Abstract
Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5)/survivin genetic microRNA (miRNA) binding site variants in the 3′ untranslated region (3′UTR) are known to be significantly associated with cancer risk. However, the roles of genetic variants in BIRC5/survivin gene 3′UTRs and post-transcriptional regulation have not been elucidated. In the present study, we revealed that rs1042489, rs1042542, rs17882360, rs2239680, rs2661694 and rs4789560 in the BIRC5/survivin 3′UTR have potential miRNA binding sites using bioinformatics analysis. However, only rs1042489 was significantly associated with BIRC5/survivin mRNA expression in lymphoblastoid cell lines (P=0.030); rs1042489 may be a putative variant mediating the post-transcriptional regulation of the target BIRC5/survivin gene. An in-depth understanding of how 3′UTR variants regulate BIRC5/survivin activity is expected to pave the way to targeting the BIRC5/survivin pathway in cancer therapy.
Keywords: genetic, microRNA, polymorphisms, baculoviral inhibitor of apoptosis repeat-containing 5, survivin, variant
Introduction
Survivin, encoded by baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5), is the smallest member of the inhibitor of apoptosis (IAP) family of proteins. It is widely distributed during the embryonic and fetal developmental stages, yet absent in terminally differentiated normal tissue (1). Previous studies have demonstrated that survivin, unlike other IAPs, is strongly expressed in the majority of neoplasms. Overexpression of survivin is also significantly correlated with a poor prognosis and decreased survival rates in breast and lung cancer, colon, bladder and Ewing sarcoma, and lymphomas (2–7). BIRC5/survivin is an essential protein involved in Ewing sarcoma cell growth and proliferation (7). In addition, BIRC5/survivin plays a significant role in the pathogenesis of malignant tumors and the progression of various types of cancer.
It is well known that the microRNA (miRNA) binding regions variants influence altered gene functions. miRNAs regulate the activity of BIRC5/survivin, and miRNA dysregulation has been associated with malignant neoplasms. Zu et al evaluated the association between genetic variants in the 3′ untranslated region (3′UTR) of cancer-related genes and risk of lung cancer in Chinese populations, and defined a 3′UTR single nucleotide polymorphism (SNP) in the human BIRC5 oncogene that may increase individual susceptibility to lung cancer, possibly by attenuating the interaction between BIRC5 and miRNA-335 (8). BIRC5/survivin directly binds to the promoter of the miRNA-335 cluster, activating its transcription, and negatively modulating the translation of BIRC5/survivin miRNAs by binding sites in their 3′UTRs (8). In addition, a number of studies have revealed that BIRC5/survivin variants may play crucial roles in carcinogenesis (2). Considering that survivin is a notable member of the IAP family, but that the role of variants in miRNA binding sites of survivin remains unknown, in the present study, we performed a bioinformatic analysis and genotype-phenotype association analysis based on the HapMap database to test our hypothesis that BIRC5/survivin 3′UTR variants are associated with its mRNA expression.
The study was approved by the Ethics Committee of the Union Hospital, Tongji Medical College of Huazhong University of Science and Technology, China.
Materials and methods
Bioinformatic analysis and selection of polymorphisms
The SNPs of BIRC5/survivin were identified in the gene region and the coding region using an online database (http://www.ncbi.nlm.nih.gov/SNP/). The bioinformatic tool SNP Function Prediction (FuncPred; http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi) was used to predict the potential functional relevance affecting the miRNA binding sites. Additionally, SNPs were limited by a minor allele frequency (MAF) of >0.05 in the HapMap population derived from Utah residents with Northern and Western European ancestry. Pairwise linkage disequilibrium (LD) values of all SNPs in the same gene were calculated, then the SNPs that were not in LD (r2<0.8) were selected, and LD maps of those SNPs in BIRC5/survivin genes were plotted with the online program http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi.
Genotype and mRNA expression data of lymphoblastoid cell lines from HapMap database
Additional data on BIRC5/survivin genotypes and mRNA levels were available online (http://app3.titan.uio.no/biotools/help.php?app=snpexp) for the genotype-phenotype association analysis (9). Genome-wide expression arrays (47,294 transcripts) from Epstein-Barr virus-transformed lymphoblastoid cell lines were used from 270 HapMap individuals (142 males and 128 females) to analyze the gene expression variation (10). The genotyping data were from the HapMap phase II release 23 data set consisting of 3.96 million SNP genotypes from 270 individuals from four populations (11). The SNPexp v1.2 tool was used for calculating and visualizing correlations between HapMap genotypes and gene expression levels (Norwegian PSC Research Center, Clinic for Specialized Surgery and Medicine, Oslo University Hospital Rikshospitalet, Norway).
Statistical analysis
Genotype and phenotype correlation was analyzed using the Chi-square test. All statistics tests were two-sided and P<0.05 was considered to indicate a statistically significance result.
Results
BIRC5/survivin 3′UTR selected variants and putative miRNA binding sites
In total, 372 SNPs were identified in the BIRC5/survivin gene region and 28 in the coding region (http://www.ncbi.nlm.nih.gov/SNP/). Among them, 62 SNPs were reported in the 3′UTR, of which only 8 SNPs (rs2239680, rs202011142, rs1042489, rs2661694, rs1042541, rs1042542, rs4789560 and rs17882360) had an available MAF value >0.05, and were predicted to affect the miRNA binding site activity according to the bioinformatics analysis, as shown in Table I. The most extensively studied putative binding sites of these SNPs include hsa-miR-877, hsa-miR-936, hsa-miR-939, hsa-miR-367, hsa-miR-493, hsa-miR-601, hsa-miR-92a, hsa-miR-1256, hsa-miR-1285, hsa-miR-34a, hsa-miR-34c-5p, hsa-miR-503, hsa-miR-612, hsa-miR-626, hsa-miR-885-3p, hsa-miR-1276, hsa-miR-335, hsa-miR-577, hsa-miR-1295, hsa-miR-24, hsa-miR-298, hsa-miR-510, hsa-miR-576-3p, hsa-miR-1254 and hsa-miR-147 (http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi). Combined with other SNPs in the 3′UTR or promoter region, the variant rs2239680 is jointly involved in cancer susceptibility (8,12).
Table I.
Selected single nucleotide polymorphisms of BIRC5/survivin 3′ untranslated region and putative microRNA binding sites.
| Name | Alleles | MAF | Putative miRNA binding sites |
|---|---|---|---|
| rs1042489 | C/T | 0.3848 | hsa-miR-877, hsa-miR-936, hsa-miR-939 |
| rs1042541 | A/G | 0.3724 | NA |
| rs1042542 | C/T | 0.3875 | hsa-miR-367, hsa-miR-493, hsa-miR-601, hsa-miR-92a |
| rs17882360 | A/T | 0.0569 | hsa-miR-1256, hsa-miR-1285, hsa-miR-34a, hsa-miR-503, hsa-miR-34c-5p, hsa-miR-612, hsa-miR-626, hsa-miR-885-3p |
| rs2239680 | (>6 bp) | 0.2319 | hsa-miR-1276, hsa-miR-335, hsa-miR-577 |
| rs2661694 | A/C | 0.2185 | hsa-miR-1295, hsa-miR-24, hsa-miR-298, hsa-miR-510, hsa-miR-576-3p |
| rs4789560 | C/T | 0.3675 | hsa-miR-1254, hsa-miR-147 |
| rs202011142 | -/T | 0.3081 | NA |
BIRC5, baculoviral inhibitor of apoptosis repeat-containing 5; MAF, minor allele frequency; NA, not available.
LD of all SNPs in the BIRC5/survivin gene calculation
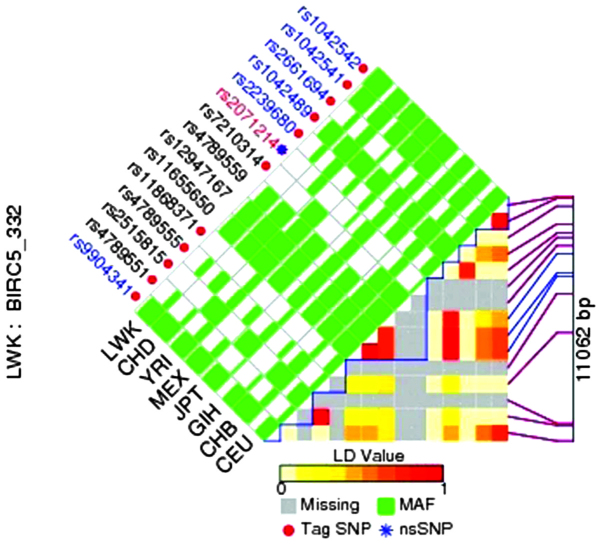
The bioinformatic tool FuncPred (http://snpinfo.niehs.nih.gov/snpfunc.htm) was used to identify the potential functional relevance of the SNPs. We calculated pairwise LD values of all SNPs in the same gene to determine the SNPs which were not in LD (r2<0.8), and plotted the LD maps of those SNPs in BIRC5/survivin genes using FuncPred (Fig. 1). The pairwise r2 correlations between the relevant two SNPs are represented by each square number. The color of each SNP spot reflects its D′ value, and when the D′ value decreases, the color changes from red to white. The haplotype blocks were estimated with the FuncPred program. The minor allele frequency of all the above alleles was greater than 0.05. rs1042489 was the predicted tag SNP in our study, and rs1042542, rs2239680 and rs2661694 in BIRC5/survivin were not included in the LD plot (Fig. 1).
Figure 1.
Linkage disequilibrium plot of the baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5)/survivin region using SNP Function Prediction (FuncPred). Each square number represents the pairwise r2 correlations between the relevant two single nucleotide polymorphisms (SNPs). The color of each SNP spot reflects its D′ value, which changes from red to white as the D′ value decreases.
BIRC5/survivin mRNA expression by genotypes in lymphoblastoid cell lines
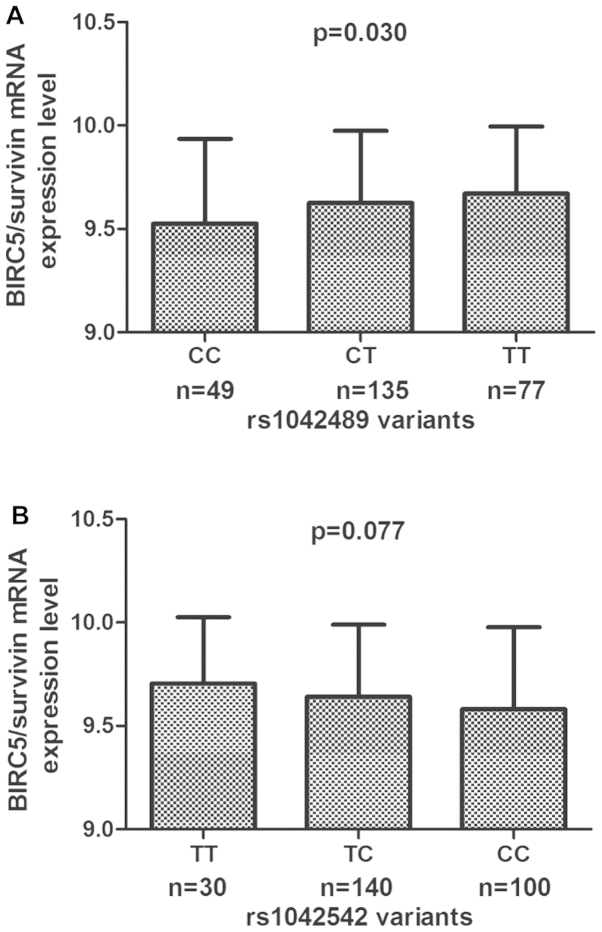
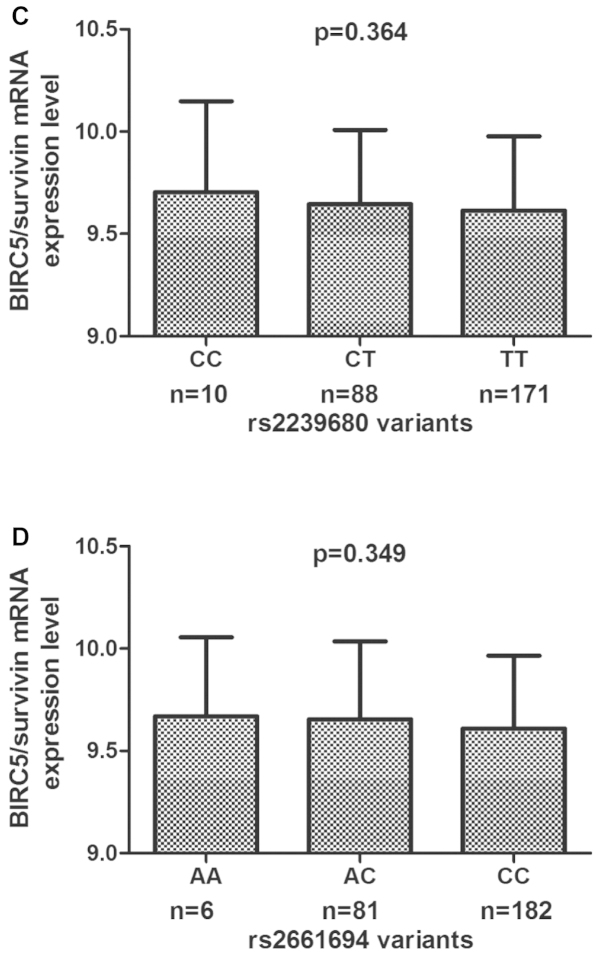
To assess the mRNA expression of the prohibitin gene in the lymphoblastoid cell lines, the available HapMap-cDNA expression database was used to analyze the correlation of the prohibitin genotype and mRNA expression in 270 HapMap lymphoblastoid cell lines. With the exception of the one cell line with unavailable values for rs1042489, 49 (0.188) cell lines with the CC genotype, 135 (0.517) cell lines with the CT genotype and 77 (0.295) cell lines with the TT genotype were identified. For rs1042542, 30 (0.111) cell lines had the TT genotype, 140 (0.519) had the TC genotype, and 100 (0.370) had the CC genotype. There were 10 (0.037) cell lines with CC, 88 (0.327) with CT, and 171 (0.636) with TT genotype for rs2239680. For rs2661694, 6 (0.022) cell lines with the AA genotype, 81 (0.301) cell lines with the AC genotype and 182 (0.677) with the CC genotype were identified. Fig. 2 shows the BIRC5/survivin mRNA expression levels of cell lines by BIRC5 genotype. The rs1042489 CC genotype was revealed to have significantly lower expression levels than the CT and TT genotypes (P=0.030; Fig. 2A), and there was no significant difference in BIRC5/survivin mRNA expression levels among cell lines carrying rs1042542 (P=0.077), rs2239680 (P=0.364) and rs2661694 (P=0.349) genotypes (Fig. 2B-D).
Figure 2.
mRNA expression level of the genotype-phenotype association analysis of baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5)/survivin variants rs1042489 (A), rs1042542 (B), rs2239680 (C) and rs2661694 (D), and mRNA expression in Epstein-Barr virus-transformed lymphoblastoid cell lines from the HapMap database.
Discussion
Human BIRC5/survivin is the smallest member of the IAP family, and encodes negative regulatory proteins that prevent apoptotic cell death. It consists of a 16.5 kDa protein encoded by a gene located on chromosome 17q25 (1). A number of studies, including tissue expression studies, animal models and clinical trials, have investigated BIRC5/survivin polymorphisms and cancer risk. The results of these studies suggested that BIRC5/survivin plays a significant role in carcinogenesis (1,4). The present study confirmed that genetic susceptibility in miRNA binding regions contributes to altered gene function. Although our findings indicate that rs1042489, rs1042542, rs17882360, rs2239680, rs2661694 and rs4789560 in the BIRC5/survivin 3′UTR have potential miRNA binding sites using bioinformatics analysis, only rs1042489 was revealed to be significantly associated with BIRC5/survivin mRNA expression in lymphoblastoid cell lines. Since rs1042489 has a significant correlation with expression, it is implied that this variant may contribute to BIRC5/survivin post-transcriptional regulation to a certain extent. The results describe the mechanism of the rs1042489 SNP functions in BIRC5/survivin regulation and provide an explanation into the tumor susceptibility associated with this SNP in chemistry research.
It is possible that the genetic variants in the BIRC5/survivin 3′UTR may modulate its expression, and that the variants in the BIRC5/survivin miRNA binding site are associated with carcinogenesis. It was suggested that carriers of rs1042489, a common polymorphism in the BIRC5/survivin gene, have a worse survival compared with the major homozygotes by breast cancer-specific survival analysis of BIRC5/survivin (13). rs1042489 was also observed to have a significant association with expression, implying that this variant may contribute in part to BIRC5/survivin post-transcriptional regulation. It may improve our understanding of the regulatory roles of miRNA variants in BIRC5/survivin 3′UTR in its mRNA expression.
In summary, BIRC5/survivin variant plays a significant role in post-transcriptional regulation, which may highlight the contribution of miRNA-mediated regulation of cancer-associated gene expression and lead to its consideration as a prognostic and diagnostic marker of malignancy. In addition, these findings improve our understanding of how the 3′UTR variants regulate BIRC5/survivin activity and may pave the way to targeting the BIRC5/survivin pathway in cancer therapy. However, this finding needs to be validated by functional analysis of the underlying mechanism involving BIRC5/survivin transcriptional activity associated with variants in the 3′UTR.
References
- 1.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 2.Waligórska-Stachura J, Jankowska A, Waśko R, Liebert W, Biczysko M, Czarnywojtek A, Baszko-Błaszyk D, Shimek V, Ruchała M. Survivin - prognostic tumor biomarker in human neoplasms - review. Ginekol Pol. 2012;83:537–540. [PubMed] [Google Scholar]
- 3.Farnebo L, Tiefenböck K, Ansell A, Thunell LK, Garvin S, Roberg K. Strong expression of survivin is associated with positive response to radiotherapy and improved overall survival in head and neck squamous cell carcinoma patients. Int J Cancer. 2013;133:1994–2003. doi: 10.1002/ijc.28200. [DOI] [PubMed] [Google Scholar]
- 4.Knizhnik AV, Kovaleva OB, Laktionov KK, Mochal'nikova VV, Komel'kov AV, Chevkina EM, Zborovskaia IB. Arf6, RalA and BIRC5 protein expression in non small cell lung cancer. Mol Biol (Mosk) 2011;45:307–315. doi: 10.1134/S0026893310061032. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Zhang X, Wang L, Zheng G, Du L, Yang Y, Dong Z, Liu Y, Qu A, Wang C. Investigation of cell free BIRC5 mRNA as a serum diagnostic and prognostic biomarker for colorectal cancer. J Surg Oncol. 2014;109:574–579. doi: 10.1002/jso.23526. [DOI] [PubMed] [Google Scholar]
- 6.Lv S, Turlova E, Zhao S, Kang H, Han M, Sun HS. Prognostic and clinicopathological significance of survivin expression in bladder cancer patients: A meta-analysis. Tumour Biol. 2014;35:1565–1574. doi: 10.1007/s13277-013-1216-y. [DOI] [PubMed] [Google Scholar]
- 7.Hingorani P, Dickman P, GarciaFilion P, WhiteCollins A, Kolb EA, Azorsa DO. BIRC5 expression is a poor prognostic marker in Ewing sarcoma. Pediatr Blood Cancer. 2013;60:35–40. doi: 10.1002/pbc.24290. [DOI] [PubMed] [Google Scholar]
- 8.Zu Y, Ban J, Xia Z, Wang J, Cai Y, Ping W, Sun W. Genetic variation in a miR-335 binding site in BIRC5 alters susceptibility to lung cancer in Chinese Han populations. Biochem Biophys Res Commun. 2013;430:529–534. doi: 10.1016/j.bbrc.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Holm K, Melum E, Franke A, Karlsen TH. SNPexp-A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics. 2010;11:600. doi: 10.1186/1471-2105-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International HapMap Consortium: The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham JM, Vierkant RA, Sellers TA, Phelan C, Rider DN, Liebow M, Schildkraut J, Berchuck A, Couch FJ, Wang X, et al. Cell cycle genes and ovarian cancer susceptibility: A tagSNP analysis. Br J Cancer. 2009;101:1461–1468. doi: 10.1038/sj.bjc.6605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Bevier M, Johansson R, EnquistOlsson K, Henriksson R, Hemminki K, Lenner P, Försti A. Prognostic impact of polymorphisms in the MYBL2 interacting genes in breast cancer. Breast Cancer Res Treat. 2012;131:1039–1047. doi: 10.1007/s10549-011-1826-2. [DOI] [PubMed] [Google Scholar]