Abstract
AIM: To investigate the association between tumor protein 53 (TP53) codon 72 polymorphisms and the risk for inflammatory bowel disease (IBD) development.
METHODS: Numerous genetic and epigenetic drivers have been identified for IBD including the TP53 gene. Pathogenic mutations in TP53 gene have only been reported in 50% of colorectal cancer (CRC) patients. A single nucleotide polymorphism (SNP) in the TP53 gene resulting in the presence of either arginine (Arg) or proline (Pro) or both at codon 72 was shown to alter TP53 tumor-suppressor properties. This SNP has been investigated as a risk factor for numerous cancers, including CRC. In this study we analyzed TP53 codon 72 polymorphism distribution in 461 IBD, 181 primary sclerosing cholangitis patients and 62 healthy controls. Genotyping of TP53 was performed by sequencing and restriction fragment length polymorphism analysis of genomic DNA extracted from peripheral blood.
RESULTS: The most frequent TP53 genotype in IBD patients was Arg/Arg occurring in 54%-64% of cases (and in only 32% of controls). Arg/Pro was the most prevalent genotype in controls (53%) and less common in patients (31%-40%). Pro/Pro frequency was not significantly different between controls and IBD patients.
CONCLUSION: The data suggests that the TP53 codon 72 Arg/Arg genotype is associated with increased risk for IBD development.
Keywords: Inflammatory bowel disease, Colorectal cancer, 72 codon single nucleotide polymorphism, Tumor protein 53, rs1042522
Core tip: Tumor protein 53 (TP53) codon 72 polymorphism distribution was analyzed by RFLP in 461 inflammatory bowel disease (IBD), 181 primary sclerosing cholangitis patents and in 62 healthy controls. The data suggests that the TP53 codon 72 Arg/Arg genotype is associated with increased risk for IBD development.
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic intestinal disease characterized by gastrointestinal (GI) inflammation resulting in abdominal pain, chronic diarrhea and weight loss[1]. It is highly prevalent across the world with North America having the highest frequency of people suffering from Crohn’s disease[2] (CD, a subtype of IBD). Moreover, about 129000 Canadians suffer from Crohn’s disease[2] with Alberta having the highest per capita increase within Canada. Chronic inflammation is associated with malignancy and the early onset of IBD increases the risk of colorectal cancer (CRC) later in life. In fact, for every 10 years with IBD, colorectal cancer risk increases by 5%[3]. Studies have shown that the risk of IBD progressing to CRC in less than 20 years once IBD diagnosis is made is about 10%-20%[4,5]. The incidence of CRC in IBD patients is six times higher than that of the healthy population, with CRC accounting for about 15% of IBD related deaths[6]. IBD subtypes include ulcerative colitis (UC) and CD. The simultaneous occurrence of primary sclerosing cholangitis (PSC, defined as inflammation and subsequent obstruction of bile ducts both inside and outside of the liver) and UC was reported to increase chances of developing CRC by up to 25% in 10 years[7,8].
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in Canada with 24400 Canadians diagnosed with colorectal cancer and 9300 expected to die from it in 2014[9]. The sporadic form of CRC and IBD-related CRC have very few clinical differences, however a two fold higher rate of mortality is reported in the later form[10,11]. In the four step pathway to CRC tumorigonesis[12], TP53 inactivation usually occurs at a later stage leading to the transition into a carcinoma and represents a major driver event in CRC[12]. The TP53 tumor suppressor gene, located at 17p13, encodes a transcription factor that regulates expression of genes involved in numerous biological processes, such as cell cycle control, DNA repair, apoptosis[13]. Mutations in the TP53 gene have been reported in 50% of CRC cases that mainly lead to the loss of transcriptional control[14]. Mutations in the TP53 gene affect mainly five “hotspot” amino acid positions: 175, 245, 248, 273 and 282.
Over 200 single nucleotide polymorphism (SNP)s have been identified in the TP53 gene[15]. The most studied is codon 72 polymorphism (rs1042522) encoding either arginine (CGC, 72Arg) or proline (CCC, 72Pro). Codon 72 encodes an amino acid located in the proline rich region of TP53 (amino acids 64-92) whereby the proline constitutes one of the five PxxP motifs important for interactions with proteins involved in apoptosis and growth suppression[16]. Arg has never been found in primates other than human[17] therefore Pro is considered the ancestral allele. The Pro allele was found to be more frequent in Africa as Arg is most prevalent in Europe suggesting that selection (most possibly UV and winter temperatures[18]) act on TP53 creating a latitudinal allele frequency gradient. Therefore, codon 72 polymorphisms will have an effect on TP53 function.
The two resulting TP53 polymorphisms (encoded by codon 72) differ in their biological and biochemical properties, such as electrophoretic mobility[16,19,20], subcellular localization[21-23], ability to induce apoptosis[16,24,25], ability to suppress transformed cell growth[16], susceptibility to HPV E6 degradation[26,27] and transcriptional activation[16,20,28,29]. Please see Table 1 for a summary of the biological properties of TP53 72Arg vs 72Pro TP53. Furthermore, the frequency of either allele (Arg or Pro) is highly affected by ethnicity. One study reported that the frequency of the proline allele increased from 17% in Swedish Saamis to 63% in Nigerian African Blacks[30]. This dictates the importance of comparing patients and controls of the same ethnic group in codon 72 polymorphism association studies.
Table 1.
Summary of biological and biochemical differences between 72Arg and 72Pro TP53 isoforms
| Property | 72Arg vs72Pro TP53 |
| Electrophoretic mobility | 72Arg migrates slightly faster on SDS[16,19,20] |
| Subcellular localization | 72Arg is cytoplasmic/mitochondrial possibly through interaction with CD2AP/Cin85 adaptor protein family[21] or with mitochondrial (mt) polymerase gamma, which may also promote mtDNA repair[22,23] 72Pro is predominantly nuclear[21,24] |
| Apoptosis induction | 72Arg is a stronger apoptosis inducer[24], with faster kinetics of cell death[16] possibly through more efficient inhibition of 72Pro by iASPP[25] or preferential localization of 72Arg to mitochondria and induction of apoptosis through a transcriptionally independent pathway[24] |
| Suppression of transformed cell growth | 72Arg is two times more active in suppressing colony formation[16] |
| Susceptibility to HPV E6 degradation | 72Arg is more susceptible to degradation[26,27] |
| Transcription activation | 72Arg associates and inactivates p73 more efficiently[20], thus inhibiting apoptosis72Pro is a stronger inducer of transcription and it has more affinity to TAFII32 and TAFII70[16] |
| Leukemia inhibitory factor transactivation | 72Arg is more active towards this function[28,29], which may be advantageous in cold climates due to increased fertility[18] |
The association between the 72 codon polymorphism and cancer risk has been studied extensively for many cancers, including CRC[31-40]. With only two papers studying association of 72 codon polymorphism and UC development[36,41], there has been no studies to associate codon 72 polymorphism with CD and PSC susceptibility. The purpose of this study is to investigate a possible link between the 72 codon polymorphism (rs1042522) and the risks of developing IBD and thus IBD-induced colorectal cancer.
MATERIALS AND METHODS
Cases and controls
Blood samples were collected at University of Alberta Adult and Pediatric Gastroenterology Clinics for IBD patients and at the University of Calgary for PSC and PSC/UC patients. The non-IBD control group was composed of 62 healthy blood donors from the same geographical region with no prior history of irritable bowel syndrome (IBS), IBD or cancer. The study was approved by the human ethics research board under the protocol No. Pro00001523 (study No. RES1598) and informed consent was obtained from all patients and controls before samples were analyzed. The diagnosis of IBD patients for this study was based on clinical, endoscopic and pathology findings as well as imaging findings on magnetic resonance or CT enterography for small bowel disease. Video capsule endoscopy was used to assess the small bowel in those with suspected IBD but have equivocal findings on endoscopy and imaging. Infectious causes were screen for and excluded. All pediatric patients were also screened for tuberculosis. In those under the age of 7, common immunodeficiency syndromes such as common immune variable deficiency, chronic granulomatous disease and mutations in IL-10 signalling pathway were screened for and excluded. The diagnosis of PSC was made on the basis of raised serum alkaline phosphatase levels and typical cholangiographic finding on ERC or MRC[42].
DNA extraction
Genomic DNA was extracted from peripheral blood collected by venous puncture. Blood was collected into PAXgene Blood DNA Tubes (QIAGEN, Country of Origin, Germany) and kept in a fridge at +4 °C until DNA extraction (1-2 d). DNA was extracted using PAXgene Blood DNA Kit (QIAGEN, Country of Origin, Germany) according to the manufacturer’s instructions. Polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP) analysis of codon 72 was used to identify TP53 genotypes.
Polymerase chain reaction
The forward primer used was P329 5’-TGCTCTTTTCACCCATCTAC-3’, and the reverse primer was P330 5’-ATACGGCCAGGCATTGAAGT-3’ (IARK protocol, 2010 update)[43]. Each PCR reaction contained 1 x OneTaq® Standard Reaction Buffer (NEB), 0.3 μmol/L each primer, 3 μmol/L of each dNTP, 100 ng genomic DNA, 0.1U OneTaq® Hot Start DNA Polymerase (NEB) in a final volume of 8 μL. The PCR conditions were as follows: denaturation at 94 °C for 4 min, followed by amplification for 40 cycles at 94 °C for 30 s, at 56 °C for 15 s, 68 °C for 40 s, with final extension at 68 °C for 10 min. The PCR products were checked on an agarose gel first 3 times and PCR appeared to work robustly, so for the rest of the analysis gel electrophoresis of PCR products was omitted.
RFLP analysis
Eight microlitres of PCR product was digested with 2 U of BstuI at 60 °C for at least 4 h. DNA fragments were run on a 2% agarose gel stained with ethidium bromide. The Arg allele was cleaved by BstUI, yielding two fragments 213 and 140 bp. The Pro allele was not cleaved by BstUI and had a single 353 bp band. Heterozygotes contained all three bands (Figure 1). RFLP results were confirmed by direct sequencing of 100 randomly chosen samples. DNA templates were amplified with P329 and P330 primers using the reaction settings described in PCR section. PCR products with sequencing P329 primer were then sent to McGill University and Génome Québec Innovation Centre Facility for Sanger sequencing.
Figure 1.
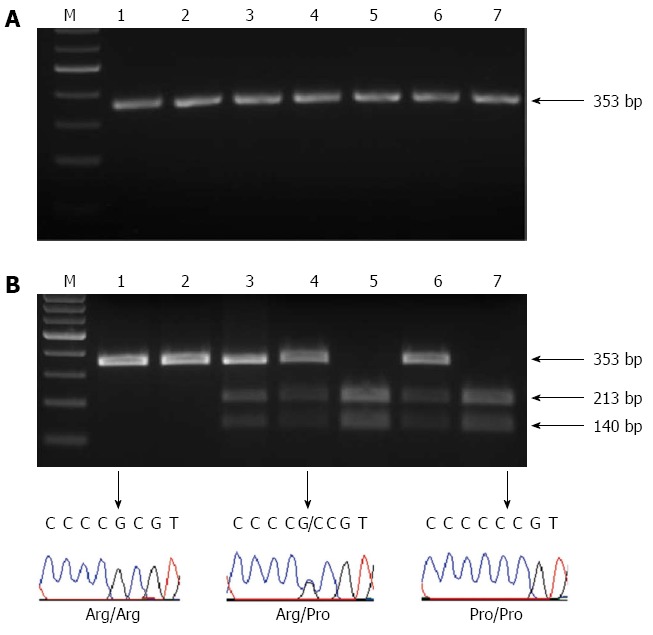
Polymerase chain reaction-restriction fragment length polymorphisms analysis. A: A representative analysis of the polymerase chain reaction (PCR) product. The product corresponded to TP53 with a single band at 353; B: The PCR-restriction fragment length polymorphisms (PCR-RFLP) analysis and sequencing results of TP53 codon 72 polymorphism. The Pro allele was not cleaved by BstUI and had a single 353 bp band (lanes 1 and 2). The Arg allele was cleaved by BstUI, yielding two fragments 213 and 140 bp (lanes 5 and 7). Heterozygotes contained all three bands (lanes 3, 4 and 6). RFLP results were confirmed by direct sequencing of 100 randomly selected samples. M: Molecular weight marker.
Statistical analysis
The χ2 test was applied to identify the deviations from the Hardy-Weinberg proportion. Odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated using logistic regression method. The statistical methods of this study were reviewed by Sung Hyun Kang from Biostatistics Service Core of Women and Children’s Health Research Institute at the University of Alberta.
RESULTS
In this study we investigated the association between TP53 codon 72 polymorphism and susceptibility to IBD. The allele and genotype frequencies of TP53 are summarized in Tables 2, 3 and Figure 2.The observed genotype frequencies did not deviate significantly from the Hardy-Weinberg equilibrium (P = 0.6543 for cases; P = 0.2856 for controls). We found that the allele distribution differed significantly between cases and controls. In particular, the frequencies of Arg and Pro alleles were 75.9% and 24.1% in IBD patients vs 58.9% and 41.1% in healthy controls (P < 0.0001, χ2 test, Table 2). This suggests that the TP53 codon 72 polymorphism may serve as a predisposing factor for IBD development.
Table 2.
Allele frequencies of TP53 72Arg/Pro polymorphism in cases and controls n (%)
| Patients/Controls |
Allele |
P value1 | |
| Arg | Pro | ||
| Cases (n = 642): | 974 (75.9) | 310 (24.1) | < 0.0001 |
| UC Adult (n = 151) | 222 (73.5) | 80 (26.5) | 0.0029 |
| CD Adult (n = 138) | 210 (76.1) | 66 (23.9) | 0.0005 |
| UC Pediatric (n = 78) | 118 (75.6) | 38 (24.4) | 0.0028 |
| CD Pediatric (n = 94) | 145 (77.1) | 43 (22.9) | 0.0006 |
| PSC (n = 42) | 67 (79.8) | 17 (20.2) | 0.0016 |
| PSC + UC (n = 139) | 212 (76.3) | 66 (23.7) | 0.0004 |
| Controls (n = 62) | 73 (58.9) | 51 (41.1) | |
P value calculated by χ2 test for comparison of patient groups with controls.
Table 3.
Genotype frequencies of TP53 polymorphism in cases and controls
| Patients/controls |
Genotype, % (n) |
OR1 | 95%CI | P value | ||
| Arg/Arg | Arg/Pro | Pro/Pro | ||||
| Cases (n = 642): | 56.9 (365) | 38.0 (244) | 5.1 (33) | 2.77 | 1.59-4.82 | 0.0001 |
| UC adult (n = 151) | 54.3 (82) | 38.4 (58) | 7.3 (11) | 2.50 | 1.34-4.65 | 0.0102 |
| CD adult (n = 138) | 55.8 (77) | 40.6 (56) | 3.6 (5) | 2.65 | 1.41-4.98 | 0.0011 |
| UC pediatric (n = 78) | 55.1 (43) | 41.0 (32) | 3.8 (3) | 2.58 | 1.29-5.17 | 0.0078 |
| CD pediatric (n = 94) | 59.6 (56) | 35.1 (33) | 5.3 (5) | 3.09 | 1.58-6.07 | 0.0023 |
| PSC (n = 42) | 64.3 (27) | 31.0 (13) | 4.8 (2) | 3.78 | 1.66-8.63 | 0.0046 |
| PSC + UC (n = 139) | 57.6 (80) | 37.4 (52) | 5.0 (7) | 2.85 | 1.52-5.35 | 0.0016 |
| Controls (n = 62) | 32.3 (20) | 53.2 (33) | 14.5 (9) | |||
OR is calculated for Arg/Arg vs Arg/Arg + Pro/Pro. CD: Crohn’s disease; UC: Ulcerative colitis; PSC: Primary sclerosing cholangitis.
Figure 2.
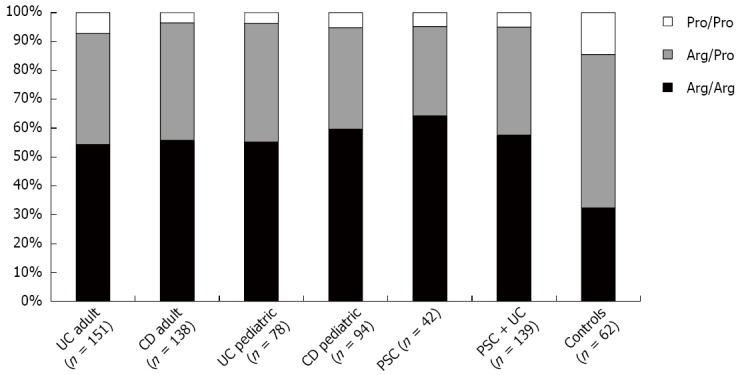
TP53 72Arg/Pro polymorphism distribution in adult and pediatric inflammatory bowel disease, primary sclerosing cholangitis (PSC), PSC/ulcerative colitis (UC) and healthy non- inflammatory bowel disease (IBD) controls. TP53 72Arg homozygosity is strongly associated with the development of IBD. Please find P values and ORs in Table 2.
The genotype for TP53 72 Arg/Arg tended to predominate in all patient groups compared to controls. In control samples utilized in this study, the genotype distribution for TP53 polymorphism revealed 32.3%, 53.2% and 14.5% for Arg/Arg, Arg/Pro and Pro/Pro genotypes, respectively. Allelic frequencies corresponded to 0.59 for the arginine and 0.41 for the proline allele (Tables 2 and 3). On the other hand, 56.9% of all cases were Arg/Arg, 38% were Arg/Pro and 5.1% were Pro/Pro. The arginine allelic frequency was 0.76 and the proline allele frequency was 0.24. Significant differences between cases and controls were found for the Arg/Arg genotype compared with grouped Arg/Pro and Pro/Pro genotypes (Table 3). Pediatric and adult IBD patients did not show statistically significant differences for the presence of TP53 SNPs. To our knowledge 72 codon polymorphism distribution has never been studied in PSC patients. Our results indicate that PSC had higher Arg/Arg percentage compared to IBD groups, but it may be due to a small sample size effect (n = 42).
DISCUSSION
The healthy population utilized in this study was mainly composed of Caucasian individuals of Eastern European origin form the Edmonton area and Northern Alberta. Surprisingly, the TP53 codon 72 genotype distribution found in our healthy controls was different from the genotype distribution observed in controls in Europe (Slovac Republic, Sweden, Italy, Germany, for references please see Figure 3). Patients from Greece had TP53 72 codon polymorphism distribution most similar to our healthy controls. These data demonstrate the importance of careful selection of controls for genetic understanding of disease states. Although these differences were observed, we have used the controls within our geographic zone for the purpose of this study.
Figure 3.
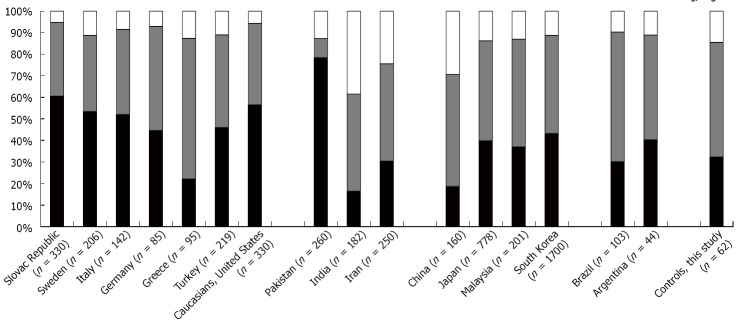
TP53 72Arg/Pro polymorphism distribution in non-inflammatory bowel disease controls in different countries and comparison to controls utilized in this study (mainly of Eastern European descent). References for indicated bar graphs: Slovak Republic[55], Sweden[30]; Italy[41], Germany[33], Greece[32], Turkey[36], Caucasians/United States[40], Pakistan[56], India[57], Iran[34], China[58], Japan[38], Malaysia[35], South Korea[39], Brazil[59], Argentina[31].
Several published articles have provided evidence that TP53 polymorphism at codon 72 may be associated with tumor development, although results are varied. Consistent with our results, it was shown that 72Arg is associated with higher CRC risk in Argentina[31], Greece[32], Germany[33] and Iran[34]. On the contrary, 72Pro has been detected as an indicator of higher risk of CRC in Malaysia[35], Turkey[36], in one ethnic Kashmiri population[37]. At the same time, studies conducted in Japan[38], Korea[39] and United States[40] failed to link 72 polymorphism to CRC development. The varied nature of these findings could be explained by the differences in ethnic distribution of 72 codon alleles through the world. Is has been shown that 72Arg frequency increases from south to north and even then, the 72 codon polymorphism distribution can differ substantially between the neighboring countries (Figure 3).
To our knowledge, no studies have been done so far on 72 codon polymorphism distribution in the CD or PSC populations although two articles have explored 72 codon polymorphism in UC. The first, an Italian study, failed to reveal an association of 72 codon polymorphism with UC development; however, 72Pro homozygosity was associated with chronic UC and family history of CRC[41]. The other study was conducted in Turkey and 72Pro was found to be associated with moderate risk of UC[36].
Our results indicate that IBD and PSC patients have significantly different TP53 codon 72 polymorphism distribution, suggesting that the 72Arg allele may predispose individuals to IBD development and an increased levels of apoptosis in the colonic area. This may lead to apoptotic degradation of the intestinal epithelial cell leading to the impairment of gut barrier - a prominent feature in IBD patients[44]. This increased ability of 72Arg allele to induce apoptosis (please see Table 1 for other isoform’s differences) may partially explain why Arg/Arg genotype is more abundant in IBD group compared to controls. Additionally, it was shown that 72Arg genotype carriers have increased risk of TP53 mutations potentially leading to further imbalances and cancer development[33,45]. Thus, if differences between 72Arg and 72Pro carriers progress through IBD to cancer development are confirmed in long-term studies, it could be useful for IBD related cancer development prediction and better screening of 72Arg carriers.
In addition to other factors predisposing to IBD, there is accumulating evidence that epigenetic changes, such as DNA methylation and histone modifications also play a role in the pathogenesis of IBD (please see following reviews for references[46-49]). One of the histone acetyltransferase-containing domain transcription co-activators, p300/CBP1, acetylates histones making DNA more accessible to transcription machinery after being recruited by sequence-specific DNA binding transcription factors, such as TP53[50]. It was shown that proline-rich domain of TP53 influences the extent of interaction between TP53 and p300/CBP and, subsequently, histone acetylation and the promoter accessibility[51]. It is tempting to speculate that p300/CBP can discriminate TP53 isoforms and associate with them differently in a similar way as CD2AP/Cin85 adaptor protein family does[21]. This differential association could lead to differential epigenetic silencing of target genes. One of such candidate target genes is the tumor suppressor RASSF1A (Ras association domain family 1A). It was demonstrated that RASSF1A promoter has a TP53 binding site and TP53 is able to bind to the RASSF1A promoter and inhibit RASSF1A expression[52]. It would be interesting to study if 72 codon polymorphism can influence this TP53 function to repress RASSF1A expression. Interestingly, it was shown that RASSF1A epigenetic silencing has a role in cancer development (for a recent review please see[53]) and we demonstrated recently that RASSF1A plays a role in controlling inflammation in dextran sodium sulphate (DSS)-induced colitis in mice, a well-known model for IBD[54].
In conclusion, our data indicate 72 Arg/Arg TP53 genotype may predispose individuals to IBD development possibly through increased efficiency of apoptosis induction. The increased risk of CRC development could also be explained by increased rate of TP53 mutations in patients carrying Arg72 genotypes[33,45]. However, more studies are needed to confirm the clinical significance of this polymorphism and better understand the effect of 72 codon polymorphism on IBD development.
ACKNOWLEDGMENTS
We would like to thank all the members of the Baksh laboratory for their helpful discussions and for Adrienne Anna Marie DeCorby for editing this manuscript. We are grateful to the support of the division of Hematology/Oncology/Palliative Care/Epidemiology under Dr. David Eisenstat. We would also like to thank Sung Hyun Kang from Biostatistics Service Core of Women and Children’s Health Research Institute at the University of Alberta for reviewing the statistical methods.
COMMENTS
Background
Inflammatory bowel disease (IBD) is a chronic intestinal idiopathic disease characterized by inflammation of the gastrointestinal (GI) area resulting in abdominal pain, chronic diarrhea, and weight loss. IBD includes Crohn’s disease and ulcerative colitis. Currently, research suggests that IBD is caused by a combination of genetic and environmental influences, intestinal microbial disruptions, epigenetic regulation and immunologic dysfunction. How all of these factors determine the appearance of IBD is an area of active research including this current report.
Research frontiers
The authors aimed to establish a relation between changes in the gene, TP53, and the risk of developing inflammatory bowel disease. This gene is commonly targeted for genetic changes in cancer but very little is known for its role in IBD.
Innovations and breakthroughs
This study demonstrated for the first time an association between codon 72 Arg/Arg TP53 polymorphism and increased susceptibility to IBD.
Applications
The authors suggest that the increased incidence of 72 Arg/Arg TP53 in IBD patients may suggest increased apoptotic efficiency of intestinal cells that may contribute to higher susceptibility to IBD due to higher erosion of the inflamed gut. Higher risks of colorectal cancer can be attributed to increased rates of TP53 mutations in patients carrying the 72 Arg phenotype.
Terminology
A single nucleotide polymorphism (SNP) is a variation in the DNA sequence which is common in a population. SNPs usually affect the susceptibility to diseases and response to treatments. The common TP53 codon 72 SNP is termed (rs1042522).
Peer-review
This article provides important insight into the association of SNPs and diseases and brings forward the importance of personalized medicine or treatments. Patients with different ethnic backgrounds and polymorphisms may respond differently. This manuscript may have potential to increase knowledge on the treatment of IBD.
Footnotes
Supported by Grants from AI-HS and The Stollery Children’s Foundation/Hair Massacure Grant (under the Department of Pediatric generously donated by the MacDonald family; to Baksh S); Queen Elizabeth II Graduate Scholarship, the Biochemistry Doctoral Recruitment Scholarship and The Stollery Children’s Foundation/Hair Massacure Grant (to Salla M).
Institutional review board statement: The study was approved by the human Ethics Research Board under the protocol No. Pro00001523 (study No. RES1598); and informed consent was obtained from all patients and controls before samples were analyzed.
Conflict-of-interest statement: The authors declare no conflict of interest with respect to competing commercial, personal, political, intellectual, or religious interests in relation to the submitted work.
Data sharing statement: Technical appendix, statistical code and dataset available from the corresponding author at sbaksh@ualberta.ca. Participants gave informed consent for data sharing.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: February 26, 2015
First decision: March 26, 2015
Article in press: July 15, 2015
P- Reviewer: Said Z S- Editor: Yu J L- Editor: A E- Editor: Zhang DN
References
- 1.Marcus SB, Strople JA, Neighbors K, Weissberg-Benchell J, Nelson SP, Limbers C, Varni JW, Alonso EM. Fatigue and health-related quality of life in pediatric inflammatory bowel disease. Clin Gastroenterol Hepatol. 2009;7:554–561. doi: 10.1016/j.cgh.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Fakhoury M, Negrulj R, Mooranian A, Al-Salami H. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res. 2014;7:113–120. doi: 10.2147/JIR.S65979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munkholm P. Review article: the incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther. 2003;18 Suppl 2:1–5. doi: 10.1046/j.1365-2036.18.s2.2.x. [DOI] [PubMed] [Google Scholar]
- 4.Harpaz N, Polydorides AD. Colorectal dysplasia in chronic inflammatory bowel disease: pathology, clinical implications, and pathogenesis. Arch Pathol Lab Med. 2010;134:876–895. doi: 10.5858/134.6.876. [DOI] [PubMed] [Google Scholar]
- 5.Azer SA. Overview of molecular pathways in inflammatory bowel disease associated with colorectal cancer development. Eur J Gastroenterol Hepatol. 2013;25:271–281. doi: 10.1097/MEG.0b013e32835b5803. [DOI] [PubMed] [Google Scholar]
- 6.Mattar MC, Lough D, Pishvaian MJ, Charabaty A. Current management of inflammatory bowel disease and colorectal cancer. Gastrointest Cancer Res. 2011;4:53–61. [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Kim HW, Kang DH, Kim MD, Lee JH, Lee JH, Kim BG, Park JH. [A case of intrahepatic cholangiocarcinoma associated with Type IV choledochal cyst] Korean J Gastroenterol. 2012;60:123–127. doi: 10.4166/kjg.2012.60.2.123. [DOI] [PubMed] [Google Scholar]
- 8.Delaunoit T, Limburg PJ, Goldberg RM, Lymp JF, Loftus EV. Colorectal cancer prognosis among patients with inflammatory bowel disease. Clin Gastroenterol Hepatol. 2006;4:335–342. doi: 10.1016/j.cgh.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 9. Available from: http://www.colorectal-cancer.ca/en/just-the-facts/colorectal/
- 10.Jensen AB, Larsen M, Gislum M, Skriver MV, Jepsen P, Nørgaard B, Sørensen HT. Survival after colorectal cancer in patients with ulcerative colitis: a nationwide population-based Danish study. Am J Gastroenterol. 2006;101:1283–1287. doi: 10.1111/j.1572-0241.2006.00520.x. [DOI] [PubMed] [Google Scholar]
- 11.Herrinton LJ, Liu L, Levin TR, Allison JE, Lewis JD, Velayos F. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology. 2012;143:382–389. doi: 10.1053/j.gastro.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 12.Raskov H, Pommergaard HC, Burcharth J, Rosenberg J. Colorectal carcinogenesis--update and perspectives. World J Gastroenterol. 2014;20:18151–18164. doi: 10.3748/wjg.v20.i48.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 14.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 15.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 16.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puente XS, Velasco G, Gutiérrez-Fernández A, Bertranpetit J, King MC, López-Otín C. Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics. 2006;7:15. doi: 10.1186/1471-2164-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Tan SJ, Zhong H, Hu W, Levine A, Xiao CJ, Peng Y, Qi XB, Shou WH, Ma RL, et al. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet. 2009;84:534–541. doi: 10.1016/j.ajhg.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris N, Brill E, Shohat O, Prokocimer M, Wolf D, Arai N, Rotter V. Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol. 1986;6:4650–4656. doi: 10.1128/mcb.6.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin MC, Jost CA, Brooks LA, Irwin MS, O’Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 21.Panni S, Salvioli S, Santonico E, Langone F, Storino F, Altilia S, Franceschi C, Cesareni G, Castagnoli L. The adapter protein CD2AP binds to p53 protein in the cytoplasm and can discriminate its polymorphic variants P72R. J Biochem. 2015;157:101–111. doi: 10.1093/jb/mvu059. [DOI] [PubMed] [Google Scholar]
- 22.Altilia S, Santoro A, Malagoli D, Lanzarini C, Ballesteros Álvarez JA, Galazzo G, Porter DC, Crocco P, Rose G, Passarino G, et al. TP53 codon 72 polymorphism affects accumulation of mtDNA damage in human cells. Aging (Albany NY) 2012;4:28–39. doi: 10.18632/aging.100425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 25.Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38:1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- 26.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 27.Zehbe I, Voglino G, Wilander E, Genta F, Tommasino M. Codon 72 polymorphism of p53 and its association with cervical cancer. Lancet. 1999;354:218–219. doi: 10.1016/S0140-6736(99)01914-5. [DOI] [PubMed] [Google Scholar]
- 28.Jeong BS, Hu W, Belyi V, Rabadan R, Levine AJ. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J. 2010;24:1347–1353. doi: 10.1096/fj.09-146001. [DOI] [PubMed] [Google Scholar]
- 29.Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR, Rosenwaks Z, Levine AJ, Hu W. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci USA. 2009;106:9761–9766. doi: 10.1073/pnas.0904280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Själander A, Birgander R, Athlin L, Stenling R, Rutegård J, Beckman L, Beckman G. P53 germ line haplotypes associated with increased risk for colorectal cancer. Carcinogenesis. 1995;16:1461–1464. doi: 10.1093/carcin/16.7.1461. [DOI] [PubMed] [Google Scholar]
- 31.Pérez LO, Abba MC, Dulout FN, Golijow CD. Evaluation of p53 codon 72 polymorphism in adenocarcinomas of the colon and rectum in La Plata, Argentina. World J Gastroenterol. 2006;12:1426–1429. doi: 10.3748/wjg.v12.i9.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dakouras A, Nikiteas N, Papadakis E, Perakis M, Valis D, Rallis G, Tzanakis N, Peros G, Tsigkris C, Kittas C, et al. P53Arg72 homozygosity and its increased incidence in left-sided sporadic colorectal adenocarcinomas, in a Greek-Caucasian population. Anticancer Res. 2008;28:1039–1043. [PubMed] [Google Scholar]
- 33.Schneider-Stock R, Boltze C, Peters B, Szibor R, Landt O, Meyer F, Roessner A. Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia. 2004;6:529–535. doi: 10.1593/neo.04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dastjerdi MN. TP53 codon 72 polymorphism and P53 protein expression in colorectal cancer specimens in Isfahan. Acta Med Iran. 2011;49:71–77. [PubMed] [Google Scholar]
- 35.Aizat AA, Shahpudin SN, Mustapha MA, Zakaria Z, Sidek AS, Abu Hassan MR, Ankathil R. Association of Arg72Pro of P53 polymorphism with colorectal cancer susceptibility risk in Malaysian population. Asian Pac J Cancer Prev. 2011;12:2909–2913. [PubMed] [Google Scholar]
- 36.Eren F, Akkiprik M, Atuğ O, Sönmez O, Tahan G, Ozdemir F, Hamzaoğlu HO, Celikel CA, Imeryüz N, Avşar E, et al. R72P polymorphism of TP53 in ulcerative colitis patients is associated with the incidence of colectomy, use of steroids and the presence of a positive family history. Pathol Oncol Res. 2010;16:563–568. doi: 10.1007/s12253-010-9255-9. [DOI] [PubMed] [Google Scholar]
- 37.Sameer AS, Shah ZA, Syeed N, Banday MZ, Bashir SM, Bhat BA, Siddiqi MA. TP53 Pro47Ser and Arg72Pro polymorphisms and colorectal cancer predisposition in an ethnic Kashmiri population. Genet Mol Res. 2010;9:651–660. doi: 10.4238/vol9-2gmr751. [DOI] [PubMed] [Google Scholar]
- 38.Joshi AM, Budhathoki S, Ohnaka K, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, et al. TP53 R72P and MDM2 SNP309 polymorphisms and colorectal cancer risk: the Fukuoka Colorectal Cancer Study. Jpn J Clin Oncol. 2011;41:232–238. doi: 10.1093/jjco/hyq200. [DOI] [PubMed] [Google Scholar]
- 39.Song HR, Kweon SS, Kim HN, Piao JM, Yun WJ, Choi JS, Hwang JE, Yoon JY, Kim HR, Park YK, et al. p53 codon 72 polymorphism in patients with gastric and colorectal cancer in a Korean population. Gastric Cancer. 2011;14:242–248. doi: 10.1007/s10120-011-0034-4. [DOI] [PubMed] [Google Scholar]
- 40.Koushik A, Tranah GJ, Ma J, Stampfer MJ, Sesso HD, Fuchs CS, Giovannucci EL, Hunter DJ. p53 Arg72Pro polymorphism and risk of colorectal adenoma and cancer. Int J Cancer. 2006;119:1863–1868. doi: 10.1002/ijc.22057. [DOI] [PubMed] [Google Scholar]
- 41.Vietri MT, Riegler G, Ursillo A, Caserta L, Cioffi M, Molinari AM. p53 codon 72 polymorphism in patients affected with ulcerative colitis. J Gastroenterol. 2007;42:456–460. doi: 10.1007/s00535-007-2026-z. [DOI] [PubMed] [Google Scholar]
- 42.Chapman R, Fevery J, Kalloo A, Nagorney DM, Boberg KM, Shneider B, Gores GJ. Diagnosis and management of primary sclerosing cholangitis. Hepatology. 2010;51:660–678. doi: 10.1002/hep.23294. [DOI] [PubMed] [Google Scholar]
- 43. Available from: http://p53.iarc.fr/ProtocolsAndTools.aspx.
- 44.Pastorelli L, De Salvo C, Mercado JR, Vecchi M, Pizarro TT. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. doi: 10.3389/fimmu.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider-Stock R, Mawrin C, Motsch C, Boltze C, Peters B, Hartig R, Buhtz P, Giers A, Rohrbeck A, Freigang B, et al. Retention of the arginine allele in codon 72 of the p53 gene correlates with poor apoptosis in head and neck cancer. Am J Pathol. 2004;164:1233–1241. doi: 10.1016/S0002-9440(10)63211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenke AC, Zilbauer M. Epigenetics in inflammatory bowel disease. Curr Opin Gastroenterol. 2012;28:577–584. doi: 10.1097/MOG.0b013e328357336b. [DOI] [PubMed] [Google Scholar]
- 47.Petronis A, Petroniene R. Epigenetics of inflammatory bowel disease. Gut. 2000;47:302–306. doi: 10.1136/gut.47.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Däbritz J, Menheniott TR. Linking immunity, epigenetics, and cancer in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:1638–1654. doi: 10.1097/MIB.0000000000000063. [DOI] [PubMed] [Google Scholar]
- 49.Karatzas PS, Gazouli M, Safioleas M, Mantzaris GJ. DNA methylation changes in inflammatory bowel disease. Ann Gastroenterol. 2014;27:125–132. [PMC free article] [PubMed] [Google Scholar]
- 50.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 51.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- 52.Tian Y, Hou Y, Zhou X, Cheng H, Zhou R. Tumor suppressor RASSF1A promoter: p53 binding and methylation. PLoS One. 2011;6:e17017. doi: 10.1371/journal.pone.0017017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 2014;588:2671–2684. doi: 10.1016/j.febslet.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 54.Gordon M, El-Kalla M, Zhao Y, Fiteih Y, Law J, Volodko N, Anwar-Mohamed A, El-Kadi AO, Liu L, Odenbach J, et al. The tumor suppressor gene, RASSF1A, is essential for protection against inflammation -induced injury. PLoS One. 2013;8:e75483. doi: 10.1371/journal.pone.0075483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zubor P, Stanclova A, Kajo K, Hatok J, Klobusiakova D, Visnovsky J, Danko J. The p53 codon 72 exon 4 BstUI polymorphism and endometrial cancer in Caucasian women. Oncology. 2009;76:173–183. doi: 10.1159/000201570. [DOI] [PubMed] [Google Scholar]
- 56.Saleem S, Azhar A, Hameed A, Khan MA, Abbasi ZA, Qureshi NR, Ajmal M. P53 (Pro72Arg) polymorphism associated with the risk of oral squamous cell carcinoma in gutka, niswar and manpuri addicted patients of Pakistan. Oral Oncol. 2013;49:818–823. doi: 10.1016/j.oraloncology.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Singhal P, Hussain S, Thakur N, Batra S, Salhan S, Bhambani S, Bharadwaj M. Association of MDM2 and p53 polymorphisms with the advancement of cervical carcinoma. DNA Cell Biol. 2013;32:19–27. doi: 10.1089/dna.2012.1718. [DOI] [PubMed] [Google Scholar]
- 58.Jiang P, Liu J, Li W, Zeng X, Tang J. Role of p53 and p21 polymorphisms in the risk of cervical cancer among Chinese women. Acta Biochim Biophys Sin (Shanghai) 2010;42:671–676. doi: 10.1093/abbs/gmq069. [DOI] [PubMed] [Google Scholar]
- 59.Contu SS, Agnes G, Damin AP, Contu PC, Rosito MA, Alexandre CO, Damin DC. Lack of correlation between p53 codon 72 polymorphism and anal cancer risk. World J Gastroenterol. 2009;15:4566–4570. doi: 10.3748/wjg.15.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]


