Abstract
Carbon fixation has a central role in determining cellular redox poise, increasingly understood to be a key parameter in cyanobacterial physiology. In the cyanobacterium Prochlorococcus—the most abundant phototroph in the oligotrophic oceans—the carbon-concentrating mechanism is reduced to the bare essentials. Given the ability of Prochlorococcus populations to grow under a wide range of oxygen concentrations in the ocean, we wondered how carbon and oxygen physiology intersect in this minimal phototroph. Thus, we examined how CO2:O2 gas balance influenced growth and chlorophyll fluorescence in Prochlorococcus strain MED4. Under O2 limitation, per-cell chlorophyll fluorescence fell at all CO2 levels, but still permitted substantial growth at moderate and high CO2. Under CO2 limitation, we observed little growth at any O2 level, although per-cell chlorophyll fluorescence fell less sharply when O2 was available. We explored this pattern further by monitoring genome-wide transcription in cells shocked with acute limitation of CO2, O2 or both. O2 limitation produced much smaller transcriptional changes than the broad suppression seen under CO2 limitation and CO2/O2 co-limitation. Strikingly, both CO2 limitation conditions initially evoked a transcriptional response that resembled the pattern previously seen in high-light stress, but at later timepoints we observed O2-dependent recovery of photosynthesis-related transcripts. These results suggest that oxygen has a protective role in Prochlorococcus when carbon fixation is not a sufficient sink for light energy.
Introduction
The marine picophytoplankter Prochlorococcus is the most numerous oxygenic marine phototroph and has the smallest genome among them (Partensky et al., 1999; Dufresne et al., 2003). It maintains access to a very large pan-genome via horizontal gene transfer, with hotspots in genomic islands, some of which contain genes that clearly have a role in niche differentiation (Coleman et al., 2006; Kettler et al., 2007; Scanlan et al., 2009). Its genomic diversity has allowed Prochlorococcus to thrive over a broad range of environmental conditions, with distinct lineages adapted to different light levels, temperatures, and nitrogen, phosphorus, oxygen, and iron availabilities (Moore et al., 1995; Moore and Chisholm, 1999; Goericke et al., 2000; Johnson et al., 2006; Coleman and Chisholm, 2007; Kettler et al., 2007; Lavin et al., 2010; Malmstrom et al., 2013). One constant across ecotypes is their relative fitness in oligotrophic waters (Partensky et al., 1999; Dinsdale et al., 2008), promoted by traits like high surface-to-volume ratio and low phosphorus demand (Dufresne et al., 2003; Van Mooy et al., 2006, 2009). In these waters, Prochlorococcus can account for as much as half of net primary production at a given location (Vaulot et al., 1995).
In oxygenic phototrophs, intracellular oxygen can reduce the efficiency of carbon fixation, because the enzyme Rubisco can catalyze the reaction of ribulose-1,5-bisphosphate with either CO2 or O2 (Bowes et al., 1971). Carboxylation leads to the Calvin cycle; the energetically wasteful oxygenation reaction typically leads to photorespiration to clear the cytotoxic product 2-phosphoglycolate from the cell. The cyanobacterial carbon-concentrating mechanism is thought to promote carboxylation by accumulating CO2 and sequestering Rubisco in O2-impermeable carboxysomes (Price et al., 2008), yet, in light, some cyanobacteria may photorespire almost constantly (Eisenhut et al., 2008). Recent work suggests that the Prochlorococcus carbon-concentrating mechanism, although minimal, is highly efficient (Hopkinson et al., 2014), compensating for the comparatively weak CO2 affinity of Prochlorococcus Rubisco (KM~260–750 μM) (Scott et al., 2007; Roberts et al., 2012; Hopkinson et al., 2014).
Carbon dioxide and oxygen are also linked in cyanobacteria by cellular redox poise. The health of the photosynthetic electron transport chain depends on the dynamic balance between oxidized and reduced cofactors. Increasingly, that balance appears to feed into cellular signaling in many cyanobacteria, contributing not just to control of photosynthesis-related gene expression but also to systems from the circadian clock to phototaxis (Li and Sherman, 2000; Wakabayashi et al., 2011; Kim et al., 2012); it is not known whether Prochlorococcus, which has only a partial circadian clock (Holtzendorff et al., 2008; Axmann et al., 2009), makes use of redox poise as a signal. Redox poise can be threatened by factors at both ends of the electron transport chain: either an excess of arriving photons or an inability to siphon off sufficient reducing equivalents through carbon fixation can create what amounts to an impedance mismatch. In such cases, absent other photoprotective strategies, excess electrons can flow to molecular oxygen to produce damaging reactive oxygen species (Niyogi, 1999). Remarkably, in the course of genome reduction, Prochlorococcus appears to have ‘outsourced' reactive oxygen species decontamination to its neighboring heterotrophic bacteria (Morris et al., 2011, 2012) and to have minimized its photoprotective strategies (Mella-Flores et al., 2012), likely limiting its ability to handle acute changes in redox poise.
To explore the relationships between oxygen and carbon dioxide physiology in Prochlorococcus, we first tracked the growth and fluorescence of the high-light-adapted Prochlorococcus strain MED4 under CO2 and O2 levels ranging from atmospheric to near-zero, with CO2:O2 ratios ranging from ~1:5250 to >4:1 (ambient: ~1:550). We then performed whole-genome transcriptional profiling of the MED4 response to acute CO2 and O2 limitation over a 24-h time course. Although Prochlorococcus would not encounter absolute CO2 limitation at the severe levels examined in this study, midday light intensities in tropical surface waters should be well above saturating cellular CO2 fixation capacity (Hopkinson et al., 2014), making a steep imbalance between photon flux and CO2 fixation a regular occurrence. Previous studies have exposed Prochlorococcus to high-light shock and to high-peak irradiances in a day:night cycle (Steglich et al., 2006; Zinser et al., 2009; Mella-Flores et al., 2012). The experiments reported here provide an opportunity to examine how Prochlorococcus responds when low CO2 rather than high light causes a photosynthetic impedance mismatch, and how O2 availability conditions this response.
Materials and methods
Growth experiment
Axenic Prochlorococcus MED4 was grown in Sargasso seawater-based Pro99 medium (Moore et al., 2007) amended with 20-mM Na-HEPES pH 8.1. The addition of HEPES at this concentration did not affect growth rate (data not shown). Standing (non-sparged) cultures were maintained in 25-mm glass tubes at 21 °C with continuous illumination at ~60 μmol photons m−2 s−1 for >30 generations before inoculation of experimental cultures. After inoculation, tubes were capped loosely and allowed to grow without bubbling for ~24 h (Shi and Xu, 2009). Tubes were then sampled, fitted with sterile cap-and-frit assemblies, and connected to gas lines. Parallel standing controls were left with their original caps. Gas mixtures were prepared and certified by Airgas. Gases were bubbled through Milli-Q water, then passed through a needle valve, a 0.2-μm filter, and, in the culture tube, a fritted glass tube (porosity C, Ace Glass, Vineland, NJ, USA). Sparging rates were maintained as steadily as possible at ~2 ml min−1 given the limitations of standard gas-cylinder regulators.
Cultures were sampled daily by withdrawing 0.3 ml with a sterile transfer pipet. Experimental cultures were immediately resealed. Samples were protected from light and rapidly subsampled for bulk fluorescence measurements (data not shown) and flow cytometry. For flow cytometry, 10- or 100-μl samples were diluted to 1 ml with filtered seawater. Glutaraldehyde (5 μl, 25%) was added and samples were mixed, then incubated under foil for ~10 min. Fixed samples were flash-frozen in liquid nitrogen. Cells were counted using the blue (488 nm) laser on an Influx flow cytometer (Cytopeia, Seattle, WA, USA) with a 2- μm fluorescent bead standard. Quantification of flow cytometric results was performed in R (v. 2.12.0) (R Development Core Team, 2010) using the package curvHDR (v. 1.0-3) (Naumann et al., 2010).
Transcriptional profiling
Axenic Prochlorococcus MED4 cultures were grown as described above at ~60 μmol photons m−2 s−1 and 21 °C for >30 generations before the shock experiment. Three parallel 1.2-l cultures were then inoculated and grown to mid-log phase in clear 2-l bottles sparged with air at ~10 ml min−1. At t=−24 h, 400 -ml fresh medium was transferred to each of 12 clear 500-ml bottles for pre-equilibration with the experimental gases. Gases (Airgas, Cambridge, MA, USA) were 360 p.p.m. CO2+<0.001% O2 (balance N2) for −O2 shock; 40 p.p.m. CO2+<0.001% O2 (balance N2) for −CO2/−O2 shock; and 40 p.p.m. CO2+21% O2 (balance N2) for −CO2 shock. Three bottles were sparged vigorously with each experimental gas, and three control bottles with air, for 24 h.
To start the experiment, each mid-log-phase 1.2-l culture was split into six and harvested by pelleting in sterile acid-washed 250-ml centrifuge bottles in a JA-14 rotor, spinning at 8000 r.p.m. for 10 min. Supernatants were immediately poured off and each pellet resuspended in 3-ml fresh medium. Cell suspensions were pooled and split evenly among four 500-ml bottles, each pre-equilibrated with a different gas. Bottles were swirled to mix, 21-ml samples were immediately (t=0) withdrawn into 30-ml centrifuge tubes, and sparging continued. A 1-ml subsample was immediately removed from each sample for bulk fluorescence and flow cytometry and protected from light; these subsamples were processed as described above. Meanwhile, the remaining 20 ml of each sample was pelleted by spinning at 12 500 r.p.m. and 4°C for 10 min in a JA-25.50 rotor. Supernatants were immediately discarded and each pellet resuspended in 500-μl resuspension buffer (200 mM sucrose, 10 mM NaOAc pH 5.2, 5 mM EDTA). Cell suspensions were transferred to 1.5-ml tubes and flash-frozen in liquid nitrogen. Sampling for each timepoint was completed within 20 min (from withdrawal of samples to flash freezing). Samples were taken at t=0, 70 min, 3 h, 6 h, 9 h, 12 h, 18 h and 24 h.
For RNA extraction, cell suspensions were thawed in small batches and processed with the Mini RNA Isolation II kit (Zymo Research, Irvine, CA, USA). The manufacturer's protocol was followed except that cell lysis was allowed to continue for 20 min on ice. DNA was removed by treating 1 μg total nucleic acids with 2-μl Turbo DNAfree (Ambion, Foster City, CA, USA) in a 50-μl reaction for 1 h, followed by DNase inactivation according to the manufacturer's protocol. RNA was concentrated by ethanol precipitation and yields were quantified with Ribogreen (Molecular Probes, Carlsbad, CA, USA). RNA amplification (using the MessageAmp II—Bacteria kit (Ambion) with 200 ng input), labeling, and hybridization to the custom Affymetrix microarray MD4-9313 (Santa Clara, CA, USA) were performed by the MIT BioMicro Center (Cambridge, MA, USA). Amplification, labeling, and hybridization were performed for two biological replicates at timepoints t=0, 70 min, 6 h, 12 h and 24 h.
Microarray data analysis was performed in R. Chip images were processed using the package Harshlight (Suárez-Fariñas et al., 2005) before RMA normalization with the package affy (Bolstad et al., 2003). Microarray data were deposited in the GEO database with accession number GSE65684. Genes showing significant differential expression were identified by Bayesian time-series analysis using the package betr (Aryee et al., 2009), controlling for false positives at α=0.02 and setting the threshold for significance at P(differential expression)>0.98 (that is, P(null)<0.02). Clustering analysis of significantly differentially expressed genes was performed using the fuzzy c-means package Mfuzz (Futschik and Carlisle, 2005) with four cluster centers and ‘fuzziness' parameter m=1.1.
Gene functional categorizations are as annotated in CyanoBase (http://genome.kazusa.or.jp/cyanobase/) as of May 2009, except where updated gene annotation or comparison with KEGG pathways prompted manual reassignment. These exceptions are marked with asterisks in Supplementary Table S1, which gives both the original and reassigned gene categories.
Results and Discussion
Growth rates under different steady-state CO2 and O2 regimes
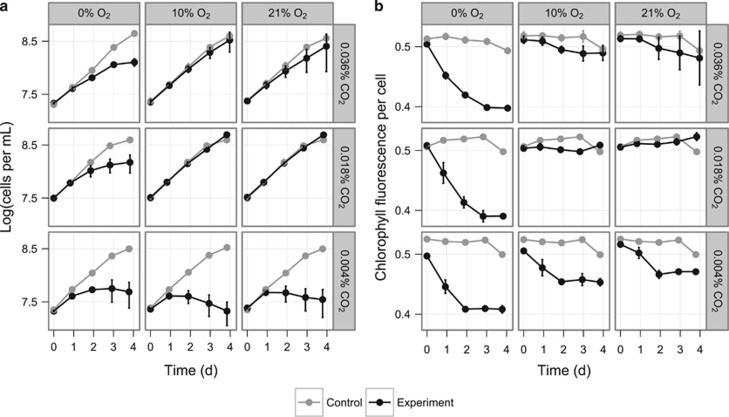
We first asked how growth is affected by CO2 and O2 availability by sparging cultures of Prochlorococcus MED4 with different gas mixtures. Three CO2 concentrations (high, 0.036% (~atmospheric); moderate, 0.018% and low, 0.004%) and three O2 concentrations (high, 21% (atmospheric); moderate, 10% and low, <0.001%) were examined in combination to provide nine growth conditions. We expected that, under moderate CO2 limitation, competition between Rubisco's carbon fixation and oxygenation reactions would cause a greater reduction in growth rate at high O2 than at moderate or low O2, whereas severe CO2 limitation would inhibit growth at any O2 level. We used flow cytometry to measure cell counts and per-cell chlorophyll fluorescence, a proxy for cellular chlorophyll concentration (DuRand et al., 2002).
Notably, growth and per-cell chlorophyll fluorescence of MED4 sparged with any combination of moderate and high CO2 and O2 was nearly indistinguishable from growth of parallel controls grown under lab air without sparging (Figure 1). By contrast, cultures grown at low O2 and either moderate or high CO2 began to show slower growth within 2 days, and approached stationary phase at lower cell densities. That is, far from releasing cells from photorespiratory pressure, suboxic conditions appeared to disrupt Prochlorococcus MED4 growth. With sufficient CO2 for continuous carbon fixation, cells grown under constant light should produce a steady stream of O2 at photosystem II (PSII). That exogenous O2 limitation nonetheless limited growth suggests that the photosynthetic O2 supply is insufficient to meet some cellular need, perhaps because of rapid diffusive loss to the suboxic medium.
Figure 1.
(a) Prochlorococcus MED4 cell concentration and (b) mean chlorophyll fluorescence per cell in cultures sparged with nine mixtures of O2 and CO2. All cultures were inoculated at t=−1 day. Experimental cultures (black) were sparged with the indicated gas mix (balanced with N2), beginning at t=0 days; control cultures (gray) were inoculated and grown in parallel but not sparged. Data points are the mean of at least three cultures. Error bars indicate 1 s.d.; where error bars are not visible, they are smaller than the plotting symbol. The drop in chlorophyll fluorescence per cell in control cultures from t=3 days to t=4 days is expected for cultures entering stationary phase. In experimental cultures, the addition of CO2 is expected to delay this transition (Moore et al., 2007).
Low CO2 severely limited growth at all O2 levels, with little or no cell division observed at t>1 day (Figure 1a). Yet flow cytometry revealed O2-dependent differences in chlorophyll fluorescence per cell among the low-CO2 conditions, with a sharper drop under low O2 than high or moderate O2 (Figure 1b). Interestingly, in contrast to the pattern for growth, chlorophyll fluorescence per cell appeared to be more strongly influenced by low O2 than low CO2, as low O2 caused a similarly steep decline at all CO2 levels. These results suggest an indispensable role for O2 under the growth conditions studied. We sought to investigate this role and its connection to CO2 levels further by studying the response at the transcriptional level.
Global transcriptional responses to acute changes in CO2 and O2 supply
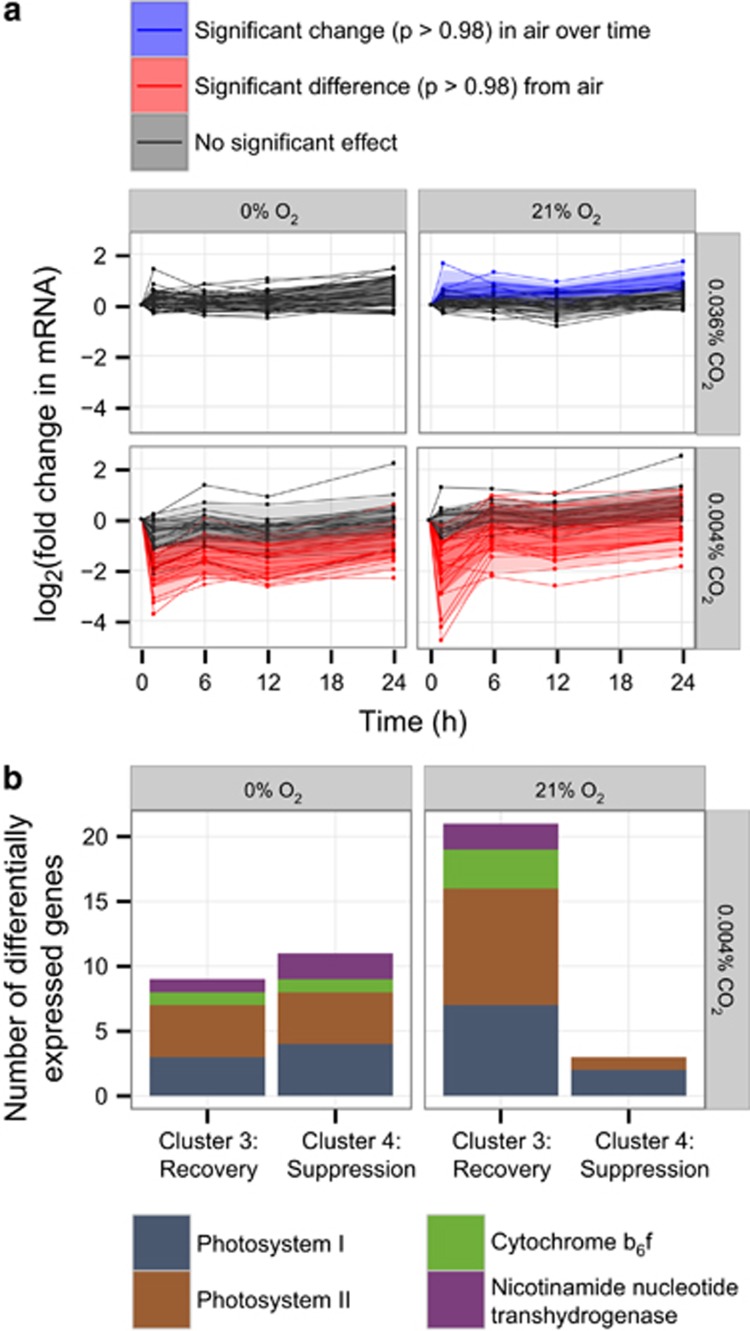
For the transcription experiment, we focused on the first 24 h after cultures were split four ways for transfer to air-sparged medium and the growth experiment's extremes: media sparged with 0.004% CO2+21% O2 (hereafter, −CO2 shock), 0.004% CO2+<0.001% O2 (−CO2/−O2 shock) and 0.036% CO2+<0.001% O2 (−O2 shock). We used an empirical Bayes approach to detect changes in expression (Aryee et al., 2009), explicitly including the time dependence of samples within each condition to identify genes whose expression profiles over the time course differed significantly (P-value threshold, 0.98; false discovery rate, 0.02) from baseline. Within the air control, the baseline was expression at t=0; for the experimental conditions, the baseline was the full profile in air.
General patterns
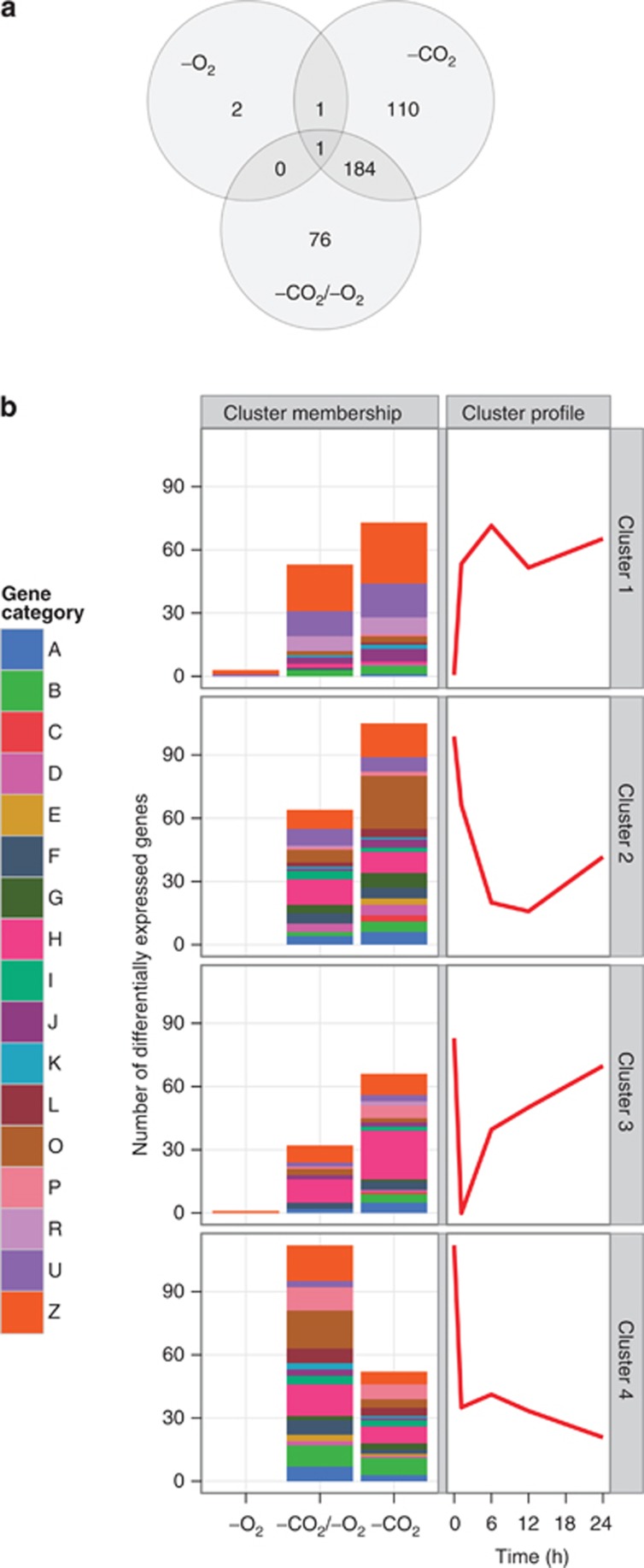
Within the air-sparged control, we observed differential expression of 188 genes, including 67 hypothetical genes (Supplementary Table S1). These changes were typically small (Supplementary Table S1) and may reflect the stress of centrifugation immediately before the t=0 timepoint. Differential expression with respect to air was minimal under −O2 shock (4 of 2048 genes probed) but widespread under −CO2 shock (296 genes) and −CO2/−O2 shock (261 genes) (Figure 2a,Supplementary Table S1). Although the latter two sets of genes overlap substantially, sharing 184 genes, clustering analysis of relative expression patterns in shock conditions with respect to air revealed an oxygen dependence to the expression profiles of many genes under carbon limitation (Figure 2b). Under −CO2/−O2 shock, sustained suppression of expression (cluster 4, 112 genes) was far more common than suppression with early recovery (cluster 3, 32 genes) among the differentially expressed genes; under −CO2 shock, sustained suppression was slightly less common than early recovery (52 vs 66 genes), raising the possibility that oxygen availability partially mitigated the effects of carbon limitation on the timescale studied here.
Figure 2.
(a) Results of whole-genome transcriptional profiling in the gas shock experiment. Venn diagram of genes showing significant differential expression with respect to air in the three experimental conditions: −O2 shock (0.036% CO2, 0% O2), −CO2 shock (0.004% CO2, 21% O2) and −CO2/−O2 shock (0.004% CO2, 0% O2). (b) Clustering analysis of changes in gene expression under gas shocks relative to changes in air. Autoscaled cluster centroids are shown at right, in red. Clusters 2 and 3 are categorized as showing suppression and subsequent recovery. At left, bar plots give the number of genes in each cluster under each gas shock condition. Gene categories: A, amino-acid biosynthesis; B, biosynthesis of cofactors, prosthetic groups, and carriers; C, cell envelope; D, cellular processes; E, central intermediary metabolism; F, energy metabolism; G, fatty acid, phospholipid, and sterol metabolism; H, photosynthesis and respiration; I, purines, pyrimidines, nucleosides, and nucleotides; J, regulatory functions; K, DNA replication, recombination, and repair; L, transcription; O, translation; P, transport and binding proteins; R, RNA; U, other categories; Z, hypothetical proteins.
As a first look at the relationship between the responses to upstream and downstream triggers of a photosynthetic imbalance, we examined the genes showing differential expression under acute high-light stress in earlier work (Steglich et al., 2006, 2008) compared with those reported here for CO2 limitation. There was very strong concordance in the direction of change: high light and –CO2/–O2 shock evoked congruent initial responses (increased high-light expression at t=45 min and gas shock cluster 1, or decreased high-light expression and gas shock cluster 2, 3 or 4; Figure 2b) in 35 of 41 shared genes, and high light and –CO2 shock did so in 44 of 46 (Supplementary Table S2). Among genes that responded to –CO2/–O2 shock, the distribution of genes across clusters 2–4 was no different for genes that did respond to light stress than for those that did not (P=0.387, Fisher's exact test). By contrast, among all genes whose expression initially dropped under –CO2 shock, those that also responded to light stress were significantly more likely to recover rapidly even as CO2 limitation continued (cluster 3; P=0.012, Fisher's exact test). That is, access to oxygen appears to reduce the extent to which CO2 limitation mimics high-light stress. As described below, this conclusion is borne out by closer examination of the groups of genes affected by –CO2 and –CO2/–O2 shock.
Hli gene family
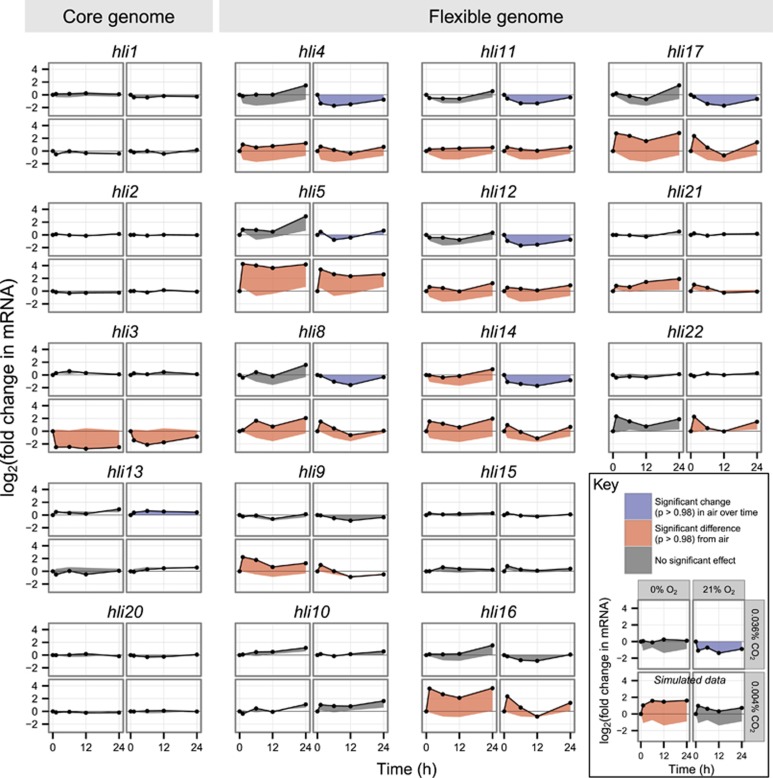
The hli gene family encodes small proteins thought to interact with the photosystems under oxidative stress conditions (He et al., 2001; Yao et al., 2007). Named for their high-light inducibility, hli genes in MED4 have been induced by phage infection, N starvation, Fe starvation and a range of light shocks (Steglich et al., 2006; Tolonen et al., 2006; Lindell et al., 2007; Thompson et al., 2011), suggesting a more general role in stress response. We found that expression of six hli genes decreased after t=0 in the air control (Figure 3), and interpret this drop as a recovery to baseline after upregulation due to sample-handling stresses (centrifugation and resuspension) just before the t=0 timepoint. By contrast, expression of these hli genes remained at or above t=0 levels throughout the –CO2/–O2 time course, suggesting that this condition did not permit recovery to a non-stressed state.
Figure 3.
Expression of high-light-inducible (hli) genes of the core (left) and flexible (right) MED4 genome in the gas shock experiment. Data points are the geometric means of two biological replicates. Fold change is relative to t=0 for each condition. All thumbnail plots share the same layout; for details, see sample plot at lower right. Data in sample plot (see Key) is simulated for illustration. The region between each time course and the baseline used for significance testing (for the air control, expression at t=0; for experimental conditions, the parallel time course in air) is shaded according to the outcome of that test: a significant change in expression in air with respect to t=0, blue; a significantly different time course in an experimental condition than in air, red; no significant effect, gray.
Under carbon limitation, the large majority of differentially expressed hli genes showed a persistent and strong increase in expression with respect to air (−CO2, 11 of 12 differentially expressed hli genes in cluster 1; −CO2/−O2, 10 of 11), but hli3 (PMM1482) was an intriguing exception. MED4's hli suite is dominated by multi-copy genes (Bhaya et al., 2002; Lindell et al., 2004), which respond to a variety of stimuli (Tolonen et al., 2006; Steglich et al., 2006; Lindell et al., 2007; Thompson et al., 2011). Their presence in genomic islands (Coleman et al., 2006) and phage suggests that they are exchanged through horizontal gene transfer, forming part of the highly niche-dependent ‘flexible' genome (Kettler et al., 2007). By contrast, the five single-copy hli genes, including hli3, belong to the core genome and have never previously been seen to respond to a stress condition in MED4 (Steglich et al., 2006; Tolonen et al., 2006; Lindell et al., 2007; Thompson et al., 2011; Kettler, personal communication). Here, both carbon limitation conditions strongly suppressed hli3 expression (Figure 3). The direction of this change substantiates the idea that hli genes in the core and flexible genomes have fundamentally different roles (Zinser et al., 2009).
Regulatory genes
Under both −CO2 and −CO2/−O2 shocks, RNA polymerase genes (rpoABC1C2) were significantly downregulated, likely contributing to a broad suppression of transcription (Figure 1,Supplementary Figure S1). The translational machinery was likewise substantially suppressed under both conditions, with roughly half the 30S and a third of 50S ribosome genes showing lowered expression (Supplementary Figure S2, Supplementary Table S1). Consistent with these changes, groEL and groEL2, components of the GroEL chaperone for newly synthesized proteins, were also expressed at lower levels (Supplementary Table S1). Despite these broad similarities, we observed some distinct regulatory responses to the stresses. The type II alternative sigma factor encoded by PMM1289 has previously been found to show differential responses to nutrient stresses, with transcript abundance increasing in the early hours of N starvation but not in response to P starvation (Martiny et al., 2006; Tolonen et al., 2006). Here, PMM1289 was significantly upregulated in −CO2 stress only (Supplementary Figure S1), suggesting that CO2 limitation in the presence of oxygen triggers invocation of a different transcriptional program than does co-limitation of CO2 and O2.
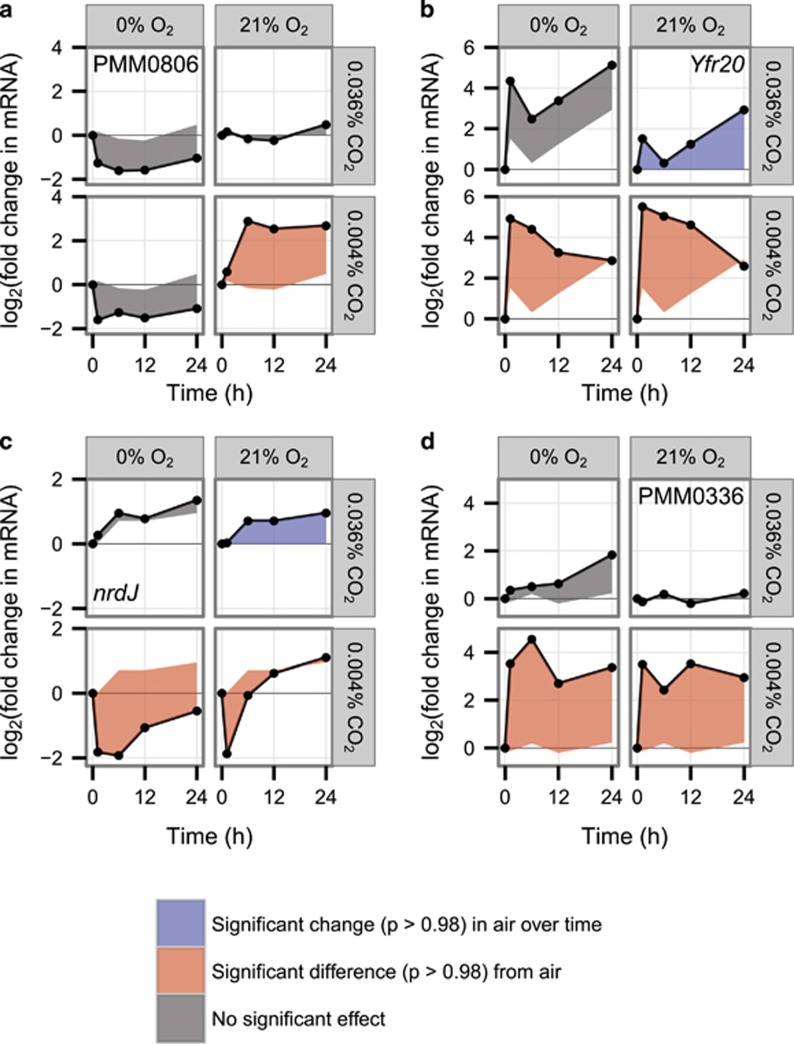
Mean expression of the Crp-family gene at locus PMM0806 showed a >4-fold increase in expression under −CO2 shock, while dropping by nearly 4-fold under both −O2 shock and −CO2/−O2 shock (Figure 4a). Although these decreases failed to meet our conservative differential expression threshold with respect to air, the difference between expression under –CO2 shock and under the oxygen limitation conditions was highly significant (P=1, false discovery rate controlled at 0.001). Crp-family proteins are transcriptional regulators, typically activators, controlled by a range of signals including cAMP, redox potential, and O2 (Körner et al., 2003). The highly oxygen-dependent transcriptional response we observe, together with previous experiments demonstrating PMM0806 induction under iron limitation (Thompson et al., 2011), strongly suggests that PMM0806 expression responds to a redox-related signal.
Figure 4.
Expression of (a) the putative CRP-family regulatory gene encoded by MED4 locus PMM0806, (b) Yfr20, (c) nrdJ, and (d) the PTOX locus PMM0336 in the gas shock experiment. Data points are the geometric means of two biological replicates. Fold change is relative to t=0 for each condition. The region between each time course and the baseline used for significance testing (for the air control, expression at t=0; for experimental conditions, the parallel time course in air) is shaded according to the outcome of that test: a significant change in expression in air with respect to t=0, blue; a significantly different time course in an experimental condition than in air, red; no significant effect, gray.
Although genome streamlining has left Prochlorococcus with a minimal complement of regulatory proteins, its complement of non-coding and antisense RNA genes is comparable to those of well-studied enterobacteria (Steglich et al., 2008) and is expected to have a major role in regulation. Here, a few antisense RNA genes were differentially expressed, but these patterns showed little clear relation to the expression of their target genes (Supplementary Figure S3). Strikingly, however, the strongest response observed for any gene was the ~30-fold upregulation of the non-coding RNA Yfr20 under both –CO2 and –CO2/–O2 shock (Figure 4b). Yfr20 is common to high-light Prochlorococcus strains and has previously been observed to be very strongly induced by high-light stress (Steglich et al., 2008).
Carbon transport and fixation
Prochlorococcus has few known transporters for inorganic carbon (Ci) (Scanlan et al., 2009). Like most open-ocean cyanobacteria, MED4 lacks inducible high-affinity Ci uptake systems, likely reflecting the relative homogeneity of Ci levels in pelagic waters as compared with coastal and lacustrine systems (Badger et al., 2006; Scanlan et al., 2009). Instead, MED4's Ci acquisition is expected to depend primarily on the Na+/HCO3- symporter BicA (PMM0214) (Badger et al., 2002; Price et al., 2004; Hopkinson et al., 2014). Although minimal, this Ci uptake machinery contributes to a highly efficient carbon-concentrating mechanism; its half-saturation point for uptake has recently been measured at 80–130 μM HCO3- (Hopkinson et al., 2014). With pH controlled at 8.1, both CO2 limitation conditions should give [HCO3-]~213 μM, theoretically within the saturation regime. That we observe marked growth limitation and a substantial transcriptional stress response at this CO2 level suggests either that the half-saturation point is higher under our culture conditions or that, even at moderate cell densities (~5 × 107 cells ml−1), photosynthesis renders bubbling insufficient to maintain pCO2 at 40 μatm (Shi and Xu, 2009) and HCO3- at saturating concentrations. Whatever the mechanism of the limitation, its effect on MED4's carbon-concentrating mechanism is remarkable: expression of bicA dropped rapidly and remained suppressed under both −CO2 and −CO2/−O2 shock (Supplementary Table S1). No other candidate cryptic Ci transporters (Harano et al., 1997; Palinska et al., 2002; Shibata et al., 2002; Scanlan et al., 2009) were induced by CO2 limitation. It is difficult to rationalize this strategy if we assume that MED4 is directly sensing Ci availability, but given MED4's extensive genome streamlining that assumption is likely false: the environmental Ci concentrations Prochlorococcus encounters should consistently be well above saturating for Ci uptake, such that direct Ci sensing is unlikely to offer a selective advantage.
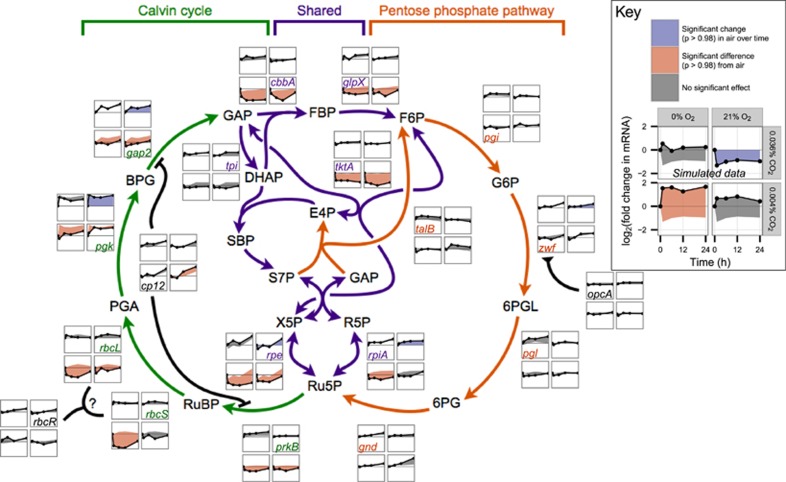
Both carbon limitation conditions also caused broad transcriptional suppression of the machinery necessary for carbon fixation. Fixation places a heavy demand for ATP production on the cell, high enough that ATP synthase expression during daytime carbon fixation exceeds expression during nighttime aerobic respiration (Zinser et al., 2009). Under both carbon limitation conditions, expression of 9 of the 10 ATP synthase complex genes (at loci PMM1438–1439 and PMM1450–1457) was significantly decreased (Supplementary Table S1), with similar time course profiles across the two conditions. We further observed decreased expression of genes encoding Calvin cycle enzymes (Figure 5), several carboxysome components (csoS1 (PMM0549), csoS2 (PMM0552), csoSCA (PMM0553), and csoS4B (PMM0553); Supplementary Table S1), and glycogen synthesis enzymes (glgABC, encoded by PMM0609, PMM0584 and PMM0769, respectively; Supplementary Table S1) under CO2 limitation. However, the availability of oxygen appears to modulate this response: while Calvin cycle gene expression was reduced under both carbon limitation conditions, the effect was generally stronger and more sustained under −CO2/−O2 shock than under –CO2 shock (Figure 5). This disparity is most striking in the Rubisco genes rbcLS. As discussed below, the minimal disruption of Rubisco expression under –CO2 shock suggests that, with carbon fixation throttled, the availability of molecular oxygen allows Prochlorococcus to buffer light stress via Rubisco's oxygenation reaction.
Figure 5.
Expression of Calvin cycle and pentose phosphate pathway genes. The Calvin cycle (green) and pentose phosphate pathway (orange) interlock; because their shared reactions (purple) run in opposite directions, the cycles cannot productively run at the same time. Gas shock expression data (thumbnail plots) for each gene in these pathways suggests that, as expected, CO2 starvation broadly downregulates the carbon-fixing Calvin cycle, while leaving the reductive pentose phosphate pathway largely intact. Data points are the geometric means of two biological replicates. Fold change is relative to t=0 for each condition. All thumbnail plots share the same layout and axis scales; for details, see sample plot at lower right. Data in sample plot (see Key) is simulated for illustration. The region between each time course and the baseline used for significance testing (for the air control, expression at t=0; for experimental conditions, the parallel time course in air) is shaded according to the outcome of that test: a significant change in expression in air with respect to t=0, blue; a significantly different time course in an experimental condition than in air, red; no significant effect, gray. Genes: cbbA, fructose-1,6-bis-P/sedoheptulose-1,7-bis-P aldolase; cp12, regulator CP12; gap2, glyceraldehyde-3-phosphate dehydrogenase; glpX, fructose-1,6/sedoheptulose-1,7-bisphosphatase; gnd, 6-phosphogluconate dehydrogenase; opcA, putative glucose-6-phosphate dehydrogenase effector; pgi, P-glucose isomerase; pgk, P-glycerate kinase; pgl, 6-P-gluconolactonase; prkB, P-ribulokinase; rbcLS, ribulose-1,5-bis-P carboxylase/oxygenase large and small subunits; rbcR, putative Rubisco transcriptional regulator; rpe, ribulose-5-P epimerase; rpiA, ribulose-5-P isomerase; talB, transaldolase; tktA, transketolase; tpi, triose-P isomerase; zwf, glucose-6-phosphate dehydrogenase. Substrates: BPG, 2,3-bis-P-glycerate; DHAP, dihydroxyacetone P; E4P, erythrose-4-P; FBP, fructose-1,6-bis-P; F6P, fructose-6-P; GAP, glyceraldehyde-3-P; G6P, glucose-6-P; PGA, 3-P-glyceric acid; R5P, ribose-5-P; RuBP, ribulose-1,5-bis-P; Ru5P, ribulose-5-P; SBP, sedoheptulose-1,7-bis-P; 6PG, 6-P-gluconate; 6PGL, 6-P-gluconolactone; S7P, sedoheptulose-7-P; and X5P, xylulose-5-P.
Photosystems
Halting carbon fixation slows NADPH consumption, threatening the photosynthetic electron transport chain with over-reduction. Two-thirds of the genes encoding proteins in the light-harvesting complex and PSI and PSII were differentially expressed under carbon limitation (–CO2/–O2 shock, 15 of 34 genes; –CO2 shock, 19; total, 22). Of these, all were initially suppressed relative to air (Supplementary Table S1); but whereas just over half the photosystem genes suppressed under −CO2/−O2 shock remained suppressed (cluster 4; Figures 2b and 6), nearly all photosystem genes affected by −CO2 shock recovered sharply and substantially (cluster 3; Figures 2b and 6). The same pattern marked cytochrome b6f and nicotinamide nucleotide transhydrogenase (pntA1, pntA2 and pntB), which couples the proton gradient to the transfer of reducing equivalents between the NAD(H) and NADP(H) pools (Figure 6b). The expression profiles seen throughout the photosynthetic electron transport chain—sustained suppression under −CO2/−O2, suppression with recovery under −CO2—strongly suggest that, in carbon-limited cells, access to oxygen affects the health of this system.
Figure 6.
(a) Expression of MED4 genes encoding PSI and PSII, cytochrome b6f and pnt genes in the gas shock experiment. Data points are the geometric means of two biological replicates. Fold change is relative to t=0 for each condition. Shock conditions causing significant differential expression in the air control with respect to time 0 are highlighted in blue; shock conditions causing significant differential expression over the time course with respect to air are highlighted in red. Ninety percent of data points lie within the outer (light) shaded bands; 70% lie within the inner (dark) bands. (b) Oxygen dependence of the cluster affiliations of the genes in panel a under carbon limitation. No differentially expressed genes in this set fell into cluster 1 (increased expression) or 2 (delayed recovery). See Figure 2b for cluster profiles.
Cyanobacterial iron physiology is closely tied to photosystem status (Thompson et al., 2011), and again, we observe divergent responses under −CO2 and −CO2/−O2 shocks. Expression of the gene encoding the iron sequestration protein ferritin (PMM0804) and of one ferric uptake regulator (Fur) gene (PMM1030) increased significantly only under −CO2, whereas the other Fur gene (PMM0637) was upregulated more strongly under –CO2 shock than –CO2/–O2 shock (Supplementary Table S1). Unusually for regulatory proteins, Fur is typically expressed at high copy number; each subunit binds one Fe(III) ion, so upregulation contributes directly to iron sequestration (Andrews et al., 2003). Oxygen-dependent differences in the expression of these genes under carbon limitation are consistent with distinct patterns of photosystem remodeling under −CO2 and −CO2/−O2 shocks.
DNA synthesis
One gene related to DNA synthesis also showed oxygen-dependent recovery under carbon limitation: the class II ribonucleotide reductase nrdJ (PMM0661) (Figure 4c). Ribonucleotide reductases convert ribonucleotides to deoxyribonucleotides, preparing the cell for DNA replication and cell division. The strong initial decrease in nrdJ transcript abundance under both −CO2 and −CO2/−O2 shocks was consistent with the expectation that severe carbon limitation would halt cell division. But why should nrdJ expression then fully recover under −CO2 shock? Intriguingly, in the obligate aerobe Streptomyces coelicolor, nrdJ activity is essential for rapid recovery after oxygen starvation (Borovok et al., 2004); ongoing deoxyribonucleotide synthesis may prepare the cell to begin DNA synthesis as soon as environmental conditions permit. If the same is true in Prochlorococcus, cultures released from −CO2 shock would be primed for recovery, whereas those released from −CO2/−O2 shock would not.
Opening safety valves under carbon limitation: PTOX and 2-phosphoglycolate
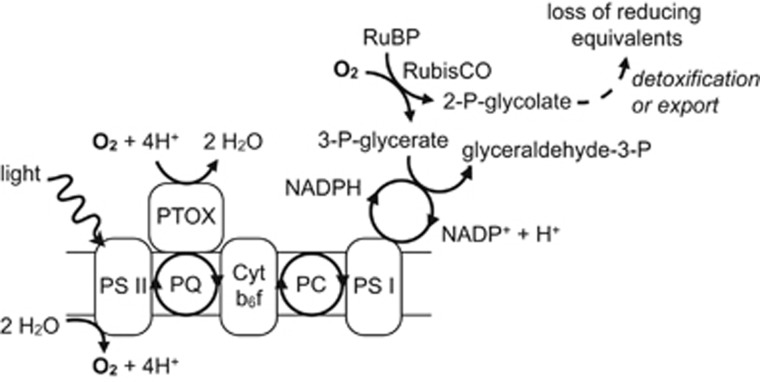
When energy cannot flow from photons to reducing equivalents to carbon–carbon bonds, the cell risks widespread damage. Of the protective strategies thought to be available to Prochlorococcus (Scanlan et al., 2009), no mechanism is yet known for non-photochemical quenching, and we find no indication at the transcriptional level that carbon limitation modulates cyclic electron flow or the Mehler reaction, with 13 of 14 ndh genes, flv1 (PMM0042), and flv3 (PMM0043) unchanged under −CO2 and −CO2/−O2 shocks. Instead, carbon-limited MED4 appears to use two photoprotective strategies, both relying on oxygen (Figure 7). These strategies may explain why light-stress-related genes whose expression initially responded to CO2 limitation were disproportionately likely to recover in cultures with access to exogenous O2.
Figure 7.
Proposed ‘safety valves' for carbon-limited Prochlorococcus. Light energy drives the photosynthetic electron transport chain, transferring reducing power from PSII via PQ to cytochrome b6f (Cyt b6f); from there via plastocyanin (PC) to PSI; and from there to the NADP(H) pool. In the presence of CO2, carbon fixation provides a final sink for these reducing equivalents; in its absence, the electron transport chain becomes over-reduced. However, so long as oxygen is still present, the electron transport chain can divert excess electrons from PQ to O2 via the PQ terminal oxidase (PTOX), or ribulose-1,5-bisphosphate (RuBP) can be oxygenated, forcing the consumption of reducing equivalents to process the products.
First, PSII can pass electrons to oxygen instead of cytochrome b6f via the plastoquinol terminal oxidase (PTOX, PMM0336), a strategy demonstrated in Synechococcus WH8102 cultures and observed in an iron-limited open-ocean cyanobacterial population dominated by Prochlorococcus (Bailey et al., 2008; Mackey et al., 2008). Previous Prochlorococcus transcriptome experiments have shown PTOX upregulation when iron limitation induced PSI downregulation (Thompson et al., 2011) and when light shocks threatened PSII and the plastoquinone (PQ) pool with over-reduction (Steglich et al., 2006). By contrast, when the PSII inhibitor DCMU was used to trap PQ in an oxidized state, PTOX transcription was significantly repressed (Steglich et al., 2006). Here, PTOX expression increased >11-fold under both −CO2 and −CO2/−O2 shocks (Figure 4d). Interpretation of this result is difficult for –CO2/–O2 shock: PSII may have continued to generate molecular oxygen, but given the steep gradient for diffusive loss to the 0% O2 medium, little of that oxygen may have been available to PTOX. With transcriptional data alone, we cannot say whether continued PTOX expression under –CO2/–O2 shock represented maintenance of a successful strategy or the futile result of signaling from an electron transport chain under continuing stress. But the broader patterns of O2-dependent transcriptional recovery suggest that any PTOX activity achieved under –CO2/–O2 shock was insufficient. If, like Synechococcus elongatus, Prochlorococcus uses PQ redox poise as a signaling input (Kim et al., 2012), different rates of PTOX activity in the presence and absence of exogenous O2 may have had a major role in triggering the different transcriptional programs observed under −CO2 and −CO2/−O2 shocks.
Second, Prochlorococcus can exploit Rubisco's oxygenation reaction to produce one 3-phosphoglycerate and one 2-phosphoglycolate. The former can be recycled through the Calvin cycle, consuming NADPH directly; the latter must be either detoxified via photorespiration or expelled. Although photorespiration appears to be essential in some cyanobacteria (Eisenhut et al., 2008), MED4, like most sequenced Prochlorococcus strains, lacks some components of the known detoxification pathways (Scanlan et al., 2009); moreover, it is known to secrete substantial quantities of glycolate even under exponential growth (Bertilsson et al., 2005). Whereas PTOX may retain some activity under –CO2/–O2 shock through use of the photosynthetic O2 generated nearby, the sequestration of Rubisco within carboxysomes makes any use of this safety valve highly unlikely when cellular O2 is low. Consistent with this, rbcLS were strongly (~4-fold) downregulated under −CO2/−O2 shock; but under −CO2 shock, rbcS was not significantly affected, and rbcL expression dropped but rapidly recovered (cluster 3, Figures 2b and 5).
We find no evidence for increased expression of reactive oxygen species scavengers under carbon limitation (data not shown), leaving cells without photoprotection vulnerable to mounting oxidative damage. In some heterotrophs, reactive oxygen species defense during starvation is necessary for recovery thereafter (McDougald et al., 2002). The effect may be the same in Prochlorococcus: the expression pattern of nrdJ, discussed above, suggests that only MED4 cells with O2 access could prepare for recovery from CO2 starvation. But why should MED4 need the ability to recover from CO2 starvation at all, when it is unlikely ever to encounter such low CO2 levels in the oligotrophic ocean? The concordance between the CO2 limitation and light stress responses, including their shared extremely strong expression of the non-coding RNA Yfr20, points to the conclusion that, on short (<1 day) timescales, the relevant factor is not the absolute CO2 concentration but the balance between CO2 and irradiance.
Threats to this central balance would be part of daily life for Prochlorococcus populations in tropical surface waters, so the ability to recover after a few hours' exposure to a photosynthetic impedance mismatch may be highly adaptive. If this is true, access to oxygen and the safety valve machinery should be most important in surface waters; conversely, exogenous oxygen may be dispensable at the very low irradiances that penetrate to oxygen minimum zones. Notably, although PTOX homologs are widespread in high-light-adapted Prochlorococcus, the only low-light-adapted Prochlorococcus ecotype known to carry a PTOX homolog is the LLI clade, whose members are abundant in some deeply mixed surface waters (Zinser et al., 2007; Scanlan et al., 2009).
Though a photosynthetic impedance mismatch may be a regular environmental stress for Prochlorococcus, a mismatch lasting continuously for >1 day is an impossibility, and the ability to withstand such a stress will not have been under selection. Here, the timing of the shifts observed in the growth experiment (Figure 1) bears further examination. From t=0 to 1 day, MED4 cells at 0.004% CO2 and 21% O2 tracked closely with the controls in both growth and per-cell chlorophyll fluorescence. Beyond 1 day in constant light, cell density increased no further, and per-cell chlorophyll fluorescence fell off. Although constant, the irradiance used in this study was too low (~60 μmol photons m−2 s−1) to provoke photoinhibition in carbon-replete and oxygen-replete MED4 (Moore and Chisholm, 1999); but lack of exogenous O2 provokes a substantial drop in per-cell chlorophyll fluorescence at all CO2 levels within 1 day of constant light, and a slowing of growth within 2 days. Oxygen-dependent safety valves may thus be essential not only for recovery from severe but transient imbalances between irradiance and carbon fixation capacity but also for the maintenance of normal photosynthetic electron transport in carbon-replete cells in moderate light.
Although the carbon limitation used in these experiments is far more extreme than Prochlorococcus would ever experience in nature, our results suggest that the resulting general stress is a familiar one, perhaps even daily for marine populations in low latitudes. Whether the photosynthetic electron transfer chain is threatened by increasing irradiance or throttling carbon fixation, the cell must dissipate an excess of reducing equivalents to survive. For Prochlorococcus MED4, the cell's capacity to respond and recover appears to hinge on oxygen. To explore this response, future work should directly measure NAD(P)/NAD(P)H balance, pigment content, and the maximum photosynthetic quantum yield (Fv/Fm) in the presence and absence of exogenous O2 across a range of CO2 levels and irradiances. The recovery phase will be of particular interest: for severely CO2-limited cells, could recovery be initiated simply by turning out the lights?
Acknowledgments
This work was supported by grants from the US Department of Energy—GTL, NSF Biological Oceanography, NSF Center for Microbial Oceanography Research and Education (C-MORE), the Seaver Foundation and the Gordon and Betty Moore Foundation. SCB was an HHMI Pre-doctoral Fellow. We thank Jessica Wiedemier Thompson, Rogier Braakman, Qinglu Zeng, Luke Thompson, and Brian Hopkinson for their helpful comments on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andrews SC, Robinson AK, Rodríguez-Quiñones F. (2003). Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237. [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Gutiérrez-Pabello JA, Kramnik I, Maiti T, Quackenbush J. (2009). An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinformatics 10: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axmann IM, Dühring U, Seeliger L, Arnold A, Vanselow JT, Kramer A et al. (2009). Biochemical evidence for a timing mechanism in Prochlorococcus. J Bacteriol 191: 5342–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Hanson D, Price GD. (2002). Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol 29: 161–173. [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD, Long BM, Woodger FJ. (2006). The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot 57: 249–265. [DOI] [PubMed] [Google Scholar]
- Bailey S, Melis A, Mackey KRM, Cardol P, Finazzi G, van Dijken G et al. (2008). Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim Biophys Acta 1777: 269–276. [DOI] [PubMed] [Google Scholar]
- Bertilsson S, Berglund O, Pullin MJ, Chisholm SW. (2005). Release of dissolved organic matter by Prochlorococcus. Vie Milieu Paris 55: 225–231. [Google Scholar]
- Bhaya D, Dufresne A, Vaulot D, Grossman A. (2002). Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol Lett 215: 209–219. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Åstrand M, Speed TP. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Borovok I, Gorovitz B, Yanku M, Schreiber R, Gust B, Chater K et al. (2004). Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol Microbiol 54: 1022–1035. [DOI] [PubMed] [Google Scholar]
- Bowes G, Ogren WL, Hageman RH. (1971). Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem Biophys Res Commun 45: 716–722. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. (2007). Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol 15: 398–407. [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L et al. (2008). Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One 3: e1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V et al. (2003). Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA 100: 10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuRand MD, Green RE, Sosik HM, Olson RJ. (2002). Diel variations in optical properties of Micromonas pusilla (Prasinophyceae). J Phycol 38: 1132–1142. [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. (2008). The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci USA 105: 17199–17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik ME, Carlisle B. (2005). Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol 3: 965–988. [DOI] [PubMed] [Google Scholar]
- Goericke R, Olson RJ, Shalapyonok A. (2000). A novel niche for Prochlorococcus sp. in low-light suboxic environments in the Arabian Sea and the Eastern Tropical North Pacific. Deep Sea Res Part 1 Oceanogr Res Pap 47: 1183–1205. [Google Scholar]
- Harano Y, Suzuki I, Maeda S, Kaneko T, Tabata S, Omata T. (1997). Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J Bacteriol 179: 5744–5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Dolganov N, Björkman O, Grossman AR. (2001). The high light-inducible polypeptides in Synechocystis PCC6803: Expression and function in high light. J Biol Chem 276: 306–314. [DOI] [PubMed] [Google Scholar]
- Holtzendorff J, Partensky F, Mella D, Lennon J-F, Hess WR, Garczarek L. (2008). Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J Biol Rhythms 23: 187–199. [DOI] [PubMed] [Google Scholar]
- Hopkinson BM, Young JN, Tansik AL, Binder BJ. (2014). The minimal CO2 concentrating mechanism of Prochlorococcus MED4 is effective and efficient. Plant Physiol 166: 2205–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-I, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. (2012). Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci USA 109: 17765–17769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner H, Sofia HJ, Zumft WG. (2003). Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev 27: 559–592. [DOI] [PubMed] [Google Scholar]
- Lavin P, González B, Santibáñez JF, Scanlan DJ, Ulloa O. (2010). Novel lineages of Prochlorococcus thrive within the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol Rep 2: 728–738. [DOI] [PubMed] [Google Scholar]
- Li H, Sherman LA. (2000). A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 182: 4268–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. (2004). Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA 101: 11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T et al. (2007). Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449: 83–86. [DOI] [PubMed] [Google Scholar]
- Mackey KRM, Paytan A, Grossman AR, Bailey S. (2008). A photosynthetic strategy for coping in a high-light, low-nutrient environment. Limnol Oceanogr 53: 900–913. [Google Scholar]
- Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, Thompson A et al. (2013). Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. ISME J 7: 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA 103: 12552–12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D, Gong L, Srinivasan S, Hild E, Thompson L, Takayama K et al. (2002). Defences against oxidative stress during starvation in bacteria. Antonie Van Leeuwenhoek 81: 3–13. [DOI] [PubMed] [Google Scholar]
- Mella-Flores D, Six C, Ratin M, Partensky F, Boutte C, Le Corguillé G et al. (2012). Prochlorococcus and Synechococcus have evolved different adaptive mechanisms to cope with light and UV stress. Front Microbiol 3: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Goericke R, Chisholm SW. (1995). Comparative physiology of Synechococcus and Prochlorococcus: Influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser 116: 259–275. [Google Scholar]
- Moore LR, Chisholm SW. (1999). Photophysiology of the marine cyanobacterium Prochlorococcus: Ecotypic differences among cultured isolates. Limnol Oceanogr 44: 628–638. [Google Scholar]
- Moore LR, Post AF, Rocap G, Chisholm SW. (2002). Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr 47: 989–996. [Google Scholar]
- Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D et al. (2007). Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods 5: 353–362. [Google Scholar]
- Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. (2011). Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One 6: e16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER. (2012). The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3: e00036–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann U, Luta G, Wand MP. (2010). The curvHDR method for gating flow cytometry samples. BMC Bioinform 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359. [DOI] [PubMed] [Google Scholar]
- Palinska KA, Laloui W, Bédu S, Loiseaux-de Goër S, Castets AM, Rippka R et al. (2002). The signal transducer PII and bicarbonate acquisition in Prochlorococcus marinus PCC 9511, a marine cyanobacterium naturally deficient in nitrate and nitrite assimilation. Microbiology 148: 2405–2412. [DOI] [PubMed] [Google Scholar]
- Partensky F, Hess WR, Vaulot D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63: 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. (2004). Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA 101: 18228–18233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. (2008). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2010), R: A language and environment for statistical computing. http://www.R-project.org/.
- Roberts EW, Cai F, Kerfeld CA, Cannon GC, Heinhorst S. (2012). Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J Bacteriol 194: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KM, Henn-Sax M, Harmer TL, Longo DL, Frame CH, Cavanaugh CM. (2007). Kinetic isotope effect and biochemical characterization of form IA RubisCO from the marine cyanobacterium Prochlorococcus marinus MIT9313. Limnol Oceanogr 52: 2199–2204. [Google Scholar]
- Shi D, Xu Y, Morel FMM. (2009). Effects of the pH/pCO2 control method on medium chemistry and phytoplankton growth. Biogeosciences 6: 1199–1207. [Google Scholar]
- Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H et al. (2002). Genes essential to sodium-dependent bicarbonate transport in cyanobacteria. J Biol Chem 277: 18658–18664. [DOI] [PubMed] [Google Scholar]
- Steglich C, Futschik M, Rector T, Steen R, Chisholm SW. (2006). Genome-wide analysis of light sensing in Prochlorococcus. J Bacteriol 188: 7796–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. (2008). The challenge of regulation in a minimal photoautotroph: Non-coding RNAs in Prochlorococcus. PLoS Genet 4: e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Pellegrino M, Wittkowski KM, Magnasco MO. (2005). Harshlight: a ''corrective make-up” program for microarray chips. BMC Bioinform 6: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Huang K, Saito MA, Chisholm SW. (2011). Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J 5: 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen AC, Aach J, Lindell D, Johnson ZI, Rector T, Steen R et al. (2006). Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol Syst Biol 2: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH. (2006). Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci USA 103: 8607–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mooy BAS, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblížek M et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458: 69–72. [DOI] [PubMed] [Google Scholar]
- Vaulot D, Marie D, Olson RJ, Chisholm SW. (1995). Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial Pacific Ocean. Science 268: 1480–1482. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Misawa Y, Mochiji S, Kamiya R. (2011). Reduction-oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 108: 11280–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Kieselbach T, Komenda J, Promnares K, Hernández Prieto MA, Tichy M et al. (2007). Localization of the small CAB-like proteins in photosystem II. J Biol Chem 282: 267–276. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. (2007). Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr 52: 2205–2220. [Google Scholar]
- Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML et al. (2009). Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One 4: e5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.