Abstract
DNA strand exchange plays a central role in genetic recombination across all kingdoms of life, but the physical basis for these reactions remains poorly defined. Using single-molecule imaging, we found that bacterial RecA and eukaryotic Rad51 and Dmc1 all stabilize strand exchange intermediates in precise three-nucleotide steps. Each step coincides with an energetic signature (0.3 kBT) that is conserved from bacteria to humans. Triplet recognition is strictly dependent on correct Watson-Crick pairing. Rad51, RecA, and Dmc1 can all step over mismatches, but only Dmc1 can stabilize mismatched triplets. This finding provides insight into why eukaryotes have evolved a meiosis-specific recombinase. We propose that canonical Watson-Crick base triplets serve as the fundamental unit of pairing interactions during DNA recombination.
Homologous recombination enables the exchange of genetic information between DNA molecules and is a driving force in evolution. During homologous recombination, a presynaptic single-stranded DNA (ssDNA) is paired with the complementary strand of a homologous double-stranded DNA (dsDNA), resulting in displacement of the noncomplementary strand (1). This strand exchange reaction plays essential roles in double-strand DNA break (DSB) repair (1, 2), the rescue of stalled or collapsed replication forks (2, 3), chromosomal rearrangements (2), horizontal gene transfer (4, 5), and meiosis (6, 7). These reactions are promoted by the Rad51/RecA family of DNA recombinases, which are adenosine triphosphate (ATP)–dependent proteins that form helical filaments on DNA (1, 8). Crystal structures of RecA-ssDNA presynaptic complexes and RecA-dsDNA postsynaptic complexes reveal that the DNA is organized into near B-form base triplets separated by ~7.1 to 8.4 Å between adjacent triplets (Fig. 1A) (9, 10); for brevity, we refer to these nucleic acids as RS-DNA (Rad51/RecA-stretched DNA).
Fig. 1. Structure of the presynaptic complex and experimental overview.
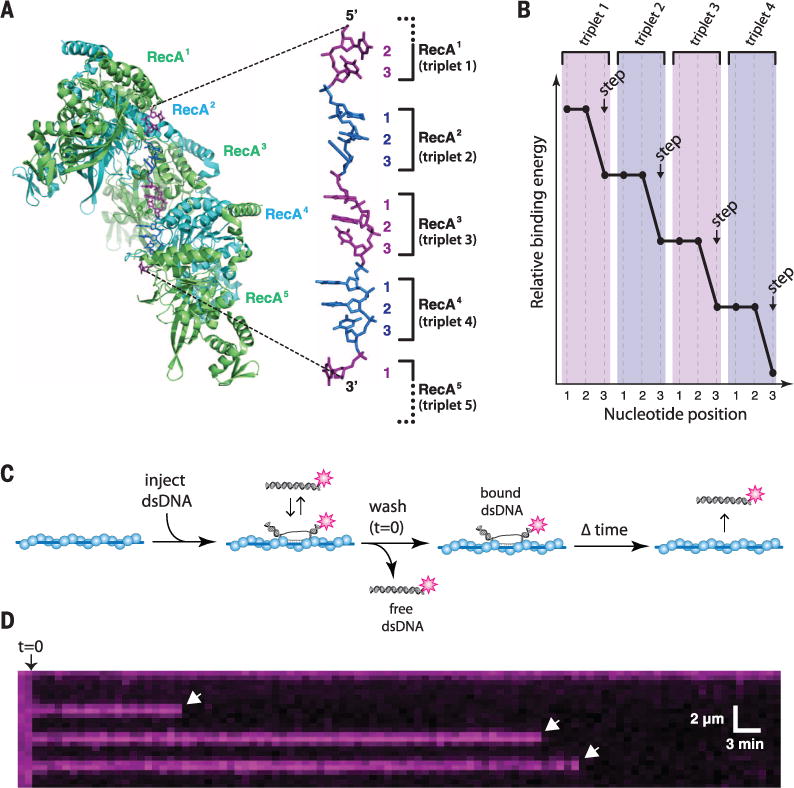
(A) Structure of the RecA-ssDNA filament highlighting the base triplet organization of the presynaptic RS-ssDNA (9). (B) Cartoon illustration of base triplet stepping for S. cerevisiae Rad51. Quantized reductions in binding energy are proposed to coincide with the third base of each triplet. (C) Experimental outline for measuring the survival probability of fluorescently tagged dsDNA oligonucleotides bound to the presynaptic complex. (D) Example of a kymograph showing the binding of single Atto565-dsDNA molecules to a ScRad51 presynaptic complex. White arrowheads highlight individual dsDNA dissociation events.
We previously developed single-molecule methods to study DNA recombination by total internal reflection fluorescence microscopy (TIRFM) (11). Using this approach, we showed that presynaptic complexes search for homology by sampling dsDNA for ≥8-nucleotide (nt) tracts of microhomology and rapidly reject sequences with ≤7 nt of microhomology (11). We have also reported that strand exchange by Saccharomyces cerevisiae Rad51 (ScRad51) occurs in 3-nt steps, presumably reflecting the base triplet organization of RS-DNA (Fig. 1, A and B) (11), and 3-nt stepping has also been proposed for bacterial RecA (12). Here, we sought to determine the underlying principles that contribute to base triplet stepping and establish how these principles influence the mechanism and fidelity of DNA recombination. To address these questions, we assembled presynaptic complexes on ssDNA curtains (Fig. 1C and fig. S1) using one of four different recombinases: Escherichia coli RecA (EcRecA) (1); ScRad51 or human Rad51 (hRad51), eukaryotic orthologs of RecA (2); or S. cerevisiae Dmc1 (ScDmc1), which is specialized for meiotic recombination (fig. S2) (6, 13).
For visualization of strand exchange intermediates, 70-base pair (bp) Atto565-dsDNA substrates were briefly incubated with the presynaptic complexes and unbound dsDNA was flushed away. Complexes were visualized by TIRFM (Fig. 1, C and D), and dissociation rates were obtained from the survival probabilities of the bound Atto565-dsDNA (fig. S3). The dsDNA substrates bore 8-to 15-nt tracts of microhomology targeted to two different regions of the presynaptic ssDNA (Fig. 2A, fig. S1B, fig. S4A, and table S1) (11). The dissociation rates for both sets of substrates scaled with microhomology length for each of the recombinases. In each instance, pronounced changes in dissociation rates coincided with recognition of the 9th, 12th, and 15th nucleotides, and similar results were observed in reactions with the nonhydrolyzable ATP analogs adenylyl imidodiphosphate (AMP-PNP) and adenosine 5′-O-(3-thiotriphosphate) (ATPγS) (Fig. 2 and fig. S4). These binding patterns demonstrate that 3-nt stepping is a broadly conserved feature of the Rad51/RecA family of DNA recombinases.
Fig. 2. Conservation of base triplet stepping among Rad51/RecA family members.

(A) Schematic of the 70-bp dsDNA substrates (set #2; see table S1). All substrates contain an internal 8- to 15-nt tract of microhomology (as indicated) flanked by nonhomologous sequence. (B) Atto565-dsDNA dissociation rates (mean ± SD) for ScRad51 plus ATP. (C) EcRecA plus ATPγS. (D) hRad51 plus ATP. (E) ScDmc1 plus ATP. (F) ScRad51 plus AMP-PNP In (B) to (F), each data point was calculated from an average of ~150 molecules (N = 70 to 250); the color-coded shading highlights each base triplet; magenta shading indicates the minimum 8 nt necessary for binding; purple shading corresponds to additional homologous nucleotides; arrows indicate stepwise reductions in dissociation rates coincident with recognition of the third base of each triplet; dashed lines report the mean rate for each step; and the free energy changes (ΔΔG‡) associated with each triplet step are indicated.
Comparison of reactions with EcRecA, ScRad51, hRad51, and ScDmc1 revealed that the free energy change (ΔΔG‡) associated with the binding of each base triplet was similar for all recombinases (Fig. 2 and fig. S4), corresponding to 0.30 ± 0.14 kBT (mean ± SD) for completion of a single triplet step (fig. S5). This result supports the conclusion that the free energy changes associated with triplet steps during DNA recombination are broadly conserved. In addition, reactions with AMP-PNP or ATPγS revealed no appreciable shift in the free energy change associated with each triplet step (Fig. 2 and fig. S4). Thus, the physical determinants governing the energetics of strand exchange have been retained during the evolution of the Rad51/RecA gene family.
Whereas base stacking dominates the stability of B-DNA (14), stacking interactions are disrupted in RS-DNA, which suggests that the structure of RS-DNA may enhance the fidelity of recombination by relying more on correct Watson-Crick pairing. Therefore, we next asked whether noncomplementary bases affect individual strand exchange steps (Fig. 3A). These experiments demonstrated that a single mismatch anywhere within a base triplet completely abolishes recognition of the entire triplet (Fig. 3B and fig. S6). All noncomplementary nucleotides abolished triplet recognition regardless of mismatch identity, and this high level of discrimination was conserved across the Rad51/RecA family (Fig. 3B and fig. S6). In addition, reactions with ATPγS or AMP-PNP revealed that ATP hydrolysis played no discernible role in mismatch discrimination at the level of a single triplet step (Fig. 3B and fig. S6, C and F).
Fig. 3. Effects of mismatches on base triplet recognition.
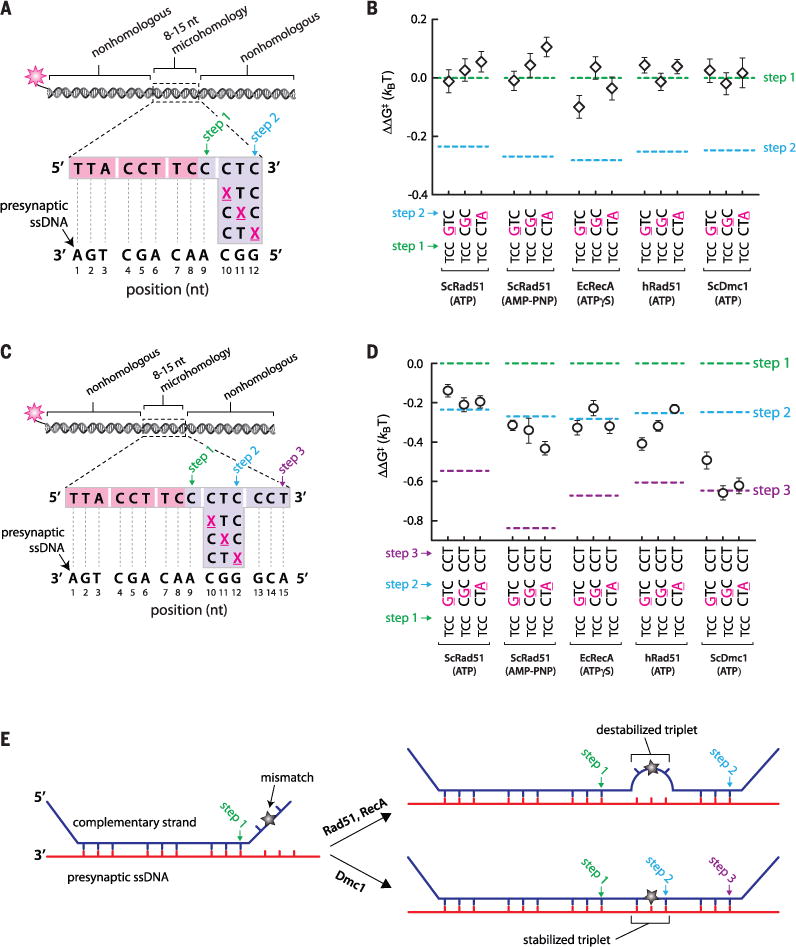
(A) Schematic highlighting the design of dsDNA substrates (set #2; see table S3) bearing mismatched bases within the fourth nucleotide triplet, corresponding to the second strand exchange step; each mismatch position is highlighted as an underlined magenta X. (B) Mismatch substrate binding for ScRad51, EcRecA, hRad51, and ScDmc1, as indicated. All indicated ΔΔG‡ values are relative to the step 1 binding data for the substrate bearing 9 nt of microhomology (see Fig. 2); each data point was calculated from an average of ~150 molecules. The green and blue dashed lines correspond to data obtained for non-mismatched substrates with each of the four different recombinases (see Fig. 2, B to F). (C) Substrate design (set #2; see table S4) for testing whether Rad51/RecA is capable of stepping over a mismatched triplet. (D) Triplet binding data illustrating how base mismatches within the fourth triplet affect recognition of the fifth triplet. (E) Schematic highlighting the different models predicted for mismatched substrates in reactions with RecA and Rad51 versus Dmc1.
We next sought to determine whether strand exchange could progress beyond a mismatch (Fig. 3C and fig. S7A). Extending the length of homology by just a single triplet allowed each recombinase to step past the mismatches, as evidenced by the corresponding reduction in binding free energy (Fig. 3D and fig. S7B). When EcRecA, ScRad51, or hRad51 stepped over a mismatched triplet, the mismatched triplets did not contribute to the binding free energy of the resulting intermediates. This conclusion is based on the observation that the corresponding reduction in binding free energy was comparable to products reflecting a second rather than a third strand exchange step (Fig. 3D and fig. S7B). This result supports a model in which these internal mismatch-bearing triplets remain destabilized (Fig. 3E). In contrast, when ScDmc1 stepped over a mismatched triplet, the stability of the resulting intermediates was comparable to expectations for completion of both the second and the third strand exchange steps (Fig. 3D and fig. S7B). The simplest interpretation of this result is that in reactions with ScDmc1, the internal mismatched triplet was either partially or fully paired with presynaptic ssDNA (Fig. 3E). To determine whether the ability to stabilize mismatched triplets is a conserved feature of the Dmc1 lineage, we next performed experiments using human Dmc1 (hDmc1). Like the other recombinases, hDmc1 strand exchange intermediates were stabilized in 3-nt steps, triplet recognition coincided with a change in free energy of ~0.3 kBT, and mismatches abolished triplet recognition (fig. S8, A to D). Like ScDmc1, hDmc1 is able to step over mismatches, and also stabilizes the mismatched triplets (fig. S8, E and F). Although Rad51, RecA, and Dmc1 can all step over mismatches, the ability to stabilize mismatched triplets embedded within longer tracts of homology is only conserved within the Dmc1 lineage.
It is not known why eukaryotes have evolved two recombinases. Rad51 is expressed ubiquitously, but Dmc1 is present only during meiosis (6, 13). A fundamental difference between mitotic and meiotic recombination is the choice of template used to direct DSB repair (6, 7, 15). Mitotic recombination is biased toward use of the identical sister chromatid to ensure accurate repair. Meiotic recombination favors use of the homolog, yielding crossovers and increasing genome diversity through reshuffling of parental alleles and meiotic gene conversion (6, 7, 15). The mechanisms that direct template choice remain poorly defined (6, 7, 15). Our findings reveal that Dmc1 can stabilize mismatches, which may play a role in guiding template choice. For example, the inability of Rad51 to stabilize mismatches could help bias mitotic recombination toward use of the sister chromatid by disfavoring interhomolog recombination. This bias may be enforced through mismatch repair (MMR) anti-recombination activity (16, 17), which could selectively disrupt mismatched intermediates arising from any attempts by Rad51 to promote interhomolog recombination. Conversely, the ability of Dmc1 to stabilize mismatches may allow recombination between polymorphic maternal and paternal alleles during meiosis by masking mismatched intermediates from premature dissolution by the MMR machinery. Dissociation of Dmc1 upon completion of strand invasion would then allow efficient gene conversion through MMR-mediated repair of the mismatched heteroduplex.
We next sought a quantitative explanation for why strand exchange takes place in 3-nt steps. Given our findings, it is conceivable that base-pairing transitions within the interior of the presynaptic complexes are governed primarily by the thermodynamic characteristics of RS-DNA. Concordant with this view is the finding that the complementary strand within the RecA-dsDNA postsynaptic complex is held in place mainly by Watson-Crick hydrogen bonds (9). However, the thermodynamic properties of RS-DNA cannot be experimentally accessed outside the context of Rad51/RecA nucleoprotein filaments. As an alternative, we used Monte Carlo and molecular dynamics simulations to explore how extension of every third phosphodiester bond may alter the melting and annealing transitions of RS-DNA These simulations used a coarse-grained model that recapitulates structural, thermodynamic, and mechanical characteristics of DNA (18). Although more complex in silico models can be envisaged, such models would require further assumptions that could only serve to confuse the general argument that the physical architecture of RS-DNA itself may be sufficient to define some mechanistic attributes of strand exchange.
Monte Carlo simulations reveal that RS-DNA annealing is highly unfavorable because of the energetic penalty associated with extension of the complementary ssDNA strand (fig. S9A). However, annealing becomes favorable when the incoming homologous ssDNA strand is constrained into base triplets to mimic the effect of the recombinases (fig. S9A). This result agrees with the expectation that Rad51/RecA must locally stretch the incoming homologous DNA to more closely conform to the extended configuration of the presynaptic RS-ssDNA (9, 10, 19). Moreover, the free energy profiles for RS-DNA reveal pronounced energetic barriers with 3-nt periodicity (fig. S9A). Molecular dynamics simulations confirm that B-DNA annealing and melting transitions occur in 1-bp increments; consistent with a 1-bp zippering mechanism (18), the lifetimes of pairing intermediates are largely independent of nucleotide position (Fig. 4, A to C, and fig. S9, B and D). In contrast, the annealing and melting transitions for RS-DNA occur in 3-nt steps (Fig. 4, D to F, and fig. S9, C and D), and partially paired RS-DNA triplets are highly transient relative to fully paired triplets (Fig. 4E and fig. S9D). These simulations reveal that partially paired RS-DNA triplets are not stable, which recapitulates a key feature of our experimental data—base triplet stepping—even though the model intentionally omits any detailed contributions of the proteins or amino acid side chains other than to stretch the DNA strands into an RS-DNA configuration. Together, the simulations and experimental work suggest that canonical Watson-Crick base triplets act as the fundamental pairing unit within RS-DNA.
Fig. 4. RS-DNA governs triplet stepping during strand exchange.
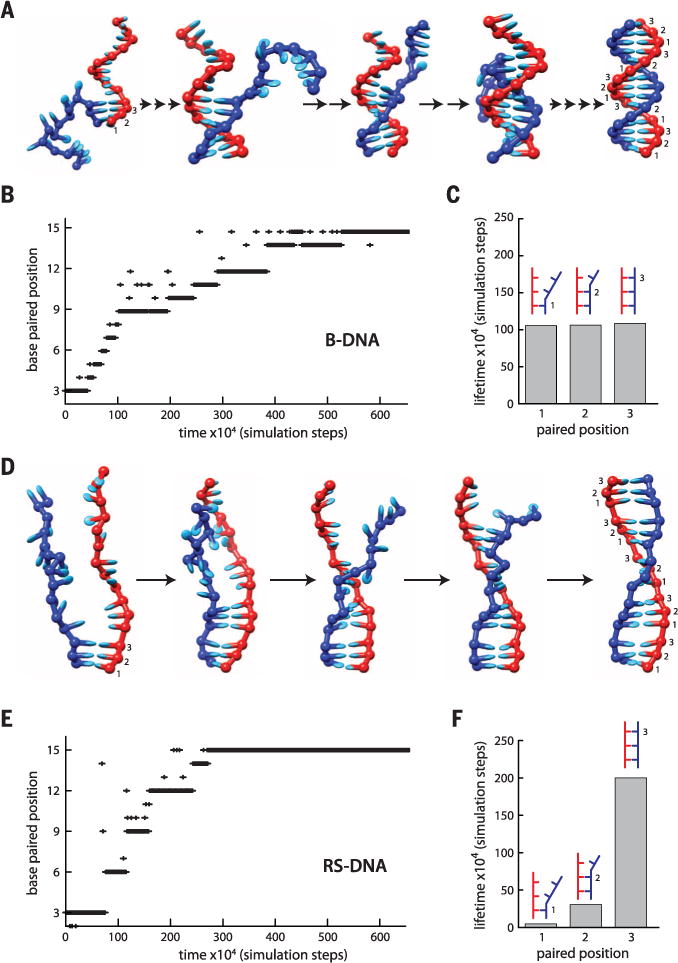
(A) Molecular dynamics snapshots of B-DNA annealing intermediates. Paired bases are numbered 1 to 3 for comparison to RS-DNA triplets. (B) Sample annealing trajectory for B-DNA. (C) Lifetimes of B-DNA intermediates calculated from 50 separate simulation runs (5 × 108 simulation steps total). (D) Molecular dynamics snapshots of RS-DNA. (E) Sample annealing trajectory for RS-DNA. (F) Lifetimes of RS-DNA intermediates calculated from 50 separate simulation runs (5 × 108 simulation steps total). In (A) and (D), the number of arrows between each image corresponds to the number of steps necessary to reach the depicted state. The y axes in (B) and (E) denote the position of the most distal paired base relative to the pre-annealed end of the DNA. The data in (C) and (F) do not include the terminal triplets.
RecA, Rad51, and Dmc1 differ in many structural, functional, and mechanistic details; they each interact with different accessory factors; and they are adapted for diverse biological functions (1, 2, 6). Our work reveals that despite these differences, unifying mechanistic principles underlie the process of strand exchange: Rad51/RecA family members all stabilize strand exchange intermediates in 3-nt steps; each step coincides with a broadly conserved energetic signature; and a single mismatch can abolish recognition of an entire base triplet. We also show that RecA, Rad51, and Dmc1 can all step over mismatches, but only Dmc1 can stabilize mismatched triplets; this distinction likely reflects the role of Dmc1 in promoting strand exchange between polymorphic alleles during meiosis. Our findings support a model in which a primary role of the Rad51/RecA family members during strand exchange is to establish the structure of RS-DNA, whereas the mechanism, energetics, and fidelity of the base-pairing transitions that take place during recombination are all governed by the physical architecture of RS-DNA itself. We propose that RS-DNA was selected at least ~2 billion years ago by an ancestral recombinase as the most energetically favorable solution for allowing efficient exchange of genetic information between related DNA molecules.
Supplementary Material
Acknowledgments
We thank R. Rothstein, L. Symington, H. Klein, M. Lichten, M. Gottesman, J. Dworkin, D. Duzdevich, and other members of the Greene and Sung laboratories for comments on the manuscript. Supported by NIH grants GM074739 (E.C.G.), RO1ES015252 (P.S.), and CA146940 (E.C.G. and P.S.); NSF grant MCB-1154511 (E.C.G.); and a Japan Society for the Promotion of Science fellowship (T.T.). E.C.G. is an HHMI Early Career Scientist. The data described in this manuscript are archived in the laboratory of E.C.G. in the Department of Biochemistry and Molecular Biophysics, Columbia University.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Bianco PR, Tracy RB, Kowalczykowski SC. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 2.Symington LS, Rothstein R, Lisby M. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox MM, et al. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 4.Fraser C, Hanage WP, Spratt BG. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeling PJ, Palmer JD. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 6.Brown MS, Bishop DK. Cold Spring Harb Perspect Biol. 2014 doi: 10.1101/cshperspect.a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neale MJ, Keeney S. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheridan SD, et al. Nucleic Acids Res. 2008;36:4057–4066. doi: 10.1093/nar/gkn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Yang H, Pavletich NP. Nature. 2008;453:489–4. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 10.Kowalczykowski SC. Nature. 2008;453:463–466. doi: 10.1038/453463a. [DOI] [PubMed] [Google Scholar]
- 11.Qi Z, et al. Cell. 2015;160:856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragunathan K, Joo C, Ha T. Structure. 2011;19:1064–1073. doi: 10.1016/j.str.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop DK, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 14.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selva EM, New L, Crouse GF, Lahue RS. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans E, Sugawara N, Haber JE, Alani E. Mol Cell. 2000;5:789–799. doi: 10.1016/s1097-2765(00)80319-6. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Jinks-Robertson S. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doye JP, et al. Phys Chem Chem Phys. 2013;15:20395–20414. doi: 10.1039/c3cp53545b. [DOI] [PubMed] [Google Scholar]
- 19.Danilowicz C, et al. Nucleic Acids Res. 2014;42:526–533. doi: 10.1093/nar/gkt867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


