Abstract
Denitrification and dissimilatory nitrate reduction to ammonium (DNRA) are competing microbial nitrate-reduction processes. The occurrence of DNRA has been shown to be effected qualitatively by various parameters in the environment. A more quantitative understanding can be obtained using enrichment cultures in a laboratory reactor, yet no successful DNRA enrichment culture has been described. We showed that a stable DNRA-dominated enrichment culture can be obtained in a chemostat system. The enrichment was based on the hypothesis that nitrate limitation is the dominant factor in selecting for DNRA. First, a conventional denitrifying culture was enriched from activated sludge, with acetate and nitrate as substrates. Next, the acetate concentration in the medium was increased to obtain nitrate-limiting conditions. As a result, conversions shifted from denitrification to DNRA. In this selection of a DNRA culture, two important factors were the nitrate limitation and a relatively low dilution rate (0.026 h−1). The culture was a highly enriched population of Deltaproteobacteria most closely related to Geobacter lovleyi, based on 16S rRNA gene sequencing (97% similarity). We established a stable and reproducible cultivation method for the enrichment of DNRA bacteria in a continuously operated reactor system. This enrichment method allows to further investigate the DNRA process and address the factors for competition between DNRA and denitrification, or other N-conversion pathways.
Introduction
Dissimilatory reduction of nitrate is a well-studied microbial process, which is embodied in three main pathways in the nitrogen cycle: denitrification, anaerobic ammonium oxidation and dissimilatory nitrate reduction to ammonium (DNRA) (Kraft et al., 2011). All three processes compete for nitrate and nitrite. In this study we will focus on the competition between denitrification and DNRA. During denitrification nitrate is reduced to nitrogen gas, whereas in DNRA ammonium is the end product. The denitrification process is very well studied and understood to a great extent (Kraft et al., 2011; Isobe and Ohte, 2014). On the other hand, little is known about the role of DNRA in the nitrogen cycle and the factors controlling its succes (Burgin and Hamilton, 2007; Kraft et al., 2011).
A number of field studies report the occurance of DNRA in soils, sediments, anoxic zones in waters and other sites (Burgin and Hamilton, 2007; Kraft et al., 2011; Rütting et al., 2011). These studies indicate that DNRA bacteria are generally found in anoxic, electron donor-rich zones with a low nitrate availability. Lab- and field studies generated several similar hypotheses on promoting conditions for DNRA. The dominant suggested selecting condition is low or limiting nitrate availability, which is mostly conveyed as a high mass ratio of available electron donor (chemical oxygen demand (COD) equivalents) over nitrate–nitrogen (COD:N) (Tiedje et al., 1982; King and Nedwell, 1985; Akunna et al., 1994; Rütting et al., 2011). However, none of the selective conditions have been experimentally substantiated and little is known about the underlying mechanisms.
Laboratory studies mainly consist of batch tests with environmental samples (for example, Sørensen (1978) and King and Nedwell (1985)), in which, as in field studies, the system and microbial community was insufficiently defined. They also include a few pure culture studies (for example, Rehr and Klemme (1989) and Strohm et al. (2007)), but in how far those represent environmental populations is unclear. These bacterial cultures have usually not been enriched and isolated on the basis of their DNRA capacity. Enrichment culture experiments specific for DNRA bacteria have not been described. Yet, performing this kind of experiments is essential to acquire better understanding of the DNRA process. Bacteria that are competitive based on their DNRA capacity are enriched and the environmental conditions can be simulated reasonably well, while the system is quantitatively defined in terms of carbon and nitrogen turnover. Nitrate-limited growth conditions can, for instance, not be achieved in a batch culture but can be easily achieved in a chemostat reactor. Recently, Kraft et al. (2014) used such an approach to study the role of DNRA in the nitrogen conversions of a marine sediment environment. Owing to the complex substrate used, a complex microbial community of fermentative denitrifying and DNRA bacteria was enriched, making it difficult to identify and study the DNRA organisms as such.
This study aimed to develop a cultivation method for the enrichment of a highly enriched population of DNRA bacteria in a mixed, open culture, the nutrient-limited chemostat. A conventional denitrifying culture was enriched from activated sludge, with acetate and nitrate as substrates. Next, based on the proposed hypotheses, the COD:N ratio in the medium was gradually increased to shift conversions from denitrification to DNRA. The enrichment culture is well suited to systematically study the DNRA process and its competition with denitrification or other N-conversion pathways.
Materials and methods
Chemostat reactor operation
A double-jacket glass bioreactor with a working volume of 2 l (Applikon, Delft, The Netherlands) was used for the cultivation of a denitrifying culture. The reactor was operated as an open continuous stirred-tank reactor (that is, a flow-controlled chemostat) and inoculated with activated sludge from the wastewater treatment plant Leiden-Noord, The Netherlands. The reactor was operated at 400 r.p.m. with a stirrer that contained two standard geometry six-blade turbines. The flow of nitrogen gas to the reactor was kept at 50 ml min−1 using a mass flow controller (Brooks Instrument, Ede, The Netherlands) and the reactor temperature was controlled at 20 °C by means of a water jacket and a cryostat bath (Lauda, Lauda-Königshofen, Germany). The concentration of dissolved oxygen in the reactor was measured using a dissolved oxygen electrode (Mettler Toledo, Tiel, The Netherlands) as percentage of air saturation. The pH of the reactor liquid was monitored with a pH electrode (Mettler Toledo) and was maintained at 7.1±0.05 using 0.5 M HCl and 0.5 M NaOH. The pH pumps and the pH were controlled by an ADI 1030 biocontroller (Applikon). MFCS/win (Sartorius Stedim Systems, Bohemia, NY, USA) was used for data acquisition of the online measurements (dissolved oxygen, pH, temperature, acid dosage and base dosage).
The dilution rate of the system was controlled at 0.026±0.0002 h−1 and the influent and effluent were pumped using peristaltic pumps (Masterflex, Vernon Hills, IL, USA). The effluent pump was controlled by a level sensor. The influent pumps, using L/S 14 mm tubes, were set to pump 26 ml h−1. The medium was supplied in two separate flows of a mineral medium (A) and substrate medium (B), thus a total of 52 ml h−1 influent was pumped in.
The culture media was autoclaved before use and sparged with a small flow of nitrogen gas while connected to the chemostat. Medium A contained per liter (day 0–271): 7.4 mmol KH2PO4, 0.41 mmol MgSO4.7H2O, 0.37 mmol NaOH, 0.02 mmol yeast extract, 4 ml trace element solution (Vishniac and Santer, 1957), with only 2.2 g ZnSO4.7H2O, and NaNO3 and NH4Cl. The concentration of NaNO3 was 6.7 mM (day 0 until 39) or 5.9 mM (from day 39). NH4Cl concentrations were 0.01 mM (day 26–68), 0.02 mM (day 1–26, 68–82 and 94–122), 0.04 mM (day 82–94 and 122–186) and was finally omitted (from day 186). Medium B contained, per liter, initially 2.8 mM NaCH3COO.3H2O; this was gradually increased to 4.4 mM (day 26 until 39), 5.1 mM (day 39 until 47), 6.3 mM (day 47 until 122) or 9.9 mM (day 122–271).
Balances were set up over the reactor conversions. The nitrogen not accounted for in ammonium, nitrate, nitrite or biomass was assumed to be converted to N2. The concentration of volatile suspended solids (VSS) was used for the biomass. For the computation of the CO2 production rate from the off-gas partial pressure, we used the molar gas volume 24.5 l mol−1. Losses by washout of dissolved CO2 and ionized species are included in the balancing.
Analytical procedures
Oxygen, carbon dioxide, nitric oxide and nitrous oxide concentrations in the headspace of the reactor were monitored in dried gas using a gas analyzer (NGA 2000, Rosemount, Chanhassen, MN, USA). To obtain a sufficient gas flow in the analyzer for quick response, gas was circulated in a closed loop between the analyzer and the head space at a rate of 400 ml min−1. The headspace volume of the reactor set up was 1 liter.
Samples taken from the reactor for analysis of acetate and nitrogen compounds were immediately filtered through a 0.45-μm pore size filters (polyvinylidene difluoride membrane, Merck Millipore, Carrigtohill, Ireland). Initially, the acetate concentration in the liquid phase was measured as COD. After 3 weeks, the acetate concentration was measured with a Chrompack CP 9001 gas chromatograph (Chrompack, Middelburg, The Netherlands). Samples were separated on a HP Innowax column (Aligent Technologies, Santa Clara, CA, USA) and compounds were detected with a flame ionization detector. An indication of the nitrite and nitrate concentration in the reactor liquid was obtained with test strips. When this was not zero, the concentrations were measured more accurately. COD, nitrate, nitrite and ammonium concentrations were determined spectrophotometrically with commercial cuvette test kits (Hach Lange, Düsseldorf, Germany).
The biomass concentration was measured by filtration and drying according to standard methods (Taras et al., 1971) for the denitrifying biomass. For the DNRA bacteria, the biomass was centrifuged (10 000 r.p.m. for 20 min) and the pellet dried at 105 °C. To compute VSS, an ash content in the biomass of 10% was assumed.
DGGE and sequence analysis of PCR amplified 16S genes
The microbial composition of the culture was analyzed by denaturing gradient gel electrophoresis (DGGE). Biomass samples were collected from the reactor, and centrifuged and stored at −20 °C. The genomic DNA was extracted using the UltraClean Microbial DNA isolation kit (MO BIO, Carlsbad, CA, USA), following manufacturer's instructions. The extracted DNA products were evaluated on 1% (w/v) agarose gel.
The extracted DNA was used as for PCR amplification of the 16S rRNA gene. The set of primers used is the 341F (containing a 40-bp GC clamp) and 907R (Schäfer and Muyzer, 2001). The used PCR thermal profile started with a pre-cooling phase at 4 °C for 1 min, followed by initial denaturation at 95 °C for 5 min, 32 cycles of 95 °C for 30 s, 55 °C for 40 s, 72 °C for 40 s, followed by an additional extension step at 72 °C for 30 min.
DGGE band isolation and DNA sequencing were performed as described by Bassin et al. (2012) for 16S rRNA. The obtained 16S rRNA gene sequences were manually corrected using the program Chromas Lite 2.1.1 (http://technelysium.com.au). The corrected sequences were compared with those stored in GenBank using the Basic Local Alignment Search Tool algorithm (http://www.ncbi.nlm.nih.gov/blast). The sequences have been deposited in the GenBank under accession number KM403199 to KM403205.
FISH and microscopic analysis of the culture
Fluorescent in situ hybridization (FISH) was performed as described by Johnson et al. (2009), using a hybridization buffer containing 35% (v/v) formamide. The applied probes are listed in Table 1. The general probe mixture EUB338 labeled with Cy5 was used to indicate all eubacteria species in the sample. No hybridization result was obtained with a probe specific for beta- (Beta42a (Manz et al., 1992)) and gammaproteobacteria (Gamma42a (Manz et al., 1992)), but was with a probe for deltaproteobacteria (Delta495) (not shown). In the shown result, we used the EUB338 (Cy5), the Beta42a probe, labeled with FLUOS (plus an unlabeled Gamma42a probe, to minimize erroneous hybridizations of Beta42a) and a probe labeled with Cy3 specifically designed for the detection of the 16S rRNA of the enriched microorganism, that is, based on the DGGE-obtained sequence under GenBank accession number KM403205. Probes were synthesized and 5′ labeled with either the FLUOS or with one of the sulfoindocyanine dyes Cy3 and Cy5 (Thermo Hybaid Interactiva, Ulm, Germany). Slides were observed with an epifluorescence microscope (Axioplan 2, Zeiss, Sliedrecht, The Netherlands), and images were acquired with a Zeiss MRM camera and compiled with the Zeiss microscopy image acquisition software (AxioVision version 4.7, Zeiss) and exported as TIFF format.
Table 1. Probes used in FISH analysis of the culture.
| Probe | Sequence (5′–>3′) | Dye | Specificity | Reference |
|---|---|---|---|---|
| EUB338mix | gcwgccwcccgtaggwgt | Cy5 | Most bacteria | Amann etal. (1990); Daims et al. (1999) |
| Beta42a | gccttcccacttcgttt | Fluos | Betaproteobacteria | Manz et al. (1992) |
| Gamma42a | gccttcccacatcgttt | None | Gammaproteobacteria | Manz et al. (1992) |
| GeoBac464 | agcctctctacacttcgtc | Cy3 | Specific for 16S of Geobacter sp. in enrichment culture | This study |
Abbreviation: FISH, fluorescence in situ hybridization.
Results
Reactor operation
A chemostat reactor was operated under non-sterile conditions, with acetate as electron donor and nitrate as electron acceptor. The reactor was kept anaerobic by flushing with 50 ml min−1 N2 gas. During the experiments, acetate concentrations in the medium were changed with respect to nitrate (COD:N mass ratio) (Table 2). The dilution rate was 0.026 h−1, which is reported as proper for growth of both denitrifiers and DNRA bacteria (Rehr and Klemme, 1989).
Table 2. List of chemostat operational conditions.
| Period | Medium | ||||||
|---|---|---|---|---|---|---|---|
| Reference to text | (No. of days) | CH3COO− (mg l−1) | NO3− (mg N per l) | COD:NO3−-N (mg per mg N) | NO3−-N to NH4+a (%) | Biomass (mg VSS l−1) | Limiting nutrient |
| A | 0–26 | 160 | 93 | 1.8 | — | 33 | Ac− |
| B | 27–38 | 265 | 82 | 3.4 | — | b | Ac− |
| C | 39–47 | 309 | 82 | 4.0 | — | 41 | Ac− |
| D | 48–122 | 375 | 82 | 4.9 | — | 60 | NO3− |
| E | 123–230 | 595 | 82 | 7.7 | 90c | 90 | NO3− |
Includes both the dissimilatory and the presumed assimilatory conversion.
No data on biomass concentration is available for period.
Ammonium was supplied in the medium. This is corrected for in the calculations.
First, a denitrifying culture was enriched to establish denitrifying conditions. Acetate-limited growth was applied and ammonium was supplied in the medium for biomass growth (period A, Table 2). When a stable culture was established, medium acetate concentrations were increased gradually, increasing the COD:N ratio (mg per mgN−1), to enrich a DNRA culture (Table 2). In period D, nitrate had become the limiting nutrient but denitrification still prevailed. The culture was stable and performed full denitrification, emitting neither NO nor N2O. There was also no nitrite accumulated in the medium.
The conversions shifted toward production of ammonium, when the COD:N ratio was further increased to 7.7 (period E, Table 2). Up to 90% of all nitrate was converted to ammonium, which includes the presumed assimilatory use of ammonium. In this steady-state culture, NO and N2O were not detectable. The biomass concentration was 84±9 mg VSS per liter (0.63±0.02 mg protein per mg VSS) and the nitrogen content of the biomass was 123±11 mg N per g VSS.
To confirm that the enrichment of the DNRA microorganisms was solely based on the culture conditions, a second reactor was started up during period E (Table 2). Applying the same high COD:N ratio conditions, a similar culture was obtained directly from an activated sludge inoculum. This confirmed that these operating conditions select for a DNRA culture, and that the role of the history in the first reactor was not important for the selection.
The conversion rates of denitrification and DNRA were averaged over a period and shown in Table 3. For denitrification period D was used and period E used for DNRA (Table 2). The biomass yields during denitrification and DNRA periods were 0.47 and 0.45, respectively (Table 6). In the denitrification steady-state reactor, the analyzed data showed a closed carbon balance, while the electron balance closed with 87±12%. For the DNRA process, the electron balance was closed, but only 86±3% of the incoming carbon was recovered in the C-balance. N2 was not measured explicitly, thus the N-compounds could not be balanced. During DNRA, 90±4% of N is recovered in ammonium and biomass; the missing fraction of nitrogen is assumed to be emitted N2, produced by a still present small fraction of denitrifiers in the community.
Table 3. Average conversion rates in the DEN and DNRA processes in the reactor (dilution rate 0.026 h −1).
| Compound conversion rates (mmol h−1) | |||||
|---|---|---|---|---|---|
| Ac− | NO3− | NH4+a | CH1.8O0.5N0.2b | CO2 | |
| DEN | −0.32±0.02 | −0.31±0.01 | −0.039±0.000 | 0.15±0.03 | 0.51±0.06 |
| DNRA | −0.40±0.03 | −0.31±0.01 | 0.25±0.01 | 0.18±0.01 | 0.51±0.02 |
Abbreviations: DEN, denitrification (day 70 till 90); DNRA, dissimilatory nitrate reduction to ammonium (day 137 till 160); n.a., not available.
During both periods no NO or N2O was emitted.
Ammonium was present in the influent, also during DNRA. This is taken into account in the calculations.
Calculated from the measured volatile suspended solids.
Microbial population
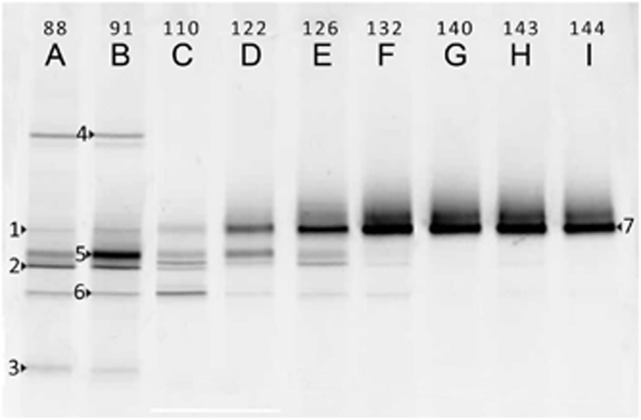
DGGE analysis of the culture (Figure 1) shows the population change over time. The lanes A and B show the culture composition in the reactor in period D (Table 2) when denitrification was dominant. The microbial population consisted of a variety of ribotypes, five of which were clearly more abundant. The samples in lanes C, D, E and F (Figure 1) cover a period of 3 weeks at the start of period E (Table 2) in which the population composition is visibly shifting. The bacteria represented in band 3 and 4 in Figure 1 disappeared quickly. Gradually, the other bands also disappeared, except one. One ribotype, which was only marginally present when denitrification was dominant (band 1), became more and more abundant (band 7). After the population shift, a stable, seemingly almost pure culture of bacteria was present in the reactor (lanes G, H, I, Figure 1). The bands were excised from the gel and sequenced. The sequence represented by band 1 in lane A was the same as the sequence of the dominant band (7) in lane G, H and I, indicating that the same ribotype was already present when denitrification prevailed in period D.
Figure 1.
Photograph of DGGE gel of bacterial 16S rRNA gene PCR products amplified from the chemostat culture. The numbers above the lanes indicate the day on which the sample was taken (Table 2).
The sequences of the PCR-amplified excised DGGE gel bands were analyzed using the NCBI BLASTn algorithm. The bacteria most closely related to the abundant denitrifiers, represented by band 2–6 (Figure 1), are shown in Table 4. During DNRA, only one bacterium appeared to be abundant on the DGGE gel (lane G, H and I, Figure 1). This ribotype (band 1 and 7, Figure 1) relates most closely (97% 16S sequence similarity) to the deltaproteobacteria G. lovleyi and Geobacter thiogenes (Table 5). The culture composition of the second chemostat was the same as that of the first, with dominance of the same ribotype (data not shown).
Table 4. BLASTn result for the 16S sequences.
| Band # | Description | Identity (%) | Isolation site | Enrichment |
|---|---|---|---|---|
| 1 | G. lovleyi SZ strain SZ | 95 | Creek sediment | PCE reduction using acetate |
| 2 | Azospira restricta SUA2 | 98 | Groundwater | General isolation |
| 3 | Bacterium GPB6 | 99 | WWTP-activated sludge | Dinitrotoluene degradation |
| 4 | Acinetobacter sp. ZH-14 | 98 | WWTP-activated sludge | Degradation of pyrethroids |
| 5 | Magnetospirillum magneticum AMB-1 | 100 | Fresh water, pond water | Magnetic+aerobic growth |
| 6 | Acidovorax caeni | 99 | Anoxic tank-activated sludge | Denitrification |
Abbreviations: PCE, tetrachloroethene; WWTP, wastewater treatment plant.
Sequences with the most similarity to those of band 1–6 indicated in Figure 1.
Table 5. BLASTn result for the 16S sequence of band 7 (Figure 1).
| Description | Identity (%) | Isolation site | Enrichment |
|---|---|---|---|
| G. lovleyi SZ strain SZ | 97 | Non-contaminated creek sediment | PCE reduction using acetate |
| G. lovleyi strain Geo7.1A | 97 | soil impacted with TCA and cis-DCE | PCE to-cis-DCE dechlorination |
| G. lovleyi strain Geo7.3B | 97 | Soil impacted with TCA and cis-DCE | PCE to-cis-DCE dechlorination |
| G. lovleyi strain Geo7.2B | 97 | Soil impacted with TCA and cis-DCE | PCE to-cis-DCE dechlorination |
| G. lovleyi strain Geo7.2A | 97 | Soil impacted with TCA and cis-DCE | PCE to-cis-DCE dechlorination |
| G. lovleyi SZ | 97 | Non-contaminated creek sediment | PCE reduction using acetate |
| G. thiogenes strain K1 | 97 | Soil leached with chlorinated chemicals | TCA dechlorination |
| G. lovleyi | 97 | Non-contaminated creek sediment | PCE reduction using acetate |
| G. lovleyi strain Geo7.3C | 96 | soil impacted with TCA and cis-DCE | PCE to-cis-DCE dechlorination |
| G. sp. IFRC128 | 96 | Uranium-contaminated ground water | Fe(III) reduction |
| G. thiogenes | 96 | freshwater sediment | Fe(III) reduction |
Abbreviations: DCE, dichloroethene; PCE, tetrachloroethene; TCA, trichloroacetate.
List of the 10 most closely related bacteria (>95%), from what environment they were isolated and on which characteristics their enrichment was based.
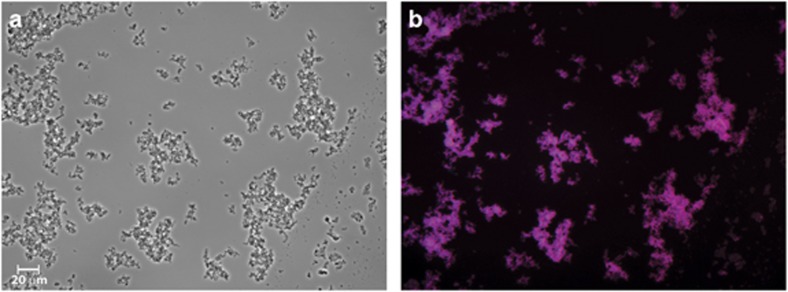
The DNRA-performing population was additionally studied with FISH analysis (Figure 2), to validate the one species dominance observed in DGGE analysis. A FISH probe was developed specific for the 16S sequence of the dominant species obtained in DGGE (band 7). In the FISH picture (Figure 2), almost all fixed bacteria are colored purple and thus hybridized with both eubacterial probe (blue) and our specific probe (red). This confirms that an almost pure culture of the Geobacter species is present in the reactor. Furthermore, the microscopic images also show that the bacteria are rod shaped and ~2 μm long.
Figure 2.
FISH microscopic photographs. (a) Image of the fixated cells of the DNRA performing culture. (b) FISH image of the DNRA culture stained with Cy5-labeled probe for bacteria (EUB338 mix, blue), fluorescein-labeled probe for most betaproteobacteria (Beta42a, green) and Cy3-labeled probe specific for the reactor species (GeoBac464, red). Blue color indicates only EUB338mix hybridized. The purple color indicates both EUB338mix and GeoBac464 hybridized. A color version of this figure is available on The ISME Journal online.
Discussion
Dissimilative nitrate reduction
We managed for the first time to cultivate a highly enriched population of DNRA bacteria in an open culture. This provides a new opportunity to study the ecophysiology of the DNRA process. This study confirms nitrate limitation, a result of high COD:N ratio, as a factor promoting nitrate conversion to ammonium. This will strengthen the insight into the competition between the denitrification and DNRA process.
The COD:N ratio of available substrates is the most suggested controlling factor in previous studies and regarded as the dominant parameter that directs the competition between DNRA and denitrification. These studies highly varied in their set-up. In batch tests with sediment or sludge samples, ammonium production for varying initial nitrate or C-source concentrations was observed, especially at higher COD:N ratios (Tiedje et al., 1982; King and Nedwell, 1985; Akunna et al., 1994). In field studies in soil and marine environments, the change in end product of nitrate reduction on addition of nitrate or C-source has been studied (Burgin and Hamilton, 2007; Rütting et al., 2011). In a chemostat reactor with a mixture of two pure cultures (Rehr and Klemme, 1989), a high COD:N ratio benefitted the DNRA culture. The observation in our chemostat enrichment culture that DNRA increases with increasing COD:N ratio of available substrates clearly confirms that this factor affects the nitrate partition. Matheson et al. (2002) argue that the change in COD:N ratio alters the oxidation state of the environment and claim that the oxidation state or prevailing redox potential is the actual key factor affecting the competition. In addition, Buresh and Patrick, (1981) state that it is the redox potential that influences the competition between DNRA and denitrification. They controlled the redox potential in sediment suspensions by sparging with different N2/O2 gas mixtures (Patrick et al., 1973) and measured a higher DNRA activity at lower potentials. The COD:N ratio is inextricably linked to the oxidation state, but oxidation state can also be influenced by the presence of reductants. To distinguish between these factors and to verify and address the importance of one or the other, further studies in well-defined enrichment cultures are required.
The nitrate limitation in our system, a result of the high COD:N feed to a chemostat reactor, promoted the success of DNRA. In many environments, nitrate is generally limiting, and hence nitrate is a growth-limiting substrate. DNRA is thought to have an advantage over denitrification under these nitrate-limiting conditions for their ability to accept eight instead of five electrons per nitrate (Tiedje et al., 1982; Kraft et al., 2014). Truly, growth-limiting conditions in the lab can only be obtained in a chemostat or fed batch system. In these systems, microorganisms compete for the uptake of the growth-limiting substrate and the important competitive trait is the substrate affinity, μmax/KS (Healey, 1980; Kuenen and Johnson, 2009). DNRA bacteria outcompeted regular denitrifiers under nitrate-limiting conditions in our system. As these bacteria have a lower μmax (Kraft et al., 2014), we have to assume that the affinity constant (KS value) for nitrate uptake is lower for DNRA organisms. An example is the KS for nitrate uptake by the denitrifier Paracocccus denitrificans, which is about 200 μM (Goddard et al., 2008), while the KS for nitrate of Escherichia coli, which performs DNRA, is estimated 15 μM (Potter et al., 1999). As described by Kuenen and Johnson (2009), the respective substrate saturation curves (Monod) of a denitrifier and a DNRA organism in the example would cross. Hence, at an adequately low-dilution-rate DNRA bacterium would be able to grow faster at the concentration of the growth-limiting nitrate. Thus, the nitrate limitation should be an effective condition to control the competition toward DNRA, as a result from high COD:N ratio, in our system.
In batch processes with high COD:N, thus relatively low, but not limiting nitrate conditions during growth, respiratory DNRA bacteria are not successful. Behrendt et al. (2014) performed denitrifying batch experiments, with a high acetate:nitrate ratio in the medium, in which no DNRA was observed. Akunna et al. (1993) performed mixed culture batch experiments for varying C-sources at similar initial amounts of COD. In both experiments, the main selective force was the μmax. Akunna et al. (1993) reported DNRA activity only when fermentative growth on glucose and glycerol occurred, but not for conversion of acetic acid, lactic acid and methanol. Possibly, the ability to ferment at high rate, using nitrate as terminal electron acceptor for excess reduction equivalents gives an advantage for DNRA over the respiratory process of denitrification. Probably, the μmax of organisms performing respiratory DNRA was not high enough to compete successfully under the nitrate excess conditions of batch cultivation. Kraft et al. (2014) also indicated that supply of fermentable substrates to a nitrate-limited system can lead to enrichment of DNRA. This underlines the requirement of nitrate limitation for successful selection of respiratory DNRA bacteria in mixed culture laboratory experiments. In the context of the work of Akunna et al. (1993), it is unclear whether the DNRA in the work of Kraft et al. (2014) was associated to fermentation or was performed by specialized DNRA bacteria, as they based their conclusions on molecular genetic analysis solely.
The yields for DNRA and denitrification are shown in Table 6. For acetic acid as a C-source, growth yields for denitrification have been limitedly reported (Strohm et al., 2007) and not at all for DNRA. Yields are theoretically correlated to the Gibbs energy released in the catabolic reaction (Heijnen, 1999). The catabolic energy gain from acetate is different when it is oxidized during DNRA and denitrification (Strohm et al., 2007; Tiedje et al., 1982). Based on the Gibbs energy values (Table 6), a higher yield for denitrification per mole of acetate has been suggested, while in this study we observe a similar yield for denitrification and DNRA per mole of acetate. The theoretical catabolic energy gain would predict yields that are similar per mole of nitrate for both processes, but they differ experimentally (Table 6). Strohm et al., (2007) observed similar deviations in the practical yields for growth on formate compared with the theoretical values for both processes. They proposed denitirifiers have a lower biomass yield on ATP. Table 6 shows the net energy gain per electron is lower in the DNRA process. However, the net energy dissipation is similar for both processes (900–1000 kJ C-mol−1 biomass produced), indicating that the growth efficiency is not influenced by the catabolic process. This would mean that a difference in growth yield is not related to a different (ATP) efficiency in the anabolism, but is due to the different energy gains in the catabolic process. For a chemostat as used here, the growth yield is not influencing the competition outcome (Gottschal and Thingstad, 1982), but in field situations with irregular (batch wise) substrate supply or growth in biofilms a higher yield on the limiting substrate would indeed lead to a better competitiveness.
Table 6. Experimental parameters and calculated Gibbs energy values for the DEN and DNRA.
| Parameter | Units | DEN | DNRA | |
|---|---|---|---|---|
| YSX | Biomass yield on acetate | (C-mol X/mol Ac−) | 0.47±0.12 | 0.45±0.07 |
| YNX | Biomass yield on nitrate | (C-mol X/mol NO3−) | 0.48±0.09 | 0.58±0.07 |
| YeX | Biomass yield on e− transferred in catabolic process | (C-mol X/e−-mol) | 0.1 | 0.07 |
| ΔGCAT01 | Catabolic energy change per mole donora | (kJ/mol Ac−) | −802 | −505 |
| ΔGCAT01 | Catabolic energy change per mole acceptor | (kJ/mol NO3−) | −501 | −505 |
| ΔGe01 | Gibbs energies per transferred electron | (kJ/e−–mol) | −100 | −63 |
Abbreviations: DEN, denitrification; DNRA, dissimilatory nitrate reduction to ammonium.
Calculated using the standard Gibbs free-energy values defined by (Thauer et al., 1977).
A high COD:N ratio of available substrates clearly affected the prevailing nitrate reduction process. The non-fermentative simple substrate acetate ensured an enrichment of specialized dissimilatory nitrate-respiring bacteria. Most likely, the nitrate limitation in combination with the adequately low dilution rate were the major factors in the selection of DNRA bacteria and the affinity for nitrate was the distinctive trait.
Microbial population
In general, it is expected that in chemostats with one limiting substrate one organism will become dominant (Kuenen and Johnson, 2009). However, during denitrification we observed several dominant species. Most likely, effectively at least two or possibly even four different limiting substrates (nitrate, nitrite, nitric oxide and nitrous oxide, respectively) are present in the nitrate-limited denitrifying chemostat, leading to the accumulation of a diverse population of partial denitrifyers (Gottschal and Thingstad, 1982). However, also perturbations in period D (data not shown) could have prevented the accumulation of one dominant organism. In this study, the role of each organism in the chemostat with denitrification was not investigated; this should be a topic of future research.
The DNRA performing culture in the reactor was an almost pure culture, as the results from DGGE and FISH analysis (lanes G, H and I, Figures 1 and 2) clearly showed. The bacteria appear to be most closely related to G. lovleyi or G. thiogenes (Table 5). Both G. lovelyi and G. thiogenes are reported rod-shaped bacteria and both can reduce nitrate to ammonium using acetate as electron donor (Nevin et al., 2007; Sung et al., 2006). However, G. thiogenes was reported to be non-motile, while G. lovleyi is a motile bacterium. Microscopic analysis of the chemostat DNRA culture showed motile cells. This indicated G. lovleyi bacteria are likely the closest relatives of our DNRA performing organism in the reactor. Geobacter species appear to have significant environmental relevance and potential practical applications. The organisms are, for example, used in bioremediation of contaminated environments, microbial fuel cells and anaerobic sludge digesters. The Geobacter species are known for their physiological capacity to couple oxidation of organic compounds to the reduction of insoluble Fe(III) minerals. Furthermore, all Geobacter species are known to use acetic acid as an electron donor, among various others, but not glucose or glycerol. Apart from reduction using Fe(III), Geobacter species are also able to conserve energy from organic matter by reduction of various other e-acceptors, such as Mn(IV) and U(VI), anthraquinone-2,6-disulfonic acid and elemental sulfur (Lovley et al., 2011). In addition, some species are capable of reductive dechlorination and reduction of nitrate to ammonium (De Wever et al., 2000; Sung et al., 2006). Furthermore, some Geobacter species produce pili that are electrically conductive, allowing them to grow on cathodes or anodes. Most of the recent attention to this group is related to their ability for direct electron transfer to minerals. DNRA capability is reported in the characterization of these organisms when found (for example, Sung et al. (2006)), but has not been further investigated.
Conclusion
We showed that a DNRA culture can be reproducibly enriched in a continuously operated reactor system. Nitrate limitation and a low dilution rate were the most important aspects in the competition between DNRA and denitrifying bacteria. The enriched culture mainly consisted of Deltaproteobacteria, closely related to G. lovleyi.
These chemostat-enrichment experiments represent the environmental selection conditions reasonably well, whereas batch enrichments are likely not selective for DNRA organisms. Future studies can use this method to further investigate the DNRA process and address the factors in its competition with denitrification.
Acknowledgments
We thank Dmitry Sorokin and Gijs Kuenen for their comments on the manuscript. The investigation was supported by the BE-Basic Foundation, project number fp0702.
The authors declare no conflicts of interest.
References
- Akunna JC, Bizeau C, Moletta R. (1993). Nitrate and nitrite reductions with anaerobic sludge using various carbon sources. Wat Res 27: 1303–1312. [Google Scholar]
- Akunna JC, Bizeau C, Moletta R. (1994). Nitrate reduction by anaerobic sludge using glucose at various nitrate concentrations: ammonification, denitrification and methanogenic activities. Environ Technol 15: 41–49. [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Envir Microbiol 56: 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassin JP, Kleerebezem R, Muyzer G, Rosado AS, van Loosdrecht MC, Dezotti M. (2012). Effect of different salt adaptation strategies on the microbial diversity, activity, and settling of nitrifying sludge in sequencing batch reactors. Appl Microbiol Technol 93: 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt A, Tarre S, Beliavski M, Green M, Klatt J, De Beer D et al. (2014). Effect of high electron donor supply on dissimilatory nitrate reduction pathways in a bioreactor for nitrate removal. Bioresour Technol 171: 291–297. [DOI] [PubMed] [Google Scholar]
- Buresh RJ, Patrick WH. (1981). Nitrate reduction to ammonium an oranic nitrogen in an estaurine sediment. Soil Biol Biochem 13: 279–283. [Google Scholar]
- Burgin AJ, Hamilton SK. (2007). Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ 5: 89–96. [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 434–444. [DOI] [PubMed] [Google Scholar]
- De Wever H, Cole JR, Fettig MR, Hogan DA, Tiedje JM. (2000). Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenes gen. nov., sp. nov. Appl Environ Microbiol 66: 2297–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard AD, Moir JW, Richardson DJ, Ferguson SJ. (2008). Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol Microbiol 70: 667–681. [DOI] [PubMed] [Google Scholar]
- Gottschal JC, Thingstad TF. (1982). Mathematical description of competition between two and three bacterial species under dual substrate limitation in the chemostat: a comparison with experimental data. Biotechnol Bioeng 24: 1403–1418. [DOI] [PubMed] [Google Scholar]
- Healey F. (1980). Slope of the Monod equation as an indicator of advantage in nutrient competition. Microb Ecol 5: 281–286. [DOI] [PubMed] [Google Scholar]
- Heijnen JJ. (1999). Bioenergetics of microbial growth. In: Flickinger MC, Drew SW (eds) Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis and Bioseperation. Wiley-Interscience: New York, pp 267–291. [Google Scholar]
- Isobe K, Ohte N. (2014). Ecological perspectives on microbes involved in N-cycling. Microbes Environ 29: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Jiang Y, Kleerebezem R, Muyzer G, van Loosdrecht MC. (2009). Enrichment of a mixed bacterial culture with a high polyhydroxyalkanoate storage capacity. Biomacromolecules 10: 670–676. [DOI] [PubMed] [Google Scholar]
- King D, Nedwell DB. (1985). The influence of nitrate concentration upon the end-products of nitrate dissimilation by bacteria in anaerobic salt marsh sediment. FEMS Microbiol Ecol 31: 23–28. [Google Scholar]
- Kraft B, Strous M, Tegetmeyer HE. (2011). Microbial nitrate respiration - genes, enzymes and environmental distribution. J Biotechnol 155: 104–117. [DOI] [PubMed] [Google Scholar]
- Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL et al. (2014). The environmental controls that govern the end product of bacterial nitrate respiration. Science 345: 676–679. [DOI] [PubMed] [Google Scholar]
- Kuenen JG, Johnson OJ. (2009). Continuous cultures (chemostats). In: Schaechter M (ed) Encyclopedia of Microbiology. Elsevier: Oxford, pp 130–147. [Google Scholar]
- Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA et al. (2011). Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv Microb Physiol 59: 1–100. [DOI] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol 15: 593–600. [Google Scholar]
- Matheson F, Nguyen M, Cooper A, Burt T, Bull D. (2002). Fate of 15N-nitrate in unplanted, planted and harvested riparian wetland soil microcosms. Ecol Eng 19: 249–264. [Google Scholar]
- Nevin KP, Holmes DE, Woodard TL, Covalla SF, Lovley DR. (2007). Reclassification of Trichlorobacter thiogenes as Geobacter thiogenes comb. nov. Int J Syst Evol Microbiol 57: 463–466. [DOI] [PubMed] [Google Scholar]
- Patrick W, Williams B, Moraghan J. (1973). A simple system for controlling redox potential and pH in soil suspensions. Soil Sci Soc Am J 37: 331–332. [Google Scholar]
- Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. (1999). Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J 344: 77–84. [PMC free article] [PubMed] [Google Scholar]
- Rehr B, Klemme JH. (1989). Competition for nitrate between denitrifying Pseudomonas stutzeri and nitrate ammonifying enterobacteria. FEMS Microbiol Ecol 62: 51–58. [Google Scholar]
- Rütting T, Boeckx P, Müller C, Klemedtsson L. (2011). Assessment of the importance of dissimilatory nitrate reduction to ammonium for the terrestrial nitrogen cycle. Biogeosciences 8: 1779–1791. [Google Scholar]
- Schäfer H, Muyzer G. (2001). Denaturing gradient gel electrophoresis in marine microbial ecology. In: John HP (ed) Methods in Microbiology. Elsevier: Oxford, pp 425–468. [Google Scholar]
- Sørensen J. (1978). Capacity for denitrification and reduction of nitrate to ammonia in a coastal marine sediment. Appl Environ Microbiol 35: 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohm TO, Griffin B, Zumft WG, Schink B. (2007). Growth yields in bacterial denitrification and nitrate ammonification. Appl Environ Microbiol 73: 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y, Fletcher KE, Ritalahti KM, Apkarian RP, Ramos-Hernandez N, Sanford RA et al. (2006). Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl Environ Microbiol 72: 2775–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taras M, Greenberg A, Hoak R, Rand M. (1971) Standard Methods for the Examination of Water and Wastewater 13th edn American Public Health Association: Washington, DC. [Google Scholar]
- Thauer RK, Jungermann K, Decker K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje JM, Sextone AJ, Myrold DD, Robinson JA. (1982). Denitrification: ecological niches, competition and survival. Antonie Van Leeuwenhoek 48: 569–583. [DOI] [PubMed] [Google Scholar]
- Vishniac W, Santer M. (1957). The Thiobacilli. Microbiol Mol Biol Rev 21: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]