Summary
Hepatotoxicity, including drug-induced liver injury, is frequently accompanied by cell death. The latter is typically driven by apoptosis or necrosis, which substantially differ based upon biochemical and morphological criteria. The present chapter describes 2 commonly used methods to probe apoptotic and necrotic activities in adherent monolayer cultures of primary hepatocytes. The apoptosis assay uses a prototypical substrate of caspase 3, the main executor of apoptotic cell death, which can be cleaved, yielding a product that can be measured fluorimetrically. The second assay relies on the disruption of the cell plasma membrane, which typically occurs in necrotic cell death and that results in the extracellular release of cytoplasmic enzymes, such as lactate dehydrogenase. The latter can be indirectly assessed by spectrophotometrically measuring the consumption of reduced nicotinamide adenine dinucleotide.
Keywords: primary hepatocyte culture, apoptosis, caspase 3, necrosis, lactate dehydrogenase
1. Introduction
Drug-induced liver injury is a ubiquitous issue in clinical settings and pharmaceutical industry. Recently, a unifying mechanistic model of drug-induced liver injury has been introduced. According to this model, drug-induced hepatotoxicity relies on 3 consecutive steps, namely an initial cellular insult that leads to the occurrence of mitochondrial permeability transition, which in turn ultimately burgeons into the onset of cell death. The latter may be manifested as apoptosis or necrosis (1-3). Apoptosis is a genetically programmed and well-orchestrated process that can be triggered via 2 routes, namely through the mitochondria-mediated intrinsic cascade or through the death receptor-mediated extrinsic pathway. Both routes rely on the proteolytic activity of cystein proteases, the caspases, which are expressed as inactive pro-enzymes that become activated upon proteolytic cleavage. As such, 2 subsets of caspases can be distinguished, namely the initiator caspases and the effector caspases. The former, including caspase 8, caspase 9 and caspase 10, mediate the cleavage and thus the activation of effector caspases. Effector caspases, such as caspase 3, caspase 6 and caspase 7, subsequently cleave a large number of cellular proteins, like major cytoplasmic and nuclear elements, which forms the biochemical basis of the apoptotic morphological phenotype. The extrinsic apoptotic pathway is initiated by binding of a specific subset of ligands, such as Fas ligand, to their corresponding receptors at the cell plasma membrane surface, resulting in the proteolytic cleavage and auto-activation of procaspase 8. Activated caspase 8 induces caspase 3, which subsequently cleaves its cellular targets. In the intrinsic apoptotic pathway, cytochrome C is released from mitochondria, a process that is mediated by members of the B-cell lymphoma-2 protein family. Cytochrome C triggers caspase 9 activation, which then induces caspase 3 (4-9). Necrosis, as opposed to apoptosis, is a rather passive and unorganized process that is caused by a plethora of external stress factors, including extremely high concentrations of xenobiotics. It usually starts with the loss of ion homeostasis, which eventually evokes cell swelling, loss of cell plasma membrane integrity, and cell lysis (4, 5, 9-12).
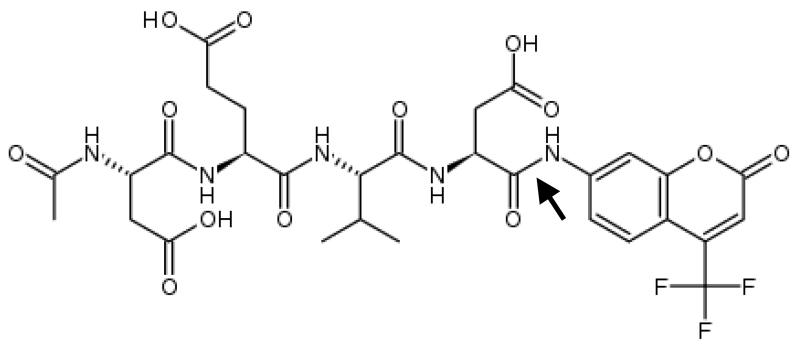
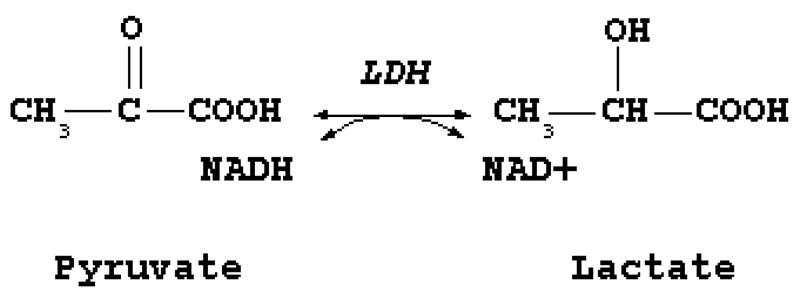
The current chapter describes 2 methods to measure apoptotic and necrotic cell death in primary hepatocyte cultures at the activity level, using an established in vitro model of Fas-mediated cell death (13). The apoptosis activity method is based upon the use of a synthetic caspase 3 substrate called acetyl-aspartic acid-glutamic acid-valine-aspartic acid-7-aminotrifluoromethylcoumarin (Ac-DEVD-AFC) (Fig. 1) (see Note 1). When the AFC label is attached to the peptide substrate, it produces a blue fluorescence upon exposure to ultraviolet light. Caspase 3 cleaves this tetrapeptide between the second aspartic acid and the AFC moiety. Released AFC produces a yellow-green fluorescence at 505 nm when exposed to ultraviolet light, which can be measured fluorimetrically. The caspase 3 activity is proportional to the amount of free AFC produced (8, 14). The assay to assess necrotic cell death relies on the disruption of the cell plasma membrane, which results in the extracellular release of cytoplasmic enzymes. Among those is lactate dehydrogenase (LDH), a stable enzyme that leaks from the cell upon cell plasma membrane damage in relatively high amounts. LDH catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of reduced (NADH) and oxidized nicotinamide adenine dinucleotide (Fig. 2). The principle of the assay described in this chapter is based upon this reaction. In particular, the consumption of NADH is spectrophotometrically assessed and serves as a measure that is proportional to LDH activity (15).
Figure 1. Chemical structure acetyl-aspartic acid-glutamic acid-valine-aspartic acid-7-aminotrifluoromethyl-coumarin (Ac-DEVD-AFC).
The arrow indicates the cleavage site for caspase 3.
Figure 2. Catalytic function of lactate dehydrogenase (LDH).
LDH catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of reduced and oxidized nicotinamide adenine dinucleotide.
2. Materials
2.1. Establishment of the in vitro model of Fas-mediated hepatocellular cell death
Hepatocyte seeding medium. William’s medium E containing 7 ng/mL glucagon, 292 mg/mL L-glutamine, 7.33 IE/mL sodium benzyl penicillin, 50 μg/mL kanamycin monosulfate, 10 μg/mL sodium ampicillin, 50 μg/mL streptomycin sulfate and 10% fetal bovine serum. Prepare in a laminar air flow cabinet and store for maximum 7 days at 4°C. Prior to use, the hepatocyte seeding medium should be placed for 30 min in a thermostated bath at 37°C.
Hepatocyte culture medium. Serum-free hepatocyte seeding medium supplemented with 25 μg/mL hydrocortisone sodium hemisuccinate and 0.5 μg/mL insulin. Prepare in a laminar air flow cabinet and store for maximum 7 days at 4°C. Prior to use, the hepatocyte culture medium should be placed for 30 min in a thermostated bath at 37°C.
Hepatocyte cell death medium. Hepatocyte culture medium supplemented with 200 ng/mL Fas ligand (Alexis, Switzerland) and 2 μg/mL cycloheximide (Sigma-Aldrich, Belgium). Prepare ex tempore in a laminar air flow cabinet. Prior to use, the hepatocyte cell death medium should be placed for 30 min in a thermostated bath at 37°C.
Incubator (37°C ± 1°C, 90% ± 5% humidity, 5% ± 1% CO2).
Laminar air flow cabinet.
Thermostated bath.
2.2. Measurement of caspase 3 activity in cultured primary hepatocytes
Phosphate-buffered saline (PBS). 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4.2H2O, 1.8 mM KH2PO4 in deionized water. Adjust to pH 7.4, sterilize by passing through a 0.22 μm filter and store for maximum 6 months at 4°C. Prior to use, PBS should be placed for 30 min at room temperature.
25× concentrated reaction buffer. 250 mM 1,4-piperazinediethanesulfonic acid, 125 mM DL-dithiothreitol, 50 mM ethylenediaminetetraacetic acid tetrasodium salt dehydrate in double distilled water. Adjust pH of this solution to 7.4 (see Note 2) and store in aliquots of 2.5 mL for maximum 6 months at −20°C.
Reaction buffer. 1 volume part 25× concentrated reaction buffer and 24 volume parts double distilled water. Prepare this buffer ex tempore and place at least 10 min at 37°C prior to use.
N-[2-hydroxyethyl]piperazine-N’[2-ethanesulfonic acid] solution. 50 mM N-[2-hydroxyethyl]piperazine-N’[2-ethanesulfonic acid] in double distilled water. Sterilize this solution by passing through a 0.22 μm filter and store for maximum 6 months at 4°C.
Ethylenediaminetetraacetic acid tetrasodium salt dihydrate solution. 500 mM ethylenediaminetetraacetic acid tetrasodium salt dihydrate solution in double distilled water. Adjust the pH of this solution to 7.4. Sterilize by passing through a 0.22 μm filter and store for maximum 6 months at room temperature.
Phenylmethylsulfonyl fluoride solution. 100 mM phenylmethylsulfonyl fluoride in isopropanol. Protect this solution from light and store for maximum 2 weeks at −20°C.
Pepstatin A solution. 1.5 mM pepstatin A in dimethyl sulfoxide. Protect this solution from light and store for maximum 4 weeks at −20°C.
Aprotinin solution. 0.15 mM aprotinin in PBS. Protect this solution from light and store for maximum for 6 months at −20°C.
Leupeptin solution. 2.1 mM leupeptin in PBS. Protect this solution from light and store for maximum 4 weeks at −20°C.
Lysis buffer. 1 mg/mL 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 5 mM DL-dithiothreitol, 20% N-[2-hydroxyethyl]piperazine-N’[2-ethanesulfonic acid] solution, 0.4% ethylenediaminetetraacetic acid tetrasodium salt dihydrate solution, 1% phenylmethylsulfonyl fluoride solution, 1% pepstatin A solution, 1% aprotinin solution, 2% leupeptin solution in double distilled water. Prepare this buffer ex tempore (see Note 3).
Ac-DEVD-AFC solution. 1 mg/mL Ac-DEVD-AFC in dimethyl sulfoxide. Protect this solution from light and store for maximum 6 months at −20°C.
AFC solution. 200 mM AFC in dimethyl sulfoxide. Protect this solution from light and store for maximum 2 weeks at room temperature.
Amber microcentrifuge tubes (VWR, Belgium).
Black 96-well plates (VWR, Belgium).
Multiplate reader.
2.3. Measurement of the extracellular release of lactate dehydrogenase in cultured primary hepatocytes
LDH reaction buffer (DiaSys, Germany). 4 volume parts of reagent 1 (i.e. NADH dissolved in 1 mM Good’s buffer, pH 9.6) and 1 part of reagent 2 (i.e. pyruvate in 800 μM phosphate buffer, pH 7.5). Prepare this buffer ex tempore and place in a thermostated bath at 25°C before use.
Plastic microcuvettes.
Spectrophotometer, computer and software.
Ultrasonicator.
3. Methods
3.1. Establishment of a monolayer culture of primary hepatocytes and induction of cell death
Use freshly isolated primary rat hepatocytes (16).
Evenly plate the hepatocytes on plastic culture dishes at a density of 0.56 × 105 cells/cm2 in hepatocyte seeding medium. Place the cell cultures in an incubator at 37°C and 5% CO2 for 4 h.
Remove the hepatocyte seeding medium and replace by identical volumes of hepatocyte culture medium. Place the cell cultures in an incubator at 37°C and 5% CO2 for 24 h.
Replace the hepatocyte culture medium. Place the cell cultures in an incubator at 37°C and 5% CO2 for 20 h.
Remove the hepatocyte culture medium and replace by identical volumes of hepatocyte cell death medium. Place the cell cultures in an incubator at 37°C and 5% CO2.
Sample at the start of cell death induction and 2, 4 and 6 h thereafter.
3.2. Measurement of caspase 3 activity in cultured primary hepatocytes
3.2.1. Sampling from cultured primary hepatocytes
Remove the hepatocyte cell death medium from 10 cm diameter culture dishes and wash the cells twice with 10 mL PBS (see Note 4).
Add 5.5 ml PBS to the culture dish and scrape the cells (see Note 5).
Transfer the cell suspension to a 15 mL centrifuge tube on ice.
Repeat steps 2 and 3.
Centrifuge for 5 min at 4332xg and 4°C.
Gently remove the supernatant and resuspend the pellet in 1 mL ice-cold PBS.
Transfer the cell suspension to a 2 mL microcentrifuge tube on ice.
Centrifuge for 5 min at 2040xg and 4°C.
Remove the supernatant and directly merge the microcentrifuge tube into liquid nitrogen.
Stock the microcentrifuge tubes, containing the dry pellets, at −80°C.
3.2.2. Preparation of cell homogenates
Defrost the microcentrifuge tubes on ice.
Add 100 μL lysis buffer to the microcentrifuge tubes and resuspend by pipetting up and down (see Note 5).
Leave the microcentrifuge tubes on ice for 20 min.
Merge the microcentrifuge tubes in liquid nitrogen and subsequently thaw at 37°C in a thermostated bath for 2 min.
Repeat step 4 twice without mixing the microcentrifuge tube between the freeze-thaw cycles.
Centrifuge for 30 min at 16065xg and 4°C.
Transfer 5 μL of the supernatant to another microcentrifuge tube, add 95 μL double distilled water and determine the protein concentration (see Note 6).
Transfer the remaining supernatant to a new microcentrifuge tube, merge into liquid nitrogen and store at −80°C.
3.2.3. Measurement of caspase 3 activity in the standards
Prepare the reaction buffer and the AFC solution.
Prepare a series of AFC standards (Table 1) in 2 mL amber microcentrifuge tubes protected from light.
Add 240 μL of each standard to a corresponding well in a black 96-well plate.
Repeat step 3 for the same standard.
Repeat steps 3 to 4 for the remaining standards.
Place the black 96-well plate in the multiplate reader and measure the fluorescence at 37°C with λex 390 nm and λem 500 nm (see Note 7).
Table 1. Preparation of the 7-amino-trifluoromethylcoumarin (AFC) standards.
| Standard number | Concentration AFC (nM) | Reaction buffer (μL) | Solution (μL) |
|---|---|---|---|
| 0 (blank) | 0 | 1000 | 0 |
| 1 | 1200 | 994 | 6 (AFC solution) |
| 2 | 600 | 500 | 500 (standard 1) |
| 3 | 300 | 500 | 500 (standard 2) |
| 4 | 150 | 500 | 500 (standard 3) |
| 5 | 75 | 500 | 500 (standard 4) |
| 6 | 37.5 | 500 | 500 (standard 5) |
| 7 | 18.75 | 500 | 500 (standard 6) |
| 8 | 9.375 | 500 | 500 (standard 7) |
3.2.4. Measurement of caspase 3 activity in the samples
For each of the samples, calculate the volume that corresponds with 50 μg protein (x μL).
Mix 230-x μL double distilled water with 10 μL 25× concentrated reaction buffer and × μL sample in a well of a black 96-well plate.
Repeat step 2 twice for the same sample.
Repeat steps 2 and 3 for the remaining samples.
In a well of the same black 96-well plate, mix 230 μL double distilled water with 10 μL 25× concentrated reaction buffer.
Repeat step 5 twice for the blank.
Incubate the black 96-well plate for 2 min at room temperature under mild shaking (i.e. 120 oscillations per minute).
Add 10 μL Ac-DEVD-AFC solution to each well using a dispenser pipette.
Slightly tap the black 96-well plate and place into the multiplate reader.
Place the black 96-well plate in the multiplate reader and perform 99 measurements of the fluorescence at 37°C with λex 390 nm and λem 500 nm) over a total time frame of 60 min (see Note 7).
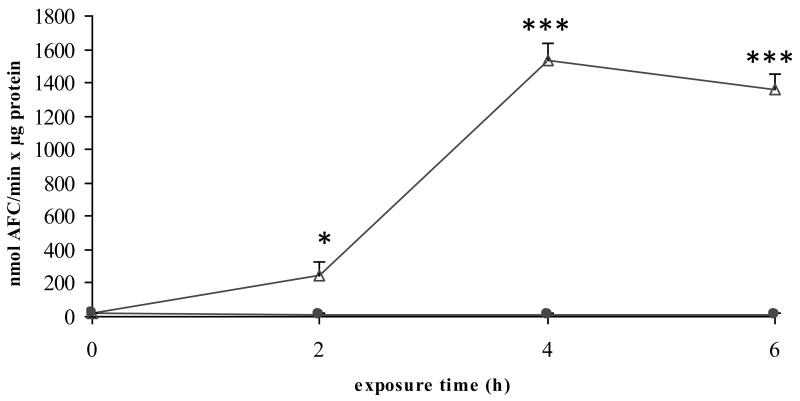
3.2.5. Processing of the results (Fig. 3) (see Note 8)
Figure 3. Measurement of caspase 3-like activity.
Freshly isolated rat hepatocytes were cultivated in a monolayer configuration and were exposed to 200 ng/mL Fas ligand and 2 μg/mL cycloheximide, starting at 44 h postplating. Samples were taken at the start of the exposure (0 h), and after 2, 4 and 6 h. Caspase 3-like activity was determined and results are expressed as nmol aminotrifluoromethylcoumarin (AFC)/min × μg protein. Results represent the means ± standard deviation of 3 independent experiments. Results were evaluated by 1-way analysis of variance followed by post hoc Bonferroni tests. ● = control; Δ = Fas ligand/cycloheximide; *p < 0.05; ***p < 0.001. Reproduced in modified form with permission from (13).
- Collect the results of the measurement of the caspase 3 activity in the standards and calculate the mathematical equation of the best-fit line:
-
Calculate the caspase 3 activity in the samples, expressed as nmol produced AFC/minute × μg protein, by completing the following equation:
With parameters:
ΔFsample = Fsample - Fblank
b = intercept of the best-fit line
250 = total reaction volume (μL)/well
a = slope of best-fit line
X = volume (μL) sample added/well
P = protein concentration (μg/μL) of the sample
60 = reaction time (min)
3.3. Measurement of the extracellular release of lactate dehydrogenase in cultured primary hepatocytes
3.3.1. Sampling from cultured primary hepatocytes
Take 500 μL cell culture medium from a 3.5 cm diameter culture dish and transfer into a recipient. Snapfreeze in liquid nitrogen and store at −20°C (see Note 9).
Scrape the cells in the remaining hepatocyte cell death medium using a policeman and transfer the cell suspension into a recipient. Snapfreeze in liquid nitrogen and store at −20°C.
3.3.2. Measurement of the lactate dehydrogenase activity
Defrost the microcentrifuge tubes on ice.
Homogenize the content of the cell culture medium and the cell suspension by ultrasonication with 20 pulses and 2 × 20 pulses with repeat duty cycle 0.5 s, respectively (see Note 10).
Mix 1 mL LDH reaction buffer with 20 μL homogenized cell suspension in a plastic microcuvette and place in the spectrophotometer (see Note 11).
After exactly 1 min, measure the decrease in absorption (ΔC/min) at 340 nm for a total of 1 min against an air blank (see Note 12).
Repeat steps 3 and 4 twice.
Repeat steps 3 to 5 for the corresponding homogenized cell culture medium.
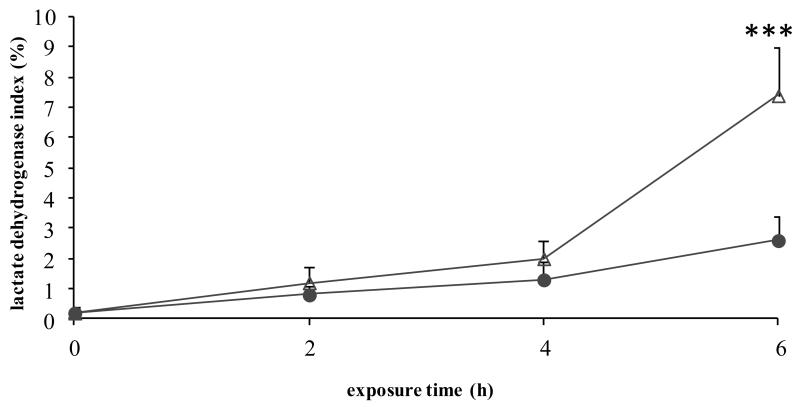
3.3.3. Processing of the results (Fig. 4) (see Note 8)
Figure 4. Measurement of lactate dehydrogenase leakage.
Freshly isolated rat hepatocytes were cultivated in a monolayer configuration and were exposed to 200 ng/mL Fas ligand and 2 μg/mL cycloheximide, starting at 44 h postplating. Samples were taken at the start of the exposure (0 h), and after 2, 4 and 6 h. Lactate dehydrogenase activity was measured and the lactate dehydrogenase index was calculated as [(100 × lactate dehydrogenase activity in supernatant)/(lactate dehydrogenase activity in cell suspension)]. Results represent the means ± standard deviation of 3 independent experiments. Results were evaluated by 1-way analysis of variance followed by post hoc Bonferroni tests. ● = control; Δ = Fas ligand/cycloheximide; ***p < 0.001. Reproduced in modified form with permission from (13).
Collect and average the 3 ΔC/min values for both the cell suspension (i.e. total LDH activity) and the cell culture medium (i.e. extracellular LDH activity).
- Calculate the LDH index by assessing the relative extracellular release of LDH, using the following equation:
Table 2. Troubleshooting of the caspase 3-like activity measurement procedure.
| Problem | Cause | Solution |
|---|---|---|
| Negative testing results | Decomposed reaction buffer and solutions | Prepare new reaction buffer and solutions and protect from light |
| Air bubbles in the wells of the black 96-well plate | Tap the black 96-well plate before incubation in the multiplate reader | |
|
| ||
| Divergent testing results | Incomplete lysis of the cells | Prepare new lysis buffer and strongly pipette up and down when lyzing cells |
| Variation in temperature | Check temperature in the multiplate reader and ensure a constant temperature of 37°C | |
| Variation in pH and crystallization of the reaction buffer | Check the pH of the reaction buffer and adjust to 7.4 | |
Table 3. Troubleshooting of the lactate dehydrogenase (LDH) assay.
| Problem | Cause | Solution |
|---|---|---|
| Negative testing results | Decomposed LDH reaction buffer | Prepare new LDH reaction buffer and store for a maximum of 5 days or 8 h at 4°C or 25°C, respectively |
| Air bubbles in the microcuvette | Shake the microcuvette upon mixing the LDH reaction buffer and the sample | |
|
| ||
| Divergent testing results | Incomplete homogenization of samples | Subject samples to an additional series of 20 ultrasonication pulses |
| Variation in temperature | Check temperature and store the LDH reaction buffer at 25°C | |
Acknowledgements
This work was financially supported by the grants of the University Hospital of the Vrije Universiteit Brussel - Belgium (Willy Gepts Fonds UZ-VUB), the University of São Paulo - Brazil (USP), the São Paulo Research Foundation - Brazil (FAPESP), the Fund for Scientific Research - Flanders (FWO-Vlaanderen), the European Research Council (project CONNECT), the European Union (FP7) and Cosmetics Europe (projects DETECTIVE and HeMiBio).
Footnotes
With respect to the specificity of the apoptosis assay, it should be kept in mind that Ac-DEVD-AFC not only is a substrate for caspase 3, but also for other caspases, including caspase 1, caspase 4, caspase 6, caspase 7, caspase 8 and caspase 10 (8). Hence, the enzymatic activity measured is not restricted to caspase 3 and thereby should be rather considered as caspase 3-like activity.
The pH of the reaction buffer is a crucial parameter and should be checked regularly. Deviation of the pH can result in crystallization of the compounds in the reaction buffer, which in turn will affect the outcome of the assay (Table 2).
Dissolve the 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate and the DL-dithiothreitol in 746 μL double distilled water when preparing a volume of 1 mL lysis buffer and subsequently add the different solutions.
This washing step could remove dead cells and therefore may be omitted.
The diameter of the culture dish used mainly depends on the purpose of the assay intended. For measurement of caspase 3 activity, it is strongly recommended to use 10 cm diameter culture dishes with hepatocytes plated at a density of 0.57 × 105 cells/cm2 (i.e. a total of 4.4 × 106 hepatocytes). When using 6 cm diameter culture dishes (i.e. a total of 1.6 × 106 hepatocytes), harvesting of the cells and preparation of cell homogenates should be performed with 3 mL PBS and 50 μL lysis buffer, respectively.
Among the different assays to assess protein concentration in biological samples, the Bradford method (17), is the most commonly used one. In this assay, which has been commercialized (Bio-Rad, Belgium), a protein sample is added to a solution of the dye Coomassie Brilliant Blue G-250 in phosphoric acid and ethanol. Under the acid conditions, the dye normally has a brownish colour, but upon binding to the protein, the blue form of the dye is produced. The optical absorbance of the solution is measured at a wavelength of 595 nm and is proportional to the amount of protein present and bound to the dye. It is a relative method, whereby bovine serum albumin is a typical standard used. Clearly, other assays, such as the bicinchoninic acid-based method (18) and the Lowry assay (19), can also be addressed to assess protein concentration.
Temperature is a critical parameter in this assay, as it strongly influences protein stability and hence caspase 3 activity. Thus, it is of the utmost importance to ensure a constant temperature of 37°C in the cabinet of the multiplate reader before and during incubation of the black 96-well plate (Table 2).
From a statistical point of view, it is strongly recommended to perform the experiments on hepatocytes isolated from the livers of 3 different animals. Results can be processed and evaluated by 1-way analysis of variance followed by post hoc Bonferroni tests.
The diameter of the culture dish used mainly depends on the purpose of the assay intended. For measurement of the extracellular release of LDH, it is sufficient to use 3.5 cm diameter culture dishes, though culture dishes with a higher diameter can also be used.
It is of the utmost importance to continuously keep samples on ice during ultrasonication, since this procedure produces heat, which may cause proteolysis and hence loss of enzyme activity.
Temperature is a critical parameter in this assay, as it strongly influences enzyme activity. Therefore, it is important to continuously store the LDH reaction buffer at 25°C and to regularly check the temperature. The procedure may also be performed at 37°C, though in this case only 10 μL homogenized sample must be mixed with 1 mL LDH reaction buffer and a correction factor of 16030 must be handled during processing of the results (see Note 12).
The assessment of the enzymatic activity relies on modifications in NADH concentration as a function of time (ΔC/min). Upon implementation of the Beer-Lambert law, this yields the equation (ΔA/min)/(ε×l), whereby the parameters ΔA, ε and l represent the decrease in absorption, the molecular absorption coefficient (cm−1mol−1l) and the optical path (1 cm), respectively. When applied to LDH activity, this gives [(ΔA/min) × 106]/(ε×l). In this equation, LDH activity is expressed in units per liter, whereby 1 unit corresponds with the quantity of LDH that catalyzes the conversion of 1 μmol NADH/min at 25°C. This formula can be simplified by taken into account an established correction factor of 8095, as experimentally established by the manufacturer. Consequently, LDH activity can be calculated as (ΔA/min) × 8095. The LDH activity is automatically calculated by the computer software upon measurement of the samples (Table 3).
References
- 1.Vinken M, Maes M, Vanhaecke T, et al. Drug-induced liver injury: mechanisms, types and biomarkers. Curr Med Chem. 2013;20:3011–3021. doi: 10.2174/0929867311320240006. [DOI] [PubMed] [Google Scholar]
- 2.Russmann S, Kullak-Ublick GA, Grattagliano I. Current concepts of mechanisms in drug-induced hepatotoxicity. Curr Med Chem. 2009;16:3041–3053. doi: 10.2174/092986709788803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinken M, Maes M, Oliveira AG, et al. Primary hepatocytes and their cultures in liver apoptosis research. Arch Toxicol. 2014;88:199–212. doi: 10.1007/s00204-013-1123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaeschke H, Gores GJ, Cederbau AI, et al. Mechanisms of hepatotoxicity. Toxicol Sci. 2002;65:166–176. doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- 5.Raffray M, Cohen GM. Apoptosis and necrosis in toxicology: a continuum or distinct modes of cell death? Pharmacol Ther. 1997;75:153–177. doi: 10.1016/s0163-7258(97)00037-5. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Lechon MJ, O’Connor E, Castell JV, et al. Sensitive markers used to identify compounds that trigger apoptosis in cultured hepatocytes. Toxicol Sci. 2002;65:299–308. doi: 10.1093/toxsci/65.2.299. [DOI] [PubMed] [Google Scholar]
- 7.Gill GH, Dive D. Apoptosis: basic mechanisms and relevance to toxicology. In: Roberts R, editor. Apoptosis in toxicology. Taylor & Francis; London: 2000. pp. 1–20. [Google Scholar]
- 8.Yin X-M, Dong Z. Essentials of apoptosis: a guide for basic and clinical research. Humana Press; New Jersey: 2003. [Google Scholar]
- 9.Feldmann G. Liver apoptosis. J Hepatol. 1997;26:1–11. doi: 10.1016/s0168-8278(97)80491-6. [DOI] [PubMed] [Google Scholar]
- 10.Decrock E, Vinken M, De Vuyst E, et al. Connexin-related signaling in cell death: to live or let die? Cell Death Differ. 2009;16:524–536. doi: 10.1038/cdd.2008.196. [DOI] [PubMed] [Google Scholar]
- 11.Malhi H, Gores GJ. Cellular and molecular mechanisms of liver injury. Gastroenterology. 2008;134:1641–1654. doi: 10.1053/j.gastro.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulze-Bergkamen H, Untergasser A, Dax A, et al. Primary human hepatocytes: a valuable tool for investigation of apoptosis and hepatitis B virus infection. J Hepatol. 2003;38:736–744. doi: 10.1016/s0168-8278(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 13.Vinken M, Decrock E, De Vuyst E, et al. Biochemical characterization of an in vitro model of hepatocellular apoptotic cell death. Altern Lab Anim. 2009;37:209–218. doi: 10.1177/026119290903700210. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz LM, Ashwell JD. Methods in cell biology volume 66: apoptosis. Academic Press; San Diego: 2001. [DOI] [PubMed] [Google Scholar]
- 15.Bergmeyer HU. Lactate dehydrogenase. In: Bergmeyer HU, editor. Methods of enzymatic analysis. Academic Press; New York: 1974. pp. 574–579. [Google Scholar]
- 16.Papeleu P, Vanhaecke T, Henkens T, et al. Isolation of rat hepatocytes. Methods Mol Biol. 2006;320:229–237. doi: 10.1385/1-59259-998-2:229. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Smith PK, Krohn RI, Hermanson GT, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 19.Lowry OH, Rosebrough NJ, Farr AL, et al. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]