Abstract
Background
The Wnt signaling pathway is abnormally activated in many human cancers. Secreted frizzled-related proteins (SFRPs) function as negative regulators of Wnt signaling and play an important role in carcinogenesis. SFRP promoter hypermethylation has often been identified in human cancers; however, the precise role of SFRPs in cutaneous squamous cell carcinoma (SCC) is unclear.
Methods
The methylation status of the SFRP family was analyzed in an age-and sex-matched case-control study, including 40 cutaneous SCC cases and 40 normal controls, using the MassARRAY EpiTYPER system.
Results
The methylation rate of SFRP1, SFRP2, SFRP4, and SFRP5 promoters was significantly higher in cutaneous SCC tissues than in adjacent tissue and normal skin samples.
Discussion
Our manuscript mainly discussed the average methylation rate of SFRPs (SFRP1, SFRP2, SFRP4, and SFRP5) promoters are significantly high in tumor tissue samples and the average CpG island methylation rate among different pathological levels of cutaneous SCC between these genes are different.
Conclusions
Our findings suggest that promoter hypermethylation of SFRPs is associated with the development of carcinoma, and could be a useful tumor marker for cutaneous SCC and other types of cancers.
Keywords: Cutaneous squamous cell carcinoma, Signaling pathway, DNA methylation, SFRP
Background
Non-melanoma skin carcinomas, comprising cutaneous squamous cell carcinoma (SCC) and basal cell carcinoma (BCC), are the most common malignancies in fair-skinned Caucasians [1]. Unlike almost all basal cell carcinomas, cutaneous SCCs are associated with a substantial risk of metastasis [2]. The etiology of cutaneous SCC is multifactorial, with both environmental and host factors being important. However, exposure to ultraviolet radiation is the most common cause [3, 4]. Other risk factors include impaired immune surveillance, as seen in organ transplant recipients, and human papillomavirus infections [5].
The Wnt family of proteins includes a wide range of secreted growth factors that regulate cell differentiation, proliferation, migration, and organogenesis during embryonic development [6]. Aberrant activation of the Wnt pathway has been observed to inhibit tumor cell apoptosis during human carcinogenesis [7, 8]. Wnt signaling is activated on binding of the Wnt ligand to the frizzled membrane receptor [9], and a family of five secreted frizzled-related glycoproteins (SFRP1-5) has been identified as modulators of Wnt signaling [10]. Moreover, the epigenetic silencing of SFRP genes, leading to oncogenic activation of the Wnt pathway and tumor progression was reported in many human cancers [11–14].
Although the involvement of Wnt signaling in carcinogenesis has been extensively studied, the associations of the SFRP family with tumorigenesis have only recently begun to be explored. Previous studies have reported SFRP downregulation in colorectal cancer, gastric cancer [15], and invasive breast tumors [16, 17]. Additionally, Wnt5a signaling was found to contribute to tissue invasion by non-melanoma skin cancer, including both SCC and BCC [18]. Although non-melanoma skin cancer is common, the incidence in China is very low, and differences have been demonstrated between Caucasians and Hong Kong Chinese patients regarding disease features [19]. In the present study, we used the Mass ARRAY EpiTYPER system to examine the methylation status of SFRP family members, including SFRP1, SFRP2, SFRP4, and SFRP5 in different tissue samples from a Chinese population of the Xinjiang Uygur Autonomous Region.
Methods
Study populations and tissue samples
To investigate the methylation status of the SFRP family, we conducted an age- and gender- matched case-control study including 40 cutaneous SCC cases and 40 normal controls. The patients were recruited from the Dermatology Department of the People’s Hospital of Xinjiang Uygur Autonomous Region (Xinjiang, China), and had received a diagnosis of histologically confirmed cutaneous SCC between January 2012 and February 2014. Controls were normal individuals from the Plastic Surgery Department of the hospital who reported no history of any type of cancer at the time of study recruitment. The basic characteristics for the participants included in this study are presented in Table 1. Cutaneous SCC tissues (n = 40) and adjacent tissues (n = 40) were obtained from the head or the hands/legs of the 40 cutaneous SCC patients. Normal skin samples (n = 40) were collected from the face or the hands/legs of the 40 controls. This study was approved by the ethics committee of the People’s Hospital of Xinjiang Uygur Autonomous Region. Written informed consent was also obtained from each participant enrolled in the study.
Table 1.
Basic characteristics of the cases and controls in this study
| Variables | Case (N = 40) | Control (N = 40) | Total | p-value |
|---|---|---|---|---|
| Sex, No. (%) | 0.749* | |||
| Male | 21 (52.50) | 22 (55.00) | 43 | |
| Female | 19 (47.50) | 18 (45.00) | 37 | |
| 40 | 40 | |||
| Grade, No. (%) | ||||
| Stage I | 21 (52.50) | |||
| Stage II | 12 (30.00) | |||
| Stage III | 7 (17.50) | |||
| Stage IV | 0 | |||
| Mean age ± SD | 67.11 ± 9.24 | 64.31 ± 8.91 | 0.707** |
*P values were calculated from two-sided chi-squared tests
**P values were calculated by Student’s t-tests
DNA extraction, sodium bisulfite modification, and PCR
Genomic DNA from tissue samples was extracted using the Tissue DNA Kit (Qiagen) according to the manufacturer’s recommendations. The DNA concentration and quality were measured using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Houston, TX). Bisulfite modification of genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s protocol. Bisulfite-treated genomic DNA was amplified in a 384-well plate using HotStar Taq Polymerase in a 5-μl reaction volume (Qiagen). PCR conditions were 94 °C for 4 min followed by 45 cycles of 95 °C for 20 s, 56 °C for 20 s, and 72 °C for 60 s, with a final extension of 72 °C for 3 min. Primer sequences are given in Table 2.
Table 2.
Primer sequences for amplifying SFRP genes
| Gene | Forward | Reverse | bp | CpGs | Tm | |
|---|---|---|---|---|---|---|
| SFRP1 | SFRP1_1 | aggaagagagTATGTGTGTTTGAGTGATGGATTTG | cagtaatacgactcactatagggagaaggctAAACAACACCTCTCCAAATAAAACC | 290 | 7 | 56 |
| SFRP1_2 | aggaagagagAGTTTGGGAGGTTAAGGTAGGAGTA | cagtaatacgactcactatagggagaaggctAAAACCTAAATCATACTTACAAACCCAT | 300 | 12 | 56 | |
| SFRP2 | SFRP2_1 | aggaagagagGGTTAGGTTTTTTTGTTTGTTGTTT | cagtaatacgactcactatagggagaaggctCACAACCAAAATTTTCTTAACCTTT | 235 | 9 | 56 |
| SFRP2_2 | aggaagagagGGGGATGAATGAGTTAATTTTAGTT | cagtaatacgactcactatagggagaaggctAAAAATCTCACCAATCACACAAAAC | 296 | 9 | 56 | |
| SFRP4 | SFRP4_1 | aggaagagagGTATGTGTGTTTGAGTGATGGATTT | cagtaatacgactcactatagggagaaggctTCCCAACTAACAAAAATTCAAAAAA | 314 | 6 | 56 |
| SFRP4_2 | aggaagagagTTTTGAATTTTTGTTAGTTGGGAAA | cagtaatacgactcactatagggagaaggctAAAACCTAAATCATACTTACAAACCCA | 390 | 36 | 56 | |
| SFRP5 | SFRP5_1 | aggaagagagGTATGTGTGTTTGAGTGATGGATTT | cagtaatacgactcactatagggagaaggctTCCCAACTAACAAAAATTCAAAAAA | 323 | 6 | 56 |
| SFRP5_2 | aggaagagagTTTTGAATTTTTGTTAGTTGGGAAA | cagtaatacgactcactatagggagaaggctAAAACCTAAATCATACTTACAAACCCA | 329 | 6 | 56 | |
SFRP_1 and SFRP_2 refer to two amplified fragments of SFRP
Quantitative DNA methylation analysis by MassARRAY EpiTyper
The PCR products were treated according to the standard protocol (Sequenom EpiTyper Assay) by SAP treatment and T-cleavage reaction. The samples were then cleaned by Resin and were dispensed to a 384 SpectroCHIP by Nanodispenser. DNA methylation levels were determined as previously described [20] using MassARRAY EpiTYPER (SEQUENOM Inc., Herston, QLD, Australia) system, which is based on MALDI-TOF MS [21]. The mass spectrum was collected by MassARRAY Spectrometer and analyzed by EpiTYPER v.1.0 software (SEQUENOM).
Statistical analysis
The SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA) and Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) were used for statistical analysis. The methylation levels in cutaneous SCC patients and normal samples were compared by nonparametric Mann-Whitney U test or the Kruskal-Wallis H test. All P-values presented in this study were two sided, and we used P < 0.05 as the cutoff value for statistical significance. The receiver operating characteristic (ROC) curve was used to estimate the specificity, sensitivity, and accuracy of the regression test.
Results
In the present study, we investigated the methylation status of SFRP1, SFRP2, SFRP4, and SFRP5 in 40 cutaneous SCC patients and 40 normal subjects. Table 1 showed the distribution of age, sex, and pathological level among cases and controls. The mean age for the case group was 67.11 ± 9.24 years and 64.31 ± 8.91 years for the control group. There were no significant differences in age and sex distribution between the case and control groups (p > 0.05).
We investigated the promoter methylation rate of SFRP1, SFRP2, SFRP4, and SFRP5 in 40 cutaneous SCC tissues, 40 adjacent tissues, and 40 normal samples using the MassARRAY EpiTYPER system. Aberrant promoter methylation of SFRP1, SFRP2, SFRP4, and SFRP5 was detected in cutaneous SCC tissues and adjacent tissues compared with normal samples. Additionally, the frequency of SFRP promoter methylation in tumors was significantly higher than in adjacent tissues and normal samples (Table 3).
Table 3.
Methylation frequency (%) comparison of SFRP CpG sites in three tissue groups
| Genes | No. of CpG islands | Frequency of promoter methylation (%) | ||
|---|---|---|---|---|
| T vs. N | T vs. A | A vs. N | ||
| SFRP1 | 17 | 82.35 (14/17) | 58.82 (10/17) | 29.41 (5/17) |
| SFRP2 | 14 | 50 (7/14) | 57.14 (8/14) | 42.86 (6/14) |
| SFRP4 | 18 | 72.22 (13/18) | 72.22 (13/18) | 61.11 (11/18) |
| SFRP5 | 11 | 54.54 (6/11) | 54.54 (6/11) | 54.54 (6/11) |
| P*-value | <0.05 | <0.05 | <0.05 | |
T tumor tissues; A adjacent tissues; N normal samples
*Nonparametric Mann-Whitney U test
We also assessed the average CpG island methylation rate of SFRPs among different pathological levels of cutaneous SCC. We observed a significant increase in the methylation rate of different SFRP CpG islands with higher pathological levels (Table 4). CpG island methylation of different SFRPs was shown to vary among different tissues. The methylation rate was also much higher in cutaneous SCC tissues than in adjacent tissues, but was lower in normal control samples (Fig. 1a–d).
Table 4.
Methylation frequency (%) of SFRP CpG islands at different cutaneous SCC pathological levels
| Gene | CpG Site | Stage I | Stage II | Stage III | H | P-value | |||
|---|---|---|---|---|---|---|---|---|---|
| N | N | N | |||||||
| SFRP1 | CpG1_5 | 0 | 0.4012 ± 0.098 | 0 | 0.4521 ± 0.115 | 9 | 0.4952 ± 0.109 | 6.157 | 0.024* |
| CpG1_7 | 9 | 0.4011 ± 0.134 | 6 | 0.4811 ± 0.024 | 2 | 0.5121 ± 0.124 | 5.103 | 0.031 | |
| CpG2_1 | 7 | 0.4141 ± 0.19 | 0 | 0.4267 ± 0.314 | 2 | 0.4013 ± 0.217 | 2.136 | 0.021 | |
| CpG2_8 | 5 | 0.4231 ± 0.824 | 7 | 0.4501 ± 0.224 | 4 | 0.5011 ± 0.523 | 8.514 | 0.017 | |
| SFRP2 | CpG1_5 | 44 | 0.3329 ± 0.101 | 29 | 0.3796 ± 0.170 | 19 | 0.4241 ± 0.183 | 6.142 | 0.015 |
| CpG2_1 | 47 | 0.4094 ± 0.036 | 48 | 0.4367 ± 0.316 | 17 | 0.4713 ± 0.107 | 6.136 | 0.023 | |
| CpG2_3.4 | 55 | 0.4017 ± 0.098 | 42 | 0.4406 ± 0.194 | 18 | 0.4891 ± 0.153 | 5.218 | 0.015 | |
| SFRP4 | CpG1_3 | 2 | 0.3029 ± 0.089 | 3 | 0.3496 ± 0.012 | 2 | 0.3867 ± 0.083 | 7.158 | 0.009 |
| CpG2_2 | 9 | 0.2994 ± 0.037 | 2 | 0.3367 ± 0.016 | 5 | 0.4113 ± 0.086 | 7.156 | 0.023 | |
| SFRP5 | CpG1_5 | 40 | 0.3014 ± 0.052 | 33 | 0.3834 ± 0.021 | 18 | 0.4345 ± 0.037 | 5.54 | 0.031 |
| CpG2_1 | 32 | 0.2941 ± 0.098 | 19 | 0.3167 ± 0.314 | 34 | 0.4013 ± 0.057 | 3.136 | 0.021 | |
N: number of samples; H: the Kruskal-Wallis H test
*p ≤ 0.05 indicates statistical significance
Fig. 1.
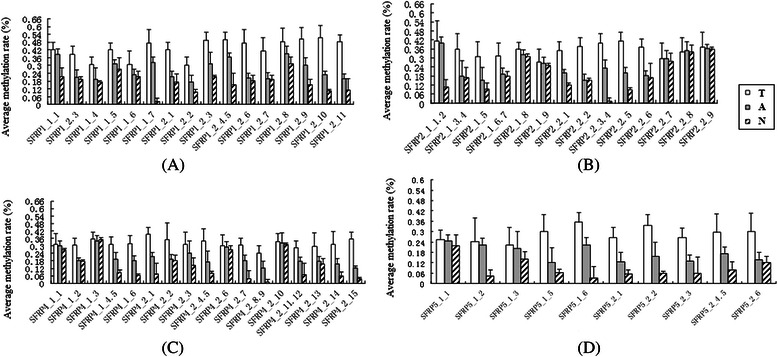
Comparison of average CpG island methylation rate in the SFRP gene family. T, tumor tissues; A, adjacent to carcinoma tissue; N, normal tissue samples
Finally, we performed ROC curve analysis of SFRP1 CpG island methylation (Fig. 2). ROC curve is a graphic presentation of the relationship between both sensitivity and specificity, and it helps to decide the optimal model through determining the best threshold for a diagnostic/predictive test [22]. The area under the curve (AUC) of the ROC curves provides a way to measure the accuracy of a diagnostic/predictive test. The larger the area, the more accurate the test is [22]. As shown in Fig. 2, all these six CpG sites showed high AUC of the ROC curves, ranging from 0.725 to 0.903. These results indicate that the detection of SFRP1 CpG methylation may be used as an early biomarker for SCC diagnosis.
Fig. 2.
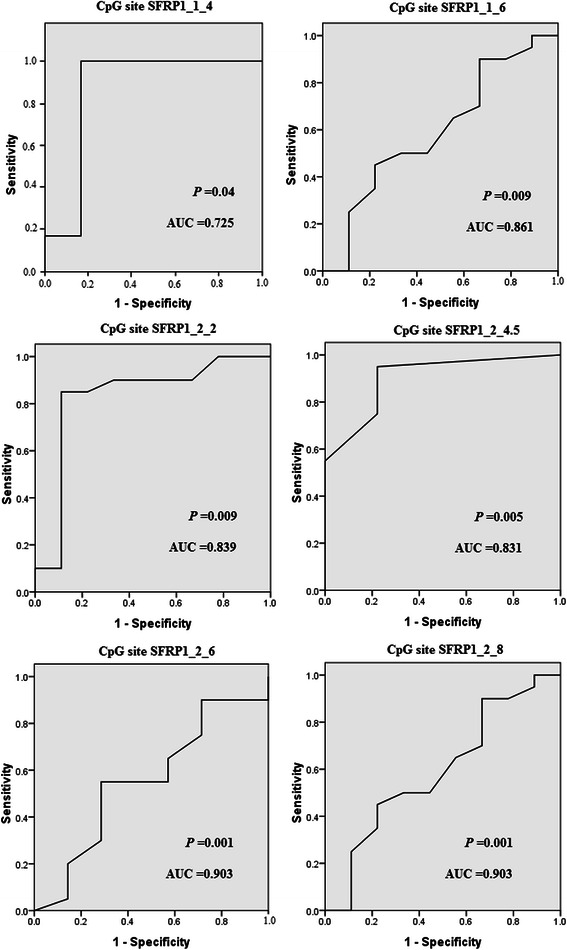
Receiver operating characteristic (ROC) curves for six CpG sites in the SFRP1 promoter. The ROC curves plot sensitivity and 1-specificity. Areas under the curve (AUC) and P values were shown in the graph
Discussion
Cutaneous SCC is the second most common cancer among Caucasians [2]. Although it has been suggested to arise through the accumulation of multiple genetic changes involving a complex multi-step process [1], the precise pathogenesis remains unclear. However, numerous studies have recently shown that the epigenetic dysregulation of tumor suppressor genes could serve as a biomarker to predict the diagnosis and prognosis of human disease and malignancy [23–25].
In the present study, we describe the aberrant DNA methylation of four members of the SFRP family, SFRP1, SFRP2, SFRP4, and SFRP5, in cutaneous SCC in a Chinese population. Methylation of these genes has been reported previously in lung cancer, prostate cancer, colorectal cancer [26], and human glioblastoma [27]. Additionally, several studies have reported the lack of methylation in nonmalignant tissues and described methylation as a tumor-restricted event [23, 28, 29]. Here, we showed that frequent promoter hypermethylation of SFRPs was significantly elevated in cutaneous SCC samples compared with adjacent tissues and control samples. Interestingly, we also observed marked differences in the levels of aberrant DNA methylation between these genes (SFRP1, SFRP2, SFRP4, and SFRP5), suggesting that hypermethylation of these SFRPs, particularly of SFRP1, could be important in the onset of cutaneous SCC.
Aberrant hypermethylation of CpG islands in gene promoters was previously shown to be a primary mechanism in the inactivation of several tumor suppressor genes [30]. We assessed the methylation status of the CpG islands in SFRP promoters in different pathological levels of cutaneous SCC tissue, and found the methylation rate to increase with the pathological level. ROC curve analysis showed that the SFRP1 CpG site is a potential biomarker of cutaneous SCC, and the further identification of genes that are methylated during cutaneous SCC carcinogenesis together with an improvement in the quantitative rather than the qualitative analysis of methylation will be of great value in the future molecular screening of cutaneous SCC.
The potential importance of SFRP silencing in cutaneous SCC relates to its involvement in the Wnt signaling pathway. This pathway is involved in the tumorigenesis of many human cancers, including those of the colon, breast, lung, and liver, as well as melanomas [7]. Chung et al. also recently demonstrated the frequent aberrant methylation of SFRPs in cervical SCC [31]. Additionally, two further studies indicated that frequent aberrant methylation of SFRPs occurs in hepatocellular carcinoma, and that restoration of SFRP1 attenuates Wnt signaling, decreases the abnormal accumulation of β-catenin in the nucleus, and suppresses cell growth [32, 33]. To date, however, the regulation of the Wnt pathway in cutaneous SCC has remained unclear. Pourreyron et al. [18] demonstrated that Wnt5a is overexpressed in non-melanoma skin cancer, which contributes to tissue invasion. Additionally, the study presented here also suggests the upregulation of SFRP1 and SFRP2 expression in cutaneous SCC, in particular SFRP2, exhibit very high constitutive expression in normal skin. The data provided by the Human Protein Atlas on cutaneous SCC (www.proteinatlas.org) showing that the SFRPs (SFRP1, SFRP2, SFRP4, and SFRP5) exhibit very low expression in cancer tissues, except SFRP1, which was slightly high. Our results showed that the SFRPs were highly methylated in cutaneous SCC, which in accordance with the data of the Human Protein Atlas on cutaneous SCC. Nevertheless, much work remains to be done to obtain a clearer understanding of the role of SFRPs in Wnt signaling and tumor pathogenesis.
Conclusions
In conclusion, our study demonstrates that promoter hypermethylation of SFRPs (SFRP1, SFRP2, SFRP4, and SFRP5) is associated with the development of cutaneous SCC. Although it is not possible to determine whether SFRPs are directly related to carcinogenesis from this study, the aberrant methylation of SFRPs may nevertheless be a useful tumor biomarker for early diagnosis. Despite the current study possessing enough statistical power, some limitations should be considered. First, we limited our investigation to only four SFRPs: SFRP1, SFRP2, SFRP4, and SFRP5. Therefore, we cannot exclude the possibility that other genes may play a role in the pathogenesis of cutaneous SCC. Second, the sample size of our study was relatively small, so our findings should be confirmed in larger case-control studies.
Acknowledgement
This work was supported by grants from the Natural Science Foundation of China (Grant number: 81260311), and the International Science and Technology Cooperation Project of Xinjiang Uyghur Autonomous Region (20146022).
Abbreviations
- SCC
Squamous cell carcinoma
- BCC
Basal cell carcinoma
- SFRPs
Secreted frizzled-related proteins
- ROC
Receiver operating characteristic
- AUC
Area under the curve
Footnotes
Junqin Liang and Xiaojing Kang contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JL, XK, and YH designed the study, carried out the molecular genetic studies, participated in the statistical analysis and drafted the manuscript. XZ, TJ, and MZ participated in the molecular genetic studies and statistical analysis. DL, YD, and YZ participated in the design of the study and performed the statistical analysis. BY and DA conceived the study, participated in its design and coordination, and helped to draft the manuscript. XP conceived the study, participated in its design and coordination, and funded the study. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Availability of data and materials
Not applicable.
Contributor Information
Junqin Liang, Email: junqinliang@163.com.
Xiaojing Kang, Email: xiaojkang@163.com.
Yilinuer Halifu, Email: yilinuerhalifu@163.com.
Xuewen Zeng, Email: xuewen_zeng@163.com.
Tianbo Jin, Email: tianbojin1973@gmail.com.
Mingxia Zhang, Email: zhmx9155@163.com.
Dong Luo, Email: lou_dong@163.com.
Yuan Ding, Email: yuandingagent@163.com.
Yunmin Zhou, Email: yunminzhou@163.com.
Buwajier Yakeya, Email: buwajierr@163.com.
Dilinuer Abudu, Email: dilinuerr@163.com.
Xiongming Pu, Phone: 86-991-8563678, Email: xiongmipu@163.com.
References
- 1.Hameetman L, Commandeur S, Bavinck JNB, Wisgerhof HC, de Gruijl FR, Willemze R, Mullenders L, Tensen CP, Vrieling H. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer. 2013;13(1):58. doi: 10.1186/1471-2407-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344(13):975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 3.Preston DS, Stern RS. Nonmelanoma cancers of the skin. N Engl J Med. 1992;327(23):1649–1662. doi: 10.1056/NEJM199212033272307. [DOI] [PubMed] [Google Scholar]
- 4.Thomas RF, Scotto J. Estimating increases in skin cancer morbidity due to increases in ultraviolet radiation exposure. Cancer Invest. 1983;1(2):119–126. doi: 10.3109/07357908309042414. [DOI] [PubMed] [Google Scholar]
- 5.Proby C, Harwood C, Neale R, Green A, Euvrard S, Naldi L, Tessari G, Feltkamp M, De Koning M, Quint W. A case-control study of betapapillomavirus infection and cutaneous squamous cell carcinoma in organ transplant recipients. Am J Transplant. 2011;11(7):1498–1508. doi: 10.1111/j.1600-6143.2011.03589.x. [DOI] [PubMed] [Google Scholar]
- 6.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11(24):3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 7.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14(15):1837–1851. [PubMed] [Google Scholar]
- 8.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275(5307):1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 9.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 10.Jones SE, Jomary C. Secreted Frizzled‐related proteins: searching for relationships and patterns. Bioessays. 2002;24(9):811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 11.Caldwell GM, Jones C, Gensberg K, Jan S, Hardy RG, Byrd P, Chughtai S, Wallis Y, Matthews GM, Morton DG. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64(3):883–888. doi: 10.1158/0008-5472.CAN-03-1346. [DOI] [PubMed] [Google Scholar]
- 12.Lee AY, He B, You L, Dadfarmay S, Xu Z, Mazieres J, Mikami I, McCormick F, Jablons DM. Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene. 2004;23(39):6672–6676. doi: 10.1038/sj.onc.1207881. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H, Watkins DN, Jair K-W, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, van Engeland M. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36(4):417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 14.Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, Tanaka Y, Kikuno N, Nakagawa M, Igawa M. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12(7):2109–2116. doi: 10.1158/1078-0432.CCR-05-2468. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31(2):141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z, Wang J, Han X, Zhou J, Linder S. Up-regulation of human secreted frizzled homolog in apoptosis and its down-regulation in breast tumors. Int J Cancer. 1998;78(1):95–99. doi: 10.1002/(SICI)1097-0215(19980925)78:1<95::AID-IJC15>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Wong SCC, Lo SFE, Cheung MT, Ng KOE, Tse CW, Lai BSP, Lee KC, Lo YD. Quantification of plasma β-catenin mRNA in colorectal cancer and adenoma patients. Clin Cancer Res. 2004;10(5):1613–1617. doi: 10.1158/1078-0432.CCR-1168-3. [DOI] [PubMed] [Google Scholar]
- 18.Pourreyron C, Reilly L, Proby C, Panteleyev A, Fleming C, McLean K, South AP, Foerster J. Wnt5a is strongly expressed at the leading edge in non-melanoma skin cancer, forming active gradients, while canonical Wnt signalling is repressed. PLoS One. 2012;7(2):e31827. doi: 10.1371/journal.pone.0031827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng S, Luk N, Chong L. Special features of non-melanoma skin cancer in Hong Kong Chinese patients: 10-year retrospective study. Hong Kong Med J. 2001;7(1):22–28. [PubMed] [Google Scholar]
- 20.Novakovic B, Wong NC, Sibson M, Ng H-K, Morley R, Manuelpillai U, Down T, Rakyan VK, Beck S, Hiendleder S. DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. Journal of Biological Chemistry. 2010;285(13):9583–9593. doi: 10.1074/jbc.M109.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampfrath T, Levinson SS. Brief critical review: statistical assessment of biomarker performance. Clin Chim Acta. 2013;419:102–107. doi: 10.1016/j.cca.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y-W, Chung M-T, Lai H-C, De Yan M, Shih Y-L, Chang C-C, Yu M-H. Methylation analysis of SFRP genes family in cervical adenocarcinoma. J Cancer Res Clin Oncol. 2009;135(12):1665–1674. doi: 10.1007/s00432-009-0613-5. [DOI] [PubMed] [Google Scholar]
- 24.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196(1):1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 25.Jones PA, Laird PW. Cancer-epigenetics comes of age. Nat Genet. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 26.Surana R, Sikka S, Cai W, Shin EM, Warrier SR, Tan HJG, Arfuso F, Fox SA, Dharmarajan AM, Kumar AP. Secreted frizzled related proteins: Implications in cancers. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2014;1845(1):53–65. doi: 10.1016/j.bbcan.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Schiefer L, Visweswaran M, Perumal V, Arfuso F, Groth D, Newsholme P, Warrier S, Dharmarajan A. Epigenetic regulation of the secreted frizzled-related protein family in human glioblastoma multiforme. Cancer Gene Ther. 2014;21(7):297–303. doi: 10.1038/cgt.2014.30. [DOI] [PubMed] [Google Scholar]
- 28.Zöchbauer-Müller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61(1):249–255. [PubMed] [Google Scholar]
- 29.Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, Walshaw M, Sheard J, Field JK, Liloglou T. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012;72(22):5692–5701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baylin SB. Mechanisms underlying epigenetically mediated gene silencing in cancer. In: Seminars in cancer biology: 2002. Elsevier; 2002: 331–337. [DOI] [PubMed]
- 31.Chung M-T, Sytwu H-K, Yan M-D, Shih Y-L, Chang C-C, Yu M-H, Chu T-Y, Lai H-C, Lin Y-W. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol Oncol. 2009;112(2):301–306. doi: 10.1016/j.ygyno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Shih YL, Shyu RY, Hsieh CB, Lai HC, Liu KY, Chu TY, Lin YW. Promoter methylation of the secreted frizzled‐related protein 1 gene SFRP1 is frequent in hepatocellular carcinoma. Cancer. 2006;107(3):579–590. doi: 10.1002/cncr.22023. [DOI] [PubMed] [Google Scholar]
- 33.Shih YL, Hsieh CB, Lai HC, Yan MD, Hsieh TY, Chao YC, Lin YW. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer. 2007;121(5):1028–1035. doi: 10.1002/ijc.22750. [DOI] [PubMed] [Google Scholar]


