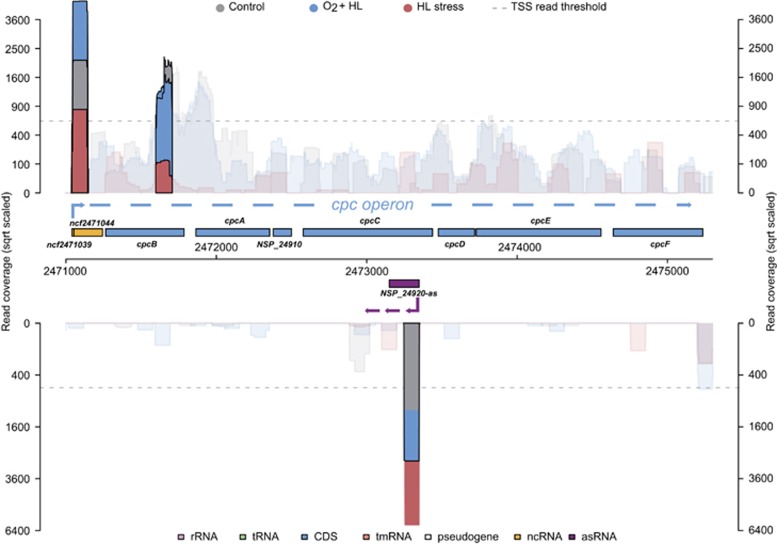
In anaerobic phototrophic bacteria, ncRNAs have been characterized as important regulators of the response to singlet oxygen (Berghoff et al., 2009). In cyanobacteria, transcriptomic changes under elevated oxygen concentrations have not been addressed to date. Therefore, we focused on the 68 aTSSs and 42 nTSSs that were induced under HL and, in particular, under HL+O2 conditions (Supplementary Table S6). For example, one aTSS that was induced by 2.5- and 5.4-fold under the HL+O2 and HL stresses, respectively, was located antisense to the cpcC gene coding for the phycocyanin-associated phycobilisome rod linker polypeptide (Figure 4). Interestingly, the gTSS-driving expression of the cpcBAC mRNA was partially inversely regulated and its read count dropped to 0.2 under HL compared with that of the control. We conclude that the asRNA to cpcC is likely functionally relevant and may contribute to the repression of cpcBAC mRNA specifically under HL. It is known from model strains that HL decreases the phycobilisome antennae size to avoid the over-reduction of PS2 (Kirilovski and Kerfeld, 2012). Interestingly, an ortholog of this asRNA also exists in Anabaena PCC7120, in which it seems to decrease cpcBAC expression under nitrogen-limiting conditions (Mitschke et al., 2011b). Thus, the functional relationship between the cpcBAC gTSS and the cpcC aTSS may not be limited to the HL response.
Figure 4.
The posttranscriptional silencing of cpcBAC mRNA accumulation may involve an asRNA induced under HL. The genes NSP_24890-NSP_24950 encompassing the genes cpcBACDEF and one very short unknown gene are shown. The read accumulation for the two TSS in this region, the gTSS in front of cpcB and for the aTSS in cpcC are given according to the three investigated conditions.
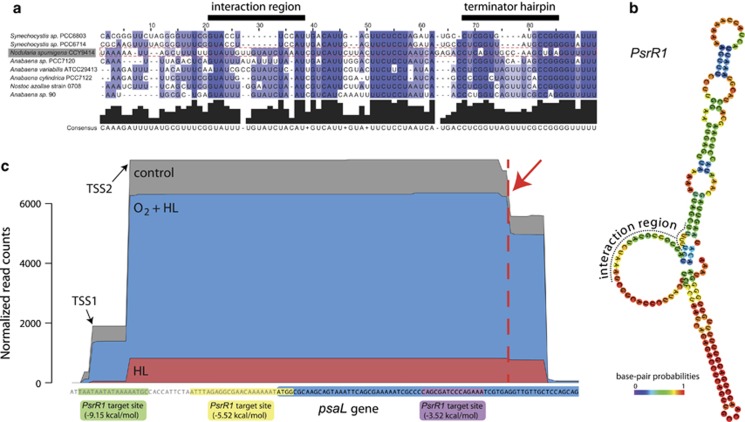
The ncRNA PsrR1 has been characterized in Synechocystis as a central regulator for the adaptation of the photosynthetic apparatus to HL (Georg et al., 2014). PsrR1 has been speculated to have similar roles in other cyanobacteria. Indeed, with the ncRNA ncr3914006, Nodularia CCY9414 possesses a likely candidate with the typical secondary structure and conserved central sequence element of PsrR1 (Figure 5). This ncRNA was induced 408- and 57-fold under the HL+O2 and HL stresses (Supplementary Table S3). The much higher induction of PsrR1 expression under HL+O2 compared with that under HL stress alone may represent an important insight, because the effect of elevated oxygen concentrations has not been analyzed to date. Our data suggest that PsrR1 may possess an even more important function than previously considered, following exposure to combined stresses. In Synechocystis, the psaL mRNA, which is one of the PsrR1 key targets, is subjected to endonucleoytic cleavage by RNase E at a specific cleavage site located 7 nt downstream of the PsrR1:psaL interaction site (Georg et al., 2014). A comparable mechanism possibly occurs in Nodularia CCY9414. The RNA sequencing analysis showed a sharp drop in transcriptome coverage at 7 nt downstream of one of three possible interaction sites (Figure 5c). However, in contrast with what has been reported in Synechocystis, this sharp drop in coverage was also observed in the control. Interestingly, two different TSSs drive the transcription of psaL in Nodularia CCY9414 compared with a single TSS in Synechocystis. As both TSSs are repressed by HL, we conclude that the light-dependent repression of psaL in Nodularia CCY9414 via PsrR1 and transcriptional repression is more pronounced than in Synechocystis.
Figure 5.
The ncRNA PsrR1 in N. spumigena CCY9414 and its major target, the psaL gene, encoding subunit XI of PSI. (a) Sequence comparison of PsrR1 with its homologs from two strains of Synechocystis and five other filamentous cyanobacteria. The regions involved in target interaction and forming a Rho-independent terminator are annotated according to Georg et al. (2014). (b) Secondary structural model for the Nodularia CCY9414 PsrR1. (c) Transcript accumulation for the psaL mRNA in control and two stress conditions. There are two closely spaced TSSs, at position 4046958 (TSS2) and 4046949 (TSS1) of the reverse strand, indicated by the black arrows. The psaL gene is highlighted by a blue box. Three possible sites for the interaction with PsrR1 are given. Sequence coverage shows a sharp drop (marked by a red arrow and vertical line), which indicates a possible processing site due to interaction with PsrR1 (Georg et al., 2014).
Several of the other ncRNAs likely have important roles as well. For example, the ncRNA that is transcribed from the nTSS at position 3867196r located 199 nt downstream of NSP_38130 is the template repeat RNA of diversity generating retroelement 1 (Voß et al., 2013). In contrast to other bacteria, in which such ncRNAs serve as templates for cDNA synthesis with subsequent recombination into protein-coding regions (Arambula et al., 2013; Pfreundt et al., 2014), this system reported here must target mainly non-coding regions, indicated by 70 nearly identical copies of diversity generating retroelement 1 (Voß et al., 2013). The ncRNA ncf2989119, which showed the sixth to seventh highest levels of expression at the nTSS, is an ortholog of ncRNA T1 in Anabaena PCC7120 (Mitschke et al., 2011b), where it is controlled by HetR, the regulator of heterocyst development. Interestingly, the expression of ncf2989119 dropped dramatically under HL, which was also confirmed independently in a time-course experiment (Figure 1d).
Conclusion
Bloom-forming cyanobacteria such as Nodularia are expected to be well adapted to stressors, including HL and/or oxidative stress, because these conditions often occur concurrent with the formation of dense scums at the water surface. Our physiological experiments supported this assumption, because the photosynthetic activity remained high even at the highest light intensities. In addition, Nodularia CCY9414 showed signs of an increase in photorespiratory flux as have been demonstrated for M. aeruginosa, another bloom-forming cyanobacterium (Meissner et al., 2014). Photorespiration cooperates with Mehler-like reactions catalyzed by flavodiiron proteins to dissipate excess absorbed energy in cyanobacteria (Hackenberg et al., 2009; Allahverdiyeva et al., 2011). Activation of this physiological reaction is consistent with our observation of many upregulated genes coding for photorespiratory enzymes and flavodiiron proteins. In addition, genes for toxin synthesis enzymes were also upregulated, implying that these compounds may specifically have an important role in the cellular acclimation to conditions of bloom formation, consistent with previous observations for Microcystis (Zilliges et al., 2011).
Our transcriptomics results suggested many additional changes in mRNA levels and also in ncRNA abundance. All mentioned changes are highly supported by statistics and key genes assessed by RT-PCR showed similar trends, validating these results. Accordingly, the RNA sequencing approach allowed the identification of many differentially expressed genes for proteins that might ensure the acclimation of Nodularia and related cyanobacteria to the harsh conditions in the surface scum. They are likely important for stress acclimation, because, by analogy, many of the annotated genes code for proteins with known functions in HL and oxidative stress acclimation in model cyanobacteria such as Synechocystis. However, we found many new stress-regulated genes that code for proteins involved in metabolism, transport, DNA stability and structure, and other still-unknown functions. These proteins are likely particularly important for successful bloom formation in Nodularia and related cyanobacteria, because they were found specifically induced under conditions characteristic for this ecological situation in Nodularia CCY9414 and not before in planktonic, non-blooming cyanobacteria. Our screen for stress-induced ncRNAs revealed several that are important to stress acclimation, including the asRNA associated with the cpc operon and PsrR1 associated with photosynthetic genes. Collectively, our physiological and transcriptomic data provide many insights into the complex adaptation of the toxic diazotrophic cyanobacterium Nodularia CCY9414 to conditions prevailing under bloom conditions in its natural habitat, the Baltic Sea.
Acknowledgments
We thank Kathrin Jahnke for technical assistance. FM was supported by a scholarship from the INF (Interdisziplinäre Fakultät, Maritime Systeme) of the University Rostock. This work was supported by the EU project MaCuMBA (Marine Microorganisms: Cultivation Methods for Improving their Biotechnological Applications; grant agreement number 311975) to WRH.
The authors declare no conflicts of interests.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allahverdiyeva Y, Ermakova M, Eisenhut M, Zhang P, Richaud P, Hagemann M et al. (2011). Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J Biol Chem 286: 24007–24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Mustila H, Ermakova M, Bersanini L, Richaud P, Ajlani G et al. (2013). Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc Natl Acad Sci USA 110: 4111–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arambula D, Wong W, Medhekar BA, Guo H, Gingery M, Czornyj E et al. (2013). Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proc Natl Acad Sci USA 110: 8212–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949). Copper enzymes in isolated chloroplasts, polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. (2009). Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol 74: 1497–1512. [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. (2008). The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci USA 105: 17199–17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelken J, Funk C, Adamska I. (2012). The extended light-harvesting complex (LHC) protein superfamily: cassification and evolutionary dynamics. In Burnap RL, Vermaas WFJ (eds). Functional Genomics and Evolution of Photosynthetic Systems. Springer: Dordrecht, The Netherlands, pp 265–284. [Google Scholar]
- Fewer DP, Jokela J, Paukku E, Österholm J, Wahlsten M, Permi P et al. (2013). New structural variants of aeruginosin produced by the toxic bloom forming cyanobacterium Nodularia spumigena. PLoS One 8: e73618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, Vermaas W. (1999). A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38: 9397–9404. [DOI] [PubMed] [Google Scholar]
- Garczarek L, Dufresne A, Blot N, Cockshutt AM, Peyrat A, Campbell DA et al. (2008). Function and evolution of the psbA gene family in marine Synechococcus: Synechococcus sp. WH7803 as a case study. ISME J 2: 937–953. [DOI] [PubMed] [Google Scholar]
- Gehringer MM, Wannicke N. (2014). Climate change and regulation of hepatotoxin production in cyanobacteria. FEMS Microbiol Ecol 88: 1–25. [DOI] [PubMed] [Google Scholar]
- Georg J, Dienst D, Schürgers N, Wallner T, Kopp D, Stazic D et al. (2014). The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 26: 3661–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg C, Engelhardt A, Matthijs HC, Wittink F, Bauwe H, Kaplan A et al. (2009). Photorespiratory 2-phosphoglycolate metabolism and photoreduction of O2 cooperate in high-light acclimation of Synechocystis sp. strain PCC 6803. Planta 230: 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Vinnemeier J, Oberpichler I, Boldt R, Bauwe H. (2005). The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol 7: 15–22. [DOI] [PubMed] [Google Scholar]
- Hayes PK, Barker GLA. (1997). Genetic diversity within Baltic Sea populations of Nodularia (Cyanobacteria). J Phycol 33: 919–923. [Google Scholar]
- He Q, Dolganov N, Bjorkman O, Grossman AR. (2001). The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J Biol Chem 276: 306–314. [DOI] [PubMed] [Google Scholar]
- Hernandez-Prieto MA, Futschik ME. (2012). CyanoEXpress: a web database for exploration and visualisation of the integrated transcriptome of cyanobacterium Synechocystis sp. PCC6803. Bioinformation 8: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. (2001). DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilovsky D, Kerfeld CA. (2012). The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim Biophys Acta 1817: 158–166. [DOI] [PubMed] [Google Scholar]
- Kopf M, Klähn S, Scholz I, Matthiessen JK, Hess WR, Voß B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res 21: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Suzuki I, Zinchenko VV, Murata N. (2008). Stress responses in Synechocystis: regulated genes and regulatory systems. In Herrero A, Flores E (eds). The Cyanobacteria - Molecular Biology, Genomics and Evolution. Caister Academic Press: Norfolk, UK, pp 117–158. [Google Scholar]
- Ludwig M, Bryant DA. (2012). Synechococcus sp. strain PCC 7002 transcriptome: acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front Microbiol 3: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur-Marzec H, Kaczkowska MJ, Blaszczyk A, Akcaalan R, Spoof L, Meriluoto J. (2013). Diversity of peptides produced by Nodularia spumigena from various geographical regions. Mar Drugs 11: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner S, Steinhauser D, Dittmann E. (2014). Metabolomic analysis indicates a pivotal role of the hepatotoxin microcystin in high light adaptation of Microcystis. Environ Microbiol e-pub ahead of print 15 July 2014 doi:10.1111/1462-2920.12565. [DOI] [PubMed]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J et al. (2011. a). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. (2011. b). Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci USA 108: 20130–20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt MC, Neilan BA. (2004). Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl Environ Microbiol 70: 6353–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möke F, Wasmund N, Bauwe H, Hagemann M. (2013). Salt acclimation of Nodularia spumigena CCY9414 - a cyanobacterium adapted to brackish water. Aquat Microb Ecol 70: 207–214. [Google Scholar]
- Mulo P, Sakurai I, Aro EM. (2012). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta 1817: 247–257. [DOI] [PubMed] [Google Scholar]
- Paerl HW, Huisman J. (2008). Blooms like it hot. Science 320: 57–58. [DOI] [PubMed] [Google Scholar]
- Paz-Yepes J, Brahamsha B, Palenik B. (2013). Role of a microcin-C-like biosynthetic gene cluster in allelopathic interactions in marine Synechococcus. Proc Natl Acad Sci USA 110: 12030–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson L, Mihali T, Moffitt M, Kellmann R, Neilan B. (2010). On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar Drugs 8: 1650–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfreundt U, Kopf M, Belkin N, Berman-Frank I, Hess WR. (2014). The primary transcriptome of the major marine diazotroph Trichodesmium erythraeum. Sci Rep 4: 6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploug H, Adam B, Musat N, Kalvelage T, Lavik G, Wolf-Gladrow D et al. (2011). Carbon, nitrogen and O2 fluxes associated with the cyanobacterium Nodularia spumigena in the Baltic Sea. ISME J 5: 1549–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdmann M, Stanier RY. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61. [Google Scholar]
- Sakurai I, Stazic D, Eisenhut M, Vuorio E, Steglich C, Hess WR et al. (2012). Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol 160: 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellner KG. (1997). Physiology, ecology, and toxic properties of marine cyanobacteria blooms. Limnol Oceanogr 42: 1089–1104. [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A et al. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255. [DOI] [PubMed] [Google Scholar]
- Sicora CI, Ho FM, Salminen T, Styring S, Aro EM. (2009). Transcription of a “silent” cyanobacterial psbA gene is induced by microaerobic conditions. Biochim Biophys Acta 1787: 105–112. [DOI] [PubMed] [Google Scholar]
- Sivonen K, Halinen K, Sihvonen LM, Koskenniemi K, Sinkko H, Rantasärkkä K et al. (2007). Bacterial diversity and function in the Baltic Sea with an emphasis on cyanobacteria. Ambio 36: 180–185. [DOI] [PubMed] [Google Scholar]
- Summerfield TC, Toepel J, Sherman LA. (2008). Low-oxygen induction of normally cryptic psbA genes in cyanobacteria. Biochemistry 47: 12939–12941. [DOI] [PubMed] [Google Scholar]
- Voß B, Bolhuis H, Fewer DP, Kopf M, Möke F, Haas F et al. (2013). Insights into the physiology and ecology of the brackish-water-adapted cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS One 8: e60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Jenkins BD, Rynearson TA, Dyhrman ST, Saito MA, Mercier M et al. (2010). Empirical bayes analysis of sequencing-based transcriptional profiling without replicates. BMC Bioinform 11: 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Vavilin D, Funk C, Vermaas W. (2004). Multiple deletions of small Cab-like proteins in the cyanobacterium Synechocystis sp. PCC 6803: consequences for pigment biosynthesis and accumulation. J Biol Chem 279: 27971–27979. [DOI] [PubMed] [Google Scholar]
- Yoshida-Takashima Y, Yoshida M, Ogata H, Nagasaki K, Hiroishi S, Yoshida T. (2012). Cyanophage infection in the bloom-forming cyanobacteria Microcystis aeruginosa in surface freshwater. Microbes Environ 27: 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP. (2011). Nitrogen fixation by marine cyanobacteria. Trends Microbiol 19: 162–173. [DOI] [PubMed] [Google Scholar]
- Zilliges Y, Kehr JC, Meissner S, Ishida K, Mikkat S, Hagemann M et al. (2011). The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of microcystis under oxidative stress conditions. PLoS One 18: e17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.