Abstract
Copy number variation is a well-known genetic variation. microRNAs (miRNAs/miRs) are non-coding RNAs that mediate gene expression by regulating target mRNAs. In the present study, copy number deletions encompassing miRNA coding regions were investigated to determine the association between the deletion of miRNA and its phenotypic effects. A total of 38 human miRNAs in copy number variants were identified and miR-650, which is functional in the human osteosarcoma MG-63 cell line, was selected. Overexpression of miR-650 decreased the expression of inhibitor of growth family member 4 (ING4) in the MG-63 cells and increased interleukin (IL)6 transcription, as well as IL6 secretion in IL1B-stimulated cells. Furthermore, miR-650 downregulated the amount of nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor α and increased the transcriptional activity of nuclear factor (NF)κB. Downregulation of ING4 also increased the production of IL6, similar to miR-650 overexpression. Taken together, these data indicate that miR-650 plays a significant role in the production of IL6 by regulating ING4 expression and NFκB signaling in IL1B-stimulated MG-63 osteosarcoma cells.
Keywords: microRNA-650, copy number variation, interleukin 6, ING4
Introduction
Since the completion of the Human Genome Project, gene alterations such as single-nucleotide polymorphisms (SNPs) and copy number variations (CNVs) have received particular attention in the field of disease etiology. Due to technical developments for high-throughput microarrays, sequencing and statistical methods, a number of gene alterations have been reported to be associated with diseases or phenotypic traits in genome-wide association studies (GWAS) (1). The majority of GWAS have examined the roles of SNPs in diseases, and curated resources of SNP-trait associations are available on the website for the GWAS Catalog (2). In addition to SNPs, structural alterations, such as CNVs, have emerged as another major reason for genetic susceptibility to human disease (3). SNPs and CNVs are responsible for 83.6 and 17.7% of the total number of detected genetic variations in gene expression, respectively. Extensive studies of these gene alterations may be effective for identifying the causes of human diseases and phenotypes (4).
Non-coding RNAs are functional RNA molecules that are not translated into functional proteins, but that may contribute to the regulation of a number of biological processes. microRNAs (miRNAs/miRs) are small non-coding RNAs of ~22 nucleotides in length that mediate gene silencing at the post-transcriptional level by targeting the 3′-untranslated region of the target mRNA (5). Since the initial discovery of the first miRNA, lin-4, from the study of post-embryonic development in Caenorhabditis elegans, numerous miRNAs have been identified and reported to be critical regulators of development, cellular physiology and malignancy (6–8). The majority of mammalian miRNA genes are located in defined transcription units (9). Copy number variable miRNA genes (CNV-miRNAs), which are miRNAs located in CNV regions, are potential functional variants in genotype-phenotype association studies (10).
miR-650 was identified from the human colorectal microRNAome (11), and its genomic association with the immunoglobulin (Ig) λ variable region gene was reported based on sequence comparisons and evolutionary approaches (12). Two studies have described the roles of miR-650 in gastrointestinal cancers such as gastric and colorectal cancer by regulating expression of its target genes, inhibitor of growth family member 4 (ING4) and N-myc downstream-regulated gene family member 2 (NDRG2), respectively (13,14). miR-650 is associated with several other tumors, including melanoma, lung adenocarcinoma, hepatocellular carcinoma and glioma (15–18). miR-650 expression is affected by Ig gene rearrangement and is associated with chronic lymphocytic leukemia by downregulation of its target genes, cyclin-dependent kinase 1, ING4 and early B-cell factor 3 in B cells (19). ING4, which is a miR-650 target gene, suppresses tumorigenesis by regulating inflammatory mediators, such as interleukin (IL)6, IL8 and nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor α (IκBα) (20). Furthermore, IL6 regulates the differentiation of osteoblasts and osteoclasts (21). However, no study has been published on the role of miR-650 in the production of IL6 in the human osteosarcoma MG-63 cell line.
In the present study, our previously reported CNV data that were obtained from population-based genome-wide approaches (22) was analyzed and candidate CNV-miRNAs with biological functions were identified. Additionally, the roles of miR-650 in the production of IL6, which is induced by IL1B in human MG-63 osteosarcoma cells, were investigated. The present study was approved by the Institutional Review Board of Korea Centers for Disease Control and Prevention (approval no. 2014-02EXP-10-1C-A).
Materials and methods
Identification of miRNAs overlapping with CNVs
To investigate CNV-miRNAs in the Korean population, CNV regions from our previously reported CNV study were analyzed (22). Briefly, 4,694 samples that are part of the Korean Genome Epidemiology Study were genotyped with the NimbleGen HD2 3×720 K comparative genomic hybridization array. A total of 9,388 CNV regions were identified in human genome build hg18. Among them, 3,601 CNV regions tagged by highly correlated SNPs were provided as the content for the database. In the present study, miRBase, a biological database of miRNA sequences and annotations, was used to obtain human miRNA genome coordinates with the human genome build hg19 (23). Next, the genome coordinates of hg19 were converted to those of hg18 using LiftOver in the University of California, Santa Cruz genome browser (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Of the 1,872 human miRNAs in miRBase, two miRNAs, hsa-mir-1273 and hsa-mir-6724, were excluded due to the absence of genome coordinate information. Moreover, as hsa-mir-511 has two different genome coordinates, both genomic positions were included in the miRNA list. Finally, 9,388 CNV regions with 1,871 miRNA regions were compared to identify CNV-miRNAs.
Cell culture and transfection
The human osteosarcoma MG-63 cell line was purchased from the American Type Culture Collection. The medium used for routine subculture was Dulbecco's modified Eagle's medium (Gibco Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml). The cells were maintained at 37°C in a humidified 5% CO2 incubator. Each 20 nM of miR-650 mimic (forward, 5′-AGGAGGCAGCGCUCUCAGGAC-3′ and reverse, 5′-GUCCUGAGAGCGCUGCCUCCU-3′; Bioneer, Daejeon, Korea) and ING4 small interfering RNA (order no. 1074590; Bioneer) were transfected into MG-63 cells with Lipofectamine RNAiMAX reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Subsequent to transfection, the cells were incubated for 48 h and then treated with 10 ng/ml IL1B (R&D Systems Inc., Minneapolis, MN, USA) prior to being harvested for further experiments.
Quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from cell lysates using the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany). Total RNA (1 µg) was mixed with the AccuPower RocketScript Cycle RT Premix (Bioneer) for cDNA synthesis according to the manufacturer's instructions. The transcribed products were used to amplify target genes. The primer sequences for PCR were as follows: miR-650 forward, 5′-AGAGGAGGCAGCGCTCT-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; ING4 (order no. P279919; Bioneer); GAPDH (order no. P267613; Bioneer); and IL6 (order no. P211161; Bioneer). Amplification was performed using the Exicycler™ 96 Real-Time Quantitative PCR System (Bioneer) in a 20 µl reaction mixture containing 2 µl cDNA template (80 ng), 2.5 µl of each primer and 13 µl distilled water with AccuPower Greenstar qPCR Premix (Bioneer), including dNTP mixture. qPCR was performed under the following conditions: Initial denaturation at 95°C for 10 min; 40 cycles of 95°C for 10 sec, 60°C for 30 sec. Exicycler 3 analysis software (version 3.55.0; Bioneer) was used to calculate cycle threshold (Ct) values for all genes. Relative expression was calculated using the 2−ΔΔCt method (24)
Immunoblotting analysis
Immunoblotting analysis was performed as previously described (25). Briefly, the cultured cells were rinsed with phosphate-buffered saline, scraped into 100 µl RIPA cell lysis buffer (Cell Signaling Technology Inc., Danvers, MA, USA) and placed on ice for 1 h. Next, the cells were centrifuged and the supernatant was harvested. Aliquots (20 µg) of soluble proteins were separated with SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were incubated overnight at 4°C with specific antibodies: Rabbit polyclonal antibody for IκBα and GAPDH, and goat polyclonal antibody for ING4 (1:1,000; Santa Cruz Biotechnology Inc., Dallas, TX, USA). The blot was then incubated with the corresponding horseradish peroxidase-conjugated anti-rabbit IgG or anti-goat IgG (1:5000; Cell Signaling Technology Inc.). The immune complex was visualized with enhanced chemiluminescence reagent (GE Healthcare Life Sciences, Chalfont, UK), and image processing was performed using an image acquisition system (Fusion FX, Vilber Lourmat, Marne-la-Vallée, France).
ELISA
The culture medium of the cells that were transfected with the miR-650 mimic was collected at 6 h post-treatment with IL1B. The level of human IL6 was determined with an ELISA kit (R&D Systems Inc.).
Reporter assay
The MG-63 cells were co-transfected with the pGL4.32 [luc2p/NF-κB-RE/Hygro] containing the nuclear factor kB (NFkB) response element, with pRL-TK expressing Renilla luciferase as an internal control (Promega, USA) and miR-650 mimic with Lipofectamine RNAiMAX reagent in OPTI-MEM (Life Technologies). Following transfection, the cells were incubated for 48 h and then treated with IL1B (10 ng/ml) prior to being harvested. Cell extracts were used for the dual-luciferase assay (Promega Corporation, Madison, WI, USA). Firefly luciferase activity as a reporter was normalized to Renilla activity to control for transfection efficiency.
Statistical analysis
Experimental results were analyzed using R software (version 3.0.2; http://www.r-project.org/). Statistical analysis was performed with a one-way analysis of variance, and data are expressed as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of miRNAs in CNV regions and selection of miR-650 for functional analysis
From the comparative analysis of 9,388 CNV regions and 1,871 miRNA regions, 38 miRNAs on CNV regions were identified (Table I). Table I shows the genome coordinates of these 38 miRNAs. When several CNV regions with different break-points (i.e., start and end positions) encompassed the same miRNA, minimal overlapping regions between CNVs were used. Among the 38 miRNAs, the target genes of three (miR-650, miR-132 and miR-212) have previously been reported (13,26). From those three miRNAs, miR-650 was selected for this functional study, as its location overlapped with the exon region of another transcript, and as its target genes have been identified in several cancer cell types, but not in osteosarcoma cells (14,16,17,20,27.
Table I.
miRNAs located in copy number-variable regions.
| ID | Chra | Start | End | CNV regionb |
|---|---|---|---|---|
| hsa-mir-6730 | 1 | 12561572 | 12561638 | chr1: 12322432–12771354 |
| hsa-mir-4256 | 1 | 112805915 | 112805978 | chr1: 112494152–113047786 |
| hsa-mir-4266 | 2 | 109296459 | 109296513 | chr2: 109295476–109299710 |
| hsa-mir-3921 | 3 | 101165848 | 101165932 | chr3: 101111409–101181644 |
| hsa-mir-4789 | 3 | 176570023 | 176570104 | chr3: 176563587–176573504 |
| hsa-mir-7978 | 4 | 21075421 | 21075479 | chr4: 21057650–21076477 |
| hsa-mir-1973 | 4 | 117440330 | 117440373 | chr4: 117260800–117551180 |
| hsa-mir-8089 | 5 | 180403009 | 180403090 | chr5: 180362646–180406960 |
| hsa-mir-6832 | 6 | 31709543 | 31709614 | chr6: 31564443–31778770 |
| hsa-mir-4646 | 6 | 31776785 | 31776847 | chr6: 31564443–31778770 |
| hsa-mir-3135b | 6 | 32825667 | 32825734 | chr6: 32734545–32828543 |
| hsa-mir-550a-3 | 7 | 29686875 | 29686969 | chr7: 29652945–29755706 |
| hsa-mir-4650-2 | 7 | 71800810 | 71800885 | chr7: 71635398–71956108 |
| hsa-mir-3674 | 8 | 1736698 | 1736765 | chr8: 1733768–1765533 |
| hsa-mir-596 | 8 | 1752804 | 1752880 | chr8: 1733768–1765533 |
| hsa-mir-876 | 9 | 28853624 | 28853704 | chr9: 28717924–28857119 |
| hsa-mir-4675 | 10 | 20880905 | 20880981 | chr10: 20871043–20896483 |
| hsa-mir-4678 | 10 | 89253618 | 89253691 | chr10: 89251878–89266538 |
| hsa-mir-3166 | 11 | 87549318 | 87549409 | chr11: 87543703–87562550 |
| hsa-mir-6763 | 12 | 131668656 | 131668720 | chr12: 131650394–131675091 |
| hsa-mir-1233-1 | 15 | 32461562 | 32461643 | chr 15: 32447266–32664650 |
| hsa-mir-1233-2 | 15 | 32607783 | 32607864 | chr 15: 32447266–32664651 |
| hsa-mir-6862-1 | 16 | 28309804 | 28309873 | chr16: 28283100–28334047 |
| hsa-mir-6862-2 | 16 | 28643074 | 28643143 | chr16: 28587048–28670623 |
| hsa-mir-132 | 17 | 1899952 | 1900052 | chr17: 1804068–1910573 |
| hsa-mir-212 | 17 | 1900315 | 1900424 | chr17: 1804068–1910573 |
| hsa-mir-6129 | 17 | 44720707 | 44720815 | chr17: 44706971–44732961 |
| hsa-mir-4524b | 17 | 64607278 | 64607392 | chr17: 64601231–64619802 |
| hsa-mir-4524a | 17 | 64607300 | 64607368 | chr17: 64601231–64619802 |
| hsa-mir-4745 | 19 | 755940 | 756001 | chr19: 726335–763296 |
| hsa-mir-1270-1 | 19 | 20371080 | 20371162 | chr19: 20367897–20508581 |
| hsa-mir-1270-2 | 19 | 20371080 | 20371162 | chr19: 20367897–20508581 |
| hsa-mir-4752 | 19 | 59477776 | 59477847 | chr19: 59477286–59499140 |
| hsa-mir-3195 | 20 | 60073253 | 60073336 | chr20: 60066989–60075374 |
| hsa-mir-650 | 22 | 21495270 | 21495365 | chr22: 21488908–21498416 |
| hsa-mir-5571 | 22 | 21558447 | 21558559 | chr22: 21551911–21565430 |
| hsa-mir-6817 | 22 | 24181613 | 24181678 | chr22: 24034085–24258990 |
| hsa-mir-6818 | 22 | 28733038 | 28733102 | chr22: 28666644–28735357 |
Chr, chromosome (chr) number
CNV region, minimal overlapping region of each copy number variation (CNV). miR/miRNA, microRNA.
Overexpression of miR-650 downregulates ING4 mRNA and protein expression in MG-63 osteosarcoma cells
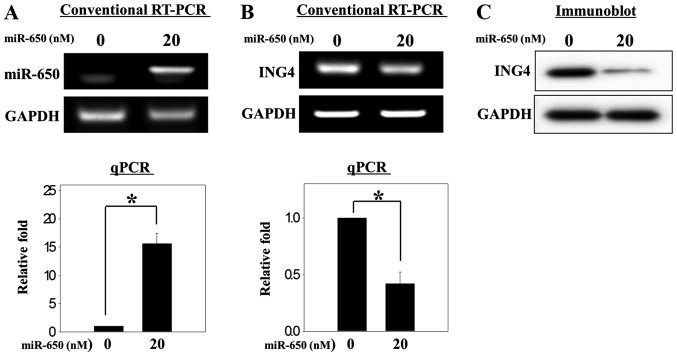
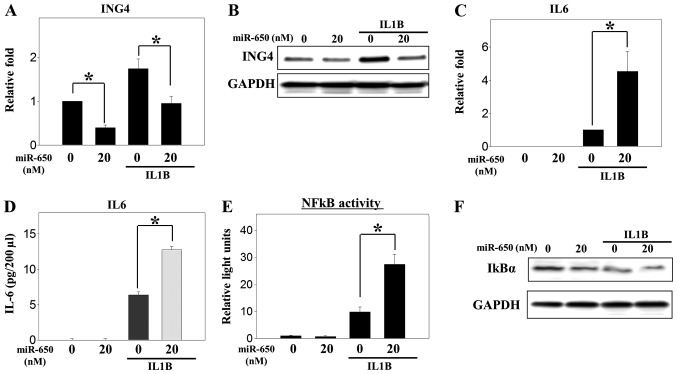
Although ING4 is a target gene of miR-650 in other cancer cell types (13,17,20), it had not been evaluated in osteosarcoma cells. Thus, a miR-650 mimic was transfected to overexpress miR-650 in the MG-63 osteosarcoma cells (Fig. 1A). Overexpression of miR-650 decreased the expression of ING4 mRNA and protein (Fig. 1B and C). As MG-63 cells are osteoblast-like osteosarcoma cells, stimulation with IL1B may accelerate the production of inflammatory cytokines, such as tumor necrosis factor-α and IL6, through the NFκB signaling pathway (28). In the present study, the overexpression of miR-650 decreased the upregulation of ING4 mRNA expression that was induced by IL1B (Fig. 2A). A similar expression pattern was observed for protein levels (Fig. 2B). These results indicate that ING4 may be a target gene of miR-650 in osteosarcoma cells, similar to previous results in other cancer cells.
Figure 1.
Overexpression of miR-650 decreases the expression of ING4 mRNA and protein in MG-63 osteosarcoma cells. (A) A total of 2.0×105 cells were plated in 6-well plates and transfected with miR-650 mimic (20 nM) to overexpress miR-650. (A) miR-650 and (B) ING4 mRNA were each measured with conventional reverse transcription polymerase chain reaction (RT-PCR) and quantitative (q)PCR. (C) Protein analysis of ING4 was performed with immunoblotting. The mRNA and protein expression of GAPDH was used as a loading control. Significant differences (*P<0.05) compared with the control were calculated with an analysis of variance. miR/miRNA, microRNA; ING4, inhibitor of growth family member 4.
Figure 2.
Overexpression of miR-650 decreases ING4 expression and increases IL6 production induced by treatment with IL1B in MG-63 osteosarcoma cells. Following transfection with the miR-650 mimic, the cells were incubated for 48 h and treated with IL1B (10 ng/ml) to induce IL6 production. (A) mRNA and (B) protein expression of ING4 and (C) mRNA expression of IL6 were detected as shown in Fig. 1. (D) Culture medium was collected and used to measure IL6 production with an ELISA. (E and F) A total of 2.0×105 cells were transfected with 200 ng pGL4.32 (luc2p/NF-κB-RE/Hygro) vector and the miR-650 mimic (20 nM). After 1 h, the cells were treated with IL1B (10 ng/ml). Firefly luciferase activity, as a reporter of NFκB activity, was normalized to Renilla activity, as a control for transfection efficiency. Protein expression of IκBα was analyzed with immunoblotting. Significant differences (*P<0.05) compared with the control were calculated with an analysis of variance. ING4, inhibitor of growth family member 4; IL6, interleukin 6; NFκB, nuclear factor κB; IκBα, nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor α.
Overexpression of miR-650 increases the production of IL6 induced by treatment with IL1B in MG-63 osteosarcoma cells
Overexpression of ING4 decreases the expression of IL6 in human umbilical vein endothelial cells (29). Therefore, we investigated the role of miR-650 in the production of IL6 induced by IL1B in MG-63 cells. As expected, IL6 mRNA and protein expression was increased by treatment with IL1B in the MG-63 cells and was more highly increased in the miR-650-overexpressing cells than in the control cells (Fig. 2C and D). ING4 may regulate the expression of IL6 by modulating NFkB activity, as reported previously in melanoma and brain tumors (29,30). Therefore, the present study investigated whether NFκB transcriptional activity is involved in the production of IL6 by miR-650 in MG-63 cells. NFκB transcriptional activity was increased by IL1B treatment and more highly increased in the miR-650-overexpressing cells than the control cells (Fig. 2E). Furthermore, IκBα protein levels were decreased by overexpression of miR-650 (Fig. 2F). These results indicate that miR-650 regulates the production of IL6 that is induced by IL1B by modulating ING4 expression and NFκB transcriptional activity in osteosarcoma cells. This is similar to the results found in other cells, such as brain tumor cells and melanoma cells (29,30).
Knockdown of ING4 expression increases the production of IL6
The present study investigated whether the effect of miR-650 on the production of IL6 induced by IL1B was mediated by ING4. The production of IL6 mRNA and protein was more highly increased in the cells in which ING4 was knocked down compared with the negative control group (Fig. 3). These results indicate that miR-650 regulates the production of IL6 by modulating its target gene, ING4.
Figure 3.
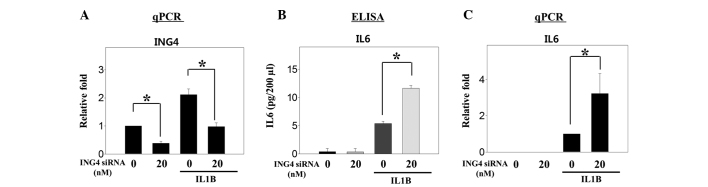
Knockdown of ING4 expression increases the production of IL6 that is induced by treatment with IL1B in MG-63 cells. A total of 2.0×105 cells were plated and transfected with small interfering (si)RNA for ING4 (20 nM). After 48 h, the cells were harvested. (A and C) mRNA expression and (B) protein expression were analyzed with quantitative polymerase chain reaction (qPCR) and ELISA, respectively. Significant differences (*P<0.05) compared with the control were calculated with an analysis of variance. ING4, inhibitor of growth family member 4; IL6, interleukin 6.
Discussion
Extensive CNV studies have shown that various human diseases, including autism, schizophrenia, epilepsy, Parkinson's disease, Alzheimer's disease, chronic pancreatitis and Crohn's disease, are associated with genomic alterations (31). miRNA signatures in a number of diseases, such as melanoma and colorectal cancer, indicate that miRNAs have significant functions in human disease through the regulation of their target genes (11,15). Gene alterations such as CNVs and SNPs in miRNA genes in the human genome are potential variants for studying the functional roles of miRNAs in human disorders (10). In the present study, 38 CNV-miRNAs were identified from the CNV discovery study of a Korean cohort using the NimbleGen HD2 3×720 K comparative genomic hybridization array. miR-650 was selected for further studies and its role was analyzed in the production of the inflammatory cytokine, IL6, in MG-63 osteosarcoma cells.
miR-650 was selected for further functional studies, as its target genes, ING4 and NDRG2, are important in gastric and colorectal cancer cells, respectively (13,14). In addition, miR-650 has previously been identified as a CNV-miRNA in another study (10). ING4 is a member of the ING family that acts as a tumor suppressor protein and is a promising candidate for the development of novel therapies in cancer research (13,17,29,30,32. In the present study, the overexpression of miR-650 decreased the expression of ING4 mRNA and protein levels (Fig. 1), and decreased the expression of the ING4 that was upregulated by treatment with IL1B to induce IL6 (Fig. 2).
A number of studies have described a role for ING4 in tumorigenesis and innate immunity by regulating the expression of p53, tumor necrosis factor α, IL6, IL8, matrix metalloproteinases, cycloxygenase-2 and IκBα. Among the genes regulated by ING4, IL6 encodes a multifunctional cytokine that activates target genes involved in a wide range of biological activities (20,27,30,33). IL6 also modulates osteoblast and osteoclast differentiation. Recently, IL6 in osteosarcoma cells was reported to promote the expression of intercellular adhesion molecule-1 and cell motility (21,34). Although roles for ING4 and IL6 in tumorigenesis and inflammation have been reported, no study has described the role of miR-650 in the production of IL6 in osteosarcoma cells. Based on previous studies, we speculated that the regulation of IL6 by ING4 may be due to the activity of miR-650. As expected, miR-650 increased the production of IL6 that was induced by IL1B by downregulating ING4 in the MG-63 osteosarcoma cells (Fig. 2).
ING4 regulates IL6 production by modulating NFκB activity (29). Therefore, the present study investigated the modulation of NFκB by miR-650 using an NFκB reporter assay, and measured the amount of IκBα protein. As shown in Fig. 2E and F, the overexpression of miR-650 increased the activity of NFκB transcriptional activity and regulated the amount of IκBα protein. Regulation of NFκB activity by ING4 may be due to a physical interaction between ING4 and p65 (RelA), a subunit of NFκB, that results in decreased activation of the canonical NFκB-responsive promoter in brain tumor cells (30). A previous study showed that ING4 promotes IκB promoter activation to suppress NFκB signaling (35). The present study did not investigate whether ING4 directly interacted with p65 or how ING4 regulates the NFκB signaling pathway to induce the differential expression of NFκB target genes. However, it may be concluded that miR-650 regulates IL6 production by modulating ING4 expression and subsequent NFκB signaling in osteosarcoma cells. Therefore, this study may be the first to elucidate the role of miR-650 in the production of IL6 in IL1B-stimulated osteosarcoma cells.
Taken together, these data indicate that the overexpression of miR-650 increases the production of IL6 induced by IL1B treatment in MG-63 osteosarcoma cells by regulating ING4 expression and subsequent NFκB transcriptional activity. Additionally, this study suggests that miR-650 may be an upstream regulator of IL6 production in MG-63 osteosarcoma cells and could be a candidate therapeutic target for IL6-related human diseases, including cancer.
Acknowledgements
This study was supported by intramural grants from the Korea National Institute of Health (nos. 2010-N73004-00 and 2013-NG73001-00). Biospecimens and data were provided by the Korean Genome Analysis Project (4845-301), the Korean Genome and Epidemiology Study (4851-302) and the Korea Biobank Project (4851-307), which were supported by the Korea Centers for Disease Control and Prevention, Republic of Korea.
References
- 1.IonitaLaza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- 4.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 7.Mendell JT. MicroRNAs: Critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 8.Lee A, McLean D, Choi J, Kang H, Chang W, Kim J. Therapeutic implications of microRNAs in pulmonary arterial hypertension. BMB Rep. 2014;47:311–317. doi: 10.5483/BMBRep.2014.47.6.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Marcinkowska M, Szymanski M, Krzyzosiak WJ, Kozlowski P. Copy number variation of microRNA genes in the human genome. BMC Genomics. 2011;12:183. doi: 10.1186/1471-2164-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S. Evolutionary origin and genomic organization of micro-RNA genes in immunoglobulin lambda variable region gene family. Mol Biol Evol. 2009;26:1179–1189. doi: 10.1093/molbev/msp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu Z, Zhang M. MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun. 2010;395:275–280. doi: 10.1016/j.bbrc.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Feng L, Xie Y, Zhang H, Wu Y. Down-regulation of NDRG2 gene expression in human colorectal cancer involves promoter methylation and microRNA-650. Biochem Biophys Res Commun. 2011;406:534–538. doi: 10.1016/j.bbrc.2011.02.081. [DOI] [PubMed] [Google Scholar]
- 15.Chan E, Patel R, Nallur S, Ratner E, Bacchiocchi A, Hoyt K, Szpakowski S, Godshalk S, Ariyan S, Sznol M, et al. MicroRNA signatures differentiate melanoma subtypes. Cell Cycle. 2011;10:1845–1852. doi: 10.4161/cc.10.11.15777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang JY, Cui SY, Chen YT, Song HZ, Huang GC, Feng B, Sun M, De W, Wang R, Chen LB. MicroRNA-650 was a prognostic factor in human lung adenocarcinoma and confers the docetaxel chemoresistance of lung adenocarcinoma cells via regulating Bcl-2/Bax expression. PLoS One. 2013;8:e72615. doi: 10.1371/journal.pone.0072615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng ZL, Li FJ, Gao F, Sun DS, Yao L. Upregulation of miR-650 is correlated with down-regulation of ING4 and progression of hepatocellular carcinoma. J Surg Oncol. 2013;107:105–110. doi: 10.1002/jso.23210. [DOI] [PubMed] [Google Scholar]
- 18.Sun B, Pu B, Chu D, Chu X, Li W, Wei D. MicroRNA-650 expression in glioma is associated with prognosis of patients. J Neurooncol. 2013;115:375–380. doi: 10.1007/s11060-013-1243-y. [DOI] [PubMed] [Google Scholar]
- 19.Mraz M, Dolezalova D, Plevova K, Stano Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B, Pavlova S, Borsky M, et al. MicroRNA-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia. Blood. 2012;119:2110–2113. doi: 10.1182/blood-2011-11-394874. [DOI] [PubMed] [Google Scholar]
- 20.Mathema VB, Koh YS. Inhibitor of growth-4 mediates chromatin modification and has a suppressive effect on tumorigenesis and innate immunity. Tumour Biol. 2012;33:1–7. doi: 10.1007/s13277-011-0249-3. [DOI] [PubMed] [Google Scholar]
- 21.Blanchard F, Duplomb L, Baud'huin M, Brounais B. The dual role of IL-6-type cytokines on bone remodeling and bone tumors. Cytokine Growth Factor Rev. 2009;20:19–28. doi: 10.1016/j.cytogfr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Moon S, Jung KS, Kim YJ, Hwang MY, Han K, Lee JY, Park K, Kim BJ. KGVDB: A population-based genomic map of CNVs tagged by SNPs in Koreans. Bioinformatics. 2013;29:1481–1483. doi: 10.1093/bioinformatics/btt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.DennisSykes CA, Miller WJ, McAleer WJ. A quantitative Western Blot method for protein measurement. J Biol Stand. 1985;13:309–314. doi: 10.1016/S0092-1157(85)80044-5. [DOI] [PubMed] [Google Scholar]
- 26.Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Xu LS, Wang ZQ, Wang KS, Li N, Cheng ZH, Huang SZ, Wei DZ, Han ZG. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004;570:7–12. doi: 10.1016/j.febslet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Yoon WJ, Heo SJ, Han SC, Lee HJ, Kang GJ, Yang EJ, Park SS, Kang HK, Yoo ES. Sargachromanol G regulates the expression of osteoclastogenic factors in human osteoblast-like MG-63 cells. Food Chem Toxicol. 2012;50:3273–3279. doi: 10.1016/j.fct.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Li G. Cell cycle regulator ING4 is a suppressor of melanoma angiogenesis that is regulated by the metastasis suppressor BRMS1. Cancer Res. 2010;70:10445–10453. doi: 10.1158/0008-5472.CAN-10-3040. [DOI] [PubMed] [Google Scholar]
- 30.Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 31.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 32.Wei Q, He W, Lu Y, Yao J, Cao X. Effect of the tumor suppressor gene ING4 on the proliferation of MCF-7 human breast cancer cells. Oncol Lett. 2012;4:438–442. doi: 10.3892/ol.2012.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–1379. doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- 34.Lin YM, Chang ZL, Liao YY, Chou MC, Tang CH. IL-6 promotes ICAM-1 expression and cell motility in human osteosarcoma. Cancer Lett. 2013;328:135–143. doi: 10.1016/j.canlet.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Coles AH, Gannon H, Cerny A, KurtJones E, Jones SN. Inhibitor of growth-4 promotes IkappaB promoter activation to suppress NF-kappaB signaling and innate immunity. Proc Natl Acad Sci USA. 2010;107:11423–11428. doi: 10.1073/pnas.0912116107. [DOI] [PMC free article] [PubMed] [Google Scholar]